Abstract
Giardia lamblia and Cryptosporidium species belong to a complex group of pathogens that cause diseases hampering development and socioeconomic improvements in the developing countries. Both pathogens are recognized as significant causes of diarrhea and nutritional disorders. However, further studies are needed to clarify the role of parasitic infections, especially asymptomatic infections in malnutrition and stunting. We developed a high-throughput multiplex quantitative polymerase chain reaction (qPCR) method for G. lamblia and Cryptosporidium spp. detection in stool samples. The sensitivity and specificity of the method were ensured by analyzing confirmed positive samples acquired from diagnostics laboratories and participating in an external quality control round. Its capability to detect asymptomatic G. lamblia and Cryptosporidium spp. infections was confirmed by analyzing stool samples collected from 44 asymptomatic 6-month-old infants living in an endemic region in Malawi. Of these, five samples were found to be positive for G. lamblia and two for Cryptosporidium spp. In conclusion, the developed method is suitable for large-scale studies evaluating the occurrence of G. lamblia and Cryptosporidium spp. in endemic regions and for clinical diagnostics of these infections.
Introduction
Giardia lamblia and Cryptosporidium species (spp.) are among the most common parasites inflicting gastroenteritis and are recognized as significant causes of diarrhea and nutritional disorders.1,2 Although numerous epidemiological studies have been conducted, there are still open questions regarding the contribution of parasitic infections, especially asymptomatic infections to malnutrition and stunting.3–7 To clarify these questions there is a clear need for a sensitive and cost-effective G. lamblia and Cryptosporidium spp. detection method suitable for analyzing a large number of clinical samples.
The traditional approach to diagnose G. lamblia and Cryptosporidium spp. infection is based on the detection of oocyst in stool samples. The most used diagnostic method is based on direct microscopy of the sample but it has downsides such as time-consuming and labor-intensive practice, high costs, and subjective interpretation of microscopic examination.8–10 Furthermore, the difficulty of detecting low numbers of oocysts limits its sensitivity. Immunoassays have also been used to detect parasites in stool samples but their sensitivity has not reached optimal levels and nonspecific cross-reactivity of detection antibodies have reduced their specificity.11 Quantitative polymerase chain reaction (qPCR) is considered to be more sensitive method compared with microscopy but needs a well-equipped laboratory to avoid cross-contaminations between samples. High sensitivity of qPCR-based detection technologies is a clear advantage in parasite diagnostics because the oocysts contain very small amount of DNA.9 However, the firm structure of the parasite stages, which are shed in stool, make them resistant to detergents creating a challenge in DNA extraction for PCR. Different stool sample pretreatment strategies have to be applied to disrupt the oocyst wall to release the DNA for extraction. Various versions of freeze-thaw cycle protocols have been used as well as proteinases, which have worked well for the oocyst wall layers of Cryptosporidium spp.12–15 Heat shock is one additional commonly used treatment, and has usually been combined with other kinds of pretreatments.10,16
In this study, we compared various pretreatment methods and finally we developed a high-throughput DNA extraction and PCR-based amplification protocol suitable for sensitive detection of G. lamblia and Cryptosporidium spp. in stool samples in a large-scale study setting.
Materials and Methods
Samples.
The method was optimized using formalin-fixed stool samples positive for G. lamblia or Cryptosporidium spp. United Kingdom Cryptosporidium Reference Unit (CRU; Public Health Wales Microbiology ABM, Singleton Hospital, Swansea, United Kingdom) provided four samples positive for Cryptosporidium spp. (one C. parvum and three C. hominis samples). On the basis of preliminary analysis, one C. hominis-positive sample was chosen for further optimization of the method. G. lamblia-positive sample was obtained from the Fimlab Laboratories (Pirkanmaa Hospital District, Tampere, Finland). In addition, the final evaluation of the optimized G. lamblia and Cryptosporidium spp. qPCR methods was done by participating in the external QCMD (Quality Control for Molecular Diagnostics: Parasitic Gastroenteritis panel 2013) quality control round, containing unfixed Cryptosporidium spp. and G. lamblia positive samples in addition to negative control and Entamoeba spp. positive samples (http://www.qcmd.org). An epidemic stool sample confirmed to be C. parvum positive by microscopy (HUSLAB, Finland) was also tested.
To find out whether the developed method could detect any asymptomatic infections in children living in high-risk region, stool samples collected from 44 6-month-old Malawian infants between January and June 2008 were analyzed. A short sedimentation step was added to the optimized protocol for these samples to prevent the clogging of extraction columns by the large particles found in these samples (sand etc.). The children were participants in the Lungwena Child Nutrition Intervention Study (registration no.: NCT00524446) evaluating the influence of dietary interventions on early childhood growth.17 The subjects were healthy 6-month-old children recruited from general population and 45% of them were male (20/44). Guardians collected a stool sample from the participants and brought it within 24 hours to the health center. The samples were then stored at −20°C and shipped later for long-term storage at −80°C. The guardians of all participating children gave a written informed consent and the trial adhered to Malawian regulatory guidelines and the principles of the Declaration of Helsinki. The trial protocol was approved by the College of Medicine's research and ethics committee (University of Malawi) and the ethical committee of the Pirkanmaa Hospital District, Finland.
Sample pretreatment.
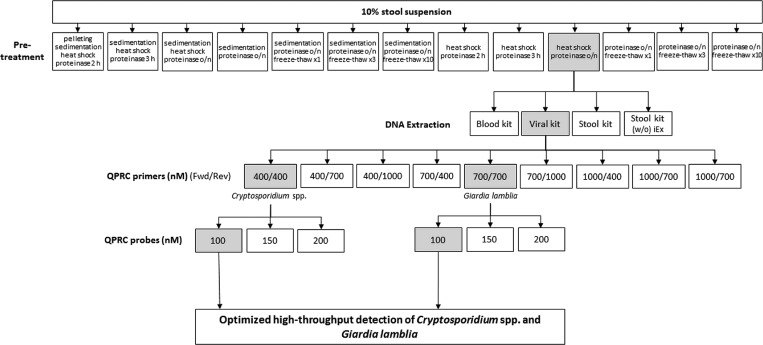
A total of 10% (w/v) stool suspensions were made in 0.2% bovine serum albumin in Hank's buffer. The sample was concentrated either by pelleting the oocysts by centrifugation followed by pellet resuspension or by sedimentation method that was based on sedimentation during a 1-hour incubation time in room temperature.14,18 In the next step, different methods were compared, which disrupt the oocysts, such as exposure to combinations of proteinase K treatment, freeze-thaw cycles, and heat shock treatment (10 minutes at +98°C). The tested sample pretreatment protocols are summarized in Figure 1.
Figure 1.
Summary of the Cryptosporidium spp. and Giardia lamblia detection method optimization. The selected options are highlighted with gray. Quantitative polymerase chain reaction (qPCR) primer and probe concentrations were tested separately for both parasites and combined into a multiplex reaction. Stool kit (w/o iEx) = stool kit without the inhibitEx tablet included in the kit; Fwd = forward (primer); Rev = reverse (primer).
DNA extraction and qPCR.
Three kits were tested for DNA extraction: QIAamp DNA Blood Mini Kit, QIAamp DNA Stool Mini Kit, and QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany). The QIAamp DNA Stool Mini Kit was also tested without the “inhibitEx” tablet provided in the kit. DNA extraction was performed from 10-fold dilution series of non-pretreated parasite positive samples together with negative control samples that were added both in the extraction and PCR stages. Parasite DNA was detected in each sample dilution using qPCR and the efficiency of amplification was evaluated based on CT-values. The primers and probe used for G. lamblia detection were designed to recognize and amplify a 62-bp fragment of the small subunit (SSU) rRNA gene (GenBank accession no. M54878) and the Cryptosporidium spp. primer-probe set was targeted against a 70-bp fragment of DNAJ-like protein gene of both C. parvum and C. hominis (GenBank accession no. AF177278.1/XM661034.1) (Table 1).11,19 QuantiTect Probe PCR kit (Qiagen, Hilden, Germany) and Applied Biosystems 7900HT were used in all qPCR analysis (LifeTech, Paisley, United Kingdom). The primer and probe concentrations were first optimized according to the qPCR kit manufacturer's instructions separately for both parasites and combined later into a multiplex reaction. In the final test format, both the DNA extraction and qPCR steps were done using 96-well plate format.
Table 1.
| Giardia lamblia | |
| Forward primer | GAC GGC TCA GGA CAA CGG TT |
| Reverse primer | TTG CCA GCG GTG TCC G |
| Probe | FAM-CCC GCG GCG GTC CCT GCT AG-MGB |
| Cryptosporidium spp. | |
| Forward primer | CTT TTT ACC AAT CAC AGA ATC ATC AGA |
| Reverse primer | TGT GTT TGC CAA TGC ATA TGA A |
| Probe | VIC-TCG ACT GGT ATC CCT ATA A-MGB |
FAM = fluorescein amidite; MGB = minor groove binder; qPCR = quantitative polymerase chain reaction; VIC is a proprietary name of Lifetech.
Results
Optimization of the method.
The ability of different stool sample pretreatments to release parasite DNA differed clearly from each other. The pretreatment protocol combining heat shock and overnight proteinase treatment, and the protocol including pelleting, heat shock, and 2-hour proteinase treatment were the most efficient ones and their sensitivity to detect parasite DNA did not significantly differ (data not shown). The protocol with heat shock followed by overnight proteinase treatment was chosen for further development (Figure 1). QIAamp Viral RNA Mini Kit was found to be the most efficient method to extract parasite DNA for qPCR whereas the QIAamp DNA Stool Mini Kit was the least efficient (Table 2). The PCR inhibitor absorbing “InhibitEx” tablet provided in the QIAamp DNA Stool Mini Kit slightly improved its performance (Table 2). The ability of the optimized methods to detect both parasites was evaluated by participating in external quality control round (QCMD). Both parasites were detected in all QCMD panel samples correctly and no wrongly positive results were obtained either (Table 3). The optimized assay detected C. parvum also in a stool sample confirmed to be positive by microscopy in a routine diagnostic laboratory (HUSLAB, Finland) (Table 3).
Table 2.
The ability of different kits to extract Giardia lamblia DNA for qPCR
| Sample dilution (log10) | Blood kit | Viral kit | Stool kit | Stool kit (NT)‡ | ||||
|---|---|---|---|---|---|---|---|---|
| CT value | SD | CT value | SD | CT value | SD | CT value | SD | |
| −1 | 27.8 | 0.2 | 26.7 | 0.6 | 32.1 | 0.3 | 35.0 | 0.3 |
| −2 | 32.5 | 0.7 | 29.2 | 0.1 | 39.0 | 1.5 | 38.2† | 3.9† |
| −3 | 37.7* | 0.1* | 34.1 | 0.8 | nd | − | nd | − |
| −4 | nd | − | 40.2* | 3.0* | nd | − | nd | − |
| −5 | nd | − | nd | − | nd | − | nd | − |
| −6 | nd | − | nd | − | nd | − | nd | − |
Blood kit = QIAamp DNA Blood Mini Kit, Viral kit = QIAamp Viral RNA Mini Kit, SD = standard deviation; Stool kit = QIAamp DNA Stool Mini Kit.
The extraction kits were tested with 10 times dilution series of G. lamblia DNA. All dilutions were processed as three parallel samples following three qPCR reactions per sample. The results are presented as mean CT values and standard deviations. For negative samples, the CT value of 45 was used for the calculations based on the number of cycles used in the PCR reaction. The mean CT and SD values are calculated from the reactions that the product was detected in.
In 33% of the reactions no product was detected.
In 66% of the reactions no product was detected.
The stool kit was also tested without the InhibitEx tablets included in the kit (NT).
Table 3.
Giardia lamblia and Cryptosporidium spp. positivity in external quality control samples (QCMD), an epidemic C. parvum sample confirmed to be positive by microscopy and samples collected from asymptomatic 6-month-old Malawian infants analyzed with the optimized qPCR method
| Sample content | G. lamblia PCR | Cryptosporidium spp. PCR |
|---|---|---|
| External quality control round (QCMD)* | ||
| Negative control | Negative | Negative |
| Entamoeba histolytica | Negative | Negative |
| C. parvum/hominis | Negative | Positive |
| C. parvum/hominis (low concentration) | Negative | Positive |
| E. dispar | Negative | Negative |
| E. histolytica | Negative | Negative |
| G. lamblia | Positive | Negative |
| G. lamblia | Positive | Negative |
| Epidemic sample positive in microscopy | ||
| C. parvum | Negative | Positive |
| 44 stool samples from endemic region (Malawi) | ||
| Prevalence | 4.5% | 11.4% |
qPCR = quantitative polymerase chain reaction, QCMD = Quality Control for Molecular Diagnostics.
QCMD: Parasitic Gastroenteritis panel 2013, QAP124154. The sample analysis was blinded.
Analysis of stool samples from Malawian infants.
Altogether five (11.4%) of the stool samples collected from asymptomatic Malawian infants were positive for Cryptosporidium spp. and two (4.5%) were positive for G. lamblia (Table 3). None of the samples was positive for both parasites.
Discussion
The aim was to develop a multiplex high-throughput qPCR method that is suitable for the detection of G. lamblia and Cryptosporidium spp. in stool samples in large-scale epidemiological studies and in diagnostic laboratories. All steps of the assay including sample pretreatment, DNA extraction, and qPCR were systematically optimized to reach maximal sensitivity and to adapt the assay to a high-throughput 96-well format. The fully optimized assay detected these parasites in both external QC samples and in clinical sample series.
Two of the tested pretreatment protocols allowed equally sensitive and reproducible detection of these parasites by qPCR. The protocol with heat shock and overnight proteinase treatment was chosen since it was more applicable to the high-throughput format compared with the other efficient protocol that included pelleting, heat shock, and 2-hour proteinase treatment. Four different DNA extraction methods were tested, and surprisingly, the QIAamp DNA Stool Mini Kit that is specifically designed for DNA extraction from stool samples and is often used as a default solution when performing stool sample DNA extraction was less efficient in purifying the parasite DNA in comparison to the other tested kits. In fact, similar results have been obtained by others as well.15,20
The high sensitivity of PCR is a clear advantage in the detection of parasites from stool samples. However, this makes PCR also prone to contamination that can happen if a positive sample contaminates negative samples that are analyzed in the same test run, or if the laboratory is contaminated by PCR products from previous amplifications. We used negative control samples in every step of the method (pretreatment, extraction, and PCR) to detect if any contamination occurred but none was found. Also, no false negative results were detected in the external quality control samples (QCMD). In addition, the quality of the detection was also confirmed by using known G. lamblia and Cryptosporidium spp. positive samples as positive control in each test run. Even if our assay was not affected by contaminations, it is important to pay attention to this issue when organizing sample collection and handling as well as the laboratory work for PCR-based parasite detection in stools.
Relatively high proportion of stool samples obtained from asymptomatic Malawian infants were found to be positive for these parasites (11.4% for Cryptosporidium spp. and 4.5% for G. lamblia). This finding, together with the result from external QCMD round, suggests that the method is sensitive. In fact, the sensitivity of PCR has been found to be superior compared with microscopy in several previous studies.11,14,19,21 The detection of several parasite positive children who had no symptoms of cryptosporidiosis or giardiasis suggests that the method can be applied for epidemiological studies in high-risk and endemic areas making it possible to estimate the background frequency and circulation of these parasites in the population. However, one limitation of this study is that we could not directly compare the detection rates by this PCR method to those using traditional microscopy. Therefore, further studies are needed to verify their relative sensitivity and specificity in different clinical materials including both symptomatic and asymptomatic patients.
The optimized protocol offers a multiplex high-throughput option for G. lamblia and Cryptosporidium spp. detection from human stool samples. The method is applicable for frozen and fixed samples, which eliminates the need to analyze the samples freshly. In addition, it is less labor-intensive compared with microscopy at least when large sample numbers need to be analyzed and the pretreatment phase is suitable for high-throughput format making it cost-effective for large screenings of G. lamblia and Cryptosporidium spp. and for clinical diagnostics. A qPCR machine is needed as a special equipment for setting up the method. The use of PCR technology also decreases the interindividual variation in the sample analysis compared with microscopy. In addition, quantification makes it possible to use the method for monitoring the efficacy of antiparasitic treatments and estimating disease severity.
In conclusion, the results suggest that this method may be suitable for clinical diagnostics of G. lamblia and Cryptosporidium spp. infections after further validation and is sensitive enough to detect even asymptomatic infections in background population in endemic regions. It offers significant advantages compared with traditional microscopic methods in terms of costs and speed of the diagnosis. Thus, this method provides an efficient tool to study the outcomes of both symptomatic and asymptomatic G. lamblia and Cryptosporidium spp. infections in clinical studies.
ACKNOWLEDGMENTS
We thank LCNI-5 study participants, their families, and the staff in Malawi, and Marie Hélou and Emily McKinney for the help in sample preparation. We also acknowledge the United Kingdom Cryptosporidium Reference Unit (CRU; Public Health Wales Microbiology ABM, Singleton Hospital, Swansea, United Kingdom), Fimlab Laboratories (Pirkanmaa Hospital District, Tampere, Finland), and HUSLAB (Finland) for providing us samples for the study.
Footnotes
Financial support: This study was financially supported by Academy of Finland (grants 200720, 108873, 111685, and 109796 [Per Ashorn]), Foundation for Pediatric Research in Finland, Nutriset Inc. (Malaunay, France), Päivikki and Sakari Sohlberg Foundation (Heikki Hyöty), Tampere Tuberculosis foundation (Heikki Hyöty). In addition, part of the study was funded by the DIABIMMUNE project (European Seventh Framework Programme HEALTH-F2-202063).
Authors' addresses: Noora Nurminen, Sami Oikarinen, and Heikki Hyöty, Department of Virology, School of Medicine, University of Tampere, Tampere, Finland, E-mails: noora.nurminen@uta.fi, sami.oikarinen@uta.fi, and heikki.hyoty@uta.fi. Rosa Juuti, EPID Research Oy, Espoo, Finland, E-mail: rosa.mattila@epidresearch.com. Yue-Mei Fan, Kirsi-Maarit Lehto, and Per Ashorn, Department for International Health, School of Medicine, University of Tampere, Tampere, Finland, E-mails: yuemei.fan@uta.fi, kirsi-maarit.lehto@uta.fi, and per.ashorn@uta.fi. Charles Mangani, Department for International Health, School of Medicine, University of Tampere, Tampere, Finland, and College of Medicine, University of Malawi, School of Public Health and Family Medicine, Blantyre, Malawi, E-mail: cmangani@medcol.mw. Kenneth Maleta, School of Public Health and Family Medicine, College of Medicine, University of Malawi, Blantyre, Malawi, E-mail: kmaleta@medcol.mw.
References
- 1.Caccio SM, Thompson RC, McLauchlin J, Smith HV. Unravelling Cryptosporidium and Giardia epidemiology. Trends Parasitol. 2005;21:430–437. doi: 10.1016/j.pt.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Savioli L, Smith H, Thompson A. Giardia and Cryptosporidium join the ‘Neglected Diseases Initiative. Trends Parasitol. 2006;22:203–208. doi: 10.1016/j.pt.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Checkley W, Gilman RH, Epstein LD, Suarez M, Diaz JF, Cabrera L, Black RE, Sterling CR. Asymptomatic and symptomatic cryptosporidiosis: their acute effect on weight gain in Peruvian children. Am J Epidemiol. 1997;145:156–163. doi: 10.1093/oxfordjournals.aje.a009086. [DOI] [PubMed] [Google Scholar]
- 4.Prado MS, Cairncross S, Strina A, Barreto ML, Oliveira-Assis AM, Rego S. Asymptomatic giardiasis and growth in young children; a longitudinal study in Salvador, Brazil. Parasitology. 2005;131:51–56. doi: 10.1017/s0031182005007353. [DOI] [PubMed] [Google Scholar]
- 5.Mondal D, Haque R, Sack RB, Kirkpatrick BD, Petri WA., Jr Attribution of malnutrition to cause-specific diarrheal illness: evidence from a prospective study of preschool children in Mirpur, Dhaka, Bangladesh. Am J Trop Med Hyg. 2009;80:824–826. [PMC free article] [PubMed] [Google Scholar]
- 6.Ajjampur SS, Sarkar R, Sankaran P, Kannan A, Menon VK, Muliyil J, Ward H, Kang G. Symptomatic and asymptomatic Cryptosporidium infections in children in a semi-urban slum community in southern India. Am J Trop Med Hyg. 2010;83:1110–1115. doi: 10.4269/ajtmh.2010.09-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veenemans J, Mank T, Ottenhof M, Baidjoe A, Mbugi EV, Demir AY, Wielders JP, Savelkoul HF, Verhoef H. Protection against diarrhea associated with Giardia intestinalis is lost with multi-nutrient supplementation: a study in Tanzanian children. PLoS Negl Trop Dis. 2011;5:e1158. doi: 10.1371/journal.pntd.0001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elwin K, Robinson G, Hadfield SJ, Fairclough HV, Iturriza-Gomara M, Chalmers RM. A comparison of two approaches to extracting Cryptosporidium DNA from human stools as measured by a real-time PCR assay. J Microbiol Methods. 2012;89:38–40. doi: 10.1016/j.mimet.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Schuurman T, Lankamp P, van Belkum A, Kooistra-Smid M, van Zwet A. Comparison of microscopy, real-time PCR and a rapid immunoassay for the detection of Giardia lamblia in human stool specimens. Clin Microbiol Infect. 2007;13:1186–1191. doi: 10.1111/j.1469-0691.2007.01836.x. [DOI] [PubMed] [Google Scholar]
- 10.Verweij JJ, Schinkel J, Laeijendecker D, van Rooyen MA, van Lieshout L, Polderman AM. Real-time PCR for the detection of Giardia lamblia. Mol Cell Probes. 2003;17:223–225. doi: 10.1016/s0890-8508(03)00057-4. [DOI] [PubMed] [Google Scholar]
- 11.Verweij JJ, Blange RA, Templeton K, Schinkel J, Brienen EA, van Rooyen MA, van Lieshout L, Polderman AM. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J Clin Microbiol. 2004;42:1220–1223. doi: 10.1128/JCM.42.3.1220-1223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robertson LJ, Gjerde BK. Effects of the Norwegian winter environment on Giardia cysts and Cryptosporidium oocysts. Microb Ecol. 2004;47:359–365. doi: 10.1007/s00248-003-0003-5. [DOI] [PubMed] [Google Scholar]
- 13.Nichols RA, Moore JE, Smith HV. A rapid method for extracting oocyst DNA from Cryptosporidium-positive human faeces for outbreak investigations. J Microbiol Methods. 2006;65:512–524. doi: 10.1016/j.mimet.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Haque R, Roy S, Siddique A, Mondal U, Rahman SM, Mondal D, Houpt E, Petri WA., Jr Multiplex real-time PCR assay for detection of Entamoeba histolytica, Giardia intestinalis, and Cryptosporidium spp. Am J Trop Med Hyg. 2007;76:713–717. [PubMed] [Google Scholar]
- 15.Adamska M, Leońska-Duniec A, Maciejewska A, Sawczuk M, Skotarczak B. PCR and real time PCR for the detection of Cryptosporidium parvum oocyst DNA. Folia Biol (Krakow) 2011;59:115–120. doi: 10.3409/fb59_3-4.115-120. [DOI] [PubMed] [Google Scholar]
- 16.Minarovicova J, Kaclikova E, Krascsenicsova K, Siekel P, Kuchta T. A single-tube nested real-time polymerase chain reaction for sensitive contained detection of Cryptosporidium parvum. Lett Appl Microbiol. 2009;49:568–572. doi: 10.1111/j.1472-765X.2009.02708.x. [DOI] [PubMed] [Google Scholar]
- 17.Mangani C, Maleta K, Phuka J, Cheung YB, Thakwalakwa C, Dewey K, Manary M, Puumalainen T, Ashorn P. Effect of complementary feeding with lipid-based nutrient supplements and corn-soy blend on the incidence of stunting and linear growth among 6- to 18-month-old infants and children in rural Malawi. Matern Child Nutr. 2013;25 doi: 10.1111/mcn.12068. doi:10.1111/mcn.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adamska M, Leonska-Duniec A, Sawczuk M, Maciejewska A, Skotarczak B. Recovery of Cryptosporidium from spiked water and stool samples measured by PCR and real time PCR. Vet Med (Praha) 2012;57:224–232. [Google Scholar]
- 19.Bruijnesteijn van Coppenraet LE, Wallinga JA, Ruijs GJ, Bruins MJ, Verweij JJ. Parasitological diagnosis combining an internally controlled real-time PCR assay for the detection of four protozoa in stool samples with a testing algorithm for microscopy. Clin Microbiol Infect. 2009;15:869–874. doi: 10.1111/j.1469-0691.2009.02894.x. [DOI] [PubMed] [Google Scholar]
- 20.Mary C, Chapey E, Dutoit E, Guyot K, Hasseine L, Jeddi F, Menotti J, Paraud C, Pomares C, Rabodonirina M, Rieux A, Derouin F, ANOFEL Cryptosporidium National Network Multicentric evaluation of a new real-time PCR assay for quantification of Cryptosporidium spp. and identification of Cryptosporidium parvum and Cryptosporidium hominis. J Clin Microbiol. 2013;51:2556–2563. doi: 10.1128/JCM.03458-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.ten Hove R, Schuurman T, Kooistra M, Moller L, van Lieshout L, Verweij JJ. Detection of diarrhoea-causing protozoa in general practice patients in the Netherlands by multiplex real-time PCR. Clin Microbiol Infect. 2007;13:1001–1007. doi: 10.1111/j.1469-0691.2007.01788.x. [DOI] [PubMed] [Google Scholar]