The crystal stucture of 3-amino-4-nitrobenzyl displays intramolecular resonance-assisted hydrogen bonding between the ortho amino and nitro groups in addition to an intermolecular network of hydrogen bonding and π-stacking.
Keywords: crystal structure, 3-amino-4-nitrobenzyl acetate, intramolecular, intermolecular, resonance-assisted hydrogen bonding, 5-amino-2-nitrobenzoic acid
Abstract
Yellow crystals of the title compound 3-amino-4-nitrobenzyl acetate, C9H10N2O4, were isolated from the reaction of acetic anhydride with (5-amino-2-nitrophenyl)methanol, prepared from reduction of commerically available 5-amino-2-nitrobenzoic acid with borane–THF. The molecule is essentially planar (r.m.s. deviation = 0.028 Å). The molecules are linked by intermolecular N—H⋯O hydrogen-bonding interactions between the carbonyl and amine groups, forming a zigzag chain along the b-axis direction lying in a plane parallel to (-102). The chains are stacked along the c axis by π–π interactions [centroid–centroid distances = 3.6240 (3) and 3.5855 (4) Å]. A strong intramolecular N—H⋯O hydrogen-bonding interaction is observed between the nitro group and the amine group [2.660 (2) Å].
Chemical Context
Often commercially available chemicals are sold with minor impurities in the range 1–5%; the user may choose to ‘use as received’ or further purify. The identities of the impurities are rarely disclosed in fine chemicals. Though these impurities may serve as benign spectators, in some cases they might hinder reactivity and/or produce undesirable by-products that are difficult to separate from the desired product. Therefore, it is important to identify these impurities to allow the users to decide if further purification is warranted. We recently purchased 5-amino-2-nitrobenzoic acid from Acros Organics© (5 g, 97%, AC33074-0050) for our ongoing studies of photo-induced decarboxylation of ortho-nitrobenzyl esters (Cabane et al., 2010 ▸; Pocker et al., 1978 ▸). The isolation of the title compound, 3-amino-4-nitrobenzyl acetate, after the reaction of crude (5-amino-2-nitrophenyl)methanol, prepared from the reduction of 5-amino-2-nitrobenzoic acid, with acetic anhydride suggests 3-amino-4-nitrobenzoic acid is an impurity in the commercially available starting material.
Structural Commentary
The asymmetric unit of the title compound (Fig. 1 ▸) displays an essentially planar molecule (r.m.s.d. 0.028 Å) with the amine, nitro and acetate groups resting in the plane of the arene. The carbonyl, C=O [1.208 (2) Å], and ester, C—O [1.3477 (19) Å], bond distances are unassuming. The nitro bond distances [O1—N1 1.2500 (16) and O2—N1 1.2401 (17) Å] are similar to those in N-(3-chlorophenyl)-3-nitropyridin-2-amine [1.222 (2) and 1.245 (2) Å] (Aznan et al., 2011 ▸). Atom O1 of the nitro group is involved in strong intramolecular hydrogen bonding [graph set S1, 1(6)] between H2B of the amine at a distance of 2.06 (2) Å, forming a rigid, thermodynamically stable six-membered ring (Fig. 1 ▸). The elongated O1—N1 bond distance, as compared to the O2—N1 distance, is consistent with resonance-assisted hydrogen bonding between O1 and H2B (Beck & Mo, 2006 ▸).
Figure 1.
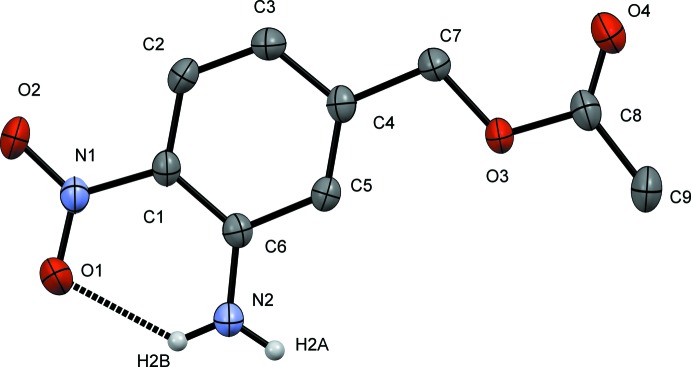
A displacement ellipsoid plot of 3-amino-4-nitrobenzyl acetate (50% probability level). C-bound H atoms have been omitted for clarity.
Supramolecular Features
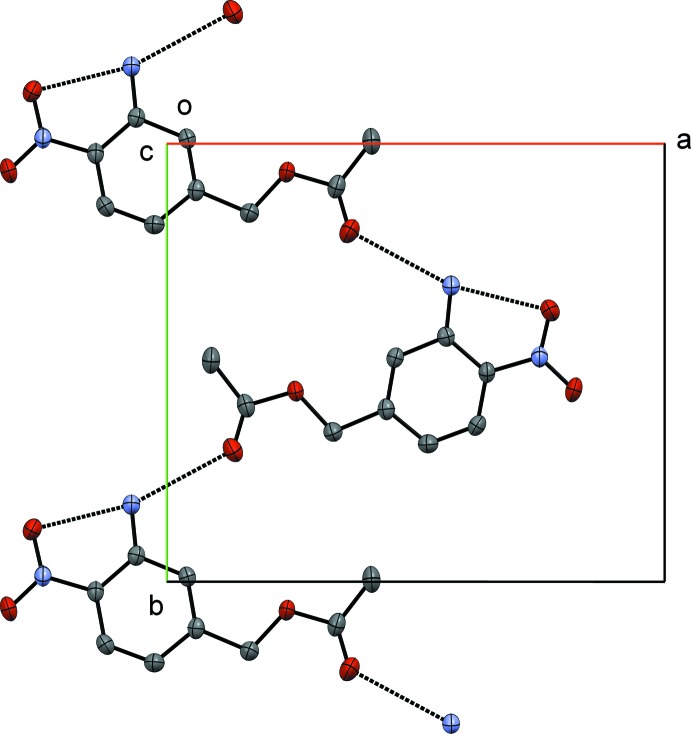
The crystal structure of 3-amino-4-nitrobenzyl acetate has interesting supramolecular features. The molecules are arranged in layers held together by intermolecular N2—H2A⋯O4 [3.005 (2) Å] hydrogen bonding [graph set C1,1(9)] interactions between the carbonyl and amine groups forming a zigzag chain along the b-axis direction (Fig. 2 ▸ and Table 1 ▸) lying in a plane parallel to ( 02). A view of a single layer along the ab plane, observed down the c axis (Fig. 2 ▸) provides a representative illustration of the hydrogen-bonding interactions of 3-amino-4-nitrobenzyl acetate. Observing the unit cell along the b-axis (Fig. 3 ▸) shows four layers along the c axis separated at a distance of 3.3163 (10) Å with the arene groups stacked one above the other. The chains stack along the c axis by π–π interactions [centroid–centroid distances = 3.6240 (3) Å (symmetry code 1 − x, 1 − y, 1 − z) and 3.5855 (4) Å (symmetry code 1 − x, y,
02). A view of a single layer along the ab plane, observed down the c axis (Fig. 2 ▸) provides a representative illustration of the hydrogen-bonding interactions of 3-amino-4-nitrobenzyl acetate. Observing the unit cell along the b-axis (Fig. 3 ▸) shows four layers along the c axis separated at a distance of 3.3163 (10) Å with the arene groups stacked one above the other. The chains stack along the c axis by π–π interactions [centroid–centroid distances = 3.6240 (3) Å (symmetry code 1 − x, 1 − y, 1 − z) and 3.5855 (4) Å (symmetry code 1 − x, y,  − z)].
− z)].
Figure 2.
A single of layer of the unit cell of 3-amino-4-nitrobenzoic acid through the ab plane (observed down the c axis), highlighting the hydrogen-bonding motif.
Table 1. Hydrogen-bond geometry (, ).
| DHA | DH | HA | D A | DHA |
|---|---|---|---|---|
| N2H2AO4i | 0.83(2) | 2.18(2) | 3.005(2) | 171.5(17) |
| N2H2BO1 | 0.84(2) | 2.06(2) | 2.6600(19) | 128.0(16) |
| N2H2BO1ii | 0.84(2) | 2.44(2) | 3.1443(19) | 142.7(16) |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Figure 3.

A displacement ellipsoid plot of the unit cell of 3-amino-4-nitrobenzoic acid observed down the b axis.
Database Survey
For a related benzyl acetate structure, see Kasuga et al. (2015 ▸). For alkyl- and aryl-3-amino-4-nitro-benzoates and benzoic acids displaying similar intramolecular hydrogen bonding between the amino and nitro groups, see: Narendra Babu et al. (2009 ▸); Abdul Rahim et al. (2010 ▸); Yoon et al. (2011 ▸); Yoon et al. (2012 ▸).
Synthesis and Crystallization
(5-Amino-2-nitrophenyl)methanol: (5-amino-2-nitrophenyl)methanol was prepared by a modified literature protocol (Yoon et al. 1973 ▸). To a solution of 5-amino-2-nitrobenzoic acid (97%, 1.5 g, 8.2 mmol) dissolved in tetrahydrofuran (10 mL), borane–THF (27.6 mL, 1.0 M in THF, 27.6 mmol) was added dropwise by dropping funnel over 30 minutes. The reaction was stirred overnight at room temperature. The reaction was quenched with aqueous potassium hydroxide (2.45 M) until pH 11 was reached and continued to be stirred for 6 h, resulting in a greenish-brown solution. The solution was treated with a saturated solution of potassium carbonate followed by treatment with hydrochloric acid until pH 1 was reached. The reaction mixture was extracted with diethyl ether three times; organic portions were collected and dried with anhydrous sodium sulfate overnight. The solution was filtered under vacuum, the filtrate was collected and all solvent removed under rotary evaporation to give a green powder (0.68 g, 49%). 1H NMR, (300 MHz, acetone-d 6) δ: 4.61 (t, 1H, –OH, 3 J HH = 5.3 Hz), 4.95 (d, 2H, CH2, 3 J HH = 5.3 Hz) , 6.03 (bs, 2H, NH2), 6.63 (dd, 1H, Ar-H, 3 J HH = 8.8 Hz, 3 J HH = 2.3 Hz), 7.07 (m, 1H, Ar-H), 8.02 (dd, 1H, 3 J HH = 9.4 Hz, 3 J HH = 3.0 Hz) (Aujard et al. 2006 ▸). Note: minor impurities were observed in the base line in the aromatic region.
3-Amino-4-nitrobenzyl acetate: (5-amino-2-nitrophenyl)methanol (10 mg, 0.0595 mmol) and triethylamine (17 µL, 0.119 mmol) were dissolved in acetonitrile-d 6 (0.7 mL) and added to an NMR tube. Acetic anhydride (11.2 µL, 0.119 mmol) was added to the tube via a syringe. The tube was held at room temperature overnight. On completion of the reaction the solvent was removed in vacuo and the residue was reconstituted in a minimum amount of methylene chloride. The sample was loaded on a column of silica and eluted with an ethyl acetate/hexane solution (70/30 v/v %). The separated solutions were allowed to slowly evaporate at room temperature. The parent compound (5-amino-2-nitrobenzyl acetate) elutes first and is isolated as a yellow powder. 1H NMR (300 MHz, CDCl3) δ: 2.10 (s, 3H, CH 3), 4.35 (bs, 2H, NH 2), 5.50 (s, 2H, CH 2), 6.55 (dd, 1H, Ar-H, 3 J HH = 8.9 Hz, 5 J HH = 2.5 Hz), 6.68 (m, 1H, Ar-H), 8.09 (dd, 1H, Ar-H, 3 J HH = 8.9 Hz, 5 J HH = 2.5 Hz) (Serafinowski et al. 2008 ▸). Yellow crystals of the title compound were isolated (less than 1 mg) in later eluate. 1H NMR (300 MHz, CDCl3) δ: 2.19 (s, 3H, CH 3), 5.53 (s, 2H, CH 2), 7.44 (bs, 2H, NH 2), 7.65 (dd, 1H, Ar-H, 3 J HH = 8.9 Hz, 5 J HH = 2.5 Hz), 7.75 (m, 1H, Ar-H), 8.15 (d, 1H, Ar-H, 3 J HH = 8.9 Hz).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. Hydrogen atoms were refined freely.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C9H10N2O4 |
| M r | 210.19 |
| Crystal system, space group | Monoclinic, C2/c |
| Temperature (K) | 173 |
| a, b, c () | 14.4803(15), 11.4054(11), 13.0936(13) |
| () | 116.341(8) |
| V (3) | 1937.9(4) |
| Z | 8 |
| Radiation type | Mo K |
| (mm1) | 0.12 |
| Crystal size (mm) | 0.25 0.25 0.10 |
| Data collection | |
| Diffractometer | Rigaku Mercury375R |
| Absorption correction | Multi-scan (REQAB; Rigaku, 1998 ▸) |
| T min, T max | 0.840, 1.000 |
| No. of measured, independent and observed [I > 2(I)] reflections | 8409, 1759, 1348 |
| R int | 0.045 |
| (sin /)max (1) | 0.601 |
| Refinement | |
| R[F 2 > 2(F 2)], wR(F 2), S | 0.037, 0.098, 1.06 |
| No. of reflections | 1759 |
| No. of parameters | 176 |
| H-atom treatment | All H-atom parameters refined |
| max, min (e 3) | 0.21, 0.17 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989015008750/pk2548sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015008750/pk2548Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989015008750/pk2548Isup3.cml
CCDC reference: 1063364
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
Acknowledgments are made to Armstrong State University and to the Donors of the American Chemical Society Petroleum Research Fund for support (or partial support) of this research (PRF No. 53848-UNI3). Additional support was provided by the NSF–STEP Program under Award No. DUE-0856593.
supplementary crystallographic information
Crystal data
| C9H10N2O4 | F(000) = 880 |
| Mr = 210.19 | Dx = 1.441 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71075 Å |
| a = 14.4803 (15) Å | Cell parameters from 513 reflections |
| b = 11.4054 (11) Å | θ = 1.6–25.4° |
| c = 13.0936 (13) Å | µ = 0.12 mm−1 |
| β = 116.341 (8)° | T = 173 K |
| V = 1937.9 (4) Å3 | Prism, yellow |
| Z = 8 | 0.25 × 0.25 × 0.10 mm |
Data collection
| Rigaku Mercury375R (2x2 bin mode) diffractometer | 1759 independent reflections |
| Radiation source: Sealed Tube | 1348 reflections with I > 2σ(I) |
| Graphite Monochromator monochromator | Rint = 0.045 |
| Detector resolution: 13.6612 pixels mm-1 | θmax = 25.3°, θmin = 2.4° |
| profile data from ω scans | h = −17→17 |
| Absorption correction: multi-scan (REQAB; Rigaku, 1998) | k = −13→13 |
| Tmin = 0.840, Tmax = 1.000 | l = −15→15 |
| 8409 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Hydrogen site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.037 | All H-atom parameters refined |
| wR(F2) = 0.098 | w = 1/[σ2(Fo2) + (0.0578P)2 + 0.2118P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.06 | (Δ/σ)max < 0.001 |
| 1759 reflections | Δρmax = 0.21 e Å−3 |
| 176 parameters | Δρmin = −0.17 e Å−3 |
| 0 restraints |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O3 | 0.74138 (8) | 0.43515 (9) | 0.74195 (10) | 0.0324 (3) | |
| O1 | 0.23008 (8) | 0.61948 (10) | 0.50492 (11) | 0.0418 (3) | |
| O2 | 0.18245 (8) | 0.43814 (10) | 0.46692 (11) | 0.0436 (3) | |
| O4 | 0.86707 (9) | 0.30050 (11) | 0.79735 (12) | 0.0456 (4) | |
| N1 | 0.25141 (9) | 0.51321 (11) | 0.50551 (11) | 0.0288 (3) | |
| N2 | 0.42892 (12) | 0.67596 (12) | 0.60612 (12) | 0.0300 (3) | |
| H2A | 0.4821 (16) | 0.7166 (16) | 0.6337 (16) | 0.036 (5)* | |
| H2B | 0.3700 (16) | 0.7046 (15) | 0.5821 (16) | 0.039 (5)* | |
| C6 | 0.43920 (11) | 0.55950 (12) | 0.60073 (12) | 0.0228 (3) | |
| C1 | 0.35704 (10) | 0.47782 (13) | 0.55271 (12) | 0.0245 (3) | |
| C4 | 0.55919 (11) | 0.39372 (13) | 0.64720 (12) | 0.0252 (3) | |
| C5 | 0.54112 (10) | 0.51152 (13) | 0.64747 (11) | 0.0228 (3) | |
| H5 | 0.5978 (13) | 0.5675 (14) | 0.6814 (13) | 0.024 (4)* | |
| C2 | 0.37680 (12) | 0.35678 (14) | 0.54982 (13) | 0.0295 (4) | |
| H2 | 0.3212 (13) | 0.3041 (14) | 0.5154 (15) | 0.031 (4)* | |
| C3 | 0.47512 (12) | 0.31496 (14) | 0.59587 (14) | 0.0312 (4) | |
| H3 | 0.4895 (13) | 0.2318 (16) | 0.5930 (15) | 0.033 (4)* | |
| C7 | 0.66619 (11) | 0.34199 (14) | 0.69873 (14) | 0.0296 (4) | |
| H7A | 0.6780 (14) | 0.2874 (16) | 0.7620 (16) | 0.040 (5)* | |
| H7B | 0.6772 (12) | 0.2957 (14) | 0.6407 (15) | 0.032 (4)* | |
| C8 | 0.84105 (11) | 0.40198 (15) | 0.79024 (13) | 0.0307 (4) | |
| C9 | 0.91100 (13) | 0.50537 (18) | 0.83137 (18) | 0.0419 (5) | |
| H9A | 0.8961 (16) | 0.5562 (19) | 0.7680 (19) | 0.055 (6)* | |
| H9B | 0.9809 (17) | 0.4806 (16) | 0.8703 (17) | 0.046 (5)* | |
| H9C | 0.8934 (17) | 0.554 (2) | 0.881 (2) | 0.067 (7)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O3 | 0.0175 (5) | 0.0288 (6) | 0.0445 (7) | 0.0021 (4) | 0.0080 (5) | −0.0018 (5) |
| O1 | 0.0242 (6) | 0.0319 (7) | 0.0621 (8) | 0.0054 (5) | 0.0128 (6) | 0.0060 (5) |
| O2 | 0.0194 (6) | 0.0411 (7) | 0.0601 (8) | −0.0073 (5) | 0.0085 (6) | −0.0033 (6) |
| O4 | 0.0271 (6) | 0.0400 (8) | 0.0605 (8) | 0.0089 (5) | 0.0111 (6) | −0.0009 (6) |
| N1 | 0.0188 (6) | 0.0314 (8) | 0.0327 (7) | 0.0002 (5) | 0.0083 (5) | 0.0031 (6) |
| N2 | 0.0199 (7) | 0.0271 (8) | 0.0379 (8) | 0.0002 (6) | 0.0082 (6) | −0.0025 (6) |
| C6 | 0.0214 (7) | 0.0266 (8) | 0.0210 (7) | 0.0013 (6) | 0.0098 (6) | 0.0011 (6) |
| C1 | 0.0180 (7) | 0.0307 (8) | 0.0234 (7) | 0.0004 (6) | 0.0078 (6) | 0.0027 (6) |
| C4 | 0.0210 (7) | 0.0312 (8) | 0.0239 (7) | 0.0010 (6) | 0.0104 (6) | 0.0002 (6) |
| C5 | 0.0198 (8) | 0.0271 (8) | 0.0212 (7) | −0.0025 (6) | 0.0086 (6) | −0.0011 (6) |
| C2 | 0.0218 (8) | 0.0283 (9) | 0.0358 (9) | −0.0063 (7) | 0.0105 (7) | −0.0029 (7) |
| C3 | 0.0279 (8) | 0.0238 (9) | 0.0406 (9) | −0.0004 (6) | 0.0142 (7) | −0.0017 (7) |
| C7 | 0.0237 (8) | 0.0262 (8) | 0.0363 (9) | 0.0007 (6) | 0.0110 (7) | −0.0024 (7) |
| C8 | 0.0211 (8) | 0.0379 (10) | 0.0305 (8) | 0.0056 (7) | 0.0091 (7) | 0.0011 (7) |
| C9 | 0.0214 (9) | 0.0483 (12) | 0.0489 (11) | −0.0017 (8) | 0.0091 (8) | −0.0015 (9) |
Geometric parameters (Å, º)
| O3—C7 | 1.4449 (19) | C4—C3 | 1.419 (2) |
| O3—C8 | 1.3477 (19) | C4—C7 | 1.509 (2) |
| O1—N1 | 1.2500 (16) | C5—H5 | 0.978 (17) |
| O2—N1 | 1.2401 (17) | C2—H2 | 0.944 (17) |
| O4—C8 | 1.208 (2) | C2—C3 | 1.362 (2) |
| N1—C1 | 1.4303 (19) | C3—H3 | 0.975 (18) |
| N2—H2A | 0.83 (2) | C7—H7A | 0.988 (19) |
| N2—H2B | 0.83 (2) | C7—H7B | 0.994 (17) |
| N2—C6 | 1.342 (2) | C8—C9 | 1.491 (3) |
| C6—C1 | 1.419 (2) | C9—H9A | 0.96 (2) |
| C6—C5 | 1.4320 (19) | C9—H9B | 0.95 (2) |
| C1—C2 | 1.414 (2) | C9—H9C | 0.97 (2) |
| C4—C5 | 1.369 (2) | ||
| C8—O3—C7 | 116.19 (12) | C3—C2—C1 | 120.94 (14) |
| O1—N1—C1 | 119.36 (12) | C3—C2—H2 | 119.5 (10) |
| O2—N1—O1 | 121.00 (12) | C4—C3—H3 | 118.7 (10) |
| O2—N1—C1 | 119.64 (13) | C2—C3—C4 | 119.75 (15) |
| H2A—N2—H2B | 122.7 (17) | C2—C3—H3 | 121.5 (10) |
| C6—N2—H2A | 118.1 (12) | O3—C7—C4 | 109.47 (13) |
| C6—N2—H2B | 119.1 (12) | O3—C7—H7A | 108.3 (11) |
| N2—C6—C1 | 125.60 (13) | O3—C7—H7B | 109.9 (9) |
| N2—C6—C5 | 118.24 (13) | C4—C7—H7A | 112.5 (10) |
| C1—C6—C5 | 116.16 (13) | C4—C7—H7B | 110.3 (10) |
| C6—C1—N1 | 122.12 (13) | H7A—C7—H7B | 106.2 (14) |
| C2—C1—N1 | 117.02 (13) | O3—C8—C9 | 111.22 (14) |
| C2—C1—C6 | 120.86 (13) | O4—C8—O3 | 122.52 (15) |
| C5—C4—C3 | 119.84 (14) | O4—C8—C9 | 126.26 (15) |
| C5—C4—C7 | 122.83 (14) | C8—C9—H9A | 108.1 (13) |
| C3—C4—C7 | 117.33 (14) | C8—C9—H9B | 110.5 (11) |
| C6—C5—H5 | 116.3 (9) | C8—C9—H9C | 110.8 (13) |
| C4—C5—C6 | 122.40 (13) | H9A—C9—H9B | 115.0 (17) |
| C4—C5—H5 | 121.3 (9) | H9A—C9—H9C | 102.2 (18) |
| C1—C2—H2 | 119.6 (10) | H9B—C9—H9C | 110.0 (17) |
| O1—N1—C1—C6 | 0.9 (2) | C5—C6—C1—N1 | 178.48 (13) |
| O1—N1—C1—C2 | −179.17 (13) | C5—C6—C1—C2 | −1.44 (19) |
| O2—N1—C1—C6 | −178.35 (13) | C5—C4—C3—C2 | −1.7 (2) |
| O2—N1—C1—C2 | 1.6 (2) | C5—C4—C7—O3 | −2.0 (2) |
| N1—C1—C2—C3 | −178.07 (14) | C3—C4—C5—C6 | 2.1 (2) |
| N2—C6—C1—N1 | −1.2 (2) | C3—C4—C7—O3 | 177.40 (13) |
| N2—C6—C1—C2 | 178.91 (14) | C7—O3—C8—O4 | 0.1 (2) |
| N2—C6—C5—C4 | 179.13 (13) | C7—O3—C8—C9 | 179.98 (14) |
| C6—C1—C2—C3 | 1.9 (2) | C7—C4—C5—C6 | −178.49 (13) |
| C1—C6—C5—C4 | −0.5 (2) | C7—C4—C3—C2 | 178.86 (15) |
| C1—C2—C3—C4 | −0.2 (2) | C8—O3—C7—C4 | −179.68 (12) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2A···O4i | 0.83 (2) | 2.18 (2) | 3.005 (2) | 171.5 (17) |
| N2—H2B···O1 | 0.84 (2) | 2.06 (2) | 2.6600 (19) | 128.0 (16) |
| N2—H2B···O1ii | 0.84 (2) | 2.44 (2) | 3.1443 (19) | 142.7 (16) |
Symmetry codes: (i) −x+3/2, y+1/2, −z+3/2; (ii) −x+1/2, −y+3/2, −z+1.
References
- Abdul Rahim, A. S., Abd Hamid, S., Narendra Babu, S. N., Loh, W.-S. & Fun, H.-K. (2010). Acta Cryst. E66, o846–o847. [DOI] [PMC free article] [PubMed]
- Aujard, I., Benbrahim, C., Gouget, M., Ruel, O., Baudin, J.-B., Neveu, P. & Jullien, L. (2006). Chem. Eur. J. 12, 6865–6879. [DOI] [PubMed]
- Aznan, A. M. A., Abdullah, Z., Ng, S. W. & Tiekink, E. R. T. (2011). Acta Cryst. E67, o3076. [DOI] [PMC free article] [PubMed]
- Beck, J. F. & Mo, Y. (2006). J. Comput. Chem. 4, 455–466.
- Cabane, E., Malinova, V. & Meier, W. (2010). Macromol. Chem. Phys. 211, 1847–1856.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Kasuga, N. C., Saito, Y., Sato, H. & Yamaguchi, K. (2015). Acta Cryst. E71, 483–486. [DOI] [PMC free article] [PubMed]
- Narendra Babu, S. N., Abdul Rahim, A. S., Abd Hamid, S., Balasubramani, K. & Fun, H.-K. (2009). Acta Cryst. E65, o2070–o2071. [DOI] [PMC free article] [PubMed]
- Pocker, Y., Davison, B. L. & Deits, T. L. (1978). J. Am. Chem. Soc. 100, 3564–3567.
- Rigaku (1998). REQAB. Rigaku Corporation, Tokyo, Japan.
- Serafinowski, P. J. & Garland, P. B. (2008). Org. Biomol. Chem. 6, 3284–3291. [DOI] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Yoon, Y. K., Ali, M. A., Choon, T. S., Loh, W.-S. & Fun, H.-K. (2011). Acta Cryst. E67, o2606. [DOI] [PMC free article] [PubMed]
- Yoon, Y. K., Manogaran, E., Ali, M. A., Arshad, S. & Razak, I. A. (2012). Acta Cryst. E68, o1684. [DOI] [PMC free article] [PubMed]
- Yoon, N. M., Pak, C. S., Krishnamurthy, S. & Stocky, T. P. (1973). J. Org. Chem. 38, 2786–2792.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989015008750/pk2548sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015008750/pk2548Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989015008750/pk2548Isup3.cml
CCDC reference: 1063364
Additional supporting information: crystallographic information; 3D view; checkCIF report