Abstract
We report the direct observation of conformational rearrangements of the ribosome during multiple rounds of elongation. Using single-molecule fluorescence resonance energy transfer, we monitor the intersubunit conformation of the ribosome – in real time – as it proceeds from codon to codon. During each elongation cycle, the ribosome unlocks upon peptide bond formation, followed by reversion to the locked state upon translocation onto the next codon. Our data reveal both the specific and cumulative effects of antibiotics on individual steps of translation, and uncover the processivity of the ribosome as it elongates. Our approach interrogates the precise molecular events occurring at each codon of the mRNA within the full context of ongoing translation.
Introduction
The ribosome is a processive molecular machine that rapidly and accurately synthesizes protein according to the genetic code stored in mRNA1. In each cycle of elongation, the ribosome selects the aminoacyl tRNA (aa-tRNA) specified by the mRNA codon in the aminoacyl-tRNA binding site (A site), catalyzes formation of a peptide bond with peptidyl-tRNA in the P site, and then steps onto the next mRNA codon, translocating the A- and P-site tRNAs into the P and E sites, respectively2–4.
Translocation requires large-scale movements of both the ribosome and its ligands. How the ribosome generates these movements while balancing the need for accuracy and processivity remains unclear. More than four decades ago, Spirin suggested a model in which ribosome movement is controlled by a series of locking and unlocking events5. Prior to peptide bond formation, the ribosomal subunits are tightly associated, to facilitate manipulation of the tRNA and preserve reading frame on the mRNA. After peptide bond formation, the ribosome unlocks. In this unlocked state, the ribosomal subunits and tRNA can move more freely, facilitating translocation of the tRNA and stepping onto the next codon of the mRNA. Full translocation returns the ribosome to the locked state, once again restricting the motion of the ribosomal subunits and tRNA. Thus, a translating ribosome would cycle between the locked state – to perform tRNA selection and catalyze peptide bond formation – and unlocked state – to translocate – at each codon of the mRNA.
Consistent with this unlocking/locking model, biochemical and structural studies have revealed a series of structural rearrangements promoted upon deacylation of the peptidyl-tRNA in the P site. Chemical probing methods have identified a hybrid tRNA configuration, in which the acceptor stems of the A- and P-site tRNAs point into the P and E sites, respectively6. Cryo-EM studies identified a ratcheted configuration of the ribosomal subunits, in which the small (30S) subunit is rotated ~3–10° counterclockwise with respect to the large (50S) subunit7–10. High-resolution crystal structures suggest that this ratcheting also involves a cocking of the 30S head domain11,12. Ribosome ratcheting upon peptide bond formation may drive formation of the tRNA hybrid state, thus preparing the ribosome for full translocation of the tRNA and stepping onto the next codon of the mRNA. Movement of the L1 stalk has also been implicated in the positioning and movement of the tRNA13.
Recent single-molecule studies have demonstrated the dynamic nature of these rearrangements, consistent with the unlocking of ribosome and tRNA motions upon peptide bond formation. FRET between P- and A-site tRNAs revealed spontaneous fluctuations between the classic and hybrid states upon peptide bond formation14–16. Similarly, Noller, Ha and coworkers observed spontaneous ratcheting in pre-translocation complexes mimicking the state of the ribosome after peptide bond formation17. In contrast, translocated ribosomes were predominantly fixed in the classical, or non-rotated state. Echoing these results, peptide bond formation also appears to release spontaneous open↔closed fluctuations of the L1 stalk, which are restricted upon binding of EF-G and translocation18–20. Taken together, these results suggest that fluctuations between the non-rotated and rotated states of the ribosome – ratcheting – are accompanied by classical↔hybrid and open↔closed fluctuations of the tRNA and L1 stalk. Consistent with this interpretation, recent cryo-EM structures suggest that hybrid state formation and ribosome ratcheting are intimately linked9,10. The observations of spontaneous ratcheting promoted by peptide bond formation is consistent with ribosome unlocking upon peptidyl transfer, and subsequent locking upon EF-G driven translocation. How these events trigger the unlocking and locking of the ribosome remains unclear.
Though the structural and dynamic behavior of the locked and unlocked states of the ribosome has been explored, the repetitive cycling of the ribosome between these states during multiple rounds of elongation has not been directly observed. Single-molecule force measurements have followed the position of the ribosome on the mRNA in real time by monitoring melting of mRNA secondary structures, but are not sensitive to ribosome conformation21. The majority of existing single-molecule FRET signals have been designed to probe ribosome dynamics within the locked and unlocked states, not the unlocking/locking event itself. Here we use single-molecule fluorescence resonance energy transfer to monitor the intersubunit conformation of the ribosome – in real time – as it proceeds from codon to codon. During each round of elongation, the ribosome cycles between two states, consistent with unlocking upon peptide bond formation and subsequent relocking after translocation. Real-time observation of this conformational cycle allows mechanistic tracking of translation at each codon of a mRNA in the full context of ongoing protein synthesis. Our results reveal the precise mechanism of antibiotic action and demonstrate the processivity of the ribosome as it elongates.
Results
Elongation from the perspective of the ribosome
We used previously characterized E. coli ribosomes that are site-specifically labeled with fluorescent dyes on the 30S and 50S subunits to yield intersubunit FRET22,23. The labeling sites on the 30S (helix 44) and 50S (helix 101) are distant from the decoding and peptidyl transferase centers of the ribosome, and from dynamic regions – the L1 stalk, regions of tRNA:ribosome interaction, the 30S head – that participate in spontaneous ratcheting. This ensures that fluorophores do not interfere with function and provide a FRET signal that is insensitive to spontaneous ratcheting. We have previously demonstrated the functionality of these labeled ribosome both in vivo and in vitro22. E. coli strains expressing h44–30S and h101–50S as their sole source of ribosomal subunits show no growth defect. Purified and labeled subunits are functionally competent, as demonstrated by bulk translocation assays.
Cy3-labeled 30S pre-initiation complexes (PICs) containing fMet-tRNAfMet were immobilized with various biotinylated mRNAs on a polyethylene glycol (PEG)-derivatized quartz surface. All mRNAs contained a naturally occurring 5′-UTR and Shine-Dalgarno sequence from T4 gene 32 and were terminated by a stop codon followed by a stretch of 12 nucleotides, to ensure that translation was not affected by arrival at the 3′ end of the mRNA (Supplementary Fig. 1). Single PICs were visualized using a prism-based total internal reflection fluorescence microscope (TIRF-M) with 532 nm excitation. Fluorescence from single molecules was filtered to separate spatially-registered images corresponding to Cy2, Cy3, or Cy5 fluorescence. Comparison of these images identified Cy2, Cy3, or Cy5 fluorescence colocalized at individual ribosomes. Data collection and analysis were performed as previously described23,24. Delivery of Cy5-labeled 50S subunits, elongator ternary complex (aa-tRNA•EF-Tu•GTP), and EF-G results in IF2-guided 70S assembly during initiation and establishment of FRET between the two ribosomal subunits (Fig. 1)23,25. One advantage of this approach is that initiation is observed in real time, and can serve as a control to eliminate non-functional translation complexes.
Figure 1. Intersubunit FRET monitors the elongation cycle at each codon during translation.
(a) Single-molecule translation assay. Cy5-labeled 50S subunits, ternary complex, and EF-G are delivered to surface-immobilized single PICs containing Cy3-labeled 30S subunits and illuminated at 532 nm; both Cy3 and Cy5 fluorescence are simultaneously detected. (b) Immobilization with an mRNA coding for 6 phenylalanines (6F) permits the observation of ribosome conformation during multiple rounds of elongation via the intersubunit FRET signal. The arrival of FRET corresponds to 50S subunit joining during initiation, and is followed by multiple cycles of high-low-high FRET, each reporting on ribosome unlocking and locking during one round of elongation.
The FRET signal from single elongating ribosomes alternates between a high-FRET state (~0.6) and a low-FRET state (~0.4). Using a series of mechanistic controls, we have previously shown that the transition from the high-FRET state to the low-FRET state occurs upon peptide bond formation, and is consistent with a conformational change signaling the unlocking of the ribosome23. This transition is blocked by either near-cognate tRNA or the antibiotic kirromycin, which prevent the accommodation of A-site tRNA. The peptidyl transferase inhibitor chloramphenicol strongly inhibits the transition. In contrast, puromycin, which participates in peptide bond formation, but does not require GTP hydrolysis by EF-Tu for accommodation, allows the transition to occur. The transition from the low-FRET state to the high-FRET state requires EF-G, and is consistent with the locking of the ribosome upon translocation23. Thus, the high-FRET state corresponds to the locked conformation of the ribosome, and the low-FRET state corresponds to the unlocked conformation of the ribosome, in which spontaneous fluctuations of the subunits and tRNA are released, but are invisible to our FRET signal. Each high-low-high FRET cycle follows the unlocking and locking of the ribosome during one round of elongation.
At the low concentrations of ternary complex and EF-G used here (20–100 nM), arrival of these factors is rate limiting for these two transitions. Consistent with this prediction, the lifetimes of the high- and low-FRET states, as determined by fitting lifetimes to a single exponential distribution, are independently sensitive to the concentration of either ternary complex or EF-G, respectively (Supplementary Fig. 2a). Linear fitting of these lifetimes as a function of TC and EF-G concentration confirms that a significant correlation is observed only between ternary complex concentration and high-FRET lifetimes, and between EF-G concentration and low-FRET lifetimes (Supplementary Fig. 2b) Thus, the cycle of high-low-high FRET represents one round of elongation from the perspective of the ribosome. The durations of the high- and low- FRET states during each unlocking/locking cycle report respectively on the waiting time to peptide bond formation and translocation at each codon of the mRNA.
In vivo, translation proceeds at a rate of ~20 amino acid additions per second26. Kinetic dissection in vitro has permitted determination of the rates for the individual steps of the elongation cycle27,28. The rates for events such as the arrival and accommodation of incoming aa-tRNA, peptide bond formation, and translocation have been determined by multiple research groups, with results differing over roughly one order of magnitude27–32. Our FRET transitions report on the events of peptide bond formation and translocation, and thus the lifetimes reported here are equivalent to kcat/Km values previously reported for these steps29,32. The rates observed here are slowed by a factor of 2–4 as compared to previous measurements made under similar conditions (Supplementary Figure 2c), consistent with previous single-molecule studies of translation17,18,21,23,25. Several factors may contribute to the observation of slower rates in single-molecule systems, with the most likely being slower bimolecular association rates due to the accessibility of ligands to surface-immobilized ribosome complexes. Recent work, which employs a single-molecule approach, but instead follows the binding of fluorescent tRNA to wild-type ribosomes, reported rates similar to those observed here33.
Multiple rounds of elongation
To confirm that each FRET cycle corresponds to a distinct round of elongation, we first followed the conformation of elongating ribosomes on mRNAs encoding 3, 6, and 12 phenylalanine codons (3F, 6F, and 12F, respectively). For each mRNA, the number of complete FRET cycles observed for hundreds of individual ribosomes can be represented as a histogram (Fig. 2). As predicted, the maximum number of cycles observed for each mRNA is controlled by the number of codons between the start (AUG) and stop (UAA) codon. Each cycle reports on one round of elongation, and these FRET cycle histograms reveal the distribution of elongation cycles performed by the ensemble of immobilized ribosomes, and can report on the efficiency with which translation proceeds on each mRNA in our system. A significant proportion of ribosomes completely decode each message, as evidenced by the accumulation of molecules showing a number of FRET cycles equal to the mRNA coding length. Full observation on longer mRNAs is challenged primarily by fluorophore photobleaching; experiments performed with increased concentrations of ternary complex and EF-G and at higher acquisition rates reveal an increased proportion of ribosomes that undergo a number of FRET cycles equal to the number of codons (Supplementary Fig. 3).
Figure 2. The number of complete FRET cycles is controlled by coding length.
Histograms of the number of complete FRET cycles observed for single ribosomes on mRNAs coding 3 (3F), 6 (6F), and 12 (12F) phenylalanines, as well as a heteropolymeric mRNA coding alternating phenylalanine and lysine residues (6FK) (139 ribosomes, 303 ribosomes, 1034, and 1145 ribosomes, respectively). Histograms are normalized to the number of ribosomes showing one FRET cycle.
FRET cycle histograms for all three mRNAs reveal a small fraction (< 2%) of ribosomes that undergo a number of FRET cycles greater than the codon length of each message. These additional events might arise from statistical errors in the identification of transitions by our analytical method. Another possibility is that these additional cycles result from non-productive unlocking/locking cycles resulting from ribosome slipping. Goldman and coworkers detected ribosome slipping on poly(U) mRNAs34, consistent with the idea that poly(U) is “slippery.” To confirm that each FRET cycle reports on a productive cycle of elongation, and not futile unlocking/locking cycles occurring on poly(U) tracts, we followed translation of a heteropolymeric mRNA encoding alternating phenylalanine and lysine codons (6FK). Like the FRET cycle histograms for 3F, 6F, and 12F, the FRET cycle histogram for 6FK is limited by the codon length of the mRNA, further suggesting the equivalence between each observed FRET cycle and one round of elongation. Interestingly, the FRET cycle histogram for 6FK shows no additional FRET cycles, supporting the interpretation that rare slipping events can result in spurious unlocking/locking events on poly(U) tracts.
Translation of the 6FK mRNA is significantly more efficient than translation of the 12F mRNA; a greater proportion of ribosomes undergoes 12 elongation cycles, signaling full translation of the mRNA (Figure 2). This effect of mRNA sequence, which underscores the ability of our approach to follow and compare translation on distinct mRNAs, may result from inefficient translation on poly(U) tracts or reflect unfavorable interactions between the synthesized poly(Phe) chain and the exit tunnel.
We next followed the intersubunit FRET signal on a three codon mRNA encoding phenylalanine-lysine-phenylalanine (FKF) (Fig. 3). This mRNA allows control over the number of codons read by the ribosome by either exclusion or inclusion of Lys-tRNALys ternary complex in the reaction mixture. Exclusion of Lys-tRNALys prevents the ribosome from proceeding beyond the first codon; accordingly, the histogram of complete FRET cycles observed shows that the majority (> 90%) of molecules undergo only one full cycle (Fig. 3b). Inclusion of Lys-tRNALys allows the ribosome to decode the full message, and the histogram of complete cycles demonstrates that individual ribosomes (> 30%) undergo three complete cycles. Thus, both coding length and codon identity control the number of FRET cycles observed, confirming that each cycle corresponds to one round of elongation at a distinct codon. Translation of FKF does not show the same efficiency gains as translation of 6FK, most likely because the shortened mRNA length does not provide a sufficient number of observable events to reveal this more subtle effect.
Figure 3. Each FRET cycle reports on one round of elongation at a distinct codon.
(a) Observing intersubunit FRET on a three codon mRNA coding phenylalanine-lysine-phenylalanine (FKF) allows control over the number of codons translated, by exclusion or inclusion of Lys-tRNALys ternary complex in the delivery mixture. (b) Histograms of the number of complete FRET cycles observed for single ribosomes on the FKF mRNA in either the absence (137 ribosomes) or presence (333 ribosomes) of Lys-tRNALys ternary complex; histograms are normalized to the number of molecules showing one FRET cycle. (c) Normalized 2D histograms of FRET and Phe-(Cy2)tRNAPhe fluorescence postsynchronized to the high-low FRET transition at each codon (27 ribosomes).
Inclusion of Cy2-labeled Phe-tRNAPhe permits real-time correlation of stable tRNA arrival with the conformational state of the ribosome during the elongation cycle. Under simultaneous illumination at 532 nm and 488 nm, the arrival of Phe-(Cy2)tRNAPhe ternary complex is signaled by a burst of blue fluorescence spatially colocalized with the intersubunit FRET signal of an elongating ribosome. The observation of individual translation complexes makes possible the arbitrary post-synchronization of both the FRET and blue fluorescence data for each ribosome, eliminating temporal averaging, and allowing temporal correlation of tRNA binding and ribosomal conformational changes.
We followed translation of the FKF mRNA in the presence of Phe-(Cy2)tRNAPhe and unlabeled Lys-tRNALys, to correlate the stable arrival of distinct tRNAs with the fluctuation of our FRET signal. The binding of Phe-(Cy2)tRNAPhe monitors the position of the ribosome on the mRNA and reports on its mechanistic progression through the elongation cycle. Post-synchronization of both FRET and blue fluorescence trajectories from individual ribosomes, according to the first, second, and third high-low FRET transitions, reveals the arrival of Phe-(Cy2)tRNAPhe exclusively during the first and third cycles, consistent with the mRNA sequence (Fig. 3c). Within the time resolution of this measurement (100ms), the arrival of Phe-(Cy2)tRNAPhe occurs simultaneously with the high-low FRET transition (Supplementary Fig. 4), confirming our prior correlation of cognate tRNA binding and peptide bond formation with the conformational cycle of the ribosome23. Thus, each high-low-high FRET cycle represents a complete, codon-dependent elongation cycle. These data also demonstrate the ability to correlate in real-time the molecular events of tRNA selection and ribosome unlocking at each codon of an mRNA.
Measuring the growing nascent chain
The number of elongation cycles performed by each ribosome provides an indirect measure of the growing polypeptide chain. We monitored ribosome unlocking and locking on two distinct mRNAs in the presence of the macrolide antibiotic erythromycin (Fig. 4), which stalls translation by binding in the exit tunnel of the 50S subunit, blocking progression of the nascent chain35. To maximize the observation of individual elongation cycles, these experiments were performed using the optimized conditions described above.
Figure 4. Erythromycin stalls single translating ribosomes by blocking the nascent chain.
Histograms of the number of complete FRET cycles for single ribosomes elongating on the 12F (left panels) and 6FK (right panels) mRNAs in the absence (blue bars) and presence (red bars) of erythromycin. Datasets are composed of 1034, 477, 1145, and 779 ribosomes for 12F, 12F plus erythromycin, 6FK, and 6FK plus erythromycin, respectively. Histograms are normalized to the number of ribosomes showing one FRET cycle.
Previous studies have reported that translation in the presence of 1 µM erythromycin results in the production of aborted polypeptides between 6–8 amino acids in length36. In our experiments, ribosomes elongating on the 12F mRNA in the presence of erythromycin do not efficiently translate the entire mRNA, as evidenced by the histogram of observed FRET cycles. Instead, erythromycin causes an accumulation (>120% increase as compared to no drug) of molecules stalled after six elongation cycles (peptide length of 7 amino acids), in agreement with translational arrest upon obstruction of the polypeptide chain in the exit tunnel. In addition, codon-resolved FRET lifetimes show the effect of this steric clash on the unlocking/locking cycle of the ribosome (Supplementary Fig. 5). As the nascent polypeptide grows, the lifetimes of both the high- and low-FRET states increase. The interaction of the nascent chain and erythromycin may block the unlocking and locking of the ribosome.
Previous studies have not observed erythromycin sensitivity for translation of poly(U) sequences37,38, suggesting that synthesized poly(Phe) does not exit the ribosome via the exit tunnel. Thus, to confirm our results, we tested the effects of erythromycin on the translation of the heteropolymeric 6FK mRNA. As with translation of the 12F mRNA, erythromycin stalls translation beyond 6 codons, drastically reducing the number of ribosomes performing additional rounds of elongation. Taken together, these results suggest that each FRET cycle reports on a productive and codon-dependent peptide synthesis event; direct observation of these elongation cycles can indirectly measure the real-time length of the nascent chain in the exit tunnel. In addition, the observation that erythromycin can stall single ribosomes translating on a poly(U) message suggests that at least some fraction of synthesized poly(Phe) peptides proceed through the exit tunnel.
Codon-resolved antibiotic effects
Our FRET approach uncovers the detailed origins of global translational effects induced by other ribosome-targeting antibiotics. We followed translation of the 12F mRNA in the presence of three additional antibiotics with distinct mechanisms: fusidic acid, viomycin, and spectinomycin (Fig. 5). All three drugs efficiently inhibit translation, as evidenced by the small number of ribosomes that successfully translate all 12 codons of the 12F mRNA in the presence of each drug (Supplementary Fig. 6). To explore the effects of these drugs more deeply, we monitored their effects on the lifetimes of the locked (high-FRET) and unlocked (low-FRET) conformations of elongating ribosomes. To eliminate the influence of photobleaching and observation time censoring, the lifetimes reported here are for productive states; only high-FRET states followed by a low-FRET state and only low-FRET states followed by a high-FRET state were included. This analysis likely dampens the observed lifetime effects of each drug, but allows more precise determination of mechanistic effects.
Figure 5. The precise mechanisms and cumulative effects of ribosome-targeting antibiotics observed at codon resolution.
(a) Comparison of mean lifetime estimations for all low-FRET states, the first high-FRET state, and subsequent high-FRET states in the absence (blue bars) or presence (red bars) of fusidic acid (301 molecules, top panel), and for all low-FRET and high-FRET states in the absence (blue bars) and presence (red bars) or either spectinomycin (middle panel) or viomycin (bottom panel) (333 and 351 molecules, respectively). All lifetimes are for productive events, defined as events followed by the next FRET state (e.g. high-FRET followed by low-FRET) (b) The relative efficiency of translation at each codon in the presence of fusidic acid, spectinomycin, and viomycin, as compared to translation in the absence of antibiotics.
Fusidic acid stabilizes EF-G•GDP on the ribosome, preventing its dissociation and thus blocking incoming ternary complex39. In the presence of 50 µM fusidic acid, the average lifetime of the high-FRET state, where the ribosome waits to select tRNA, is lengthened approximately 30% (11.2 ± 1.4 s vs. 8.6 ± 0.4 s without drug), while the low-FRET state lifetime is unaffected (6.8 ± 0.9 s vs. 6.5 ± 0.3 s). However, EF-G•GDP is only present after the initial round of elongation, and thus fusidic acid should only inhibit these subsequent rounds of peptide synthesis. Comparison of the high-FRET lifetime at the first codon (8.2 ± 1.6 s) and subsequent codons (13.9 ± 2.5 s) reveals that fusidic acid affects only the high-FRET state of the ribosome beyond the first codon.
In contrast to fusidic acid, the antibiotics spectinomycin and viomycin are thought to function by blocking conformational rearrangements of the ribosome that drive elongation. Our method follows these rearrangements in real-time and can thus dissect the precise mechanism of these drugs. Spectinomycin binds to helix 34 in the 30S subunit, and stabilizes its conformation to block translocation40. Accordingly, 100 µM spectinomycin more than doubles the lifetime of the low-FRET state (13.3 ± 1.4 s vs. 6.5 ± 0.3 s without drug), while having no effect on the high-FRET state (9.2 ± 0.9 s vs. 8.6 ± 0.4 s). Structural data suggest that spectinomycin locks the 30S head in the “cocked” state adopted in the ratcheted ribosome41. The stabilization of the low FRET state observed here further links 30S head and ratcheting dynamics with the unlocked conformation of the ribosome.
Viomycin restricts the movement of tRNA on the ribosome, blocking translocation42. Here, 500 µM viomycin lengthens both the high- (20.4 ± 2.4 s vs. 8.6 ± 0.4 s) and low-FRET (13.3 ± 1.7 s vs. 6.5 ± 0.3 s) state lifetimes. Viomycin, which is thought to interact at the ribosomal subunit interface, may function by raising the energy barrier to ribosome unlocking and locking. Of the antibiotics tested here, viomycin exhibits the most severe affects, inhibiting translation more than 80% (Fig. 5b) and significantly increasing the number of events whose lifetime cannot be determined as a result of censoring (Supplementary Fig. 7a). This latter result suggests that viomycin slows translation even more drastically than indicated by the effect on productive event lifetimes. In addition, viomycin increases the pool of ribosomes that do not elongate after initiation (68 %) and significantly lengthens the first high-FRET state, as compared to other codons (Supplementary Figure 7b–c). Recent studies have demonstrated that viomycin can induce reverse translocation43, raising the possibility that the FRET cycles observed in the presence of viomycin report on both functional elongation events, as well as futile cycles of unlocking and locking.
Previous bulk and single-molecule studies have suggested that viomycin may act by stabilizing tRNA in the hybrid state15,17,44,45. Our data suggest that viomycin blocks translation by inhibiting the unlocking/locking cycle of the ribosome. A recent crystal structure of viomycin bound to the ribosome provides a structural explanation for both observations46. In this structure, viomycin binds at the subunit interface, interacting with bridge B2a at helix 44 of the 30S subunit and helix 69 of the 50S subunit. This interaction with helix 44 affects the position of residues A1492 and A1493, which are involved in the decoding of A-site tRNA. Binding of viomycin may stabilize the hybrid conformation of the tRNA and disrupt communication at the subunit interface necessary to mediate ribosome unlocking/locking.
The magnitude of antibiotic effects on translation at individual codons observed here appears modest. Fusidic acid, spectinomycin, and viomycin slow the individual steps of the ribosome elongation cycle at most by slightly more than a factor of two. As discussed above, these modest effects are partly due to our analytical approach, which excludes censored events to focus on mechanistic effects. Moreover, in our experiments, each drug is co-delivered with initiation factors, elongation factors, tRNA, and 50S subunits, and is not pre-incubated with immobilized complexes. This scenario mimics the competition of ligands for the ribosome likely to exist in the cell, and may limit the effects of each drug at early codons, particularly in comparison to bulk systems in which antibiotics are pre-incubated with ribosome complexes. In addition, the exquisite sensitivity of single-molecule methods allows detection of rare translation events that may occur even under efficient inhibition.
These apparently modest effects are magnified through multiple rounds of peptide synthesis. In the presence of fusidic acid, spectinomycin, or viomycin, translation beyond six codons is inhibited up to ~80% as compared to no drug (Fig. 5b). This underscores the importance that subtle effects on the efficiency of individual steps can have on the overall efficiency of translation.
The origins of ribosome processivity
The ribosome processively synthesizes polypeptides. However, there has been no direct observation of this phenomenon. We directly measure the unlocking/locking cycle of the ribosome, at each codon of the mRNA, within the full context of ongoing protein synthesis (Fig. 6). Comparison of the lifetimes of the high-FRET state at each of the first ten codons of the 12F mRNA shows no discernible codon dependence for this state. In contrast, the lifetimes of the low-FRET state, or unlocked ribosome, decrease sharply from the first to the second codon, and continue to decay as the ribosome penetrates deeper into the message; this leads to a greater than twofold increase in the rate of translocation by the time the ribosome arrives at the sixth codon (7.9 ± 0.7 s vs. 3.64 ± 0.6 s).
Figure 6. Monitoring the elongation cycle during multiple rounds of translation reveals increased translocation rates beyond initial codons.
Mean lifetime estimates for the high-FRET (locked) and low-FRET (unlocked) states of the ribosome at the first ten codons of the 12F mRNA (1034 molecules). Error bars are 95% confidence intervals from single-exponential fits. The schematics above each panel represent the global conformational transition represented by each set of FRET state lifetimes.
Ribosome processivity, defined as increasing rates of translation as compared to the rate of ribosome dissociation from mRNA, may originate from increased rates of translocation beyond initial codons, and not from more rapid tRNA selection. This trend is also observed on the 6FK mRNA, and on mRNAs with distinct Shine-Dalgarno sequences, suggesting that neither sequence context nor disruption of the Shine-Dalgarno interaction are the origin of the observed increase in processivity (Supplementary Fig. 7). Notably, the rate of translocation is increased most dramatically between the first and second codon, consistent with increased processivity upon loading of the E site. It will be interesting to probe the role of EF-P, which binds in the E site47, or of the lengthening peptide in the exit tunnel, in controlling processivity.
Discussion
Our results further confirm the general role of ribosome unlocking/locking during translation. At each codon, the ribosome undergoes a cycle of high-low-high FRET, consistent with a global conformational change that unlocks spontaneous fluctuations of the ribosomal subunits and tRNA after peptide bond formation. Upon translocation, the ribosome reverts to the original, locked, state. Ribosome unlocking likely involves the rearrangement of RNA:RNA intersubunit bridges11,12, thus facilitating other dynamic motions, such as ratcheting and hybrid state formation. In particular, helix 44 of the small subunit, which participates in the conserved bridge B2a and projects into the decoding center, might function as a switch signaling the unlocking of the ribosome. High-resolution structural snapshots of the ribosome in intermediate states of ratcheting reveal a movement of helix 44 and remodeling of bridge B2a12. Our FRET signal, which employs a label on helix 44, may thus be sensitive to conformational changes that signal the unlocking and locking of the ribosome. Interestingly, viomycin, which binds at the subunit interface and interacts with bridge B2a, blocks transitions between the high- and low-FRET states in our experiments, consistent with the idea that this region is involved in unlocking/locking and that our signal is sensitive to these events. Other studies have demonstrated the role of spontaneous ratcheting, as well as L1 and L7/L12 stalk dynamics in tRNA positioning17–20,48. However, the placement of our labels suggests that our FRET signal is blind to these fluctuations.
We propose a mechanism in which a global conformational change – ribosome unlocking – facilitates these dynamic events, encouraging tRNA movement and formation of the hybrid state (Fig. 7). Single-molecule force measurements suggest that peptide bond formation may also weaken the ribosome:mRNA interaction, facilitating translocation of the ribosome onto the next codon49. During translocation, the ribosome returns to the locked state, restricting these dynamic fluctuations to preserve the proper positioning of both the tRNA and the ribosome on the mRNA. In fact, recent structural evidence supports a model in which ribosome conformation may modulate the dynamic nature of specific elements, such as the L1 stalk50, consistent with previous single-molecule observations18.
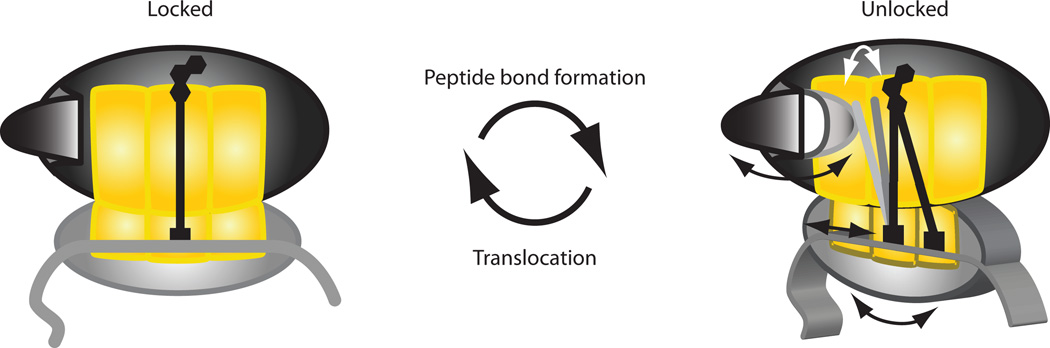
Figure 7. A general model for ribosome dynamics and function.
Interpretation of single-molecule data within the current locking/unlocking model of ribosome function. In the locked state, tRNA are held in the classical configuration, and the L1 stalk is kept open. The stability of this conformation may preserve reading frame and allow precise manipulation of tRNA during selection and catalysis. Ribosome unlocking upon peptide bond formation permits classical↔hybrid and open↔closed fluctuations of the tRNA and L1 stalk, respectively, and promotes spontaneous ratcheting. Unlocking may also loosen the ribosome:mRNA interaction. These motions might facilitate movement of tRNA and mRNA during translocation or initiation. The action of EF-G returns the ribosome to the locked state. L1 opening during this transition may facilitate dissociation of the E-site tRNA.
By following ribosome unlocking/locking in real time during translation, we observe and correlate many of the detailed molecular events of elongation on a codon by codon basis. In addition to this molecular detail, our data provide a global perspective that reports on the real-time progress of ongoing protein synthesis. Here, we have demonstrated the steps at which antibiotics inhibit translation, and have shown how translocation becomes more rapid as the ribosome elongates at early codons. The approach outlined here should allow further correlation of molecular-level translational events with mRNA context, reveal the origins and nature of rare or transient translational events, and allow parallel analysis of many mRNAs within the full context of translation. Future experiments might also explore the role of specific intersubunit bridges in the unlocking/locking process.
Methods
Single-molecule translation experiments
We prepared fluorescently-labeled 30S (Cy3 or Cy3B) and 50S (Cy5) subunits, translation factors, S1, mRNA, labeled (Cy2) and unlabeled tRNA as described previously14,22,23. To assemble 30S PICs, we mixed 0.25 µM Cy3-30S pre-incubated with stoichiometric S1, 1 µM IF2, 1 µM fMet-tRNAfMet, 1 µM biotinylated mRNA, and 4 mM GTP in a previously described Tris-based polymix buffer system without reducing agents23, and subsequently incubated this mixture at 37 °C for 5 minutes. The Mg2+ concentration in all buffers was 5 mM.
Prior to surface immobilization, we diluted assembled PICs in polymix buffer containing 1 µM IF2, 1 µM fMet-tRNAfMet, and 4 mM GTP23. We next immobilized these diluted PICs on a neutravidin-derivatized quartz slide, and washed with polymix buffer containing 1 µM IF2, 1 µM fMet-tRNAfMet, 4 mM GTP, 1 mM Trolox (for Cy5 stabilization), and an oxygen-scavenging system (2.5 mM 3,4 dihydroxybenzoic acid and 250 nM protocatechuate dioxygenase24). To initiate translation, we delivered 50 nM Cy5-50S, 1 µM IF2, 20–100 nM EF-G, 20–100 unlabeled nM ternary complex, 20 nM Phe-(Cy2)tRNAPhe (where applicable), and antibiotics (where applicable, in specified concentrations) in the polymix wash buffer using a controlled syringe pump. We prepared Phe-tRNAPhe and Lys-tRNALys ternary complexes immediately prior to use as previously described23.
Instrumentation and analysis
We performed all single-molecule fluorescence measurements using a prism-based total internal reflection instrument described previously24. For FRET measurements, we employed a diode-pumped solid-state 532 nm laser at 1 kW cm2 −1 intensity, as measured at the prism. For three-color experiments were employed dual-illumination at 532 nm and 488 nm at intensities of 1 kW (cm2)−1 and 400 W (cm2)−1, respectively. A Quad-View device (Photometrics) separated fluorescent emission into distinct color channels, corresponding to Cy2, Cy3, and Cy5 fluorescence and projected this emission signal onto an EMCCD camera (Andor Technology). We performed image acquisition using the MetaMorph software package (Molecular Devices) and subsequent analysis using scripts written in MATLAB (The Mathworks). We assigned FRET and blue fluorescence states using a hidden Markov Model approach as previously described24, and confirmed these assignments by visual inspection. For lifetime analysis, we selected only productive FRET states (followed by the next FRET state, e.g. low-FRET followed by high-FRET and vice versa, and not terminated by photobleaching or blinking), to eliminate the influence of spurious photophysical events. All lifetime estimates are the result of maximum likelihood parameter estimation for single-exponential distributions composed of between 301–1145 molecules.
Supplementary Material
Acknowledgements
This work was funded by grants to J.D.P. from NIH.
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website
Author Contributions
C.E.A. performed experiments and data analysis. C.E.A. and J.D.P discussed results and wrote the manuscript.
Competing Interests Statement
The authors declare no competing financial interests.
References
- 1.Green R, Noller HF. Ribosomes and translation. Annu. Rev. Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 2.Wintermeyer W, et al. Mechanisms of elongation on the ribosome: dynamics of a macromolecular machine. Biochem Soc Trans. 2004;32:733–737. doi: 10.1042/BST0320733. [DOI] [PubMed] [Google Scholar]
- 3.Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr Opin Chem Biol. 2008;12:674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spirin AS. A model of the functioning ribosome: locking and unlocking of the ribosome subparticles. Cold Spring Harb Symp Quant Biol. 1969;34:197–207. doi: 10.1101/sqb.1969.034.01.026. [DOI] [PubMed] [Google Scholar]
- 6.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 7.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 8.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 9.Agirrezabala X, et al. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Julian P, et al. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci U S A. 2008;105:16924–16927. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 12.Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 14.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci U S A. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim HD, Puglisi JD, Chu S. Fluctuations of transfer RNAs between classical and hybrid states. Biophys J. 2007;93:3575–3582. doi: 10.1529/biophysj.107.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munro JB, Altman RB, O'Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 19.Fei J, et al. Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proc Natl Acad Sci U S A. 2009;106:15702–15707. doi: 10.1073/pnas.0908077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornish PV, et al. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci U S A. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wen JD, et al. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorywalska M, et al. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res. 2005;33:182–189. doi: 10.1093/nar/gki151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marshall RA, Dorywalska M, Puglisi JD. Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci U S A. 2008;105:15364–15369. doi: 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall RA, Aitken CE, Puglisi JD. GTP hydrolysis by IF2 guides progression of the ribosome into elongation. Mol Cell. 2009;35:37–47. doi: 10.1016/j.molcel.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang ST, Xu YC, Dennis P, Bremer H. mRNA composition and control of bacterial gene expression. J Bacteriol. 2000;182:3037–3044. doi: 10.1128/jb.182.11.3037-3044.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savelsbergh A, et al. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Molecular Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 29.Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 31.Bilgin N, Claesens F, Pahverk H, Ehrenberg M. Kinetic properties of Escherichia coli ribosomes with altered forms of S12. J Mol Biol. 1992;224:1011–1027. doi: 10.1016/0022-2836(92)90466-w. [DOI] [PubMed] [Google Scholar]
- 32.Johansson M, Bouakaz E, Lovmar M, Ehrenberg M. The kinetics of ribosomal peptidyl transfer revisited. Mol Cell. 2008;30:589–598. doi: 10.1016/j.molcel.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 33.Uemura S, et al. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. doi: 10.1038/nature08925. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vanzi F, Takagi Y, Shuman H, Cooperman BS, Goldman YE. Mechanical studies of single ribosome/mRNA complexes. Biophys J. 2005;89:1909–1919. doi: 10.1529/biophysj.104.056283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlunzen F, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 36.Tenson T, Lovmar M, Ehrenberg M. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J Mol Biol. 2003;330:1005–1014. doi: 10.1016/s0022-2836(03)00662-4. [DOI] [PubMed] [Google Scholar]
- 37.Vazquez D. Antibiotics affecting chloramphenicol uptake by bacteria. Their effect on amino acid incorporation in a cell-free system. Biochim Biophys Acta. 1966;114:289–295. doi: 10.1016/0005-2787(66)90310-8. [DOI] [PubMed] [Google Scholar]
- 38.Karahalios P, et al. On the mechanism of action of 9-O-arylalkyloxime derivatives of 6-O-mycaminosyltylonolide, a new class of 16-membered macrolide antibiotics. Mol Pharmacol. 2006;70:1271–1280. doi: 10.1124/mol.106.026567. [DOI] [PubMed] [Google Scholar]
- 39.Bodley JW, Zieve FJ, Lin L. Studies on translocation. IV. The hydrolysis of a single round of guanosine triphosphate in the presence of fusidic acid. J Biol Chem. 1970;245:5662–5667. [PubMed] [Google Scholar]
- 40.Brink MF, Brink G, Verbeet MP, de Boer HA. Spectinomycin interacts specifically with the residues G1064 and C1192 in 16S rRNA, thereby potentially freezing this molecule into an inactive conformation. Nucleic Acids Res. 1994;22:325–331. doi: 10.1093/nar/22.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borovinskaya MA, Shoji S, Holton JM, Fredrick K, Cate JH. A steric block in translation caused by the antibiotic spectinomycin. ACS Chem Biol. 2007;2:545–552. doi: 10.1021/cb700100n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liou YF, Tanaka N. Dual actions of viomycin on the ribosomal functions. Biochem Biophys Res Commun. 1976;71:477–483. doi: 10.1016/0006-291x(76)90812-3. [DOI] [PubMed] [Google Scholar]
- 43.Shoji S, Walker SE, Fredrick K. Reverse translocation of tRNA in the ribosome. Mol Cell. 2006;24:931–942. doi: 10.1016/j.molcel.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ermolenko DN, et al. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat Struct Mol Biol. 2007;14:493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 45.Feldman MB, Terry DS, Altman RB, Blanchard SC. Aminoglycoside activity observed on single pre-translocation ribosome complexes. Nat Chem Biol. 2010;6:54–62. doi: 10.1038/nchembio.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stanley RE, Blaha G, Grodzicki RL, Strickler MD, Steitz TA. The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nat Struct Mol Biol. 2010;17:289–293. doi: 10.1038/nsmb.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blaha G, Stanley RE, Steitz TA. Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science. 2009;325:966–970. doi: 10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diaconu M, et al. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 49.Uemura S, et al. Peptide bond formation destabilizes Shine-Dalgarno interaction on the ribosome. Nature. 2007;446:454–457. doi: 10.1038/nature05625. [DOI] [PubMed] [Google Scholar]
- 50.Gao YG, et al. The Structure of the Ribosome with Elongation Factor G Trapped in the Posttranslocational State. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.