Abstract
Monoclonal antibodies that bind the native conformation of proteins are indispensable reagents for the development of immunoassays, production of therapeutic antibodies and delineating protein interaction networks by affinity purification-mass spectrometry. Antibodies generated against short peptides, protein fragments, or even full length recombinant proteins may not bind the native protein form in biological fluids, thus limiting their utility. Here, we report the application of immunocapture coupled with selected reaction monitoring measurements (immunocapture-SRM), in the rapid screening of hybridoma culture supernatants for monoclonal antibodies that bind the native protein conformation. We produced mouse monoclonal antibodies, which detect in human serum or seminal plasma the native form of the human testis-expressed sequence 101 (TEX101) protein—a recently proposed biomarker of male infertility. Pairing of two monoclonal antibodies against unique TEX101 epitopes led to the development of an ELISA for the measurement of TEX101 in seminal plasma (limit of detection: 20 pg/ml) and serum (limit of detection: 40 pg/ml). Measurements of matched seminal plasma samples, obtained from men pre- and post-vasectomy, confirmed the absolute diagnostic specificity and sensitivity of TEX101 for noninvasive identification of physical obstructions in the male reproductive tract. Measurement of male and female serum samples revealed undetectable levels of TEX101 in the systemic circulation of healthy individuals. Immunocapture-SRM screening may facilitate development of monoclonal antibodies and immunoassays against native forms of challenging protein targets.
Monoclonal antibodies that bind the native form of a protein are indispensable for the development of sensitive immunoassays, production of therapeutic antibodies and for studying protein interaction networks by affinity purification-mass spectrometry (1, 2). Large-scale purification of native proteins from biological samples may be challenging, so recombinant proteins or protein fragments are often used for antibody production. Antibodies produced against short peptides, protein fragments, or even full length recombinant proteins, however, may not bind the native protein conformation present in biological fluids, thus limiting the utility of antibodies. Rapid screening of antibody-producing hybridoma clones for native protein binders requires highly specific and sensitive assays, performed under nondenaturing conditions. Here, we report the capability of an immunocapture-SRM assay to facilitate fast screening of hybridoma cultures for monoclonal antibodies that recognize the native conformation of testis-expressed sequence 101 (TEX101)1 protein in biological fluids.
Recently, we discovered, verified, and validated two proteins, testis-specific protein TEX101 and epididymis-specific protein ECM1, as biomarkers for the differential diagnosis of azoospermia (3, 4). Combination of TEX101 and ECM1 proteins measured in seminal plasma could differentiate between normal spermatogenesis, obstructive azoospermia (OA), and nonobstructive azoospermia (NOA) with very high diagnostic sensitivity and specificity. TEX101 levels in seminal plasma also facilitated classification of NOA subtypes of hypospermatogenesis, maturation arrest and Sertoli cell-only syndrome (5). A clinical laboratory test for TEX101 in seminal plasma may confirm the success of vasectomy or vasovasostomy, eliminate diagnostic testicular biopsies, and predict the success of sperm cell retrieval for assisted reproduction.
Human TEX101 is a membrane GPI-anchored protein encoded by the TEX101 gene, located in the 19q13.31 region of chromosome 19. According to the Human Protein Atlas, TEX101 expression is restricted to testicular tissue and male germ cells, with no evidence of expression in any other human tissue or cell type (6). Investigation of the function of mouse TEX101 demonstrated its direct role in fertilization (7–9).
We initially measured TEX101 levels in seminal plasma by mass spectrometry-based selected reaction monitoring (SRM) and immuno-SRM assays, with limits of detection of 120 and 5 ng/ml, respectively (4, 5). However, because of the ultra-wide range of TEX101 concentrations in seminal plasma of infertile and healthy men (0.5 ng/ml to 50,000 ng/ml) and theoretically zero levels for some azoospermic patients, a sensitive TEX101 immunoassay is required to develop a clinical laboratory test. In addition to immunoassay, monoclonal antibodies against native TEX101 would allow investigating its interactome and revealing its functional role in spermatogenesis and male fertility. Because TEX101 may emerge as a novel biomarker of male infertility, in this work we focused on the development of an ELISA for sensitive measurement of TEX101 in seminal plasma and serum.
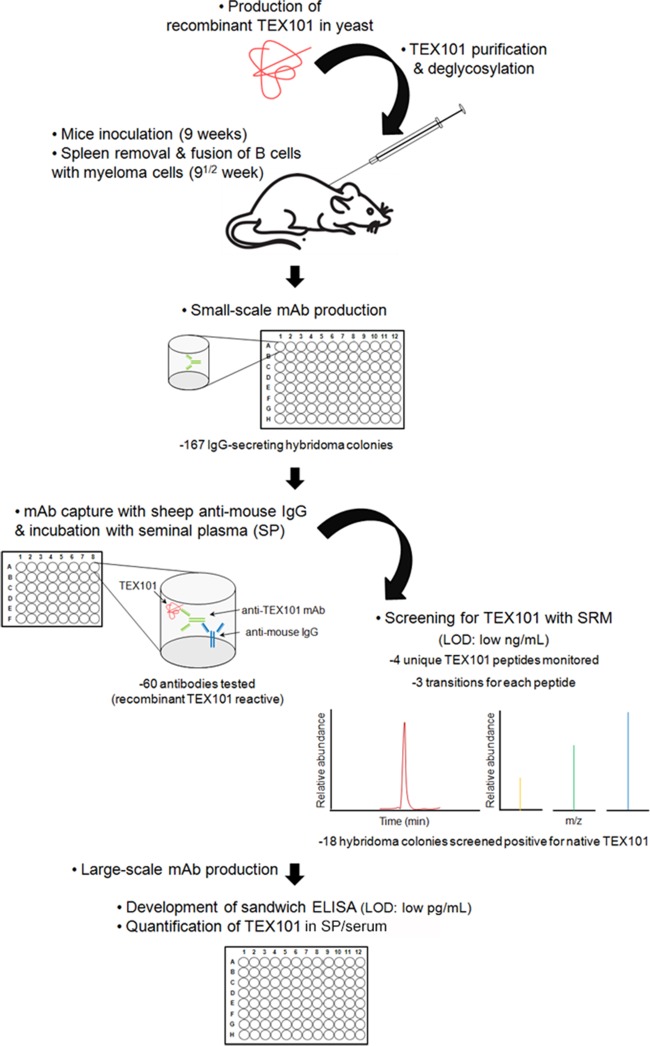
Our initial efforts to develop a TEX101 immunoassay using commercially available polyclonal antibodies were not successful. We found that commercial antibodies recognized only the denatured form of TEX101 and were useful for immunohistochemistry and Western blots, but not for the analysis of native TEX101 in seminal plasma. Here, we describe the production of mouse monoclonal antibodies against native TEX101, screening of antibody-producing clones by the two-step immunocapture and SRM assay, development of a sensitive ELISA and measurement of TEX101 in seminal plasma and serum (Fig. 1).
Fig. 1.
Pipeline for the production of mouse monoclonal anti-TEX101 antibodies and screening of colonies using two-step immunocapture-SRM assay. Screening included the coating of microtiter plates with sheep anti-mouse IgG antibodies, the addition of hybridoma cell supernatants, incubation with seminal plasma containing the native form of TEX101 followed by trypsin digestion and SRM analysis. Two-step immunocapture followed by SRM detection facilitated rapid screening of antibody-producing colonies and provided the following advantages: no requirement for previously developed TEX101 antibodies, small scale antibody production on 96-well plates, screening of low amounts of the newly-produced antibodies and direct selection of antibodies against the native form of TEX101. Eventually, all positive clones were expanded and a sensitive immunofluorescent assay for TEX101 was developed in seminal plasma and serum.
EXPERIMENTAL PROCEDURES
Cloning of TEX101 cDNA into the Yeast Expression Vector
A commercial Pichia Expression Kit (Invitrogen, Waltham, MA) was used for production of recombinant TEX101. Based on the published TEX101 cDNA sequence (transcript variant 2, NM_001130011.1), a set of oligonucleotide primers (forward 5′-GAAGAAGGGGTATCTCTCGAGAAAGACTGTATTGTCAAAAGGGTCTGTCCAT-3′ and reverse 5′-TAGGGAATTCTTAATGGTGATGGTGATGATGATTTTCAGTCTTTCGAGGTTGA-3′) were designed for PCR amplification of the fragment coding for the mature form of TEX101 present in seminal plasma (aa 26–222). Primers facilitated the generation of compatible restriction ends for ligation into the pPIC9 vector, as well as the incorporation of a C-terminus polyhistidine tag for protein purification (supplemental Fig. S1). TEX101 human cDNA ORF Clone (RC225319; Origene, Rockville, MD) was used as a template. PCR was performed in a 20 μl reaction mixture, supplemented with 0.4 μl of cDNA (5 ng/μl final concentration), 4 μl of 5x Phusion GC Buffer, which contained 7.5 mm MgCl2, and provided 1.5 mm MgCl2 in the final reaction, 200 μm deoxynucleoside triphosphates, 250 nm of the primers and 0.4 U of Phusion High-Fidelity DNA polymerase (New England BioLabs, Ipswich, MA) on an Eppendorf Mastercycler thermal cycler. The PCR conditions were 98 °C for 30 s, followed by 26 cycles of 98 °C for 10 s, 68 °C for 20 s, and 72 °C for 20 s, with a final extension at 72 °C for 7 min. In-frame cloning of the PCR product into the yeast expression vector pPIC9 was accomplished through double digestion, using XhoI and EcoRI restriction enzymes, and ligation of the two DNA fragments (supplemental Fig. S1). The sequence of the construct was confirmed by DNA sequencing.
Production of Human TEX101
Prior to transformation of yeast cells, pPIC9 vector containing the TEX101 cDNA was linearized with SacI restriction enzyme to favor the integration of the construct in P. pastoris genome via homologous recombination. The linearized construct was introduced into the yeast strains GS115 and KM71 by electroporation. A stable clone was selected from the GS115 strain according to the manufacturer's recommendations (Invitrogen). Stable yeast clones were grown in the buffered complex glycerol medium (BMGY) until the culture reached log-phase (OD600 = 2–6). Following that, the cell pellet was resuspended in the buffered complex methanol (BMMY) to an OD600 of 1.0 and was grown at 30 °C with shaking. TEX101 production was induced with 10 ml/L methanol over 4 days. Yeast culture containing secreted TEX101 was centrifuged and the supernatant was concentrated 100-fold initially by positive pressure ultrafiltration in an AmiconTM stirring chamber (Millipore, Billerica, MA) with a 10-kDa cutoff regenerated cellulose membrane (Millipore), followed by AmiconTM centrifugal filter tubes Ultracel 3K (Millipore). A rabbit polyclonal anti-TEX101 antibody HPA041915 (Sigma-Aldrich, St. Louis, MO) and a mouse monoclonal anti-His antibody (Cat# A00186–100, GenScript, Piscataway, NJ) were used to monitor TEX101 production by Western blot analysis.
Purification of Human TEX101 with Immobilized Metal Ion Affinity Chromatography
The recombinant TEX101 was purified from yeast culture supernatants by immobilized metal ion affinity chromatography. HIS-Select Nickel Affinity gel (Sigma-Aldrich) selective for recombinant proteins with histidine tags was used to purify TEX101 from yeast culture according to the manufacturer's recommendations. In summary, the nickel affinity gel was first washed with 1–2 volumes of de-ionized water to remove ethanol, and then equilibrated with 3–5 volumes of equilibration buffer (10 mm imidazole in 50 mm NaH2PO4, 0.3 m NaCl, pH 8.0). Prior to application on the affinity gel, the recombinant protein sample was clarified by centrifugation to obtain a pH between 7.0 and 8.0. Recombinant protein solution was incubated with affinity gel, which was subsequently washed with equilibration buffer. TEX101 was eluted with 250 mm imidazole in 50 mm NaH2PO4, 0.3 m NaCl, pH 8.0 at room temperature. The presence of TEX101 in various fractions was determined with Western blotting by using rabbit polyclonal anti-TEX101 antibody HPA041915 (Sigma-Aldrich). The purity and the molecular mass of TEX101 were determined by SDS-PAGE stained with Coomassie Blue. The purified TEX101 protein concentration was determined by the bicinchoninic acid assay (Pierce Biotechnology, Rockford, IL).
Analysis of Human Recombinant TEX101 by Mass Spectrometry
Following SDS-PAGE analysis, all visible gel bands were excised and analyzed by LC-MS/MS. An in-gel digestion protocol was followed, as described elsewhere (10). In all cases, peptides were extracted from solution using C18 OMIX tips (Varian Inc., Lake Forest, CA) and eluted in 5 μl of elution buffer B (65% acetonitrile, 0.1% formic acid). Buffer A (80 μl of 0.1% formic acid) was added to sample tubes and transferred to a 96-well microplate (Axygen, Union City, CA). Using a 96-well microplate autosampler, 40 μl of each sample was loaded onto a 3 cm C18 trap column (inner diameter 150 μm; New Objective, Woburn, MA) that was packed in-house with 5 μm Pursuit C18 (Varian Inc.). An increasing concentration of Buffer B (0.1% formic acid in acetonitrile) was used to elute the peptides from the trap column onto a resolving analytical 5-cm PicoTip Emitter Column (inner diameter 75 μm, 8 μm tip; New Objective). This column was packed in-house using 3 μm Pursuit C18 (Varian Inc.). The EASY-nLC system (Proxeon Biosystems, Odense, Denmark) was coupled online to an LTQ-Orbitrap XL hybrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA) and a nanoelectrospray ionization source (Proxeon Biosystems) was used with a spray voltage of 2 kV and temperature of 160 °C. A data-dependent mode was used to analyze samples and a full MS1 scan was acquired from 450–1450 m/z in the mass analyzer (resolution of 60,000). This was followed by MS2 scan acquisition of the top six parent ions in the LTQ mass analyzer. The subsequent parameters were enabled: dynamic exclusion, charge state screening, and monoisotopic precursor selection. Ions with charge states of +1, ≥ +4 and unassigned charge states did not undergo MS2 fragmentation.
For protein identification and data analysis, XCalibur software (v. 2.0.5; Thermo Fisher Scientific) was used to generate RAW files of each MS run. RAW files were subsequently used to generate Mascot Generic Files (MGF) on Mascot Daemon (version 2.2.2). Once generated, MGFs were searched with Mascot (Matrix Science, London, UK; version 2.2). Protein searches were performed against the nonredundant human UniProtKB/Swiss-Prot database (version 10, October 2013) using the following parameters: fully tryptic cleavages, 7 ppm precursor ion mass tolerance, 0.4 Da fragment ion mass tolerance, allowance of one missed cleavage and fixed modifications of carbamidomethylation of cysteines. Variable modifications included oxidation of methionine, pyro-Glu from glutamine of the N terminus-carbamoylmethylcystein cyclization at N terminus, deamidation of glutamine, oxidation of tryptophan, and acetylation of the N terminus.
Assessment of TEX101 Glycosylation
The TEX101 protein glycosylation was assessed by treatment of purified recombinant TEX101 protein with the deglycosylation enzyme PNGase F (Roche, Mannheim, Germany). The mixture was incubated at 37 °C for 3 h. PNGase F treated and nontreated TEX101 were subjected to SDS-PAGE stained with Coomassie Blue.
Animal Handling and Somatic Cell Fusion for Monoclonal Antibody Production
Female BALB/c mice were obtained from the Toronto Centre for Phenogenomics (TCP). All animal research (Animal Use Protocol# 14–04-0119a-H) was approved by TCP Animal Care Committee. Mice were inoculated subcutaneously with 100 μg of deglycosylated recombinant TEX101 protein, mixed (1:1) with Sigma Adjuvant System (Sigma-Aldrich). Two subsequent booster injections with 25 μg of antigen in adjuvant were performed at 3-week intervals. Final boost was an intraperitoneal injection of 25 μg of antigen in phosphate-buffered saline (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4). Three days later, mouse spleen was excised aseptically and homogenized. Extracted spleen cells were fused with NSO murine myeloma cells (5:1 ratio) using polyethylene glycol (Sigma-Aldrich). Successfully fused cells were selected using HAT media (Invitrogen), supplemented with 20% fetal bovine serum (Hyclone, Thermo Fisher Scientific, Waltman, MA).
Screening for IgG Secreting Clones
Cell culture supernatants were screened for the presence of IgG and IgM antibodies by using the following immunoassay protocol. 96-well microtiter plates were coated with goat anti-mouse IgG+IgM (H+L) antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) diluted (1:1,000) in sodium carbonate-bicarbonate buffer (0.2 m Na2CO3, 0.2 m NaHCO3, pH 9.2). Plates were washed twice with PBST (0.05% Tween 20 in 137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, pH 7.4) and 100 μl of 5% milk in PBST were added per well. Plates were incubated for 1 h at room temperature (RT). After washing (3×), 100 μl of each hybridoma supernatant and the appropriate controls were added in two adjacent wells and incubated for 1 h at room temperature. Following one more round of washing (3×), 100 μl of HRP-conjugated Goat anti-mouse IgG (Fc Fraction; Jackson ImmunoResearch) and 100 μl of goat anti-mouse IgM (μ chain) (Jackson ImmunoResearch) antibodies (in 1% milk/PBST), were added on each one of the paired wells, respectively. Following a final wash (3×), 100 μl of 3,3,5,5′-tetramethylbenzidine substrate solution were added and plates were incubated for 15 min at 37 °C with gentle shaking. Fifty microliters of stop solution (2 m H2SO4) were added on top. Absorbance was measured with the Wallac EnVision 2103 Multilabel Reader (Perkin Elmer, Waltham, MA) at 450 nm, with a reference wavelength of 620 nm. IgG positive colonies were transferred in 48-well culture plates.
Screening for Immunogen Reacting Clones
IgG positive clones were screened for reaction with the immunogen using an indirect immunoassay protocol. Recombinant TEX101 protein was immobilized on 96-well microtiter plates (100 ng per well) diluted in coating buffer. Plates were washed (2×) and 100 μl of 5% milk in PBST were added per well. Plates were incubated for one hour at RT, followed by wash (3×). One hundred microliters of hybridoma supernatant and the appropriate controls were added and incubated for 1 h at RT. Then, plates were washed (3×) and 100 μl of HRP-conjugated goat anti-mouse IgG (Fc Fraction) (Jackson ImmunoResearch) antibody (in 1% milk in PBST) were added on the plates. Following final wash (3×), 100 μl of 3,3,5,5′-tetramethylbenzidine substrate solution were added and plates were incubated for 15 min at 37 °C with gentle shaking. Fifty microliters of stop solution (2 m H2SO4) were added on top. Absorbance was measured with the Wallac EnVision 2103 Multilabel Reader (Perkin Elmer) at 450 nm, with a reference wavelength of 620 nm.
Immunocapture-SRM Screening for Clones Producing Antibodies Against Native TEX101 in Seminal Plasma
White 96-well microtiter plates were coated with 500 ng/well of sheep anti-mouse IgG-Fcγ fragment-specific antibody (Jackson ImmunoResearch) in TBS buffer. Plates were washed twice with PBS (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, pH 7.4). Cell culture supernatants at two dilutions in 1% (w/v) BSA in PBS buffer were applied to the plates and incubated for 2 h at RT with gentle shaking. A protein G-purified mouse polyclonal anti-TEX101 antibody (ab69522; Abcam, Cambridge, MA) was used as a positive control for the assay. Plates were washed (6×) with PBS and 100 μl of 100-fold diluted seminal plasma, from a normal donor, in 6% (w/v) BSA and PBS buffer were added. Following 2 h of incubation at RT with gentle shaking, wells were once again washed 3× with PBS and 3× with 50 mm ammonium bicarbonate. Fifty millimoles ammonium bicarbonate, 50 mm dithiothreitol (Sigma-Aldrich), 50 fmoles of heavy isotope-labeled TEX101 proteotypic peptide GALCQETILIIK tagged with a trypsin-cleavable tag (SpikeTides™_TQL, JPT Peptide Technologies GmbH, Berlin, Germany) and 0.05% RapiGest SF (Waters, Milford, MA) were mixed and ninety four μl of this mix were added to each well and kept for 15 min at RT. Then, 5 μl of 100 mm iodoacetamide were added and samples were kept for 40 min in the dark at RT. Samples were then digested by addition of 5 μl of 0.05 μg/μl of sequencing-grade modified porcine trypsin (Promega Cat# V5111, Madison, WI) in 50 mm ABC. Trypsin inactivation and cleavage of RapiGest SF was achieved with the addition of 1% trifluoroacetic acid. C18 microextraction and desalting of peptides was done as described above. EASY-nLC system (Proxeon Biosystems) was coupled online to a Quantiva triple-quadrupole mass spectrometer (Thermo Fisher Scientific) using a nanoelectrospray ionization source. All transitions being monitored were scheduled within 1.5-min intervals during a 30-min LC gradient. Four unique TEX101 peptides were monitored with the scheduled SRM mode (supplemental Table S1), with one used for quantification and the rest used for qualitative analysis. Relative abundance of TEX101 in each sample was estimated as a ratio to the spiked-in tagged heavy isotope-labeled peptide internal standard GALC[cm]QETILIIK*-JPTtag. The SRM method had the following parameters: optimized collision energy (CE) values; mass/charge ratio (m/z) scan width, 0.010; scan time, 0.015 to 0.040 s; FWHM resolution of the first quadrupole (Q1), 0.4; FWHM resolution of the third quadrupole (Q3), 0.7; pressure of the second quadrupole, 1.5 mtorr; tuned S-lens values; declustering voltage, +1 V. RAW files recorded for each sample were analyzed with the Pinpoint software, and peptide areas were used to calculate light-to-heavy peptide ratios and protein concentrations in each sample.
Clone Screening by Western blot
We confirmed results of immunocapture-SRM screening by Western blot. Seminal plasma samples (10 μg per well) were prepared in 50 mm DTT and 31.25 mm Tris-HCl pH 6.8, 12.5% glycerol, 1% SDS, 0.005% Bromphenol Blue (Laemmli sample buffer; BioRad Cat# 161–0737, Hercules, CA) and applied to 4–15% Mini-PROTEAN® TGX™ Precast Gels Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (BioRad Cat# 456–1021). Gel electrophoresis was performed at 200 V for ∼30 min. Gels were then stained with SimplyBlueTM SafeStain Coomassie® G-250 stain (Invitrogen). For Western blotting, gels were transferred using a Trans-Blot® Turbo Blotting System (BioRad) and a Trans-Blot® Turbo Transfer Pack (0.2 μm PVDF membrane, BioRad). After blocking in 5% milk in TBST (0.05% Tween 20 in 50 mm Tris, 150 mm NaCl, pH 7.5) for ∼2 h at 22 °C, membranes were attached to a Mini-PROTEAN® II Multiscreen Apparatus (BioRad Cat# 456–1021). Hybridoma culture supernatants were added on each lane and membranes were incubated for 1.5 h at room temperature. A rabbit polyclonal anti-TEX101 antibody HPA041915 (Sigma-Aldrich) was used as a positive control. After incubation, the membrane was washed (3x) in TBST, followed by the addition of alkaline phosphatase-conjugated AffiniPure goat anti-rabbit or anti-mouse IgG (0.03 μg/ml in 1% milk in TBST; Jackson ImmunoResearch) and incubation at 22 °C for 45 min. The membrane was then extensively washed with TBST, dried and 125 μl of chemiluminescence substrate (Siemens, Los Angeles, CA) per square centimeter was added. The membrane was then placed into an autoradiography cassette and exposed and developed using Radiomat™ B Plus-Full Speed Blue sensitive x-ray film (8 × 10 inches, AGFA X-Ray Film, Mortsel, Belgium).
Expansion of Clones and Purification of Anti-human TEX101 Monoclonal Antibodies
Subsequently, cells were further grown and transferred in serum-free medium (CD-1 medium; Invitrogen), containing 8 mm l-Glutamine. Supernatants were collected and purified using a protein G column, according to the manufacturer's protocol (GammaBind Plus, GE Healthcare, Little Chalfont, Buckinghamshire, UK). Culture supernatants were diluted twofold with the binding buffer (10 mm Na2HPO4/NaH2PO4, 150 mm NaCl, 10 mm EDTA, pH 7.0) and loaded on the column. The column was then washed with binding buffer and antibodies were eluted with 0.5 m acetic acid at pH 3.0.
Development of Immunoassay
White 96-well ELISA plates were coated with 500 ng/well of mouse monoclonal anti-TEX101 antibody 23-ED-616 in 50 mm Tris buffer, pH 7.8. Plates were washed (2x) with 0.05% Tween 20 in 20 mm Tris, 150 mm NaCl, pH 7.4). Treated TEX101 calibrators and samples (see below for treatment) were added into each well (100 μl/well) and incubated for 2 h with gentle shaking. Plates were then washed (2×) with the washing buffer. A biotinylated mouse monoclonal anti-TEX101 antibody 23-ED-155, diluted in a solution containing 60 g/L BSA, 25 ml/L normal mouse serum, 100 ml/L normal goat serum, and 10 g/L bovine IgG in 50 mm Tris, pH 7.8, were added (250 ng of antibody per 100 μl of solution per well) and incubated for 1 h. Plates were washed (6×) and alkaline phosphatase-conjugated streptavidin was added in the wells (100 μl per well). Incubation was for 20 min at RT with gentle shaking, followed by a final wash (6×). Diflunisal phosphate (DFP) solution was prepared in substrate buffer (0.1 m NaCl, 1 mm MgCl2 in 0.1 m Tris, pH 9.1), added on the plate (100 μl per well) and incubated for 10 min at RT with gentle shaking. Subsequently, the developing solution (1 m Tris, 0.4 m NaOH, 2 mm TbCl3, and 3 mm EDTA) was added on top and mixed for 1 min. Time-resolved fluorescence was measured with the Wallac EnVision 2103 Multilabel Reader (Perkin Elmer), as previously described (11).
Seminal Plasma and Serum Samples
Matched pre- and post-vasectomy seminal fluid samples (n = 9), as well as male (n = 17) and female (n = 17) serum samples, were obtained after informed consent and institutional review board approval (Mount Sinai Hospital, Toronto, ON, Canada). Seminal fluid was allowed to liquefy at room temperature for 1 h after collection, aliquoted in 1-ml portions and centrifuged (3×) at 13,000 × g for 15 min at room temperature to separate plasma from cells and cellular debris. The supernatant seminal plasma was then frozen at −80 °C until use.
Several seminal plasma samples were pooled and mixed to prepare the calibrators. TEX101 concentration in the pool was calculated using a quantitative SRM method, similar to the aforementioned one. Heavy isotope-labeled peptide GALC[cm]QETILIIK with a trypsin-cleavable JPT tag (JPT Peptide Technologies GmbH) was used as the internal standard for the absolute quantification of endogenous TEX101 protein. Calibration curve was prepared by spiking increasing amounts of the internal standard (0.1 to 3,000 fmoles) into 10 μl of 10-fold diluted seminal plasma pool (1 μl equivalent), before proteomic sample preparation and trypsin digestion. In parallel, different volumes of the seminal plasma pool (10-fold diluted 0.6, 1, 2 and 6 μl of seminal plasma) were supplemented with 600 fmoles of the internal standard and digested with three full-process replicates and measured by SRM with two injections each. Dilution-adjusted ratios were used to calculate TEX101 concentrations in the seminal plasma pool. Furthermore, numerous aliquots (volume of 20 μl each) of the pool were prepared and stored at −20 °C.
Calibrators for the seminal plasma assay were prepared by mixing (1:1) one aliquot of SP with reagent mix (16 μl 7.7 m guanidine-HCl, 2 μl 2 m NaOH, and 2 μl dH20). The mix was incubated for 1 h at RT, and then diluted (50×) with assay diluent (60 g/L BSA, 25 ml/L normal mouse serum, 100 ml/L normal goat serum, and 10 g/L bovine IgG in 50 mm Tris, pH 7.8). Serial dilutions of the treated calibrator sample were prepared in a twofold dilution step (ranging from ∼ 47 ng/ml to 0 ng/ml) and added on ELISA plate. Seminal plasma samples from individuals were mixed (1:1) with the reagent mix, followed by 1-h incubation at room temperature. Samples were further diluted 10-fold with assay diluent before loading on the plate.
Calibrators for the serum assay were prepared by diluting SP in a female serum pool (1:10) and 60 μl of this mixture was supplemented with 10% sodium deoxycholate [5% final]. The mixture was incubated for 1 h at 63 °C and further diluted (5×) with assay diluent (60 g/L BSA, 25 ml/L normal mouse serum, 100 ml/L normal goat serum, and 10 g/L bovine IgG in 50 mm Tris, pH 7.8). Once again, serial dilutions of the treated calibrator sample were prepared in a twofold dilution step (ranging from ∼ 47 ng/ml to 0 ng/ml) and added on ELISA plate. Serum samples from individuals were mixed (1:1) with 10% sodium deoxycholate, followed by 1-h incubation at 63 °C. Samples were further diluted three- to sixfold with assay diluent before loading on the plate.
Statistical Analysis
To assess assay's linearity, a linear regression model was built using the log-transformed values of endogenous TEX101 concentration and sample dilutions, within CV≤15%. Statistical analysis and plots were prepared using R statistical software v 2.15.2 (available from http://www.Rproject.org).
RESULTS
Production, Purification, and Analysis of Recombinant Human TEX101 Protein
The cDNA coding for mature TEX101 protein (amino acids 26 to 222) was cloned into a methylotrophic P. pastoris yeast expression system and secreted TEX101 was purified by nickel affinity chromatography from the yeast culture supernatant. Yeast mainly expressed two forms of TEX101 protein (∼30 kDa and ∼90 kDa), as assessed by SDS-PAGE (supplemental Fig. S2) and mass spectrometry (Supplemental Table S2). Higher molecular weight forms were produced because of the variable glycosylation of recombinant proteins in P. pastoris (12). Treatment of recombinant TEX101 with PNGase F reduced their molecular weight to the expected 27–35 kDa (supplemental Fig. S3).
Production and Screening of Monoclonal Antibodies
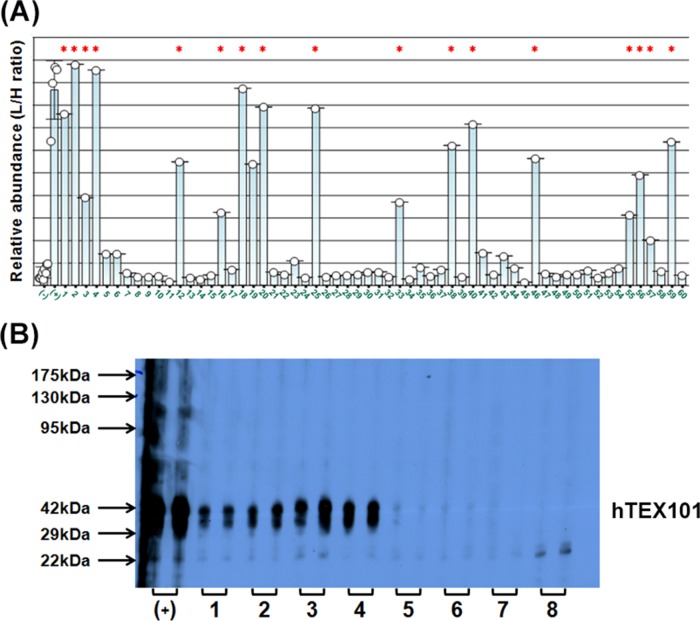
Deglycosylated recombinant TEX101 protein was used as an immunogen to generate mouse monoclonal antibodies. The fusion of murine splenocytes with murine myeloma cells resulted in the generation of 167 IgG-secreting hybridoma colonies, with 60 antibodies reacting with recombinant TEX101. These 60 antibodies were further screened for reaction with the native TEX101 protein in seminal plasma. Immunocapture-SRM revealed that 18 colonies produced antibodies that could bind to the native form of TEX101 present in seminal plasma (Fig. 2A). Western blot, however, showed positivity of 26 colonies for denatured seminal plasma TEX101 (Fig. 2B). Seventeen of the 18 aforementioned colonies were also identified by Western blot, whereas one colony was identified exclusively by the immunocapture-SRM. Thus, 18 positive hybridoma colonies were eventually expanded in the serum-free media and purified using protein G columns.
Fig. 2.
Screening of hybridoma colonies by immunocapture-SRM assay and Western blot. A, Relative abundance of the native form of TEX101 immunocaptured from seminal plasma using 60 hybridoma colonies, as measured by a TEX101 SRM assay. Lanes (-) and (+) denote the negative control (no mouse anti-TEX101 antibodies) and the positive control (anti-TEX101 mouse polyclonal antibody ab69522), respectively. Lanes 1–60 denote supernatants from 60 IgG-secreting hybridoma colonies. Asterisks mark the clones that were also positive on Western blot. B, Representative Western blot of colonies screened for reaction with TEX101 in seminal plasma. Lane (+) represents the positive control (rabbit polyclonal antibody HPA041915), whereas lanes 1–8 include 8 representative colonies measured in duplicates.
Immunoassay Development
Two mouse monoclonal antibodies (23-ED-616 and 23-ED-155) targeting different TEX101 epitopes were used to develop an immunofluorometric assay. Even though both antibodies could capture the native form of TEX101 in seminal plasma, we thoroughly investigated a wide variety of conditions that would further increase ELISA signal and thus allow for measuring TEX101 concentrations in the low pg/ml range. Incubation of seminal plasma with guanidine hydrochloride at pH 12 or treatment with sodium deoxycholate at 63 °C emerged as the most efficient procedures. We suspect that only a fraction of endogenous TEX101 is present in seminal plasma in the soluble form, whereas another fraction is either bound to other proteins, embedded into membranous vesicles such as epididymosomes, or encapsulated into membrane fragments of destroyed spermatozoa. For instance, it has been previously reported that the majority of plasma membranes of the disrupted spermatozoa heads are released in the form of unilamellar vesicles (13). These vesicles are typically pelleted by centrifugation at 285,000 × g (13) and thus most probably are present in our seminal plasma samples prepared by centrifugation of semen at 13,000 × g. Opening of such membrane vesicles and dissociation of intravesicular proteins requires high pH treatment, such as incubation with 100 mm Na2CO3 at pH 11.6 (14). Thus, treatment of seminal plasma with guanidinium at pH 12 or deoxycholate at 63 °C may lead to the release of TEX101 from such vesicles, making it available for ELISA measurements.
Assay calibrators were prepared by mixing seminal plasma samples obtained from several dozen healthy fertile men. The concentration of TEX101 protein in the pool was assessed by a quantitative SRM assay. The endogenous proteotypic peptide and spike-in internal standard were measured in four different volumes of seminal plasma, with three full process replicates for each volume and two injections for each replicate. Assuming that one mole of the proteotypic peptide represents one mole of protein, the mean TEX101 protein concentration was derived as 4.7 ± 1.5 μg/ml (supplemental Fig. S4).
ELISA Performance and TEX101 Measurement in Seminal Plasma
Initially, the assay was developed for TEX101 measurements in seminal plasma, the fluid of choice for diagnosis of male infertility. Two monoclonal antibodies (23-ED-616 and 23-ED-155) were paired in the sandwich immunoassay. The endogenous TEX101 protein present in pooled seminal plasma was used to calibrate the assay. Because limit of detection (LOD) of the immunoassay without seminal plasma pretreatment was not high enough, we examined literature (15–25) to identify conditions that would improve the assay sensitivity in seminal plasma. Thus, seminal plasma was subjected to pretreatment with the following detergents, salts and solvents: 1% SDS, 2–5% Triton X-100, 1% CHAPS, 2% sodium deoxycholate, 1 m urea, 3 m guanidine hydrochloride, 10% dimethyl sulfoxide, 10% dimethylformamide, 10% glycerol, 10% methanol, 10% acetonitrile. Additional treatments included heating between 40–70 °C and the use of high pH (10–12). As a result, the use of guanidinium at pH 12 and room temperature allowed a significant increase of ELISA signal and pg/ml sensitivity of TEX101 in seminal plasma (supplemental Fig. S5A).
The limit of blank (LOB) of the optimized immunoassay, measured with 2% (w/v) BSA (n = 7) was established as 6.5 pg/ml. The corresponding within-run limit of detection (LOB + 1.64*S.D.) was determined as 20.1 pg/ml, with a linear range spanning from 33 pg/ml to 19.4 ng/ml (regression coefficient β1 = 0.900, p < 0.0001) (supplemental Fig. S5B). Aliquots of seminal plasma were measured over 4 days in order to calculate within-run (n = 7 aliquots, each in seven replicates) and total (n = 4 aliquots, each in two replicates) imprecision. According to results, within-run and total imprecisions were generally low for the broad range of TEX101 concentrations (<5 and 11%, respectively), only getting worse when reaching the LOD (supplemental Table S3). Within-run limit of quantification (LOQ) was set at the TEX101 concentration showing CV≤20%—which in this case was close to the assay's LOD (<30.0 pg/ml). Stability of TEX101 was assessed by daily measurements of TEX101 concentrations in seminal plasma stored at 4 °C and 22 °C for 7 days. As a result, concentration of TEX101 in the untreated seminal plasma changed marginally (supplemental Fig. S6). When measured in the treated seminal plasma stored at 4 °C, TEX101 concentration slightly increased after 24 h from 50.25 ± 1.91 to 67.84 ± 0.33 ng/ml and then remained stable (supplemental Fig. S7).
TEX101 was measured in nine pairs of pre- and post-vasectomy seminal plasma samples. TEX101 concentrations in prevasectomy samples ranged from 37.1 ± 0.05 μg/ml to 0.82 ± 0.01 μg/ml, whereas TEX101 in all post-vasectomy samples was below the lowest calibrator (0.047 ng/ml) loaded on plate (Fig. 3).
Fig. 3.
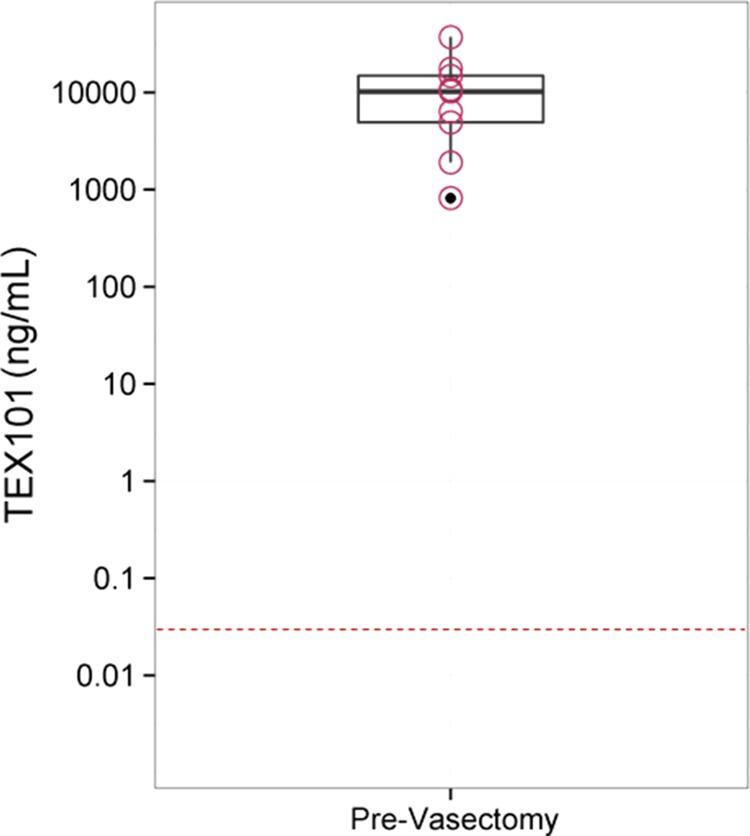
Box plot for TEX101 levels in matched seminal plasma samples (n = 9), pre- and post-vasectomy. Measured TEX101 levels in pre-vasectomy samples ranged from 37.1 ± 0.05 μg/ml to 0.82 ± 0.01 μg/ml. TEX101 levels in all post-vasectomy samples were below the lowest calibrator (0.047 ng/ml) represented by a dashed line.
ELISA Performance and TEX101 Measurement in Serum
To investigate TEX101 presence in the systemic circulation, we modified our protocol to allow for TEX101 measurements in male and female serum samples. TEX101 calibrators were prepared by diluting aliquots of seminal plasma in female serum. Treatment of serum with 10% sodium deoxycholate (5% final) at 63 °C was found to be more effective than treatment with guanidine hydrochloride at pH 12. A representative calibration curve for the TEX101 assay in female serum is shown in supplemental Fig. S5C. Within-run LOB of the optimized immunoassay measured with female serum samples (n = 7) was established as 20.7 pg/ml. The corresponding within-run LOD (LOB + 1.64*S.D.) was determined as 40.0 pg/ml, with a linear range spanning from 45.7 pg/ml to 28.9 ng/ml (regression coefficient β1 = 1.034, p < 0.0001) (supplemental Fig. S5D). Within-run (n = 4 aliquots, each in seven replicates) and total (n = 4 aliquots, each in two replicates) imprecisions assessed over 4 days were found low for the broad range of TEX101 concentrations (<6 and 13%, respectively), only deteriorating when reaching assay's LOD (Supplemental Table S4). Within-run limit of quantification (LOQ) was set at the LOD (40.0 pg/ml).
Recovery of endogenous TEX101 from serum was assessed by spiking seminal plasma pool (of TEX101 concentration: 4.7 ± 1.5 μg/ml) into female serum samples. According to results, endogenous TEX101 recovery was calculated as 90%, by comparing the measured value with the theoretical one.
Measurement of serum samples from 17 healthy females and 17 healthy males revealed undetectable levels of TEX101 in the systemic circulation (< 40 pg/ml). The apparent absence of TEX101 in serum can be explained by its high tissue specificity and the stringency of testis-blood barriers in healthy males.
DISCUSSION
Despite rapid progress in proteomics and mass spectrometry, monoclonal antibodies remain indispensable analytical tools for the ultrasensitive analysis of proteins in biological fluids. Likewise, antibody-based ELISAs are still the assays of choice for clinical laboratory diagnostics and large-scale biomarker validation studies because of their simplicity, high sensitivity and specificity, high throughput, and low cost (26). One of the most important aspects of ELISA development is the availability of antibodies that bind the native form of proteins.
We developed an immunoassay for TEX101 protein, because of the importance of this analyte for diagnosis of male infertility (5). Our initial efforts to produce an immunoassay using six different commercially available antibodies were not successful. Immunohistochemistry, prototype sandwich assays, Western blots and immuno-SRM assays revealed that commercial antibodies recognized only the denatured form of TEX101, but they could not bind the native form present in seminal plasma. Only a mouse polyclonal antibody (ab69522 from Abcam) could capture the native form of TEX101, as revealed by immuno-SRM (5). Due to its low purity and high cost, ab69522 was suitable as a positive control for colony screening, but not for ELISA development. Thus, we proceeded with production of new TEX101 monoclonal antibodies.
To obtain native immunogen, we first attempted to purify TEX101 from seminal plasma. Multistep chromatography included anion exchange, size exclusion, and reverse phase separations of seminal plasma from fertile men, with TEX101 being monitored by shotgun and SRM mass spectrometry. Multistep separations enriched TEX101 from the initial abundance of ∼0.01% (5 μg/ml) to ∼5% of total protein. Such purity, however, was not sufficient for successful mouse immunization. We also attempted to produce TEX101 in E. coli; however, expression levels were low and most of TEX101 was lost because of protein aggregation. Expression in the P. pastoris yeast system was more successful, and the recombinant TEX101 was used for mouse immunization. Because of TEX101 hyperglycosylation in P. pastoris, however, the produced antibodies recognized only recombinant TEX101, but not the endogenous TEX101 in seminal plasma, as confirmed by prototype sandwich ELISA and immuno-SRM assays (supplemental Fig. S8). The deglycosylated form of recombinant TEX101 elicited a strong immune response and led to dozens of hybridoma colonies. Screening by immunocapture-SRM assay revealed 18 colonies that produced antibodies against the native form of TEX101. In order to further improve the sensitivity of the developed sandwich immunoassay, we tested a variety of sample treatment reagents, with 3 m guanidine hydrochloride at pH 12 and room temperature or 5% sodium deoxycholate at 63 °C being particularly effective for seminal plasma.
Assay calibration was accomplished with the endogenous TEX101 protein present in a pool of seminal plasma samples (concentration of TEX101: 4.7 ± 1.5 μg/ml). We also tried using the deglycosylated recombinant TEX101 diluted in PBS, whose performance matched the endogenous following treatment with guanidinium at pH 12 (supplemental Fig. S9). Recombinant TEX101 was also recovered from seminal plasma samples (min. 112% and max. 191%) and female serum samples (min. 102% and max. 243%), with results indicating potential interference within both treated matrices; unlike the endogenous TEX101 protein.
Until recently, screening of antibody clones was accomplished either by Western blot, under denaturing conditions, or by solid-phase ELISA. Mass spectrometry- and proteomics-based approaches can revolutionize antibody production through the rapid screening of hybridoma clones (27). Immunocapture-mass spectrometry detection was previously demonstrated by Schoenherr et al., who proposed a SISCAPA-based pipeline for the screening of hybridoma supernatants for anti-peptide antibodies (28). To our knowledge, the use of two-step immunocapture followed by SRM detection to select hybridoma clones recognizing the native form of proteins present in biological fluids has not been previously demonstrated.
Two-step purification using sheep anti-mouse antibodies is a crucial step of our approach. Since dozens of IgG-producing colonies have to be tested, rapid screening is feasible only if colonies are grown in the FBS-supplemented media using a small-scale cell culture. To screen supernatants produced by each colony, mouse antibodies against TEX101 are first purified with sheep anti-mouse antibodies and then incubated with seminal plasma containing the native form of TEX101. Such two-step immunocapture, followed by SRM quantification, allows identification of hybridoma colonies that secrete antibodies against the native form of TEX101.
Our approach can be extended to develop high-quality antibodies against a variety of proteins with unknown function (29). For example, many testis- and prostate-specific proteins have not as yet been characterized and some of them have not been previously identified, because of their sequestration from the systemic circulation by blood-tissue barriers. Even though the Human Protein Atlas initiative (30) produced polyclonal antibodies against denatured forms of many testis- and prostate-specific proteins, monoclonal antibodies against the native protein forms of such proteins and corresponding immunoassays are still not available (31).
Coupling high-affinity antibodies with mass spectrometry-based quantitative analysis could complement the traditional immunoassays in the biomarker verification process (32–34). There are numerous studies on immunocapture-MS/MS or SRM, using either anti-peptide antibodies in complex digests (35, 36) or antibodies that capture the intact protein (37, 38), with quantification reaching low ng/ml. According to these studies, the advantages of immunocapture MS/MS or SRM over ELISA are associated with their high specificity and its great potential for multiplex analyses, thus proving to be a powerful alternative to immunoassays. We believe that 0.5 ng/ml or even lower LOD for TEX101 can be achieved by immunoaffinity-SRM assay with a sample volume as large as 1,000 μl of seminal plasma (reasonable volume from the clinical perspective). In future, if executed with a high throughput (>100 samples per day), a multiplex immunoaffinity-SRM assay for TEX101 and ECM1 markers may eventually replace ELISA assays for differential diagnosis of azoospermia.
To conclude, we here propose that immunocapture-SRM screening can facilitate the development of monoclonal antibodies and immunoassays against the native forms of challenging protein targets, such as membrane-bound testis-specific proteins. With more rigorous clinical validation, the developed TEX101 ELISA may emerge as a novel noninvasive clinical laboratory test for the differential diagnosis of male infertility. Native protein capturing antibodies will allow mapping of the human TEX101 interactome and reveal its functional role in reproduction and male infertility.
Supplementary Material
Acknowledgments
We thank Apostolos Dimitromanolakis for assistance with R statistical software.
Footnotes
Author contributions: E.P.D. and K.J. designed the research project; D.K., D.B. and C.S. performed the experiments; A.P.D. performed immunocapture-SRM experiments; D.K. wrote the manuscript, and all authors contributed to the revision of the manuscript.
* This work was supported by the MaRS Innovation Industry Access Program.
 This article contains supplemental Figs S1 to S9 and Tables S1 to S4.
This article contains supplemental Figs S1 to S9 and Tables S1 to S4.
1 The abbreviations used are:
- TEX101
- testis-expressed sequence 101 protein
- ABC
- Ammonium bicarbonate
- BCA
- Bicinchoninic acid
- BMGY
- Buffered complex glycerol medium
- BMMY
- Buffered complex methanol
- DFP
- Diflunisal phosphate
- DOC
- Deoxycholate
- DTT
- Dithiothreitol
- ELISA
- Enzyme-linked immunosorbent assay
- Fc
- constant region of immunoglobulin
- FWHM
- Full width at half maximum
- GPI
- glycosylphosphatidylinositol
- HAT
- Hypoxanthine-aminopterin-thymidine medium
- HRP
- Horseradish peroxidase
- KLK
- Kallikrein
- MS
- Mass spectrometry
- NOA
- Non-obstructive azoospermia
- OA
- Obstructive azoospermia
- PBS
- Phosphate-buffered saline
- PSA
- Prostate-specific antigen
- PV
- Post-vasectomy
- SP
- seminal plasma
- SRM
- Selected reaction monitoring
- TBS
- Tris-buffered saline.
REFERENCES
- 1. Weiner L. M., Surana R., Wang S. (2010) Monoclonal antibodies: versatile platforms for cancer immunotherapy. Nat. Rev. Immunol. 10, 317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lambert J. P., Ivosev G., Couzens A. L., Larsen B., Taipale M., Lin Z. Y., Zhong Q., Lindquist S., Vidal M., Aebersold R., Pawson T., Bonner R., Tate S., Gingras A. C. (2013) Mapping differential interactomes by affinity purification coupled with data-independent mass spectrometry acquisition. Nat. Methods 10, 1239–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Batruch I., Lecker I., Kagedan D., Smith C. R., Mullen B. J., Grober E., Lo K. C., Diamandis E. P., Jarvi K. A. (2011) Proteomic analysis of seminal plasma from normal volunteers and postvasectomy patients identifies over 2000 proteins and candidate biomarkers of the urogenital system. J. Proteome Res. 10, 941–953 [DOI] [PubMed] [Google Scholar]
- 4. Drabovich A. P., Jarvi K., Diamandis E. P. (2011) Verification of male infertility biomarkers in seminal plasma by multiplex selected reaction monitoring assay. Mol. Cell. Proteomics 10, M110 004127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Drabovich A. P., Dimitromanolakis A., Saraon P., Soosaipillai A., Batruch I., Mullen B., Jarvi K., Diamandis E. P. (2013) Differential diagnosis of azoospermia with proteomic biomarkers ECM1 and TEX101 quantified in seminal plasma. Sci. Transl. Med. 5, 212ra160. [DOI] [PubMed] [Google Scholar]
- 6. Djureinovic D., Fagerberg L., Hallstrom B., Danielsson A., Lindskog C., Uhlen M., Ponten F. (2014) The human testis-specific proteome defined by transcriptomics and antibody-based profiling. Mol. Hum. Reprod. 20, 476–488 [DOI] [PubMed] [Google Scholar]
- 7. Fujihara Y., Okabe M., Ikawa M. (2014) GPI-anchored protein complex, LY6K/TEX101, is required for sperm migration into the oviduct and male fertility in mice. Biol. Reprod. 90, 60. [DOI] [PubMed] [Google Scholar]
- 8. Fujihara Y., Tokuhiro K., Muro Y., Kondoh G., Araki Y., Ikawa M., Okabe M. (2013) Expression of TEX101, regulated by ACE, is essential for the production of fertile mouse spermatozoa. Proc. Natl. Acad. Sci. U.S.A. 110, 8111–8116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li W., Guo X. J., Teng F., Hou X. J., Lv Z., Zhou S. Y., Bi Y., Wan H. F., Feng C. J., Yuan Y., Zhao X. Y., Wang L., Sha J. H., Zhou Q. (2013) Tex101 is essential for male fertility by affecting sperm migration into the oviduct in mice. J. Mol. Cell. Biol. 5, 345–347 [DOI] [PubMed] [Google Scholar]
- 10. Kuzmanov U., Jiang N., Smith C. R., Soosaipillai A., Diamandis E. P. (2009) Differential N-glycosylation of kallikrein 6 derived from ovarian cancer cells or the central nervous system. Mol. Cell. Proteomics 8, 791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Christopoulos T. K., Diamandis E. P. (1992) Enzymatically amplified time-resolved fluorescence immunoassay with terbium chelates. Anal. Chem. 64, 342–346 [DOI] [PubMed] [Google Scholar]
- 12. Romanos M. A., Scorer C. A., Clare J. J. (1992) Foreign gene expression in yeast: a review. Yeast 8, 423–488 [DOI] [PubMed] [Google Scholar]
- 13. Flesch F. M., Voorhout W. F., Colenbrander B., van Golde L. M., Gadella B. M. (1998) Use of lectins to characterize plasma membrane preparations from boar spermatozoa: a novel technique for monitoring membrane purity and quantity. Biol. Reprod. 59, 1530–1539 [DOI] [PubMed] [Google Scholar]
- 14. Kasvandik S., Sillaste G., Velthut-Meikas A., Mikelsaar A. V., Hallap T., Padrik P., Tenson T., Jaakma U., Koks S., Salumets A. (2015) Bovine sperm plasma membrane proteomics through biotinylation and subcellular enrichment. Proteomics, Epub 2015 Jan 20 [DOI] [PubMed] [Google Scholar]
- 15. Bassett M. E., Thornton D. J., Sheehan J. K., Nieduszynski I. A. (1987) An enzyme-linked immunosorbent assay (ELISA) of denatured cartilage link protein. Biochim. Biophys. Acta 925, 347–355 [DOI] [PubMed] [Google Scholar]
- 16. Geumann C., Gronborg M., Hellwig M., Martens H., Jahn R. (2010) A sandwich enzyme-linked immunosorbent assay for the quantification of insoluble membrane and scaffold proteins. Anal. Biochem. 402, 161–169 [DOI] [PubMed] [Google Scholar]
- 17. Lechtzier V., Hutoran M., Levy T., Kotler M., Brenner T., Steinitz M. (2002) Sodium dodecyl sulphate-treated proteins as ligands in ELISA. J. Immunol. Methods 270, 19–26 [DOI] [PubMed] [Google Scholar]
- 18. Bumgarner G. W., Zampell J. C., Nagarajan S., Poloso N. J., Dorn A. S., D'Souza M. J., Selvaraj P. (2005) Modified cell ELISA to determine the solubilization of cell surface proteins: Applications in GPI-anchored protein purification. J. Biochem. Biophys. Methods 64, 99–109 [DOI] [PubMed] [Google Scholar]
- 19. Lee J. K., Ahn K. C., Park O. S., Kang S. Y., Hammock B. D. (2001) Development of an ELISA for the detection of the residues of the insecticide imidacloprid in agricultural and environmental samples. J. Agric. Food Chem. 49, 2159–2167 [DOI] [PubMed] [Google Scholar]
- 20. Rehan M., Younus H. (2006) Effect of organic solvents on the conformation and interaction of catalase and anticatalase antibodies. Int. J. Biol. Macromol. 38, 289–295 [DOI] [PubMed] [Google Scholar]
- 21. Russell A. J., Trudel L. J., Skipper P. L., Groopman J. D., Tannenbaum S. R., Klibanov A. M. (1989) Antibody-antigen binding in organic solvents. Biochem. Biophys. Res. Commun. 158, 80–85 [DOI] [PubMed] [Google Scholar]
- 22. Doucet J., Canadi J., Kalis C., Valentin M. A., Marrony S., Deckert-Salva F., Legay F., Avrameas A. (2009) Reduction of matrix interferences by the combination of chaotropic salt and DMSO in a broadly applicable target-based ELISA for pharmacokinetic studies of therapeutic monoclonal antibodies. J. Pharm. Biomed. Anal. 50, 924–931 [DOI] [PubMed] [Google Scholar]
- 23. Tybor P. T., Bryant J. N., Dill C. W., Landmann W. A. (1970) Heat denaturation of blood serum proteins measured in saturated sodium chloride. J. Agric. Food Chem. 18, 629–631 [DOI] [PubMed] [Google Scholar]
- 24. Verdoliva V., Senatore C., Polci M. L., Rossi S., Cordella M., Carlucci G., Marchetti P., Antonini-Cappellini G., Facchiano A., D'Arcangelo D., Facchiano F. (2013) Differential denaturation of serum proteome reveals a significant amount of hidden information in complex mixtures of proteins. PLoS One 8, e57104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Erdile L. F., Smith D., Berd D. (2001) Whole cell ELISA for detection of tumor antigen expression in tumor samples. J. Immunol. Methods 258, 47–53 [DOI] [PubMed] [Google Scholar]
- 26. Drabovich A. P., Martinez-Morillo E., Diamandis E. P. (2014) Toward an integrated pipeline for protein biomarker development. Biochim. Biophys. Acta, Epub 2014 Sept 11 [DOI] [PubMed] [Google Scholar]
- 27. Cheung W. C., Beausoleil S. A., Zhang X., Sato S., Schieferl S. M., Wieler J. S., Beaudet J. G., Ramenani R. K., Popova L., Comb M. J., Rush J., Polakiewicz R. D. (2012) A proteomics approach for the identification and cloning of monoclonal antibodies from serum. Nat. Biotechnol. 30, 447–452 [DOI] [PubMed] [Google Scholar]
- 28. Schoenherr R. M., Zhao L., Whiteaker J. R., Feng L. C., Li L., Liu L., Liu X., Paulovich A. G. (2010) Automated screening of monoclonal antibodies for SISCAPA assays using a magnetic bead processor and liquid chromatography-selected reaction monitoring-mass spectrometry. J. Immunol. Methods 353, 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Drabovich A. P., Saraon P., Jarvi K., Diamandis E. P. (2014) Seminal plasma as a diagnostic fluid for male reproductive system disorders. Nat. Rev. Urol. 11, 278–288 [DOI] [PubMed] [Google Scholar]
- 30. Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S., Wernerus H., Bjorling L., Ponten F. (2010) Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 28, 1248–1250 [DOI] [PubMed] [Google Scholar]
- 31. Alm T., von Feilitzen K., Lundberg E., Sivertsson A., Uhlen M. (2014) A chromosome-centric analysis of antibodies directed toward the human proteome using Antibodypedia. J. Proteome Res. 13, 1669–1676 [DOI] [PubMed] [Google Scholar]
- 32. Ackermann B. L., Berna M. J. (2007) Coupling immunoaffinity techniques with MS for quantitative analysis of low-abundance protein biomarkers. Expert Rev. Proteomics 4, 175–186 [DOI] [PubMed] [Google Scholar]
- 33. Nelson R. W., Borges C. R. (2011) Mass spectrometric immunoassay revisited. J. Am. Soc. Mass Spectrom. 22, 960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yassine H., Borges C. R., Schaab M. R., Billheimer D., Stump C., Reaven P., Lau S. S., Nelson R. (2013) Mass spectrometric immunoassay and MRM as targeted MS-based quantitative approaches in biomarker development: potential applications to cardiovascular disease and diabetes. Proteomics Clin. Appl. 7, 528–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anderson N. L., Anderson N. G., Haines L. R., Hardie D. B., Olafson R. W., Pearson T. W. (2004) Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA). J. Proteome Res. 3, 235–244 [DOI] [PubMed] [Google Scholar]
- 36. Whiteaker J. R., Zhao L., Zhang H. Y., Feng L. C., Piening B. D., Anderson L., Paulovich A. G. (2007) Antibody-based enrichment of peptides on magnetic beads for mass-spectrometry-based quantification of serum biomarkers. Anal. Biochem. 362, 44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Keshishian H., Addona T., Burgess M., Kuhn E., Carr S. A. (2007) Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics 6, 2212–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nicol G. R., Han M., Kim J., Birse C. E., Brand E., Nguyen A., Mesri M., FitzHugh W., Kaminker P., Moore P. A., Ruben S. M., He T. (2008) Use of an immunoaffinity-mass spectrometry-based approach for the quantification of protein biomarkers from serum samples of lung cancer patients. Mol. Cell. Proteomics 7, 1974–1982 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.