Abstract
Schizophrenia is one of the most prevalent psychiatric disorders with complex genetic etiology. Accumulating evidence suggests that energy metabolism and oxidative stress play important roles in the pathophysiology of schizophrenia. Dysfunction of mitochondrial respiratory chain and altered expression of complex I subunits were frequently reported in schizophrenia. To investigate whether nuclear-encoded core subunit genes of mitochondrial complex I are associated with schizophrenia, we performed a genetic association study in Han Chinese. In total, 46 tag single nucleotide polymorphisms (SNPs) from 7 nuclear-encoded core genes of mitochondrial complex I were genotyped in 918 schizophrenia patients and 1042 healthy controls. We also analyzed these SNPs in a large sample mainly composed of Europeans through using the available GWAS datasets from the Psychiatric Genomics Consortium (PGC). No significant associations were detected between these SNPs and schizophrenia in Han Chinese and the PGC data set. However, we observed nominal significant associations of 2 SNPs in the NDUFS1 gene and 4 SNPs in the NDUFS2 gene with early onset schizophrenia (EOS), but none of these associations survived the Bonferroni correction. Taken together, our results suggested that common SNPs in the nuclear-encoded core subunit genes of mitochondrial complex I may not confer genetic susceptibility to schizophrenia.
Schizophrenia is one of the most common psychiatric disorders with a heritability as high as 80%1. Though significant progress has been made during the past decades, the etiology and pathophysiology of schizophrenia remains largely unknown. Accumulating evidence implies that mitochondrial dysfunction may play an important role in schizophrenia2.
Human brain is the largest energy consumer among all organs (it consumes about 20% energy used by the human body)3, which makes it more susceptible to disrupted cellular energy metabolism. Most of the energy used by the human body is produced by mitochondrion, a complex intracellular organelle with many components. As the energy factory, mitochondrion plays a key role in maintaining the normal function of brain and mitochondrial dysfunction has been frequently reported in patients with brain diseases, including neurodegenerative diseases and psychiatric disorders2,4.
Several lines of evidence support the dysfunction of mitochondria in schizophrenia. First, defect of mitochondrial oxidative phosphorylation and altered expression of mitochondria-related genes were reported in brains from patients with schizophrenia5,6,7. Second, data from transcriptomics, proteomics and metabolomics revealed aberrant brain metabolism and oxidative stress in schizophrenia patients8. Third, perturbations in mitochondrial network dynamics and in complex I dependent cellular respiration were also reported in schizophrenia9. Besides, genetic variations of mitochondrial DNA (mtDNA) were also reported to be susceptible to schizophrenia and other neurological disorders2,10,11, although with controversies12,13,14,15. Despite the significant progress in recent years, the role of mitochondria in the pathophysiology of schizophrenia remains elusive, and further studies using more rigorous methodological and statistical standards are necessary to avoid false positive results.
Among the mitochondrial complexes, NADH ubiquinone oxidoreductase (complex I) may have a role in schizophrenia. Human mitochondrial complex I, which contains 45 subunits, is the largest and most complicated component of the respiratory chain and plays a central role in electron transportation16. The 14 “core” subunits of complex I are conserved from bacteria to human and are sufficient for catalysis17,18, suggesting their importance in energy metabolism. Seven of the 14 “core” subunits were nuclear-encoded which ligate the flavin mononucleotide and the iron-sulfur clusters19. Due to its pivotal role in energy metabolism, it has been speculated that mitochondrial complex I may be involved in the pathophysiology of schizophrenia. Consistent with this speculation, previous studies showed aberrant expression of mitochondrial complex I subunits or altered complex I activity in schizophrenia6,20,21,22. In addition, several “core” genes of mitochondrial complex I have been reported to be functionally related to schizophrenia23,24,25.
To further explore the potential association between mitochondrial complex I and schizophrenia, we conducted a comprehensive genetic association study by genotyping 46 tag SNPs from seven nuclear-encoded genes of mitochondrial complex I in Han Chinese with and without schizophrenia and reanalysis of the Psychiatric Genetics Consortium (PGC) dataset26. In addition, we also performed stratified analyses to test if genetic variants of the seven nuclear-encoded genes of mitochondrial complex I were associated with early onset schizophrenia (EOS) in Han Chinese.
Results
In total, 46 tag SNPs were successfully genotyped in our samples. Considering an average population minor allele frequency (MAF) of 0.1, the power to detect an odds ratio as low as 1.5 for a risk allele/genotype/haplotype was above 90% for the comparison between schizophrenia patients and controls in our sample set (Supplementary Figure 1).
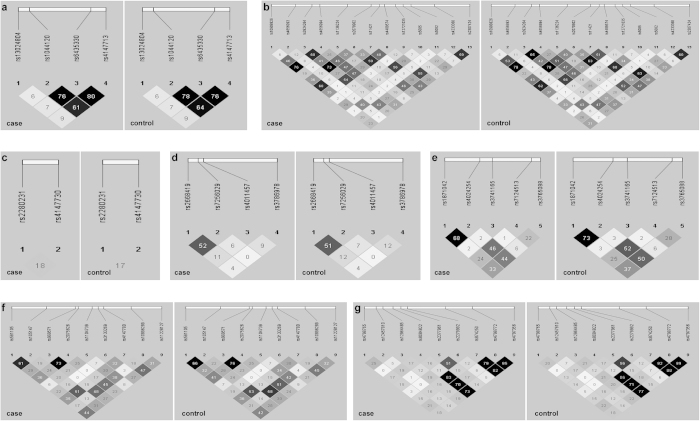
None of the 46 SNPs was deviated from HWE in both case and control samples (Supplementary Table 1). The linkage disequilibrium (LD) structures of the 46 SNPs showed high degrees of similarity between cases and controls (Fig. 1). Allelic and genotypic association analyses revealed no significant association between these 46 tag SNPs and schizophrenia (Table 1). In addition, haplotype-based analysis also showed no significant association between schizophrenia cases and healthy controls (Supplementary Table 2). Further stratified association analysis revealed that two SNPs (rs4147713, P = 0.019; and rs1044120, P = 0.049) in the NDUFS1 gene and 4 SNPs (rs3924264, P = 0.004; rs1136224, P = 0.013; rs2070902, P = 0.006; and rs4233368, P = 0.038) in the NDUFS2 gene were nominally associated with EOS (onset age ≤ 18 years) (Table 2). Nevertheless, none of the six SNPs survived for multiple testing corrections.
Figure 1. The linkage disequilibrium (LD) structures of the NDUFS1 gene (a), the NDUFS2 gene (b), the NDUFS3 gene (c), the NDUFS7 gene (d), the NDUFV1 gene (e), the NDUFS8 gene (f) and the NDUFV2 gene (g) in Han Chinese with and without schizophrenia.
The value in each square refers to r2 × 100. The blacker square represented the higher LD. The individual square showed the r2 × 100 value for each SNP pair.
Table 1. Association results of the 46 SNPs in Hunan sample and the PGC sample.
| SNP ID | A12a | Freq.b |
P-valuec |
OR (s.e.)d |
||
|---|---|---|---|---|---|---|
| Hunan | PGC | Hunan | PGC | |||
| NDUFS1 | ||||||
| rs4147713 | G/T | 0.325 | 0.570 | 0.165 | 1.039 (0.068) | 0.985 (0.011) |
| rs6435330 | T/G | 0.280 | 0.357 | 0.077 | 1.067 (0.071) | 1.019 (0.011) |
| rs1044120 | T/G | 0.243 | 0.892 | 0.106 | 0.990 (0.075) | 1.018 (0.011) |
| rs13024804 | G/A | 0.174 | 0.174 | 0.277 | 0.889 (0.086) | 1.022 (0.020) |
| NDUFS2 | ||||||
| rs10908826 | T/C | 0.346 | 0.319 | 0.028 | 0.935 (0.068) | 1.033 (0.015) |
| rs4656993 | A/G | 0.100 | 0.110 | 0.274 | 1.179 (0.103) | 0.988 (0.011) |
| rs3924264 | A/G | 0.493 | 0.628 | 0.314 | 1.031 (0.064) | 1.011 (0.011) |
| rs4656994 | A/G | 0.393 | 0.588 | 0.016 | 0.965 (0.066) | 1.031 (0.013) |
| rs1136224 | C/T | 0.304 | 0.892 | 0.105 | 0.991 (0.070) | 1.024 (0.015) |
| rs2070902 | T/C | 0.455 | 0.509 | 0.100 | 0.958 (0.064) | 0.980 (0.012) |
| rs11421 | C/T | 0.384 | 0.312 | 0.059 | 0.935 (0.066) | 0.973 (0.015) |
| rs4489574 | T/C | 0.474 | 0.826 | 0.057 | 1.014 (0.064) | 0.979 (0.011) |
| rs12721035 | A/G | 0.146 | 0.579 | 0.406 | 1.051 (0.090) | 1.018 (0.022) |
| rs5085 | C/G | 0.305 | 0.347 | 0.015 | 0.936 (0.070) | 0.967 (0.014) |
| rs5082 | C/T | 0.088 | 0.149 | 0.585 | 1.171 (0.109) | 1.006 (0.011) |
| rs4233368 | A/C | 0.398 | 0.306 | 0.066 | 0.935 (0.066) | 0.978 (0.012) |
| rs2307424 | C/T | 0.491 | 0.885 | 0.017 | 0.991 (0.064) | 1.027 (0.011) |
| NDUFS3 | ||||||
| rs2280231 | T/C | 0.278 | 0.846 | 0.167 | 0.986 (0.072) | 0.984 (0.012) |
| rs4147730 | A/G | 0.318 | 0.721 | 0.311 | 1.025 (0.069) | 1.015 (0.015) |
| NDUFS7 | ||||||
| rs2668419 | A/G | 0.438 | 0.873 | 0.383 | 1.010 (0.064) | 1.017 (0.020) |
| rs7256029 | G/A | 0.406 | 0.881 | 0.354 | 0.990 (0.065) | 1.017 (0.018) |
| rs4011457 | C/G | 0.145 | 0.987 | 0.143 | 1.002 (0.091) | 0.960 (0.028) |
| rs3786978 | C/T | 0.395 | 0.836 | 0.196 | 1.014 (0.065) | 1.044 (0.033) |
| NDUFS8 | ||||||
| rs581105 | G/T | 0.390 | 0.285 | 0.585 | 1.072 (0.065) | 1.006 (0.011) |
| rs105147 | C/T | 0.465 | 0.462 | 0.865 | 1.049 (0.064) | 1.002 (0.011) |
| rs999571 | A/G | 0.183 | 0.685 | 0.053 | 0.967 (0.083) | 0.969 (0.017) |
| rs2075626 | C/T | 0.222 | 0.594 | 0.408 | 1.042 (0.077) | 1.010 (0.013) |
| rs1104739 | C/A | 0.235 | 0.940 | 0.747 | 0.994 (0.076) | 0.996 (0.011) |
| rs3133269 | C/T | 0.202 | 0.826 | 0.675 | 1.018 (0.080) | 1.005 (0.012) |
| rs4147780 | C/T | 0.428 | 0.821 | 0.277 | 1.015 (0.065) | 1.012 (0.011) |
| rs10896289 | A/C | 0.143 | 0.897 | 0.055 | 0.988 (0.092) | 0.973 (0.015) |
| rs11228127 | A/G | 0.317 | 0.760 | 0.098 | 0.979 (0.069) | 0.973 (0.016) |
| NDUFV1 | ||||||
| rs1871042 | T/C | 0.172 | 0.267 | 0.266 | 0.909 (0.086) | 0.987 (0.012) |
| rs4024254 | C/T | 0.207 | 0.380 | 0.365 | 0.932 (0.080) | 1.010 (0.011) |
| rs3741165 | G/A | 0.134 | 0.638 | 0.390 | 1.045 (0.093) | 1.047 (0.053) |
| rs7124513 | T/C | 0.127 | 0.061 | 0.425 | 0.830 (0.100) | 1.009 (0.012) |
| rs3765088 | G/A | 0.326 | 0.692 | 0.950 | 1.027 (0.068) | 1.000 (0.011) |
| NDUFV2 | ||||||
| rs4798765 | T/C | 0.323 | 0.051 | 0.623 | 0.873 (0.069) | 1.006 (0.011) |
| rs12457810 | G/T | 0.107 | 0.980 | 0.035 | 0.997 (0.104) | 0.958 (0.021) |
| rs12964485 | T/C | 0.394 | 0.967 | 0.019 | 0.997 (0.066) | 0.975 (0.011) |
| rs8084822 | T/A | 0.250 | 0.327 | 0.786 | 0.929 (0.075) | 0.996 (0.014) |
| rs2377961 | C/T | 0.345 | 0.071 | 0.019 | 1.128 (0.067) | 0.973 (0.012) |
| rs2279992 | G/A | 0.275 | 0.119 | 0.107 | 1.117 (0.071) | 0.982 (0.011) |
| rs874250 | A/G | 0.261 | 0.250 | 0.538 | 0.919 (0.074) | 1.008 (0.014) |
| rs4798772 | G/A | 0.299 | 0.342 | 0.507 | 0.935 (0.071) | 1.008 (0.012) |
| rs4797356 | A/T | 0.283 | 0.497 | 0.163 | 0.953 (0.072) | 1.015 (0.011) |
aA12, minor allele and major allele.
bMinor allele frequency (Freq.) of control samples in Hunan.
cP-value < 0.05 was marked in bold.
dOdds ratio (OR) estimates and standard errors (s.e.).
Table 2. Association results of the 46 SNPs in 189 early onset schizophrenia patients and 1042 controls.
| SNP ID | A12a |
Frequency of minor allele |
OR | 95% CI | P-valueb | |
|---|---|---|---|---|---|---|
| EOS | control | |||||
| NDUFS1 | ||||||
| rs4147713 | G/T | 0.265 | 0.325 | 0.746 | 0.583-0.954 | 0.019 |
| rs6435330 | T/G | 0.246 | 0.280 | 0.838 | 0.651-1.079 | 0.171 |
| rs1044120 | T/G | 0.196 | 0.243 | 0.760 | 0.579-0.999 | 0.049 |
| rs13024804 | G/A | 0.204 | 0.174 | 1.213 | 0.922-1.596 | 0.168 |
| NDUFS2 | ||||||
| rs10908826 | T/C | 0.373 | 0.346 | 1.122 | 0.894-1.409 | 0.319 |
| rs4656993 | A/G | 0.130 | 0.100 | 1.336 | 0.958-1.863 | 0.087 |
| rs3924264 | A/G | 0.426 | 0.507 | 0.721 | 0.578-0.899 | 0.004 |
| rs4656994 | A/G | 0.434 | 0.393 | 1.184 | 0.948-1.477 | 0.136 |
| rs1136224 | C/T | 0.241 | 0.304 | 0.727 | 0.564-0.937 | 0.013 |
| rs2070902 | T/C | 0.378 | 0.455 | 0.729 | 0.582-0.913 | 0.006 |
| rs11421 | C/T | 0.429 | 0.384 | 1.204 | 0.964-1.503 | 0.101 |
| rs4489574 | T/C | 0.434 | 0.474 | 0.852 | 0.683-1.062 | 0.154 |
| rs12721035 | A/G | 0.161 | 0.146 | 1.130 | 0.837-1.525 | 0.425 |
| rs5085 | C/G | 0.336 | 0.305 | 1.152 | 0.913-1.454 | 0.234 |
| rs5082 | C/T | 0.087 | 0.088 | 0.994 | 0.674-1.465 | 0.974 |
| rs4233368 | A/C | 0.341 | 0.398 | 0.784 | 0.623-0.987 | 0.038 |
| rs2307424 | C/T | 0.439 | 0.491 | 0.812 | 0.651-1.012 | 0.064 |
| NDUFS3 | ||||||
| rs2280231 | T/C | 0.315 | 0.278 | 1.194 | 0.942-1.514 | 0.142 |
| rs4147730 | A/G | 0.304 | 0.318 | 0.939 | 0.741-1.191 | 0.605 |
| NDUFS7 | ||||||
| rs2668419 | A/G | 0.460 | 0.438 | 1.094 | 0.878-1.363 | 0.424 |
| rs7256029 | G/A | 0.413 | 0.406 | 1.028 | 0.823-1.285 | 0.806 |
| rs4011457 | C/G | 0.124 | 0.145 | 0.835 | 0.601-1.160 | 0.281 |
| rs3786978 | C/T | 0.400 | 0.395 | 1.019 | 0.815-1.275 | 0.868 |
| NDUFS8 | ||||||
| rs581105 | G/T | 0.378 | 0.390 | 0.951 | 0.759-1.192 | 0.665 |
| rs105147 | C/T | 0.455 | 0.465 | 0.961 | 0.771-1.198 | 0.724 |
| rs999571 | A/G | 0.156 | 0.183 | 0.824 | 0.611-1.111 | 0.204 |
| rs2075626 | C/T | 0.201 | 0.222 | 0.881 | 0.671-1.156 | 0.361 |
| rs1104739 | C/A | 0.217 | 0.235 | 0.903 | 0.693-1.176 | 0.447 |
| rs3133269 | C/T | 0.217 | 0.202 | 1.097 | 0.840-1.433 | 0.497 |
| rs4147780 | C/T | 0.426 | 0.428 | 0.992 | 0.794-1.237 | 0.940 |
| rs10896289 | A/C | 0.122 | 0.143 | 0.834 | 0.598-1.162 | 0.282 |
| rs11228127 | A/G | 0.307 | 0.317 | 0.953 | 0.752-1.208 | 0.692 |
| NDUFV1 | ||||||
| rs1871042 | T/C | 0.169 | 0.172 | 0.979 | 0.732-1.311 | 0.889 |
| rs4024254 | C/T | 0.201 | 0.207 | 0.965 | 0.735-1.268 | 0.799 |
| rs3741165 | G/A | 0.143 | 0.134 | 1.078 | 0.787-1.477 | 0.639 |
| rs7124513 | T/C | 0.101 | 0.127 | 0.767 | 0.536-1.099 | 0.147 |
| rs3765088 | G/A | 0.347 | 0.326 | 1.095 | 0.869-1.379 | 0.441 |
| NDUFV2 | ||||||
| rs4798765 | T/C | 0.275 | 0.323 | 0.794 | 0.622-1.013 | 0.063 |
| rs12457810 | G/T | 0.101 | 0.107 | 0.933 | 0.649-1.341 | 0.707 |
| rs12964485 | T/C | 0.384 | 0.394 | 0.955 | 0.763-1.197 | 0.691 |
| rs8084822 | T/A | 0.228 | 0.250 | 0.886 | 0.683-1.149 | 0.361 |
| rs2377961 | C/T | 0.386 | 0.345 | 1.197 | 0.955-1.501 | 0.118 |
| rs2279992 | G/A | 0.309 | 0.275 | 1.178 | 0.928-1.497 | 0.179 |
| rs874250 | A/G | 0.238 | 0.261 | 0.887 | 0.687-1.146 | 0.358 |
| rs4798772 | G/A | 0.271 | 0.299 | 0.875 | 0.684-1.119 | 0.287 |
| rs4797356 | A/T | 0.263 | 0.283 | 0.907 | 0.706-1.164 | 0.441 |
aA12, minor allele and major allele.
bP-value < 0.05 was marked in bold.
OR - odds ratio; 95% CI – 95% confidence interval
We further examined the genetic association between these 46 SNPs and schizophrenia in the PGC data26. Overall, 35,476 schizophrenia cases and 46,839 controls were included for association analysis. We observed marginally significant associations with schizophrenia of rs10908826, rs4656994, rs5085 and rs2307424 in the NDUFS2 gene and rs12457810, rs12964485 and rs2377961 in the NDUFV2 gene. However, none of these SNPs showed significant association after correcting for multiple testing (Table 1).
We further performed interaction analysis to test whether there is an interaction between SNPs in both case-control samples and the EOS-control samples. Our results showed that although there were many marginally significant interactions, none of them could survive the Bonferroni correction (Supplementary Tables 3-4).
Discussion
Mitochondrial dysfunction has been frequently reported in schizophrenia8,27. Previous studies have suggested that genes related to energy metabolism and oxidative stress may be responsible for mitochondrial dysfunction in schizophrenia8. As the largest component of the three membrane-bound enzymes, mitochondrial complex I plays a vital role in energy metabolism and altered activity of mitochondrial complex I was repeatedly reported in schizophrenia20,22,28,29. Though multiple studies have reported that mitochondrial dysfunction may be involved in schizophrenia pathogenesis, most of the conclusions were based on gene expression6,21. We previously analyzed the NDUFS7 gene in Han Chinese with and without schizophrenia, with an intention to discern the effect of the complex I genes on this disorder30. In this study, we analyzed other complex I core subunit genes including NDUFS1, NDUFS2, NDUFS3, NDUFS8, NDUFV1 and NDUFV2, as well as four more common SNPs of the NDUFS7 gene in a larger sample set. We systematically investigated the potential association between these nuclear-encoded mitochondrial complex I genes and schizophrenia in a Chinese case-control sample. Our results revealed no significant association between genetic variants from the seven selected genes and schizophrenia, suggesting that these genes are unlikely to confer risk of schizophrenia in Han Chinese population. Consistent with the finding in Han Chinese, we also found no robust association between these SNPs and schizophrenia in the PGC data26. It should be mentioned that if we did not consider the effect of multiple corrections, SNPs of the NDUFS2 gene would show (marginally) significant association with schizophrenia in both Han Chinese and the PGC data26.
It is interesting that we observed nominal significant association between genetic variants in the NDUFS1 and NDUFS2 genes and EOS in Han Chinese. The nominal significance would not survive from adjustment and we cannot rule out the possibility of false positive association caused by relatively small sample size of EOS used in the analyses. It should be mentioned that genetic variant in the NDUFS1 gene was reported to be associated with schizophrenia and negative symptoms in Han Chinese from Eastern China, albeit the samples size was also relatively small31. We also noticed that comparing with whole samples (odds ratio [OR] > 1), 2 of the 6 nominal significant SNPs (rs4147713 and rs3924264) were associated with EOS in reverse style (OR < 1). This observation indicated population stratification may exist in the EOS-subpopulation.
Considering that mitochondrial dysfunction was repeatedly reported in schizophrenia, it is amazing that none of the selected SNPs showed significant association with schizophrenia. One of the possible explanations is that other genes but not the seven selected core genes of mitochondrial complex I contribute to schizophrenia susceptibility. Moreover, possibility of rare variant(s) in these genes contributing to schizophrenia susceptibility needs to be studied as well. In addition, given that schizophrenia has a strong genetic heterogeneity, it is also possible that these genes would have a strong effect in other populations but not in Han Chinese23,24.
There are several limitations in this study. First, the sample size is relatively modest in this study. As a result, it may be difficult to detect a robust significant association. Second, we only analyzed seven nuclear-encoded genes in the mitochondrial complex I in this study, we could not exclude the possibility that other genes of complex I are associated with schizophrenia.
In summary, we detected no significant association between the genetic polymorphisms of the nuclear-encoded core subunit genes of mitochondrial complex I and schizophrenia by using a rigorous statistical standard so as to avoid false positive results. Further work is needed to test if the expression and rare variants, but not common variants of these genes, contribute to schizophrenia.
Materials and methods
Subjects
A total of 1960 subjects, including 918 unrelated patients with schizophrenia (561 males: mean age ± SD, 38.5 ± 13.6 years; 357 females: 38.4 ± 16.8 years) and 1042 healthy controls (631 males: mean age ± SD, 38.5 ± 14.2 years; 411 females: 46.0 ± 8.2 years) were recruited. All of the individuals are of Han Chinese origin from Hunan Province of South Central China. Among the 918 patients, 189 were EOS (first age of onset ≤ 18 years old). The patients were clinically diagnosed according to Diagnostic and Statistical Manual of Mental Disorders IV (DSM-IV) and had at least two-year history of schizophrenia. Diagnosis and review of schizophrenia cases were independently checked and verified by two senior psychiatrists prior to blood sample collection. The healthy controls were collected from local hospitals and assessed by experienced psychiatrists. Individuals with psychiatric history, alcohol dependence, drug abuse, or family history of psychiatric disorders were excluded. All of the schizophrenia patients and healthy controls have been reported in our previous studies11,32. Written informed consents conforming to the tenets of the Declaration of Helsinki were obtained from all participants or the appointed guardians of the patients (for those who were unable to provide informed consent at the time of blood collection) prior to this study. The experimental methods were carried out in accordance with the approved guidelines. All experimental protocols of this study were approved by the institutional review board / Ethics Committee of Kunming Institute of Zoology, Chinese Academy of Sciences.
SNP selection and genotyping
Genomic DNA of all participants was extracted from peripheral blood using the AxyPrepTM Blood Genomic DNA Miniprep Kit (Axygen, USA) according to the manufacturer’s instruction. Seven nuclear-encoded genes of mitochondrial complex I (NDUFS1, NDUFS2, NDUFS3, NDUFS7, NDUFS8, NDUFV1 and NDUFV2) were chosen in this study. To select the tag SNPs, we retrieved genotypic data of Han Chinese (CHB) from the HapMap database ( http://hapmap.ncbi.nlm.nih.gov/) and defined LD blocks using the Haploview 4.233. The gene region and potential regulatory sequences (20 kb of both upstream and downstream regions) were taken into consideration during the selection of tag SNPs. In total, 51 tag SNPs were selected based on the following criteria: minor allele frequency (MAF) ≥ 0.1 and r2 ≥ 0.8 (Supplementary Figures 2-5). Three tag SNPs (rs2074896, rs2074897 and rs2074898) in the NDUFS7 gene were analyzed in our previous study30, therefore were not included in this study. The remaining 48 tag SNPs were divided into four panels (12 SNPs for each panel) according to their compatibility in multiplex PCR. Genotyping of each panel was conducted by SNaPshot assays reported in our previous studies34,35. The GeneMarker software was utilized to read the genotyping results36.
Power calculation and statistical analysis
Among the 48 tag SNPs, two (rs12798346 and rs3751084) were failed to be genotyped in our samples. Therefore, these two SNPs were excluded from our statistical analysis. The genotyping call rate of each SNP was above 99.0% in 1960 individuals. LD plot of the genotyped SNPs of each gene was constructed using Haploview 4.2 program (version 4.2). We tested deviation from the Hardy-Weinberg equilibrium (HWE), individual SNP association, haplotype comparison and SNP-SNP interaction by using PLINK37. Quanto software38 was used for power analysis under the gene only hypothesis and log additive model and following parameters: risk allele frequency from 0.1 to 0.5 in increments of 0.1; overall disease risk in the general population = 0.01; sample size = 918 cases vs. 1042 controls; range of OR from 1.0 to 2.0 in increments of 0.1; two-sided type I error rate = 0.05.
PGC data analysis
To further explore if the studied SNPs are associated with schizophrenia, we extracted the genetic association data from the Psychiatric Genomics Consortium (PGC, http://www.broadinstitute.org/mpg/ricopili/)39 and reanalyzed this data set as an independent validation sample. In brief, 35,476 schizophrenia cases and 46,839 controls were included in the PGC dataset. The genotyping of each primary GWAS study composing the PGC data was performed by Affymetrix or Illumina array and the genetic association analysis was conducted by PLINK37 under an additive logistic regression model. More detailed information about the PGC can be found in the original publication26.
Additional Information
How to cite this article: Li, X. et al. Do nuclear-encoded core subunits of mitochondrial complex I confer genetic susceptibility to schizophrenia in Han Chinese populations? Sci. Rep. 5, 11076; doi: 10.1038/srep11076 (2015).
Supplementary Material
Acknowledgments
We are grateful to all of the participants in this study. This study was supported by the Strategic Priority Research Program (B) of the Chinese Academy of Sciences (XDB02020000), the Ministry of Science and Technology of China (2011CB910900), National Natural Science Foundation of China (31171225) and the West Light Foundation of the Chinese Academy of Sciences.
Footnotes
Author Contributions W.Z. and Y.G.Y. designed the study; X.C., J.T. and L.T. collected the samples and clinical information; X.L. carried out the experimental procedures; X.L., W.Z., X.j.L. and Y.G.Y. analyzed data; X.L., W.Z. and Y.G.Y. drafted the manuscript. All authors contributed to and have approved the final manuscript.
References
- Sullivan P. F., Kendler K. S. & Neale M. C. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry 60, 1187–1192 (2003). [DOI] [PubMed] [Google Scholar]
- Shao L. et al. Mitochondrial involvement in psychiatric disorders. Ann Med 40, 281–295 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mink J. W., Blumenschine R. J. & Adams D. B. Ratio of central nervous system to body metabolism in vertebrates: its constancy and functional basis. Am J Physiol 241, R203–212 (1981). [DOI] [PubMed] [Google Scholar]
- Zsurka G. & Kunz W. S. Mitochondrial involvement in neurodegenerative diseases. IUBMB Life 65, 263–272 (2013). [DOI] [PubMed] [Google Scholar]
- Maurer I., Zierz S. & Moller H. Evidence for a mitochondrial oxidative phosphorylation defect in brains from patients with schizophrenia. Schizophr Res 48, 125–136 (2001). [DOI] [PubMed] [Google Scholar]
- Karry R., Klein E. & Ben Shachar D. Mitochondrial complex I subunits expression is altered in schizophrenia: a postmortem study. Biol Psychiatry 55, 676–684 (2004). [DOI] [PubMed] [Google Scholar]
- Iwamoto K., Bundo M. & Kato T. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Hum Mol Genet 14, 241–253 (2005). [DOI] [PubMed] [Google Scholar]
- Prabakaran S. et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry 9, 684-697 643 (2004). [DOI] [PubMed] [Google Scholar]
- Rosenfeld M., Brenner-Lavie H., Ari S. G., Kavushansky A. & Ben-Shachar D. Perturbation in mitochondrial network dynamics and in complex I dependent cellular respiration in schizophrenia. Biol Psychiatry 69, 980–988 (2011). [DOI] [PubMed] [Google Scholar]
- Verge B. et al. Mitochondrial DNA (mtDNA) and schizophrenia. Eur Psychiatry 26, 45–56 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang W. et al. A matrilineal genetic legacy from the last glacial maximum confers susceptibility to schizophrenia in han chinese. J Genet Genomics 41, 397–407 (2014). [DOI] [PubMed] [Google Scholar]
- Bandelt H. J., Yao Y. G. & Kivisild T. Mitochondrial genes and schizophrenia. Schizophr Res 72, 267–269 (2005). [DOI] [PubMed] [Google Scholar]
- Fachal L. et al. No evidence of association between common European mitochondrial DNA variants in Alzheimer, Parkinson, and migraine in the Spanish population. Am J Med Genet B Neuropsychiatr Genet 168B, 54–65 (2015). [DOI] [PubMed] [Google Scholar]
- Mosquera-Miguel A. et al. No evidence that major mtDNA European haplogroups confer risk to schizophrenia. Am J Med Genet B Neuropsychiatr Genet 159B, 414–421 (2012). [DOI] [PubMed] [Google Scholar]
- Torrell H. et al. Mitochondrial DNA (mtDNA) variants in the European haplogroups HV, JT, and U do not have a major role in schizophrenia. Am J Med Genet B Neuropsychiatr Genet 165B, 607–617 (2014). [DOI] [PubMed] [Google Scholar]
- Brandt U. Energy converting NADH:quinone oxidoreductase (complex I). Annu Rev Biochem 75, 69–92 (2006). [DOI] [PubMed] [Google Scholar]
- Yip C. Y., Harbour M. E., Jayawardena K., Fearnley I. M. & Sazanov L. A. Evolution of respiratory complex I: “supernumerary” subunits are present in the alpha-proteobacterial enzyme. J Biol Chem 286, 5023–5033 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov R. G. & Sazanov L. A. The coupling mechanism of respiratory complex I - a structural and evolutionary perspective. Biochim Biophys Acta 1817, 1785–1795 (2012). [DOI] [PubMed] [Google Scholar]
- Hirst J., Carroll J., Fearnley I. M., Shannon R. J. & Walker J. E. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim Biophys Acta 1604, 135–150 (2003). [DOI] [PubMed] [Google Scholar]
- Dror N. et al. State-dependent alterations in mitochondrial complex I activity in platelets: a potential peripheral marker for schizophrenia. Mol Psychiatry 7, 995–1001 (2002). [DOI] [PubMed] [Google Scholar]
- Taurines R. et al. Expression analyses of the mitochondrial complex I 75-kDa subunit in early onset schizophrenia and autism spectrum disorder: increased levels as a potential biomarker for early onset schizophrenia. Eur Child Adolesc Psychiatry 19, 441–448 (2010). [DOI] [PubMed] [Google Scholar]
- Ben-Shachar D. et al. Increased mitochondrial complex I activity in platelets of schizophrenic patients. Int J Neuropsychopharmacol 2, 245–253 (1999). [DOI] [PubMed] [Google Scholar]
- Ben-Shachar D. & Karry R. Sp1 expression is disrupted in schizophrenia; a possible mechanism for the abnormal expression of mitochondrial complex I genes, NDUFV1 and NDUFV2. PLoS One 2, e817 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washizuka S. et al. Association of mitochondrial complex I subunit gene NDUFV2 at 18p11 with schizophrenia in the Japanese population. Am J Med Genet B Neuropsychiatr Genet 141B, 301–304 (2006). [DOI] [PubMed] [Google Scholar]
- Ben-Shachar D. & Karry R. Neuroanatomical pattern of mitochondrial complex I pathology varies between schizophrenia, bipolar disorder and major depression. PLoS One 3, e3676 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shachar D. Mitochondrial dysfunction in schizophrenia: a possible linkage to dopamine. J Neurochem 83, 1241–1251 (2002). [DOI] [PubMed] [Google Scholar]
- Ben-Shachar D. et al. Cerebral glucose utilization and platelet mitochondrial complex I activity in schizophrenia: A FDG-PET study. Prog Neuropsychopharmacol Biol Psychiatry 31, 807–813 (2007). [DOI] [PubMed] [Google Scholar]
- Gubert C. et al. Mitochondrial activity and oxidative stress markers in peripheral blood mononuclear cells of patients with bipolar disorder, schizophrenia, and healthy subjects. J Psychiatr Res 47, 1396–1402 (2013). [DOI] [PubMed] [Google Scholar]
- Ma L. et al. No association between genetic polymorphisms of the NDUFS7 gene and schizophrenia in Han Chinese. Psychiatr Genet 23, 29–32 (2013). [DOI] [PubMed] [Google Scholar]
- Zhu Y. et al. Genetic variant in NDUFS1 gene is associated with schizophrenia and negative symptoms in Han Chinese. J Hum Genet 60, 11–16 (2015). [DOI] [PubMed] [Google Scholar]
- Ma L. et al. Evaluating risk loci for schizophrenia distilled from genome-wide association studies in Han Chinese from Central China. Mol Psychiatry 18, 638–639 (2013). [DOI] [PubMed] [Google Scholar]
- Barrett J. C., Fry B., Maller J. & Daly M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005). [DOI] [PubMed] [Google Scholar]
- Bi R. et al. No association of the LRRK2 genetic variants with Alzheimer’s disease in Han Chinese individuals. Neurobiol Aging 35, 444 e445–e449 (2014). [DOI] [PubMed] [Google Scholar]
- Li X. et al. No association between genetic variants of the LRRK2 gene and schizophrenia in Han Chinese. Neurosci Lett 566, 210–215 (2014). [DOI] [PubMed] [Google Scholar]
- Holland M. M. & Parson W. GeneMarker(R) HID: A reliable software tool for the analysis of forensic STR data. J Forensic Sci 56, 29–35 (2011). [DOI] [PubMed] [Google Scholar]
- Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauderman W. J. Sample size requirements for matched case-control studies of gene-environment interaction. Stat Med 21, 35–50 (2002). [DOI] [PubMed] [Google Scholar]
- Sullivan P. F. The psychiatric GWAS consortium: big science comes to psychiatry. Neuron 68, 182–186 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.