The packing of the title compound features N—H⋯Cl hydrogen bonds and π–π stacking interactions, which form one-dimensional chains of molecules parallel to [001] further linked via N—H⋯O interactions.
Keywords: crystal structure, trans-platinum(II) complexes, hydrogen bonding
Abstract
In the title complex, [PtCl2(C5H11N)(C6H6N2O2)], the PtII metal atom displays a slightly distorted trans-PtN2Cl2 square-planar coordination geometry. The dihedral angle between the mean plane of the benzene and piperidine rings is 89.03 (3)°. In the crystal structure, inversion dimers are formed via N—H⋯Cl hydrogen-bond interactions, resulting in chains parallel to the [001] direction. The benzene rings within the chains show π–π stacking interactions [centroid-to-centroid distances of 3.801 (3) Å] and neighbouring chains interact via N—H⋯O hydrogen bonds.
Chemical context
The title compound is one of many complexes which have been synthesized for the purpose of potential medical applications (Klein & Hambley, 2009 ▸; Wilson & Lippard, 2014 ▸; Peng et al., 2014 ▸). It is notable that according to the procedure used for the synthesis of complexes of the type cis-[PtCl2(piperidine)(another amine)] (piperidine hereafter denoted Pip) (Dinh & Da, 2003 ▸; Nguyen Thi Thanh et al., 2014 ▸), the reaction between K[PtCl3(Pip)] and p-nitroaniline under appropriate conditions gave no cis complex, as expected, but instead gave the trans-[PtCl2(p-nitroaniline)(Pip)] derivative, (I).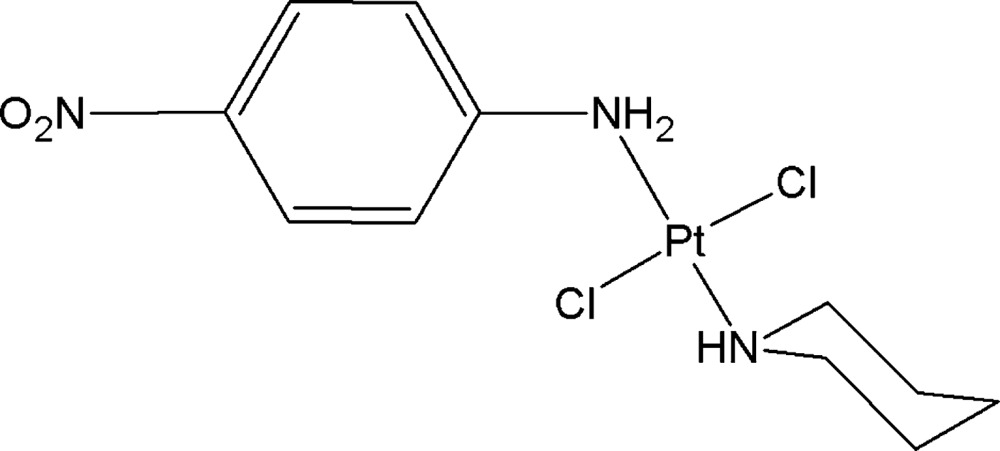
To explain this we suppose that p-nitroaniline first coordinates with PtII via the N atom of the amino group to form cis-[PtCl2(p-nitroaniline)(Pip)] based on the trans effect. Then, in the reaction solution, the cis complex converts into the trans complex and the thermodynamics of this conversion are currently under investigation by us.
The anticancer activity of the title compound was tested according to the method described by Skehan et al. (1990 ▸) against four human cancer cell lines (HepG2, RD, MCF7 and Fl). The IC50 values calculated based on OD values taken on an Elisa instrument at 515–540 nm are >10, 4.86, >10 and 8.25 µg ml−1, respectively.
Structural commentary
The molecular structure of the title compound is illustrated in Fig. 1 ▸ and surprisingly shows a trans arrangement of the two Cl atoms [Cl8—Pt1—Cl9 = 177.84 (4)°]. The piperidine ring adopts the usual chair conformation, with the N2—Pt1 bond in the equatorial position. The piperidine ring is oriented nearly perpendicular to the coordination plane of the PtII atom, thereby reducing the van der Waals repulsion; the dihedral angle between the least-squares mean planes through the piperdine ring and the four atoms coordinated to the Pt atom is 89.6 (2)°. One short intramolecular contact is observed, i.e. H7B⋯Cl8 = 2.83 Å. The mean planes through the piperidine ring and the benzene ring make a dihedral angle of 89.0 (3)°. The dihedral angle between the mean planes of the nitro substituent and the benzene ring is 16.6 (3)°.
Figure 1.
The molecular structure of the title compound, with displacement ellipsoids drawn at the 50% probability level.
Supramolecular features
In the crystal, inversion dimers are formed via N—H⋯Cl interactions between the aniline N atom and both Cl atoms, resulting in chains of molecules along the [001] direction (Fig. 2 ▸ and Table 1 ▸). Within these chains, π–π interactions occur between the aromatic rings [Cg⋯Cg
iv = 3.801 (3) Å; Cg is the centroid of the C11–C16 ring; symmetry code: (iv) −x, y, −z +  ; Fig. 2 ▸]. Neighbouring chains are linked via N—H⋯O hydrogen bonds between the piperidine N atom and a nitro O atom (Fig. 2 ▸ and Table 1 ▸).
; Fig. 2 ▸]. Neighbouring chains are linked via N—H⋯O hydrogen bonds between the piperidine N atom and a nitro O atom (Fig. 2 ▸ and Table 1 ▸).
Figure 2.
Partial packing diagram of the title compound, showing a chain of molecules formed parallel to the [001] direction via N—H⋯Cl interactions (green dotted lines) and π–π interactions (grey dotted line). Neighbouring chains interact via N—H⋯O hydrogen bonds (red dotted line).
Table 1. Hydrogen-bond geometry (, ).
| DHA | DH | HA | D A | DHA |
|---|---|---|---|---|
| N2H2O18i | 0.93 | 2.27 | 3.182(6) | 165 |
| N10H10ACl8ii | 0.92 | 2.32 | 3.198(4) | 158 |
| N10H10BCl9iii | 0.92 | 2.37 | 3.255(4) | 161 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Database survey
A search of the Cambridge Structural Database (Version 5.36; last update February 2015; Groom & Allen, 2014 ▸) for Pt complexes with Pt coordinated to exactly two Cl atoms and two N atoms gave 713 hits. The majority of these Pt complexes display a cis coordination of the Cl atoms (474 structures), with the remaining 239 structures showing a trans coordination. There is no difference in the Pt—Cl distances between both configurations. The average Pt—Cl distances are 2.300 (15) and 2.299 (12) Å for the cis and trans arrangements, respectively, and correspond to the observed distances of 2.3039 (11) and 2.2917 (12) Å for Pt1—Cl8 and Pt1—Cl9, respectively.
Synthesis and crystallization
The starting complex K[PtCl3(piperidine)] (0.425 g, 1 mmol), prepared according to the synthetic procedure of Da et al. (2001 ▸) with slight modifications, was dissolved in water (10 ml) and filtered to afford a clear solution. To this solution, p-nitroaniline (1 mmol) in ethanol (10 ml) was added gradually while stirring at 413–318 K. After 1 h, a brown powder appeared and the reaction mixture was then stirred further for 24 h until all the precipitate was completely dissolved. The solvent was removed in vacuo to give a brown–yellow product. The product was washed consecutively with a 0.1 M HCl solution (2 × 2 ml), warm water (2 × 2 ml) and diethyl ether (2 × 2 ml). The yield was 80%. Single crystals suitable for X-ray determination were obtained by slow evaporation within 12 h from an acetone solution at room temperature. IR (KBr, cm−1): 3199, 3113 (νNH); 3070, 2927, 2862 (νCH); 1596, 1525, 1479 (νC=C arom); 1342, 1325 (νNO); 1H NMR (CDCl3, 500 MHz): δ 8.21 (2H, d, 3 J = 9.0 Hz, Ar-H), 7.47 (2H, d, 3 J = 9.0 Hz, Ar-H), 5.49 (2H, br, O2NC6H4NH2), 3.66 (1H, br, C5H10NH), 3.26 (2Hα e, d, 2 J ae = 13.0 Hz, C5 H10NH), 2.99 (2Hα a, q, 2 J ae, 3 J aa, 3 J aa(NH) = 13.0 Hz, C5 H10NH), 1.69–1.43 (4Hβ, 2Hγ, ov, C5 H10NH). 13C{1H} NMR (125 MHz, CDCl3): δ 149.6, 125.1, 124.2 (O2NC6H4NH2), 54.0, 27.2, 24.3 (C5H10NH).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. All H atoms were placed at idealized positions and refined in riding mode, with U iso(H) values assigned as 1.2U eq of the parent atoms, with C—H distances of 0.95 (aromatic) and 0.99 Å (methylene), and N—H distances of 0.93 (NH) and 0.92 Å (NH2).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | [PtCl2(C5H11N)(C6H6N2O2)] |
| M r | 489.27 |
| Crystal system, space group | Monoclinic, C2/c |
| Temperature (K) | 100 |
| a, b, c () | 15.8763(11), 18.5394(11), 10.8707(6) |
| () | 103.119(7) |
| V (3) | 3116.1(3) |
| Z | 8 |
| Radiation type | Mo K |
| (mm1) | 9.35 |
| Crystal size (mm) | 0.35 0.15 0.1 |
| Data collection | |
| Diffractometer | Agilent SuperNova (single source at offset, Eos detector) |
| Absorption correction | Multi-scan (CrysAlis PRO; Agilent, 2012 ▸) |
| T min, T max | 0.538, 1.000 |
| No. of measured, independent and observed [I > 2(I)] reflections | 8156, 3109, 2713 |
| R int | 0.044 |
| (sin /)max (1) | 0.625 |
| Refinement | |
| R[F 2 > 2(F 2)], wR(F 2), S | 0.032, 0.072, 1.10 |
| No. of reflections | 3109 |
| No. of parameters | 172 |
| H-atom treatment | H-atom parameters constrained |
| max, min (e 3) | 2.75, 1.82 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989015009196/rz5159sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015009196/rz5159Isup2.hkl
CCDC reference: 1400786
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors thank the Vietnamese Ministry of Education (project B2013-17-39) and VLIR–UOS (project ZEIN2014Z182) for financial support and the Hercules Foundation for supporting the purchase of the diffractometer through project AKUL/09/0035.
supplementary crystallographic information
Crystal data
| [PtCl2(C5H11N)(C6H6N2O2)] | F(000) = 1856 |
| Mr = 489.27 | Dx = 2.086 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 15.8763 (11) Å | Cell parameters from 3549 reflections |
| b = 18.5394 (11) Å | θ = 3.4–28.8° |
| c = 10.8707 (6) Å | µ = 9.35 mm−1 |
| β = 103.119 (7)° | T = 100 K |
| V = 3116.1 (3) Å3 | , brown |
| Z = 8 | 0.35 × 0.15 × 0.1 mm |
Data collection
| Agilent SuperNova (single source at offset, Eos detector) diffractometer | 3109 independent reflections |
| Radiation source: SuperNova (Mo) X-ray Source | 2713 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.044 |
| Detector resolution: 15.9631 pixels mm-1 | θmax = 26.4°, θmin = 2.8° |
| ω scans | h = −19→15 |
| Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2012) | k = −17→23 |
| Tmin = 0.538, Tmax = 1.000 | l = −13→13 |
| 8156 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.032 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.072 | H-atom parameters constrained |
| S = 1.10 | w = 1/[σ2(Fo2) + (0.0296P)2 + 0.5839P] where P = (Fo2 + 2Fc2)/3 |
| 3109 reflections | (Δ/σ)max = 0.003 |
| 172 parameters | Δρmax = 2.75 e Å−3 |
| 0 restraints | Δρmin = −1.82 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C3 | 0.2646 (4) | 0.9772 (3) | 0.4633 (5) | 0.0233 (13) | |
| H3A | 0.2439 | 1.0130 | 0.5173 | 0.028* | |
| H3B | 0.2335 | 0.9859 | 0.3748 | 0.028* | |
| C4 | 0.3620 (4) | 0.9877 (3) | 0.4750 (5) | 0.0243 (13) | |
| H4A | 0.3815 | 0.9562 | 0.4132 | 0.029* | |
| H4B | 0.3734 | 1.0383 | 0.4551 | 0.029* | |
| C5 | 0.4128 (4) | 0.9694 (3) | 0.6083 (5) | 0.0267 (13) | |
| H5A | 0.4755 | 0.9727 | 0.6122 | 0.032* | |
| H5B | 0.3985 | 1.0045 | 0.6691 | 0.032* | |
| C6 | 0.3905 (4) | 0.8933 (3) | 0.6443 (6) | 0.0255 (14) | |
| H6A | 0.4109 | 0.8580 | 0.5892 | 0.031* | |
| H6B | 0.4210 | 0.8833 | 0.7326 | 0.031* | |
| C7 | 0.2938 (4) | 0.8839 (3) | 0.6313 (5) | 0.0220 (13) | |
| H7A | 0.2816 | 0.8332 | 0.6501 | 0.026* | |
| H7B | 0.2746 | 0.9151 | 0.6937 | 0.026* | |
| C11 | −0.0526 (3) | 0.8256 (3) | 0.5180 (4) | 0.0175 (11) | |
| C12 | −0.0838 (4) | 0.8538 (3) | 0.6181 (5) | 0.0193 (11) | |
| H12 | −0.0832 | 0.9044 | 0.6328 | 0.023* | |
| C13 | −0.1159 (3) | 0.8069 (3) | 0.6957 (5) | 0.0205 (12) | |
| H13 | −0.1370 | 0.8248 | 0.7649 | 0.025* | |
| C14 | −0.1167 (4) | 0.7338 (3) | 0.6710 (5) | 0.0225 (12) | |
| C15 | −0.0857 (4) | 0.7057 (3) | 0.5722 (5) | 0.0266 (14) | |
| H15 | −0.0870 | 0.6552 | 0.5571 | 0.032* | |
| C16 | −0.0528 (4) | 0.7522 (3) | 0.4957 (5) | 0.0218 (12) | |
| H16 | −0.0303 | 0.7339 | 0.4280 | 0.026* | |
| Cl8 | 0.10439 (9) | 0.97037 (6) | 0.62798 (11) | 0.0190 (3) | |
| Cl9 | 0.12310 (9) | 0.80109 (7) | 0.32660 (11) | 0.0212 (3) | |
| N2 | 0.2441 (3) | 0.9028 (2) | 0.5016 (4) | 0.0160 (9) | |
| H2 | 0.2629 | 0.8713 | 0.4469 | 0.019* | |
| N10 | −0.0192 (3) | 0.8739 (2) | 0.4368 (4) | 0.0157 (9) | |
| H10A | −0.0439 | 0.9185 | 0.4409 | 0.019* | |
| H10B | −0.0373 | 0.8576 | 0.3552 | 0.019* | |
| N17 | −0.1522 (3) | 0.6855 (3) | 0.7523 (4) | 0.0294 (12) | |
| O18 | −0.1616 (3) | 0.7083 (2) | 0.8546 (3) | 0.0316 (10) | |
| O19 | −0.1721 (4) | 0.6237 (2) | 0.7150 (4) | 0.0464 (14) | |
| Pt1 | 0.113556 (13) | 0.887195 (10) | 0.474086 (16) | 0.01473 (9) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C3 | 0.021 (3) | 0.032 (3) | 0.019 (3) | −0.008 (2) | 0.007 (2) | 0.001 (2) |
| C4 | 0.024 (4) | 0.030 (3) | 0.022 (3) | −0.003 (2) | 0.010 (2) | 0.000 (2) |
| C5 | 0.021 (3) | 0.031 (3) | 0.029 (3) | −0.001 (3) | 0.007 (3) | −0.003 (3) |
| C6 | 0.022 (4) | 0.026 (3) | 0.028 (3) | 0.000 (2) | 0.005 (3) | 0.000 (2) |
| C7 | 0.018 (3) | 0.026 (3) | 0.022 (3) | −0.001 (2) | 0.003 (2) | 0.003 (2) |
| C11 | 0.013 (3) | 0.022 (3) | 0.016 (2) | −0.002 (2) | 0.000 (2) | 0.000 (2) |
| C12 | 0.019 (3) | 0.019 (3) | 0.020 (3) | −0.001 (2) | 0.006 (2) | −0.004 (2) |
| C13 | 0.013 (3) | 0.034 (3) | 0.014 (2) | −0.003 (2) | 0.004 (2) | −0.002 (2) |
| C14 | 0.025 (3) | 0.024 (3) | 0.021 (3) | −0.004 (2) | 0.010 (2) | 0.005 (2) |
| C15 | 0.040 (4) | 0.017 (3) | 0.021 (3) | −0.001 (3) | 0.004 (3) | −0.002 (2) |
| C16 | 0.025 (3) | 0.021 (3) | 0.020 (3) | −0.001 (2) | 0.008 (2) | 0.001 (2) |
| Cl8 | 0.0217 (8) | 0.0195 (6) | 0.0164 (6) | 0.0017 (5) | 0.0054 (5) | −0.0009 (5) |
| Cl9 | 0.0220 (8) | 0.0239 (6) | 0.0174 (6) | 0.0023 (6) | 0.0039 (5) | −0.0042 (5) |
| N2 | 0.012 (3) | 0.020 (2) | 0.016 (2) | −0.0003 (18) | 0.0033 (18) | −0.0026 (18) |
| N10 | 0.013 (3) | 0.019 (2) | 0.015 (2) | 0.0011 (18) | 0.0022 (19) | −0.0009 (18) |
| N17 | 0.031 (3) | 0.030 (3) | 0.028 (3) | −0.003 (2) | 0.009 (2) | 0.006 (2) |
| O18 | 0.035 (3) | 0.044 (2) | 0.021 (2) | −0.007 (2) | 0.0163 (18) | 0.0022 (19) |
| O19 | 0.077 (4) | 0.026 (2) | 0.046 (3) | −0.006 (2) | 0.034 (3) | 0.003 (2) |
| Pt1 | 0.01469 (15) | 0.01670 (13) | 0.01350 (12) | 0.00081 (7) | 0.00464 (9) | 0.00004 (7) |
Geometric parameters (Å, º)
| C3—H3A | 0.9900 | C12—H12 | 0.9500 |
| C3—H3B | 0.9900 | C12—C13 | 1.387 (7) |
| C3—C4 | 1.535 (8) | C13—H13 | 0.9500 |
| C3—N2 | 1.497 (6) | C13—C14 | 1.381 (7) |
| C4—H4A | 0.9900 | C14—C15 | 1.382 (7) |
| C4—H4B | 0.9900 | C14—N17 | 1.458 (7) |
| C4—C5 | 1.528 (8) | C15—H15 | 0.9500 |
| C5—H5A | 0.9900 | C15—C16 | 1.380 (7) |
| C5—H5B | 0.9900 | C16—H16 | 0.9500 |
| C5—C6 | 1.526 (7) | Cl8—Pt1 | 2.3039 (11) |
| C6—H6A | 0.9900 | Cl9—Pt1 | 2.2917 (12) |
| C6—H6B | 0.9900 | N2—H2 | 0.9300 |
| C6—C7 | 1.520 (8) | N2—Pt1 | 2.046 (4) |
| C7—H7A | 0.9900 | N10—H10A | 0.9200 |
| C7—H7B | 0.9900 | N10—H10B | 0.9200 |
| C7—N2 | 1.492 (7) | N10—Pt1 | 2.068 (4) |
| C11—C12 | 1.396 (7) | N17—O18 | 1.231 (5) |
| C11—C16 | 1.382 (7) | N17—O19 | 1.232 (6) |
| C11—N10 | 1.440 (6) | ||
| H3A—C3—H3B | 107.9 | C13—C12—H12 | 120.5 |
| C4—C3—H3A | 109.3 | C12—C13—H13 | 120.5 |
| C4—C3—H3B | 109.3 | C14—C13—C12 | 119.0 (5) |
| N2—C3—H3A | 109.3 | C14—C13—H13 | 120.5 |
| N2—C3—H3B | 109.3 | C13—C14—C15 | 122.2 (5) |
| N2—C3—C4 | 111.8 (4) | C13—C14—N17 | 118.2 (5) |
| C3—C4—H4A | 109.5 | C15—C14—N17 | 119.6 (5) |
| C3—C4—H4B | 109.5 | C14—C15—H15 | 120.6 |
| H4A—C4—H4B | 108.1 | C16—C15—C14 | 118.9 (5) |
| C5—C4—C3 | 110.8 (4) | C16—C15—H15 | 120.6 |
| C5—C4—H4A | 109.5 | C11—C16—H16 | 120.1 |
| C5—C4—H4B | 109.5 | C15—C16—C11 | 119.7 (5) |
| C4—C5—H5A | 109.6 | C15—C16—H16 | 120.1 |
| C4—C5—H5B | 109.6 | C3—N2—H2 | 106.2 |
| H5A—C5—H5B | 108.1 | C3—N2—Pt1 | 111.5 (3) |
| C6—C5—C4 | 110.2 (5) | C7—N2—C3 | 112.2 (4) |
| C6—C5—H5A | 109.6 | C7—N2—H2 | 106.2 |
| C6—C5—H5B | 109.6 | C7—N2—Pt1 | 113.9 (3) |
| C5—C6—H6A | 109.3 | Pt1—N2—H2 | 106.2 |
| C5—C6—H6B | 109.3 | C11—N10—H10A | 108.0 |
| H6A—C6—H6B | 107.9 | C11—N10—H10B | 108.0 |
| C7—C6—C5 | 111.7 (5) | C11—N10—Pt1 | 117.0 (3) |
| C7—C6—H6A | 109.3 | H10A—N10—H10B | 107.3 |
| C7—C6—H6B | 109.3 | Pt1—N10—H10A | 108.0 |
| C6—C7—H7A | 109.3 | Pt1—N10—H10B | 108.0 |
| C6—C7—H7B | 109.3 | O18—N17—C14 | 118.7 (4) |
| H7A—C7—H7B | 108.0 | O18—N17—O19 | 122.8 (5) |
| N2—C7—C6 | 111.5 (4) | O19—N17—C14 | 118.5 (5) |
| N2—C7—H7A | 109.3 | Cl9—Pt1—Cl8 | 177.84 (4) |
| N2—C7—H7B | 109.3 | N2—Pt1—Cl8 | 91.59 (12) |
| C12—C11—N10 | 119.4 (4) | N2—Pt1—Cl9 | 88.59 (12) |
| C16—C11—C12 | 121.2 (5) | N2—Pt1—N10 | 176.94 (15) |
| C16—C11—N10 | 119.4 (4) | N10—Pt1—Cl8 | 89.56 (12) |
| C11—C12—H12 | 120.5 | N10—Pt1—Cl9 | 90.37 (12) |
| C13—C12—C11 | 119.0 (5) | ||
| C3—C4—C5—C6 | 54.8 (6) | C12—C11—N10—Pt1 | 97.9 (5) |
| C3—N2—Pt1—Cl8 | −66.5 (3) | C12—C13—C14—C15 | 0.7 (9) |
| C3—N2—Pt1—Cl9 | 115.7 (3) | C12—C13—C14—N17 | −178.9 (5) |
| C4—C3—N2—C7 | 54.8 (6) | C13—C14—C15—C16 | 0.0 (9) |
| C4—C3—N2—Pt1 | −176.0 (3) | C13—C14—N17—O18 | −16.4 (8) |
| C4—C5—C6—C7 | −55.5 (6) | C13—C14—N17—O19 | 163.1 (6) |
| C5—C6—C7—N2 | 55.4 (6) | C14—C15—C16—C11 | −0.9 (8) |
| C6—C7—N2—C3 | −54.8 (6) | C15—C14—N17—O18 | 164.0 (5) |
| C6—C7—N2—Pt1 | 177.4 (3) | C15—C14—N17—O19 | −16.5 (8) |
| C7—N2—Pt1—Cl8 | 61.7 (3) | C16—C11—C12—C13 | −0.3 (8) |
| C7—N2—Pt1—Cl9 | −116.1 (3) | C16—C11—N10—Pt1 | −81.8 (5) |
| C11—C12—C13—C14 | −0.6 (8) | N2—C3—C4—C5 | −54.9 (6) |
| C11—N10—Pt1—Cl8 | −82.4 (3) | N10—C11—C12—C13 | 180.0 (5) |
| C11—N10—Pt1—Cl9 | 95.5 (3) | N10—C11—C16—C15 | −179.2 (5) |
| C12—C11—C16—C15 | 1.1 (8) | N17—C14—C15—C16 | 179.6 (5) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2···O18i | 0.93 | 2.27 | 3.182 (6) | 165 |
| N10—H10A···Cl8ii | 0.92 | 2.32 | 3.198 (4) | 158 |
| N10—H10B···Cl9iii | 0.92 | 2.37 | 3.255 (4) | 161 |
Symmetry codes: (i) x+1/2, −y+3/2, z−1/2; (ii) −x, −y+2, −z+1; (iii) −x, y, −z+1/2.
References
- Agilent (2012). CrysAlis PRO. Agilent Technologies, Yarnton, Oxfordshire, England.
- Da, T. T., Vu, D. B. & Dinh, N. H. (2001). J. Pharm. Sci. (Vietnam), 6, 6–8.
- Dinh, N. H. & Da, T. T. (2003). J. Coord. Chem. 41, 683–689.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Groom, C. R. & Allen, F. H. (2014). Angew. Chem. Int. Ed. 53, 662–671. [DOI] [PubMed]
- Klein, A. V. & Hambley, T. W. (2009). Chem. Rev 109, 4911–4920. [DOI] [PubMed]
- Nguyen Thi Thanh, C., Nguyen Bich, N. & Van Meervelt, L. (2014). Acta Cryst. C70, 297–301. [DOI] [PubMed]
- Peng, Y., Zhong, H., Chen, Z.-F., Liu, Y.-C., Zhang, G.-H., Qin, Q.-P. & Liang, H. (2014). Chem. Pharm. Bull. 62, 221–228. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Skehan, P., Storeng, R., Scudiero, D., Monks, A., McMahon, J., Vistica, D., Warren, J. T., Bokesch, H., Kenney, S. & Boyd, M. R. (1990). J. Natl. Cancer Inst. 82, 1107–1112. [DOI] [PubMed]
- Wilson, J. J. & Lippard, S. J. (2014). Chem. Rev 114, 4470–4495. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989015009196/rz5159sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015009196/rz5159Isup2.hkl
CCDC reference: 1400786
Additional supporting information: crystallographic information; 3D view; checkCIF report




