Abstract
The HIV epidemic in Russia, one of the world's fastest growing, has been concentrated mostly among people who inject drugs (PWID). We sought to explore the epidemiology of the epidemic in St. Petersburg by sampling from the highest risk groups of PWID and men who have sex with men (MSM) and use viral sequencing data to better understand the nature of the city's epidemic. Serological testing confirmed an HIV prevalence among PWID in excess of 40%. All but 1 of 110 PWID whose blood samples were tested for genetic diversity were infected by subtype A virus, specifically by the AFSU strain. The remaining person was infected with a CRF-06cpx recombinant. Analysis of pairwise genetic distance among all PWID studied revealed an average of 3.1% sequence divergence, suggesting clonal introduction of the AFSU strain and/or constraints on sequence divergence. The HIV prevalence was less than 10% among MSM. All 17 sequences from HIV-infected MSM were found to be a clade B virus with a much higher average sequence diversity of 15.7%. These findings suggest two independent epidemics with little overlap between the two highest at-risk populations, which will require different HIV prevention approaches.
Introduction
Beginning in 1996, Russia has witnessed a rapidly expanding HIV epidemic. The epidemic began in the westernmost part of Russia and expanded rapidly.1,2 Newly diagnosed cases reported to the Russian Federal AIDS Center (RFAC) increased every year between 1996 and 2001, totaling more than 87,000 cases.3,4 Since then, an average of 40,000 new cases have been reported annually, with the number of new diagnoses exceeding 700,000 at the end of 2012. This makes Russia the only country outside of sub-Saharan African with the highest population prevalence of HIV (RFAC data, http://hivrussia.ru/files/bul_38.pdf). Throughout this period, unsafe injection drug use has been the predominant mode of HIV transmission.5 Although there appears to be some expansion of the epidemic beyond people who inject drugs (PWID), two-thirds of newly diagnosed infections since 1997 were among people officially identified as PWID.6
The city of St. Petersburg has been among those most heavily affected, with around 55,000 cases being diagnosed (RFAC data, http://hivrussia.ru/files/bul_38.pdf). MSM HIV transmission in St. Petersburg has been observed since the beginning of the epidemic in Russia and initially represented intermittent introductions of HIV subtype B from Western Europe or the United States.1,7,8 Limited data are available about the HIV prevalence in MSM. One study of 237 MSM recruited from gay clubs in St. Petersburg found a prevalence of 3.8%,9 and another study of 401 subjects showed the prevalence of about 6%.10 HIV transmission among PWID was first noted in 1998 and currently more than three-quarters of all registered infections in St. Petersburg (totaling approximately 40,000) are among the estimated 83,000 PWID.11 The HIV prevalence among PWID increased linearly from 2% in 1998 to 30% in 2003, and reached 59% in the most recent studies.12–16 Respondent-driven sampling (RDS), a chain referral sampling method commonly used to reach hidden populations, has become the most popular method to recruit samples of PWID and MSM in Russia.17,18 These populations have been studied separately; however, little is known about crossover between these groups.15
The goal of this particular analysis was to explore the molecular epidemiology of HIV-1 among PWID and MSM who were recruited simultaneously using a dual high-risk, peer-driven chain referring sampling scheme.19 By allowing PWID to recruit MSM and vice versa, we maximized the likelihood of detecting HIV-1 transmission between these two groups.
Materials and Methods
Clinical samples
The Sexual Acquisition and Transmission of HIV-Cooperative Agreement Program (SATH-CAP) was an NIDA-funded, cross-sectional study of high-risk groups (PWID, MSM, and sex partners of both groups) recruited in 2006–2008 by a modified form of RDS.19 Individuals who belong to the core populations of drug users and MSM and met the inclusion criteria for the study were recruited as seeds and recruitment continued to create chains of individuals in the core risk group populations that expand beyond the seed's own drug using or MSM network. In addition, the members of the core risk groups were instructed and encouraged to recruit their sexual partners who were not members of the core risk groups into the study.
All study procedures were approved by institutional review boards at The Biomedical Center, Yale University, and RAND Corporation.
Recruited participants completed structured interviews and answered detailed questions about their drug use and sexual behaviors, their HIV testing history, and their social and drug using networks. The participants were classified into several major categories: (1) people who inject drugs (PWID), (2) people who use noninjectable drugs (PWUD), (3) men who have sex with men (MSM), (4) combined groups (MSM/PWUD and MSM/PWID), and (5) sex partners (non PWID/PWUD/MSM).
After pretest counseling, blood samples were taken to ascertain their HIV status by an enzyme-linked immunoassay (EIA) that was confirmed by Western blot. Among those individuals who were found to be HIV positive, the incidence of recent infection cases was estimated using a BED Capture Enzyme Immunoassay (Calypte Inc.). However, since the BED assay is known for overestimation of the incidence cases, we adjusted the BED results using participants' self-reported data. Those individuals who were aware of their HIV-positive status for 6 months and longer were removed from the group of BED-determined recent infection cases.
The participants who knew their HIV-positive status for 1 year or longer were assigned to the subgroup of chronically infected cases.
Genotyping and phylogenetic analysis
Aliquots of blood were stored at −80°C until viral RNA was extracted from serum samples using a Viral RNA Extraction kit (Qiagen). Reverse transcriptase polymerase chain reactions (RT-PCR) were carried out to amplify the env V3 region. For this purpose, we used either a one-step or two-step protocol, with a One-Step RT-PCR kit (Qiagen) or SuperScript III and Platinum Taq polymerases (Invitrogen), respectively. Viral RNA was extracted from 240 μl of sera. For the one-step reaction, we took 25 μl of extracted RNA. For cDNA synthesis in the two-step protocol, we used 15 μl of RNA. As a template for the first round of PCR, we took 1 μl of cDNA. The second round of PCR was carried out only for the env region; 1 μl of the first round mix was used as a template. The following primer sequences were used: for the first round of PCR—ES7-1 (5′-AATGTCAGCACAGTACARTGYACNCA-3′, HXB2 6945–6970) and ES8-1 (5′-CCATAGTRCTTCCTGCTGYTCCYAAGA-3′, HXB2 7789–7815); for the second round of PCR and sequencing—ES7 (5′-CTGTTAAATGGCAGTCTAGC-3′, HXB2 7002–7021) and ES8 (5′-CACTTCTCCAATTGTCCCTCA-3′, HXB2 7648–7668).
PCR fragments were subjected to direct sequencing using MegaBACE1000 sequencer (Amersham Biosciences) using the DYEnamic ET Dye Terminator Cycle Sequencing Kit (Amersham Biosciences) according to the manufacturer's protocol. The sequence data were analyzed using Sequencher 4.7 and nucleotide sequences were edited and assembled using the BioEdit 7.0.5.2 software (www.mbio.ncsu.edu/BioEdit/bioedit.html). Our sequences were aligned with the subtype-specific reference sequences obtained from the Los Alamos HIV Database (www.hiv.lanl.gov).
Phylogenetic analysis was conducted using MEGA 5.2.20 Neighbor-joining trees were tested by the bootstrap method (500 bootstrap replications) using the Kimura two-parameter model. A viral cluster with a bootstrap value of 60 or more was considered as a potential transmission cluster.
The nucleotide and amino acid diversity was calculated using the p-distance model in MEGA. The differences between the groups were assessed using the Mann–Whitney test and plotted in Prism 6.0 (GraphPad Software, La Jolla, CA).
All HIV-1 env nucleotide sequences were submitted to the GenBank database under accession numbers FJ233960, FJ233974-FJ233991, FJ233993, FJ233997, FJ233999-FJ234004, FJ234006, FJ234008-FJ234009, FJ234011-FJ234016, FJ234018-FJ234020, FJ234022-FJ234024, FJ234026-FJ234028, FJ234031-FJ234062, KP319348-KP319379 for subtype A and KP100401-KP100417 for subtype B.
Results
The recruitment of the study subjects was conducted in 2006–2008. The HIV prevalence in the PWID group was 45.2% and in the MSM group was 8%. Although allowing for cross referrals between the MSM and PWID groups, we found them to be an infrequent event. Only 1.6% of PWID were recruited by MSM and 4.0% of MSM were recruited by PWID. Furthermore, only 22 individuals (2.4%) were jointly PWID and MSM and 10 of them (45.5%) were HIV positive (Table 1).
Table 1.
Demographic Characteristics of the Study Participants and HIV Testing Results
| Risk groups | Number of subjects | Number of HIV+ subjects (prevalence) | Gender | Age, mean (range) |
|---|---|---|---|---|
| Men who have sex with men (MSM) | 224 | 18 (8.0%) | M (100%) | 26 (18–58) |
| People who use noninjectable drugs (PWUD) | 18 | 3 (16.7%) | F (45%) M (55%) |
24 (18–42) |
| People who inject drugs (PWID) | 684 | 309 (45.2%) | F (26.9%) M (73.1%) |
29 (18–53) |
| MSM/PWUD | 7 | 0 (0.0%) | M (100%) | 22 (19–26) |
| MSM/PWID | 7 | 3 (42.9%) | M (100%) | 28 (23–36) |
| Sex partners, non-MSM/PWUD/PWID | 83 | 9 (10.8%) | F (63.9%) M (36.1%) |
28 (18–58) |
| Total | 1023 | 342 (33.4%) | F (24%) M (76%) |
28 (18–58) |
We genotyped viral strains from 127 individuals (110 from PWID and 17 from MSM) using a portion of the env gene in the V3 region. The final sets of sequences included the fragments with HXB2 coordinates of 7068–7564 bp for subtype A and 7137–7598 bp for subtype B. The number of analyzed MSM samples was significantly lower due to dramatic differences in prevalence (Table 1). Eight MSM blood samples came from the study participants recruited through RDS, and nine were obtained from the unrelated MSM patients of the City AIDS Center who did not report the use of injectable drugs. Two samples came from RDS-recruited men who reported both drug use (one person reported injectable and one reported single use of noninjectable drugs) and sex with other men. Broadly speaking, the sequence data revealed two distinct and nonoverlapping epidemics of HIV subtypes A and B.
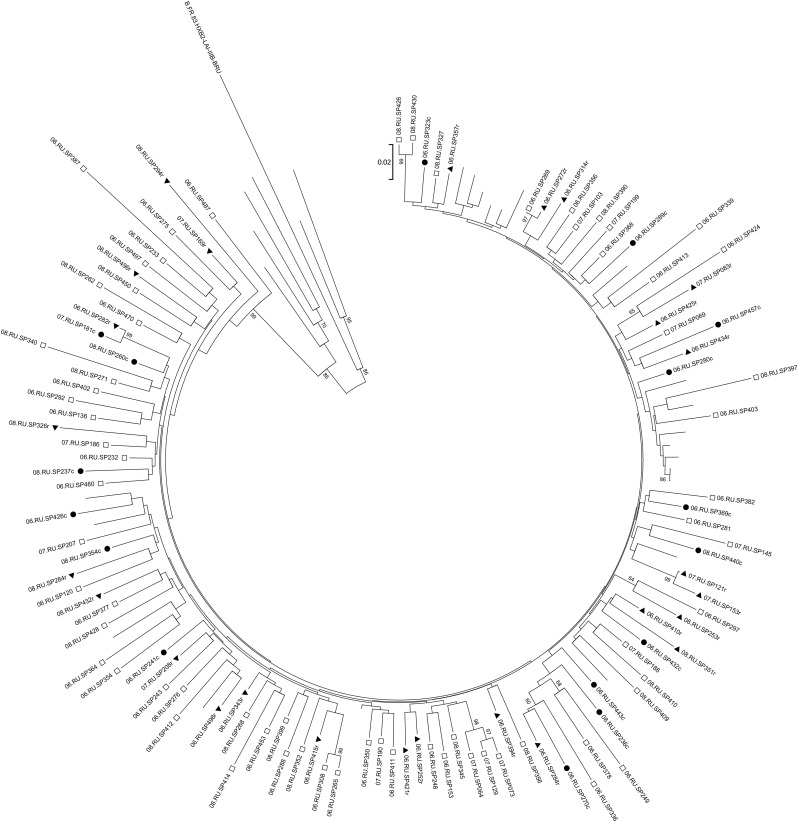
Among samples obtained from PWID, all but one individual harbored HIV of subtype A1 commonly observed in former Soviet Union (FSU) countries (Fig. 1). The one non-A1 sequence belonged to the family of complex recombinant CRF06_cpx and its subsequent recombinant with A1, CRF06-cpx/A1, and was analyzed for full-length env gene elsewhere.21 Bootstrapping analysis of the env sequences revealed nine pairs of sequences from PWID that were very closely related. Using the coupon system of RDS, we discerned that four of these pairs represented individuals who reported either sharing drugs or having sex with each other. In addition, three pairs were part of the same recruitment network.
FIG. 1.
Phylogenetic analysis of the plasma-derived env fragments of HIV-1 subtype A. Closed triangles mark the sequences obtained from blood samples of people who inject drugs (PWID) who were HIV positive for less than 6 months (recent infection cases); closed circles show the sequences obtained from blood samples of PWID who were HIV positive for 12 month or more (chronic infection cases); open squares represent the strains obtained from PWID with an unknown duration of infection. For convenience, all titles of SATH-CAP sequences start with the two digits of the sampling year (2006, 07, or 08); titles of recent and chronic infection cases end with r or c, respectively. Unlabeled branches show the subtype A reference strains.
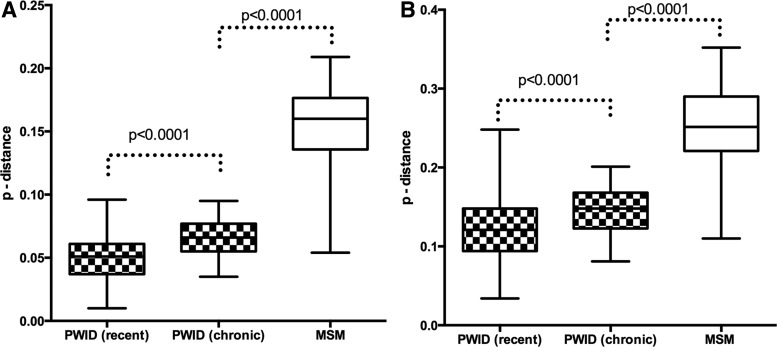
From among the 110 PWID whose HIV was amenable to sequencing, we used two methods—self-reported data on HIV test history and the BED assay that detects individuals recently infected with HIV22 with high probability—to identify 27 individuals who by both assays were recently infected. All but one of the env sequences belonged to the AFSU subtype; the other one was the CRF06-cpx/A1 strain. The median pairwise genetic diversity for all subtype A sequences studied was 3%; the average was 3.1% (range 0–12.9%). Within the subgroup of 26 individuals recently infected with subtype AFSU the nucleotide sequence diversity ranged from 1% to 9.6% with the median value of 5.1%. In contrast, 16 individuals who reported being infected with AFSU at least 365 days before providing the sample we analyzed (subgroup of chronic infection) had pairwise genetic diversity that ranged from 3.5% to 9.5% with the higher median value of 6.6% (Fig. 2A). The median amino acid diversity for the recent infection subgroup was 12.1% (range 3.4–24.8%) and for the chronic infection subgroup was 14.8% (range 8.1–20%) (Fig. 2B). All the differences between the subgroups were statistically significant.
FIG. 2.
Nucleotide and amino acid diversity of HIV-1 subtype A and B env fragments. (A) Genetic diversity of recently transmitted subtype A strains is lower than that of persistent subtype A and B strains. Patterned bars represent the p-distance values obtained for the nucleotide sequences derived from two subgroups of subtype A: infected PWID and recent and chronic infection cases; open bar represents the values obtained for the nucleotide sequences of subtype B-infected men who have sex with men (MSM). (B) Amino acid diversity of recently transmitted subtype A strains is lower than that of persistent subtype A and B strains. Patterned bars represent the p-distance values obtained for the amino acid sequences derived from two subgroups of subtype A-infected PWID (recent and chronic infection cases); open bar represents the values obtained for the nucleotide sequences of subtype B-infected MSM.
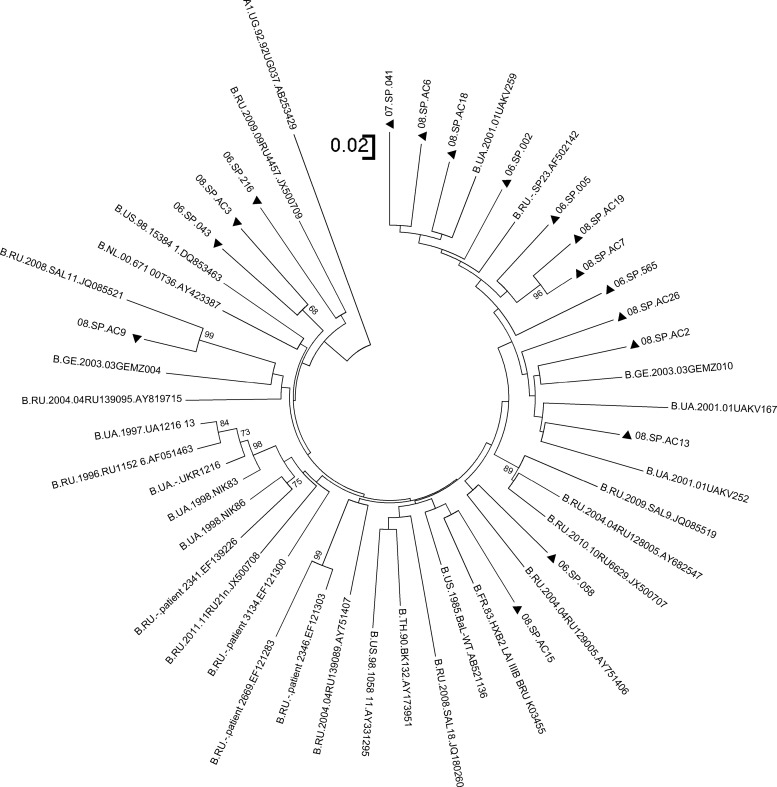
The epidemiological profile of the HIV-1 epidemic among MSM in St. Petersburg differs from the injection-related epidemic. All 17 blood samples obtained from MSM harbored HIV-1 strains of subtype B (Fig. 3). The pairwise genetic diversity of subtype B sequences ranged from 5.4% to 20.9% with a median value of 16% (Fig. 2). The median amino acid diversity for the MSM group was 25.1% (range 11–35.2%) (Fig. 2B).
FIG. 3.
Phylogenetic analysis of the plasma-derived env fragments of HIV-1 subtype B. Closed triangles indicate the sequences obtained from the blood samples of MSM within our study; other branches represent the subtype B reference strains and subtype A outgroup. For convenience, all titles of SATH-CAP sequences start with the two digits of the sampling year (2006, 07, or 08).
Of the two men who reported both having sex with other men and a history of drug use, one of them harbored the strains of subtype AFSU commonly observed in PWID while the other had HIV strains of subtype B.
No sequence comparisons for sexual partners or for individuals within the same recruitment chain were possible due to the low number of HIV-positive subjects among the recruited individuals.
Discussion
Our data suggested that PWID and MSM constitute largely, but not completely, separate populations. The low frequency of cross-referrals and the low number of men who reported both injection drug use and sex with other men have kept the two epidemics of HIV-1 largely separate. The limited genetic data from the two MSM who used drugs do suggest that if the overlap were greater, more mixing would have occurred.
The genetic diversity of the AFSU sequences remained relatively low even among individuals who had been, according to their reported testing histories, HIV positive for many years. HIV generally evolves rapidly once infection has been established,23 but that does not seem to be the case for the AFSU strain. In this study low genetic diversity was detected in cases of recent and chronic infection using the sequencing method that reveals only the predominant sequence within an infected individual. The genetic bottleneck was also observed studying the individual strains of transmitted virus in acutely infected PWID, when the analysis focused on the single copies of the full-length env gene.21
While we do not have sufficient information on the biological properties of the viruses to explain the low diversity, the behavioral patterns may shed some light on this phenomenon. Notably, the analysis of the history of drug use of our participants did not reveal any correlations between PWID experience (years of drug use) and recent HIV infection acquisition. Analyzing these data together with the genetic bottleneck phenomenon we may speculate that the PWID risk network structure may contribute to the likelihood of infection. Acute HIV-1 infection is associated with a high viral load and thus an increased likelihood of transmission. It is known that within the network PWID in St. Petersburg do share syringes and injection paraphernalia frequently,15,24,25 allowing the infection of the PWID network members to occur very rapidly after HIV-1 acquisition and viral load spike of a single member of this network. The homogeneous acute infection as a source of the epidemic may explain the low diversity of the virus circulating in the population.
Conclusions
The sequence data of the env genomic region demonstrate that two parallel epidemics of HIV exist within the city of St. Petersburg. The subtype found among MSM was subtype B unless they also reported injected drug use, in which case some species were subtype A, suggestive of transmission through unsafe injection, and others were subtype B, suggestive of transmission through unsafe sex. Given the high prevalence among PWID and the limited resources for prevention, prevention programs must first concentrate on the PWID epidemic. Indeed, epidemic modeling of the SATH-CAP data published elsewhere concludes that if transmission among PWID were stopped, there would be no further expansion into the heterosexual population of St. Petersburg.16 HIV-1 transmission within the acute infection window, which is characterized by high viremia and a high probability of transmission, should be studied in the context of PWID risk networks, and the specific surveillance systems for detection of acute HIV-1 infection that exists in many countries should be implemented in the health system of St. Petersburg. Programs for both primary prevention—to keep uninfected individuals from becoming infected—and secondary prevention—to reduce the transmission from those already infected, including people with acute HIV infection—need to be expanded to contain this growing epidemic.
Acknowledgments
This work was supported by grants from the U.S. National Institute on Drug Abuse (5U01DA017387). Support from the Fogarty International Center at the U.S. National Institutes of Health provided training for Dr. Dukhovlinova and Ms. Meringof (5D43TW001028 and 5U2RTW006893, respectively).
All the recruitment and laboratory work were conducted at the Biomedical Center. The data analysis was performed at Yale University. During the study period E.D., A.M., O.T., M.M., and T.S. were the staff members of the Biomedical Center and/or trainees at Yale University.
We thank the SATH-CAP participants and the Recruitment and Follow-Up group of the psychologists, social workers, and nurses of the Biomedical Center. We thank the St. Petersburg City AIDS Center and specifically Dr. Galina Volkova for providing additional blood samples of MSM.
We also acknowledge the assistance of the University of North Carolina Center for AIDS Research (P30 AI50410) in the preparation of the manuscript, and personally thank Prof. Ronald Swanstrom for his helpful comments and continuous support.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Kozlov AP, Volkova GV, Malykh AG, et al. : Epidemiology of HIV infection in St. Petersburg, Russia. J Acquir Immune Defic Syndr 1993; 6(2):208–212 [PubMed] [Google Scholar]
- 2.Bobkov A, Kazennova E, Khanina T, et al. : An HIV 1 subtype A strain of low genetic diversity continues to spread among injecting drug users in Russia: Study of the new local outbreaks in Moscow and Irkutsk. AIDS Res Hum Retroviruses 2001;17:257–261 [DOI] [PubMed] [Google Scholar]
- 3.Nabatov AA, Masharsky AE, Verevochkin SV, et al. : The rate of epidemiological and virological changes during the transition from nascent to concentrated HIV epidemic stage in the former Soviet Union countries. AIDS Res Hum Retroviruses 2007;23(2):183–192 [DOI] [PubMed] [Google Scholar]
- 4.AIDS Foundation East/West: HIV statistics for Belarus, Moldova, Russia, and Ukraine, 2008. (www.afew.org/knowledge-centre/hiv-statistics.)
- 5.UNAIDS Report on the Global AIDS Epidemic 2012: UNAIDS, Geneva, Switzerland [Google Scholar]
- 6.Goliusov AT, Dementyeva LA, Ladnaya NN, et al. : Federal Service for Surveillance of Consumer Rights Protection and Human Well-Being of the Russian Federation, UNAIDS; Moscow: 2010. Country Progress Report of the Russian Federation on the Implementation of the Declaration of Commitment on HIV/AIDS. Reporting period: January 2008–December 2009 [Google Scholar]
- 7.Bobkov AF, Kazennova EV, Selimova LM, et al. : Temporal trends in the HIV-1 epidemic in Russia: Predominance of subtype A. J Med Virol 2004;74(2):191–196 [DOI] [PubMed] [Google Scholar]
- 8.Thomson MM, de Parga EV, Vinogradova A, et al. : New insights into the origin of the HIV type 1 subtype A epidemic in former Soviet Union's countries derived from sequence analyses of preepidemically transmitted viruses. AIDS Res Hum Retroviruses 2007;23(12):1599–1604 [DOI] [PubMed] [Google Scholar]
- 9.World Health Organization: HIV prevalence and risks among men having sex with men in Moscow and St. Petersburg, 2007. (www.euro.who.int/__data/assets/pdf_file/0004/78547/E90854.pdf.)
- 10.Baral S., Sifakis F., Peryshkina A. et al. : Risks for HIV infection among gay, bisexual and other men who have sex with men in Moscow and St. Petersburg, Russia. AIDS Res Hum Retroviruses 2012;28:874–879 [DOI] [PubMed] [Google Scholar]
- 11.Heimer R. and White E: Estimation of the number of injection drug users in St. Petersburg, Russia. Drug Alcohol Depend 2010;109(1–3):79–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaboltas AV, Toussova OV, Hoffman IF, et al. : HIV prevalence, sociodemographic and behavioral correlates and recruitment methods among injection drug users in St. Petersburg, Russia. J AIDS 2006;41:657–663 [DOI] [PubMed] [Google Scholar]
- 13.Niccolai LM, Toussova OV, Verevochkin SV, et al. : High HIV prevalence, suboptimal HIV testing, and low knowledge of HIV-positive serostatus among injection drug users in St. Petersburg, Russia. AIDS Behav 2010;14(4):932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niccolai LM, Shcherbakova IS, Toussova OV, et al. : The potential for bridging of HIV transmission in the Russian Federation: Sex risk behaviors and HIV prevalence among drug users (DUs) and their non-DU sex partners. J Urban Health 2009;86(Suppl 1):131–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eritsyan K, Heimer R, Barbour R, et al. : Individual-level, network-level and city-level factors associated with HIV prevalence among people who inject drugs in eight Russian cities: A cross-sectional study. BMJ Open 2013;3(6):e002645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mills HL, White E, Colijn C, et al. : HIV transmission from drug injectors to partners who do not inject, and beyond: Modeling the potential for a generalized heterosexual epidemic in Saint Petersburg, Russia. Drug Alcohol Depend 2013;133:242–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heckathorn DD: Respondent-driven sampling; a new approach to the study of hidden populations. Soc Problems 1997;44:174–199 [Google Scholar]
- 18.Semaan S, Lauby J, and Liebman J: Street and network sampling in evaluation studies of HIV risk-reduction interventions. AIDS Rev 2002;4:213–223 [PubMed] [Google Scholar]
- 19.Iguchi MY, Ober AJ, Berry SH, et al. : Simultaneous recruitment of drug users and men who have sex with men in the United States and Russia using respondent-driven sampling: Sampling methods and implications. J Urban Health 2009;86(Suppl 1):5–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura K., Peterson D., Peterson N. et al. : Mega 5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011;28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masharsky AE, Dukhovlinova EN, Verevochkin SV, et al. : A substantial transmission bottleneck among newly and recently HIV-1-infected injection drug users in St. Petersburg, Russia. J Infect Dis 2010;201(11):1697–1702 [DOI] [PubMed] [Google Scholar]
- 22.Parekh BS, Kennedy MS, Dobbs T, et al. : Quantitative detection of increasing HIV type 1 antibodies after seroconversion: A simple assay for detecting recent HIV infection and estimating incidence. AIDS Res Hum Retroviruses 2002;18:295–307 [DOI] [PubMed] [Google Scholar]
- 23.Woodman Z. and Williamson C: HIV molecular epidemiology: Transmission and adaptation to human populations. Curr Opin HIV/AIDS 2009;4:247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozlov AP, Shaboltas AV, Toussova OV, et al. : HIV incidence and factors associated with HIV acquisition among injection drug users in St. Petersburg, Russia. AIDS 2006;20:901–916 [DOI] [PubMed] [Google Scholar]
- 25.Gyarmathy A, Li N, Tobin KE, et al. : Correlates of unsafe equipment sharing among injecting drug users in St. Petersburg, Russia. Eur Addict Res 2009;15(3):163–170 [DOI] [PMC free article] [PubMed] [Google Scholar]