Abstract
Understanding the regulatory landscape of the human genome is a central question in complex trait genetics. Most single-nucleotide polymorphisms (SNPs) associated with cancer risk lie in non-protein-coding regions, implicating regulatory DNA elements as functional targets of susceptibility variants. Here, we describe genome-wide annotation of regions of open chromatin and histone modification in fallopian tube and ovarian surface epithelial cells (FTSECs, OSECs), the debated cellular origins of high-grade serous ovarian cancers (HGSOCs) and in endometriosis epithelial cells (EECs), the likely precursor of clear cell ovarian carcinomas (CCOCs). The regulatory architecture of these cell types was compared with normal human mammary epithelial cells and LNCaP prostate cancer cells. We observed similar positional patterns of global enhancer signatures across the three different ovarian cancer precursor cell types, and evidence of tissue-specific regulatory signatures compared to non-gynecological cell types. We found significant enrichment for risk-associated SNPs intersecting regulatory biofeatures at 17 known HGSOC susceptibility loci in FTSECs (P = 3.8 × 10−30), OSECs (P = 2.4 × 10−23) and HMECs (P = 6.7 × 10−15) but not for EECs (P = 0.45) or LNCaP cells (P = 0.88). Hierarchical clustering of risk SNPs conditioned on the six different cell types indicates FTSECs and OSECs are highly related (96% of samples using multi-scale bootstrapping) suggesting both cell types may be precursors of HGSOC. These data represent the first description of regulatory catalogues of normal precursor cells for different ovarian cancer subtypes, and provide unique insights into the tissue specific regulatory variation with respect to the likely functional targets of germline genetic susceptibility variants for ovarian cancer.
Introduction
Understanding the regulatory landscape of the human genome is a central question in complex trait genetics. The surge of genome-wide association studies (GWASs) over the past decade has revealed that ∼90% of common trait-associated variants are located outside protein coding regions (http://www.genome.gov/gwastudies/). This suggests a major role for non-protein coding DNA elements such as enhancers and non-coding RNAs regulating the expression of target susceptibility genes, affecting complex phenotypes. The Encyclopedia of DNA Elements (ENCODE) project has catalogued genome-wide profiles of non-coding regulatory features of cell lines for multiple different cancer types. Evaluating ENCODE data with respect to GWAS data reveals that common variant risk regions are enriched for regulatory elements (1,2). However, ENCODE data are limited with respect to cell types that have been profiled, and particularly the normal precursor cells of different cancers.
The two main challenges in elucidating the functional mechanisms by which common variants modulate risk are identifying the disease causing SNPs and the target susceptibility genes they regulate. This is complicated by linkage disequilibrium (LD) in the human genome, varying by racial–ethnic group. The causal SNP at a given risk locus will usually be correlated with many surrogate variants; the 1000 Genomes Project data show that for every trait-associated variant in the NHGRI GWAS database, there are, on average, 56 correlated variants at r2 ≥ 0.53, which may represent the causal SNP. Another issue is tissue specificity. ENCODE data show that the genome-wide regulatory architecture is highly cell-type specific. This suggests that only tissues relevant to a disease's development should be evaluated in order to identify interactions between putative causal SNPs and their regulatory targets.
Epithelial ovarian cancer (EOC) is a heterogeneous disease comprising several histological subtypes, each with distinct biological characteristics. Emerging evidence suggests that the different subtypes have different genetic hallmarks. The most common subtype, high-grade serous ovarian cancer (HGSOC), is characterized by somatic TP53 mutations and genomic instability caused by defects in double strand DNA break repair pathways (3,4), whereas clear cell ovarian carcinomas (CCOCs) are characterized by mutations in ARID1A and deregulation of the phosphoinositide 3-kinase signaling pathway (5,6). Different subtypes also have different proposed cellular origins. Although it remains a matter of debate, HGSOCs are thought to derive from fallopian tube secretory epithelial cells (FTSECs) and/or ovarian surface epithelial cells (OSECs) (7–9). Given that endometriosis is a known risk factor for CCOC, it is likely that this subtype derives from endometriosis epithelial cells (EECs) (10). Different histological subtypes also show variations in the underlying genetic susceptibility. Germline BRCA1 and BRCA2 mutations usually lead to HGSOC (11,12), while the mismatch repair genes predispose women to the endometrioid subtype of ovarian cancer, in addition to endometrial and colorectal cancers (13). GWAS have so far discovered 17 common variant susceptibility regions at genome-wide levels of significance (P < 5 × 10−8) conferring risk to HGSOC, two of which also confer risk CCOC (14–21).
Currently, there are few data, either in ENCODE or the literature, describing the regulatory architecture of ovarian cancer precursor tissues. This significantly limits the ability to understand the contribution of the non-coding genome to the development of epithelial ovarian cancer, and to identify the causal genetic variants at ovarian cancer susceptibility loci. The goals of the current study were 3-fold: (i) to catalogue the tissue-specific genome-wide architecture of non-coding DNA regulatory elements in OSECs, FTSECs and EECs based on regions of nucleosome depletion and surrounding histone-3-lysine-4-monomethylation (H3K4me1) and histone-3-lysine-27-acetylation (H3K27ac) post-translational modifications; (ii) to integrate these data with risk-associated common variants at ovarian cancer susceptibility loci and identify the putative functional regulatory elements coinciding with SNPs at these loci; (iii) to use risk-associated epigenetic profiles at susceptibility loci to evaluate the cellular origins of different ovarian cancer histological subtypes.
Results
Genome-wide regulatory profiling of epithelial ovarian cancer precursor tissues
We generated genome-wide profiles of DNA regulatory marks in five different cell lines representing the possible precursor cells for HGSOC and CCOC. This included two normal ovarian epithelial cell lines (IOE4 and IOE11) (22,23) and two fallopian tube secretory epithelial cell lines (FT33 and FT246) (24,25), the possible precursors of HGSOCs, and one EEC line (EEC16) (26), the likely precursor of CCOC. We used FAIRE-seq (Formaldehyde-Assisted Isolation of Regulatory Elements) to catalogue localized regions of nucleosome depletion, and ChIP-seq (Chromatin Immunoprecipitation sequencing) to profile H3K4me1 and H3K27ac histone modifications consistent with poised and active/engaged enhancers, respectively. Putative enhancer sites were defined as regions where localized nucleosome depletion corresponded with surrounding histone peaks having either/both H3K4me1 and H3K27ac modifications.
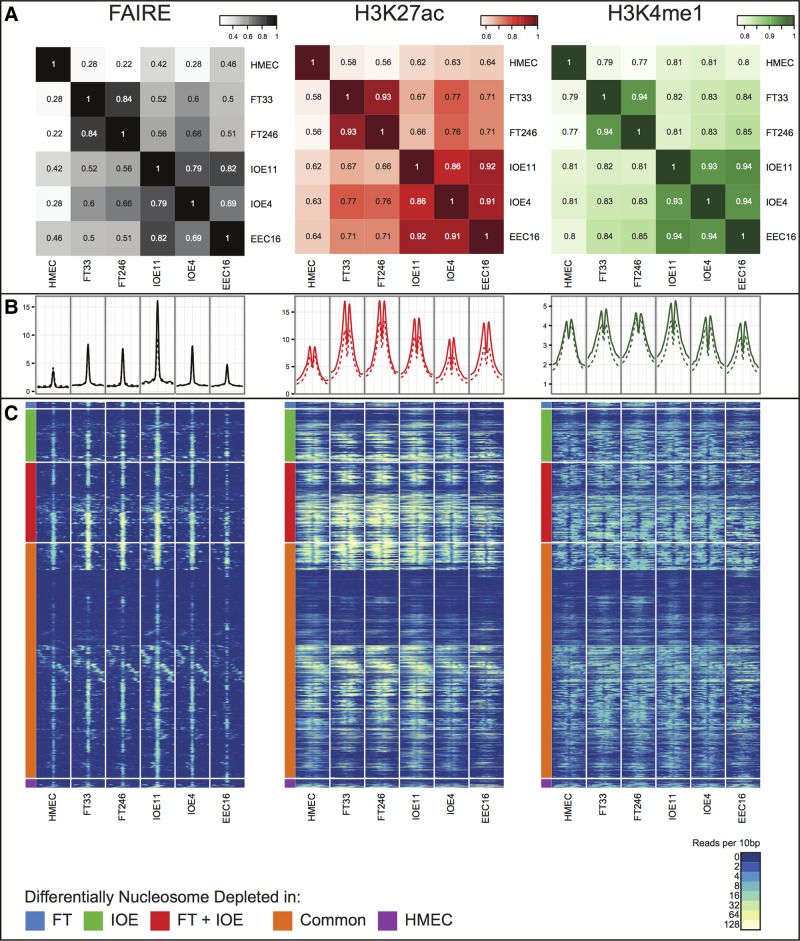
We compared the genome-wide architecture of regulatory marks by cell type, and to similar data generated previously for human mammary epithelial cells (HMECs) (27). By analyzing a global similarity matrix of affinity scores across all common peaks (shared by at least two samples), we determined a genome-wide correlation coefficient of the five cell types. We observed similar positional patterns of global enhancer signals across the five EOC precursor cell lines. For OSECs and FTSECs, where we profiled cells from two different individuals, the biological replicates showed greatest similarity to each other compared to any other cell type. By cell type, OSEC and EEC lines showed the greatest similarity to each other, but all three cell types of gynecological origin showed greater similarity to each other compared to HMECs (Fig. 1A). Taken together, these data suggest that these gynecological tissues share a common underlying regulatory biology that is distinct from non-gynecological epithelial cells, even though there are clearly unique epigenetic signatures between gynecological tissues reflecting the tissue-specific regulatory landscape of each cell type.
Figure 1.
(A) A global similarity matrix displaying the correlation (R2 values) of affinity scores between cell types for FAIRE, H3K27ac and H3K4me1 regulatory marks across the entire genome. The affinity scores are calculated from the values of the read counts across all samples with the set of common peaks (those that occurred in at least two samples) and then TMM marks normalized (using edgeR), using ChIP/FAIRE read counts minus Control read counts and Full Library size. (B) Enrichment profiles of FAIRE, H3K27ac and H3K4me1 normalized signal around the center of 4929 FAIRE peaks within the fine-mapped regions (solid lines) and 634 976 FAIRE peaks genome-wide (dashed lines). (C) Heatmap representing FAIRE, H3K27ac and H3K4me1 normalized signal in a ±1 kb window around the center of all nucleosome depleted regions as defined in Figure 1B. Normalized signal intensity represents read density within 10 bp windows. Hierarchical clustering was performed, supervised on differentially nucleosome depleted regions.
Regulatory architecture at epithelial ovarian cancer susceptibility loci
We conditioned our regulatory analysis on 17 different regions of the genome identified by genome-wide association studies, which predispose to invasive epithelial ovarian cancer. Fifteen of these regions are genome-wide significant risk loci for HGSOC (1p34; 2q31; 3q25; 4q26; 5p15; 6p22; 8q21; 8q24; 9p22; 9q34; 10p12; 17q12; 17q21.31; 17q21.32; 19p13). Two of these loci are also independently associated with CCOC risk (8q24; 17q12). Two other regions (1p36; 17q11) are borderline genome-wide significant associations for HGSOC (P-value 5.7 × 10−8 and 3.9 × 10−7, respectively) but are genome-wide significant for all invasive epithelial ovarian cancer subtypes combined (Table 1). Representative examples of browser images showing the quality of the regulatory data are shown in Supplementary Material, Figure S1, for the 1p34 and 17q21.31 loci.
Table 1.
Confirmed common variant susceptibility regions identified for epithelial ovarian cancer, by subtype, and annotated for regulatory biofeatures identified in ovarian cancer precursor tissues
| Locus | Subtypea | Top SNP | Mapped window (kb) | Number 1:100 SNPs | OR (95%CI) | P-value¶ | Nearest gene | Number predicted TF motifs | Motifs |
|---|---|---|---|---|---|---|---|---|---|
| 1p36.12 | Invasive | rs56318008 | 152 | 34 | 1.11 (1.07–1.16) | 7.6 × 10−9 | WNT4 | 9 | CTCF, TCF12, ZEB1, MEF2, RFX5, HSF1, TAL1, RUNX1, ETS1, TEAD1, CREB, SREBF1, CEBPB |
| 1p34.3 | HGSOC | rs58722170 | 33 | 14 | 1.08 (1.04–1.12) | 2.7 × 10−12 | RSPO1 | NA | NA |
| 2q31 | HGSOC | rs2072590 | 22 | 16 | 1.14 (1.10–1.19) | 3.7 × 10−13 | HOXD | 7 | AP2, UA3, UA12, UA3, UA8, ZEB1, UA9, BARHL2, UA8, EBF1, RUNX1 |
| 3q25 | HGSOC | rs7651446 | 176 | 63 | 1.59 (1.48–1.70) | 1.5 × 10−38 | TiPARP | 30 | — |
| 4q26 | Invasive | rs17329882 | 13 | 3 | 1.09 (1.06–1.13) | 1.4 × 10−8 | SYNPO2 | 2 | AP1, BARHL2, UA8, BARHL2, CEBPB, ETS1 |
| 5p15.15 | HGSOC | rs10069690 | 5 | 8 | 1.14 (1.10–1.19) | 7.6 × 10−11 | TERT | 0 | NA |
| 6p22.1 | HGSOC | rs1161331104 | 91 | 17 | 0.93 (0.91–0.97) | 3.0 × 10−8 | GPX6 | NA | NA |
| 8q21 | HGSOC | rs11782652 | 60 | 9 | 1.24 (1.16–1.32) | 5.6 × 10−11 | CHMP4C | 1 | BARHL2, PRDM1, SOX2 |
| 8q24 | HGSOC | rs10088218 | 32 | 49 | 0.77 (0.73–0.82) | 1.6 × 10−20 | MYC/ PVT1 | 19 | — |
| CCOC | rs2165805 | 18 | 2 | 1.21 (1.05–1.40) | 8.4 × 10−9 | NA | NA | ||
| 9p22 | HGSOC | rs3814113 | 10 | 17 | 0.79 (0.76–0.82) | 2.7 × 10−34 | BNC2 | 1 | TAL1 |
| 9q34.2 | Invasive | rs635634 | 224 | 15 | 1.11 (1.07–1.16) | 4.4 × 10−9 | ABO | 0 | NA |
| 10p12 | HGSOC | rs1243180 | 210 | 48 | 1.10 (1.06–1.14) | 1.2 × 10−9 | MLLT10 | 12 | |
| 17q11.2 | Invasive | chr17:29181220:I5 | 270 | 17 | 0.91 (0.88–0.94) | 2.6 × 10−9 | ATAD5 | 0 | NA |
| 17q12 | CCOC | rs11651755 | 14 | 16 | 0.80 (0.72–0.88) | 2.9 × 10−8 | HNF1B | 4 | YY1, CTCF, ETS1, ETS1 |
| HGSOC | rs757210 | 16 | 9 | 1.11 (1.07–1.15) | 8.2 × 10−9 | 1 | ETS1 | ||
| 17q21.31 | HGSOC | rs183211 | 113 | 148 | 1.11 (1.07–1.16) | 1.6 × 10−7 | PLEKHM1 | 30 | |
| 17q21.32 | HGSOC | rs9303542 | 162 | 95 | 1.14 (1.10–1.19) | 4.0 × 10−12 | SKAP1 | 8 | SOX2, TEAD1, ELF1, ELK4, ETS1, GABP, BHLHE40, MYC, AP2, UA3, UA8, SOX2, TAL1, TBP, TCF7L2, ETS1, RUNX1, SOX2, TCF12, UA9, ZEB1, SOX2 |
| 19p13 | HGSOC | rs4808075 | 20 | 13 | 1.18 (1.13–1.23) | 2.9 × 10−14 | ANKLE1 | 2 | ELF1, ELK4, GABP, GATA3 |
aHigh-grade serous ovarian cancer (HGSOC); ovarian clear cell cancer (OCCC); All invasive ovarian cancer subtypes combined (Invasive).
¶P-value for the strongest association listed by subtype. Where all invasive ovarian cancers gave the strongest P-value, the association for HGSOC was the subtypes with the strongest effect with the following P-values: 5.7 × 10−8 at 1p36.12, 1.6 × 10−8 at 4p26, 4.2 × 10−8 at 9q34.2 and 3.9 × 10−7 at 17q11.2.
We compared the average signal intensity of the chromatin signatures in all five normal gynecological cell lines, centered on the peak of the FAIRE signals to establish if regulatory signals differed between ovarian cancer risk regions (Supplementary Material, Table S1) and genome-wide profiles. We observed no differences in average peak positioning or intensity at putative enhancer sites in risk region verses whole genome regulatory architecture (Fig. 1B). Neither did we find evidence of a difference in regulatory peak intensity between different risk regions. However, we did observe some differences in FAIRE and ChIP-seq signal intensity between gynecological tissues and HMECs at peaks in risk regions consistent with the tissue-specific differences we observed in the genome-wide patterns of regulatory marks between these cell types. For example, within the risk regions there are 617 FAIRE peaks with significant nucleosome depletion [False Discovery Rate (FDR) < 0.05] unique to OSECs, 80 unique to FTSEC lines; but combining these cell types, there are 960 FAIRE peaks that were significantly different to HMECs (Fig. 1C, Supplementary Material, Table S2).
Functional annotation of risk-associated SNPs at ovarian cancer susceptibility loci
We next integrated risk-associated genetic variants in the 17 ovarian cancer risk regions with regulatory biofeatures and predicted functional motifs from in silico analyses to identify SNP-biofeature intersections. Imputation of variants from the 1000 Genomes Project data (28) identified a total of 9053 common variants in the 17 regions. Within each region, we identified all SNPs with a 1-in-100 chance of being a causal variant based on their association with the most risk-associated SNP at each locus (Table 1). These regions varied in size depending on the extent of linkage disequilibrium and the span of risk-associated SNPs. To annotate the transcriptional architecture of these regions, we extended each risk region to one megabase to encompass putative target susceptibility genes that may be regulated at long distances from causal susceptibility SNPs. In total, 676 SNPs were highly correlated with the most significant risk-associated SNP at each locus and calculated to be the most likely causal variant (21). These 676 variants were mapped with respect to the following biofeatures: (i) nucleosome-depleted sites defined by FAIRE signals defined earlier, and with ENCODE DNaseI hypersensitivity data from other cell types (HMEC and LNCaP); (ii) H3K4me1 and H3K27ac signals identified in tissues in the current study; (iii) previously defined gene regions (RefGene), their promoter-proximal regions (i.e. −1 kb to +100 bp of a transcription start site), exons and untranslated regions containing known microRNA target sites.
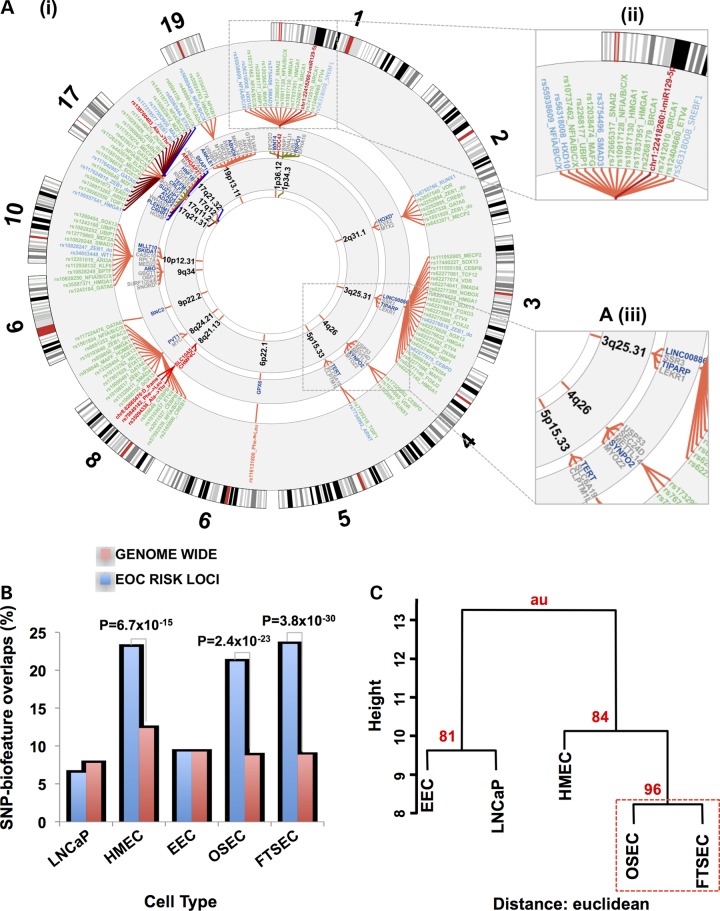
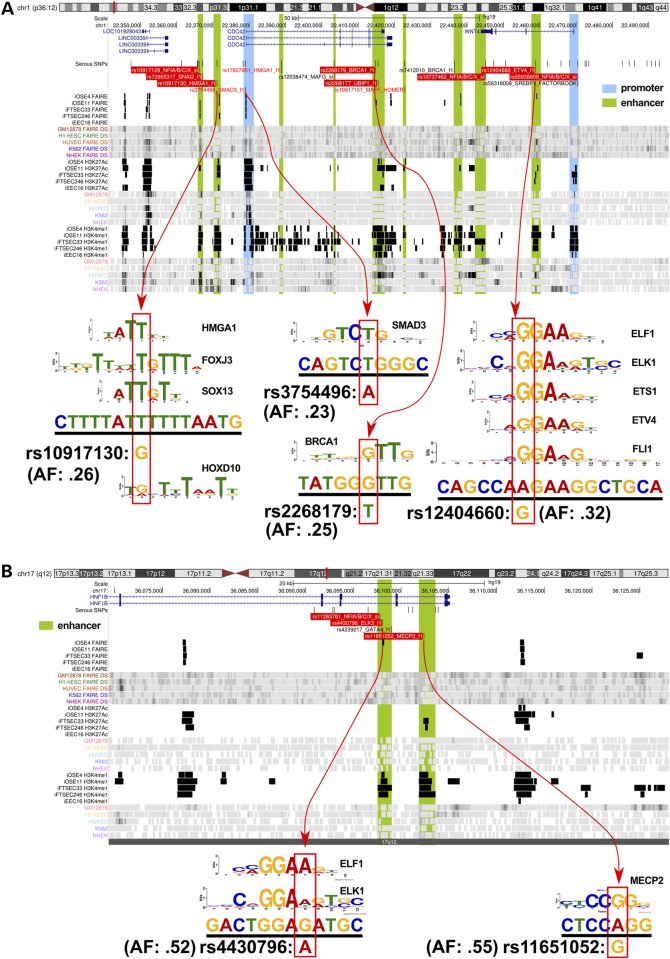
Of the 676 associated risk variants, 202 (30%) mapped to at least one of the functional categories (Fig. 2). The vast majority of risk SNPs mapped to non-coding features; only 12 (6%) mapped to coding exons of which four were non-synonymous substitutions and one was a frameshift variant (Supplementary Material, Table S3). One hundred and fifty-three SNPs mapped to 96 distinct enhancer regions (summarized in Table 2). We predicted the allele-specific impact of these SNPs on modulating transcription factor (TF) DNA binding motifs (Supplementary Material, Table S4). To specifically relate screened TF motifs to ovarian cancer, we restricted our analysis to the top 95% of expressed TFs in ovarian cancer precursor cells as determined by RNA-sequencing (Supplementary Material, Table S5). Of the 138 out of 153 enhancer SNPs that had designations in dbSNP, 103 (75%) modulated a TF motif. An additional 31 SNPs mapped to 11 distinct promoter or promoter-proximal enhancer regions. Of these 31 promoter SNPs, 18 were predicted to modulate TF motifs (Supplementary Material, Table S6). There was a single insertion variant in a putative microRNA target sequence (miR-129-5p) in the 3′ untranslated region of CDC42 at 1p36. The intersection between risk-associated SNPs and regulatory biofeatures at all 17 risk loci are illustrated in Figure 2, while more detailed illustrations for two loci, 1p34 and 17q12 regions, are given in Figure 3.
Figure 2. (.
A) (i) The ovarian cancer ‘riskome’ is displayed as a circular representation of the genome, excluding chromosomes that lack ovarian cancer risk regions. Concentric circles show, progressively from the inside-out, the chromosome band of the susceptibility region. The fine mapped SNPs (and inset A iii) in each region color-classified according to their putative functionality. Promotors are colored blue, putative enhancers green, coding variants red and miR targets burgundy. PWM motif-disruptions are included in the label with the SNP rsname (see Supplementary Material, Table S3 for full description). For some loci (e.g. chromosome 17) linking spanners have been assigned alternate colors to make the SNPs distinguishable when they are crowded. For physical disruptions (coding and miR target sequence) the gene name and spanners are also highlighted with the color corresponding to function. Genes (and inset A iii) within 100 kb of the top ranked risk SNP with the nearest gene in dark blue. (B) Analysis of SNP-regulatory biofeature intersections in gynecological tissues stratified by cell type, a human mammary epithelial cell (HMEC) line and the LNCaP prostate cancer cell line. The proportion of risk-SNPs intersecting regulatory biofeatures in the 17 ovarian cancer susceptibility regions was compared with the genome-wide distribution of SNPs intersecting regulatory biofeatures in the same tissues. (C) Bootstrap hierarchical cluster analysis for different cell types conditioned on the SNP-regulatory biofeatures interactions at the 17 confirmed HGSOC susceptibility regions.
Table 2.
Enrichment of risk SNPs at ovarian cancer susceptibility loci coinciding with regulatory biofeatures
| Cell Type | 17 HGSOC risk regions |
Genome-wide |
P-value | ||||
|---|---|---|---|---|---|---|---|
| Total number of regulatory biofeatures | Number of SNP-regulatory overlaps | Proportion of SNP-regulatory overlaps, % | Total number of regulatory biofeatures | Number of SNP-regulatory overlaps | Percentage SNP-regulatory overlaps, % | ||
| FTSEC | 676 | 159 | 23.5 | 39 706 715 | 3 547 703 | 8.9 | 3.77 × 10−30 |
| OSEC | 676 | 143 | 21.2 | 39 706 715 | 3 480 272 | 8.8 | 2.38 × 10−23 |
| EEC | 676 | 63 | 9.3 | 39 706 715 | 3 681 106 | 9.3 | 0.449 |
| HMEC | 676 | 156 | 23.1 | 39 706 715 | 4 931 084 | 12.4 | 6.71 × 10−15 |
| LNCaP | 676 | 44 | 6.5 | 39 706 715 | 3 087 796 | 7.8 | 0.878 |
Figure 3.
Illustration of SNP-functional annotation of two susceptibility regions. (A) The 1p34 regions, which predisposes to high-grade serous ovarian cancer (HGSOC) and (B) the 17q12 region which predisposes to both HGSOC and ovarian clear cell carcinoma. Each browser view highlights the position of genes within the region (top row annotation), followed by all 100:1 fine-mapped and motif-breaker SNPs for HGSOC. The following 30 tracks show the positions of significant peaks from this study (black) and other public data sources (colored). Labeled SNPs in the 2nd track (under ‘Serous SNPs’) encode for disruptions of major transcription factor motifs described by position weight matrices (PWM) (see diagrams underneath). Most SNPs break several motifs, but only the top-scoring match is shown in the browser view (see Supplementary Material, Table S1 for details). Black SNPs meet the minimum quality cutoff PWM match 85% of maximum score; red SNPs = 90%; red highlight = 95%. The diagrams underneath each browser view (indicated with red arrows) show the motif logo representing the PWM that matches to the genomic sequence (underlined) surrounding the SNP. The position of the SNP is indicated below the match sequence with the red outlined box, along with its allele frequency.
Tissue-specific enrichment of risk SNP-regulatory intersections
We evaluated whether there was enrichment at the 17 ovarian cancer susceptibility loci for those regulatory biofeatures that intersect risk SNPs compared with genome-wide SNP-regulatory biofeature intersections that may occur by chance. Of the 676 mapped risk SNPs in these regions, 184 (27.2%) were located in regulatory or promoter sites across the five different gynecological cell types. Genome-wide, we catalogued 312 245 regulatory marks (FAIRE, H3K4me1 and H3K27ac) in these cell types spanning 380 177 570 bp (12.3% of the genome). When we integrated these data with the genome-wide catalogue of common variants from the 1000 Genome Project, we found that 647 343 SNPs coincide with these regulatory biofeatures (14.2%). Thus, SNPs associated with HGSOC risk are twice as likely to fall within regulatory features identified in gynecological epithelial cell types than expected by chance (P = 4.65 × 10−19).
We performed the same analysis after stratifying by cell type. In gynecological tissues, we found that enrichment of the overlaps between SNPs and regulatory biofeatures in ovarian cancer risk regions was driven by signals from FTSECs (P = 3.8 × 10−30) and OSECs (P = 2.4 × 10−23), but not EECs (P = 0.45) (Fig. 2B and Table 2). We also found enrichment of risk SNP-regulatory biofeature intersections in HGSOC risk regions in HMECs (P = 6.7 × 10−15) but not in cells unrelated to ovarian cancer, the prostate cancer cell line LNCaP for which data on genome-wide DNaseI, H3K4me1 and H3K27ac were available (P = 0.88)16 (Fig. 2B and Table 3).
Table 3.
A summary of the numbers of SNPs located at sites of putative active and poised enhancers in ovarian cancer precursor tissues
| Cell type | SNPs in active enhancersa (n =) | SNPs in poised enhancersa (n =) | FAIRE only |
|---|---|---|---|
| IOE4 | 71 | 34 | 3 |
| IOE11 | 67 | 46 | 6 |
| FT33 | 53 | 88 | 0 |
| FT246 | 32 | 79 | 3 |
aDesignations are putative enhancers only.
We eliminated the possibility that the risk-associated enrichments at these loci are the result of bias due to a greater density of regulatory biofeatures in the risk regions compared with the rest of the genome. There are 7667 SNPs within the 17 risk regions covering 0.8% of the human genome. These regions encompass 17 Mb of which 4 688 907 bases coincide with regulatory marks identified in gynecological tissues (27.6% coverage). This figure is quite different from the genome-wide coverage of enhancer regulatory marks (16.6%). However, we could find no evidence for differences in enrichment between the cell types at the risk regions. For example, coverage of enhancer marks at the risk regions in EEC, OSEC and FTSEC was 17.9%, was 20.1 and 15.3%, respectively, compared with 9.4, 8.9 and 9.0% genome-wide. Similarly, HMEC has 16.7% enhancer coverage at the risk regions compared with 11.6% genome-wide. Only in LNCaP cells was coverage in these regions comparable to the genome-wide distribution (6.9 versus 4.4%). Thus, whereas regional enrichment for enhancers could explain a lack of enrichment in LNCaP, it does not account for the observed differences between EEC and HMECs, OSECs and FTSECs. Put together, these data suggest that the enrichment of disease associated SNPs in enhancers of specific cell types is not a function of regional enrichment of enhancer regulatory marks.
Hierarchical cluster analysis was used to compare the patterns of enrichment between OSEC, FTSEC, EEC, HMEC and LNCaP. Euclidean distance was used to evaluate the relationships between SNP-biofeature intersections detected in the different cell types. We chose complete linkage clustering to construct the hierarchies, which minimizes the maximum differences of cluster members, although we obtained similar trees using average and Ward minimum variance techniques (which is related to complete linkage). The closest relationship was observed for OSECs and FTSECs, which was highly supported by the data using a bootstrapping technique (Fig. 2iii). Ninety-six percent of randomly sampled subsets of the data (n = 10000) provide support for the same cluster. When we used correlation as a distance metric, we recapitulated the earlier relationships we found in the analysis of regulatory regions before conditioning based on risk SNPs (Fig. 1), because the majority of the SNPs that overlap with EEC regulatory marks are also common to OSECs, FTSECs and HMECs. Thus, the clustering analysis reveals that a large fraction of SNPs mapping to functional elements in OSECs, FTSECs and HMECS are not shared with EECs.
Enrichment for transcription factor response elements
We investigated whether risk SNPs coinciding with putative enhancers were enriched for known transcription factor binding motifs with respect to the biology and known pathways associated with ovarian cancer. We simulated draws (n = 1000) of 184 SNPs from the 1000 Genomes Project, or 184 SNPs located within enhancer regulatory marks (n = 1000) to distinguish bias from the source tissue. We confined these analyses to transcription factors that are expressed in ovarian cancer precursor tissues. Comparing the actual position weight matrix (PWM) disruption counts with the distribution in the simulation, we evaluated the significance based on the fraction of times we obtained as many motif disruptions or better from random draws of 184 the SNPs. We found that four response elements had a significance threshold of P < 0.05: SOX13 (17 SNPs, P = 0.003), ZNF384 (11 SNPs, P = 0.002), NFIA/B/C/X (11 SNPs, P = 0.030) and ETS2 (9 SNPs, P = 0.020). However, after correction for multiple comparisons none of these observations remained significant. We also considered how likely it would be to obtain four or more such motif enrichments from a single data set; in 1000 simulations of random SNPs, we observed enrichment of four or more motifs in 33% of simulated data sets at a P-value threshold <0.05 and in 11% of data sets at a P-value threshold <0.01. Thus, there was no evidence for significant enrichment of response elements for any specific factor. Finally, we performed ontology analyses on the ranked list of transcription factor response elements (based on P-value) and on various lists (with different ranking cutoffs) of disrupted PWMs versus all tested PWMs. We found no convincing evidence for factor/pathway enrichment.
Discussion
It is now widely recognized that the non-coding fraction of the human genome is densely populated with regulatory elements that play major roles in the initiation and development of common complex traits. In the current study, we catalogued the global architecture of regulatory chromatin marks that define poised and active enhancers, in the cell types postulated to be precursors of different subtypes of epithelial ovarian cancer: FTSECs and OSECs, both of which are the debated precursors of HGSOC (7,9,29); and EECs that are the likely precursors of CCOC (30–33). To our knowledge, this is the first time that profiles of active and poised enhancers have been described for these epithelial tissues.
The Encyclopedia of DNA Elements (ENCODE) project has shown unequivocally that the regulatory architecture of the human genome is highly tissue and cell-type specific (34,35). The data from the present study supports this; while the three gynecological tissues we studied show broad similarities in their genome-wide regulatory profiles, they also demonstrate clear differences between each other that likely reflect their underlying biology. In contrast, the regulatory architecture of these tissues is distinct from that of HMECs, breast cancer precursor cells. This is perhaps not surprising given that FTSECs, OSECs and EECs (which likely derive from uterine epithelial tissues) share a common embryological origin.
The molecular relationships between these different cell types changes when we base our analyses on the 17 genomic regions that are associated with susceptibility to ovarian cancer. Regulatory biofeatures within these regions may represent the functional mediators of tumor initiation, given that they coincide with common germline genetic variants associated with disease risk. The 17 regions are all significantly associated with risk of HGSOC, and consistent with this, we found highly statistically significant enrichment for regulatory biofeatures coinciding with risk-associated SNPs in FTSECs and OSECs. In a hierarchical cluster analysis based on intersections between risk SNPs and regulatory biofeatures, these tissues were more highly related to each other than any other cell type.
It was long thought that the ovarian surface epithelium was the cell of origin for HGSOCs, supported by both in vitro and in vitro data (7). More recently, FTSECs have emerged as a probable source of at least a proportion of HGSOCs following the observation of occult invasive serous intraepithelial carcinoma lesions in the fallopian tube in patients carrying germline BRCA1 and BRCA2 mutations (36–39); and in vivo studies have described the ability of FTSECs to develop into serous epithelial carcinomas in a background of mutant TP53 and PTEN, or SV40 (40–42). However, the close similarity between these two cell types based on their regulatory architecture at risk loci implies that both could be cells of origin for HGSOC.
We found no enrichment for SNP-regulatory biofeature interactions at HGSOC risk loci in EECs, but a highly statistically significant enrichment in HMECs. It is perhaps not surprising that endometriosis cells show few similarities with OSEC/FTSEC at these risk loci. Epidemiological studies find no evidence that endometriosis is associated with HGSOC (30); and from what is known about the development of the different ovarian cancer subtypes, the underlying biology and somatic genetic changes associated with HGSOC and CCOC are very different. The enrichment of enhancers at HGSOC risk loci in HMECs, and the close relationship between this cell type and OSECs/FTSECs, may also be unsurprising. Breast and ovarian cancers share a common etiology and an underlying genetic predisposition conferred by germline mutations in the BRCA1 and BRCA2 genes. GWASs have also identified multiple susceptibility loci that confer risk to both breast and ovarian cancer including 5p15, 8q24 and 19p13 (15–17,43,44). Of the 17 HGSOC loci evaluated in the current study, 7 (41%) are also genome-wide significant risk loci for breast cancer (P.D.P. Pharoah, personal communication). Taken together, these data suggest that similar gene regulation pathways underlie both breast cancer and HGSOC. These data are also consistent with other studies reporting genomic regions (e.g. at 8q24 and 5p15) associated with a multiple different human traits and common diseases (45–48).
One of the goals of this study was to identify the putative regulatory elements that coincide with risk-associated genetic variants at ovarian cancer susceptibility loci, that may be functional mediators of allele-specific effects of SNPs. Additional follow-up will be required to identify the disease-causing SNPs. Due to the nature of linkage disequilibrium throughout the genome, our observation that over 25% of the ‘candidate causal’ risk SNPs overlap regulatory biofeatures must be treated with caution. It is likely that only a proportion of these SNP-regulatory feature overlaps will represent true functional enhancer-associated alleles, with the remainder being chance associations. Similarly, we would have missed SNPs that intersect with other types of regulatory element in these tissue types because we only annotated risk-associated SNPs at regions of open chromatin/nucleosome depletion and histone marks indicative of active and poised enhancers. These represent a fraction of the non-coding regulatory biofeatures that are present in these tissues. There are multiple other histone modifications that regulate a range of biological processes such as gene expression, DNA repair, apoptosis, chromosome condensation, replication and meiosis, which we have not characterized. Neither did we interrogate the long non-coding RNA (lncRNA) transcriptomes of these cell types; lncRNAs are also known to show highly tissue-specific expression, and could also be effectors of risk-associated variants.
A previous study found evidence of common transcription factor binding sites located at several breast cancer susceptibility loci (49). This implies that there may be common mechanisms underlying breast cancer development. Thus, we might have expected to identify regulatory features that are common across multiple HGSOC loci. While we found evidence that the 17 risk loci may be enriched for the response elements for SOX13, ZNF384, NFIA/B/C/X and ETS2, these associations were not statistically robust when adjusted for multiple testing. This may imply that different regulatory mechanisms drive disease development at each HGSOC risk locus. Alternatively, it could be that common regulatory mechanisms exist but, as suggested earlier, we have not been able to identify them because first, our analyses were underpowered as a result of the relatively small number of risk loci indentified for HGSOC, and secondly, because we have not yet annotated all possible biofeatures in these tissue types.
In summary, we have catalogued the architecture of epigenetic marks and regions of open chromatin in three different gynecological cell types. We find evidence of tissue specificity when we compare the regulatory features between cell types and, when conditioned on risk-associated common variants, a highly statistically significant enrichment of disease-associated SNP-biofeature intersections in the proposed precursor cells of HGSOCs. These data provide a unique catalogue of epigenetic data for future studies aimed at understanding the genetics of gene regulation for different histological subtypes of invasive epithelial ovarian cancer.
Materials and Methods
Cell lines
Immortalized OSEC lines (IOE4, IOE11), derived from normal ovaries and immortalized using ectopic expression of TERT, have been previously described (23) and were cultured in medium 199:MCDB105 (mixed in a 1:1 ratio, both Sigma) supplemented with l-glutamine (Lonza) and 15% fetal bovine serum (Hyclone). FTSEC lines (FT33, FT246) were immortalized using TERT, a mutant CDK4 allele, and a shRNA targeting TP53 (24,25). FTSECs were cultured in DMEM/F12 (Lonza) supplemented with 2% Ultroser G (Crescent Chemicals). An EEC cell line (EEC16), derived from primary endometriosis epithelial cell culture (26), has been previously described and was grown in normal ovarian epithelial cell complete medium (50).
Capture of regulatory chromatin marks
For each cell line, four 15 cm culture dishes of ∼90% confluent cells were cross-linked in 1% formaldehyde for 10 min at room temperature before lysing cells in SDS buffer and sonicating to shear genomic DNA to ∼100–500 bp fragments. Insoluble cell debris was discarded, and the supernatant from pairs of dishes combined to represent two biological replicates per cell line. The material was divided into aliquots for input, FAIRE and ChIP. Immunoprecipitations were performed overnight at 4°C using antibodies against H3K27ac (Abcam ab4729) and H3K4me1 (Abcam ab8895) followed by incubation with protein A/G agarose beads (Pierce). Following washing and elution from beads, the DNA was ethanol precipitated, and the pellet washed with 70% ethanol. Recovered DNA was re-suspended in water for sequencing. Purification and recovery of FAIRE material was performed as previously described (27) in parallel with ChIP to ensure that annotation of open chromatin/nucleosome depletion was performed from the same cell lysate sample as annotation of histone acetylation and methylation marks. All ChIPseq and FAIREseq data generated in this study are deposited in the Gene Expression Omnibus (www.ncbi.nlm.nig.gov/geo/) under accession # GSE68104.
High-throughput sequencing and cell line data set generation
FAIRE and ChIP-recovered DNA was sequenced on the Illumina Hi-Seq 50 Single End read platform, and data processed by the Illumina analysis pipeline and aligned to the Human Reference Genome (assembly hg19) using BWA version 0.6.1-r104 (51). ‘Reads were processed by marking duplicates with Picard version 1.95 (http://broadinstitute.github.io/picard/) and by removing those with alignment quality scores less than 20. Marked duplicates were excluded from downstream analyses’. Enriched regions of the genome were identified by comparing ChIP and FAIRE samples using the MACS2 peak caller version 2.0.10 with default settings (52).
Analysis of differential nucleosome depletion and histone modification enrichment
Using bioconductor package DiffBind (52), a common set of peaks was generated across each set of experiments (FAIRE, ChIP-seq for H3K27ac and H3K4me1, respectively). In order to normalize across biological replicates, we retained peaks which overlapped at least once across replicates and cell types (setting minOverlaps to 2 in the dba.count function). To identify significantly differentially bound peaks, we first counted the number of reads within each experimental sample overlapping the identified peak while subtracting the number of overlapping reads from the corresponding input. Differential analysis was performed using the DESeq2 method within DiffBind. Differential scores were computed by setting the score argument to ‘DBA_SCORE_TMM_MINUS_FULL’ within the dba.count function. Briefly, this method calculates the tagwise dispersions which are subject to an exact test after normalization by computing the trimmed mean of M values (TMM). Resulting P-values were adjusted using Benjamini–Hochberg multiple testing correction to compute FDR; only sites found to be differential between cell types with an FDR <0.05 were retained for downstream analysis. Supplementary Material, Table S7 lists the read counts for each file before and after filtering for base quality.
Genome-wide regulatory fingerprinting
Using the defined nucleosome depleted regions, a set of peaks unique to OSEC, FTSEC, OSEC + FTSEC and HMEC [from previously published data (27) and ENCODE] were used to interrogate the read density of each experimental condition. A window of ±1 kb was taken from each summit, and the sequencing library densities relative to these specific positions were calculated by extending tags by their estimated ChIP-fragment length using default settings defined by Homer's annotatePeaks.pl script (53).
Germline genetic susceptibility data
Genotyping data from GWAS and follow-up replication analysis have been performed in epithelial ovarian cancer cases and controls by two different consortia: The ovarian cancer association consortium (OCAC) and the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA). For OCAC, data were available for three population-based ovarian cancer GWAS (Stage 1 data). These included 2165 cases and 2564 controls from a GWAS from North America (‘US GWAS’) (54), 1762 cases and 6118 controls from a UK-based GWAS (‘UK GWAS’) (14), and 441 cases and 441 controls from the U19 GWAS. Furthermore, 11 069 cases and 21 722 controls from 41 OCAC studies were genotyped using the iCOGS array (‘OCAC-iCOGS’ stage data). Overall, 43 studies from 11 countries provided data on 15 347 women diagnosed with invasive epithelial ovarian cancer, 9627 of whom were diagnosed with serous ovarian cancer, and 30 845 controls from the general population. From CIMBA, data were available on 15 252 BRCA1 mutation carriers and 8211 BRCA2 mutation carriers of which 2462 BRCA1 and 631 BRCA2 carriers diagnosed with invasive epithelial ovarian cancer. Samples from both consortia were genotyped for 211 155 replication SNPs as part of the collaborative oncological gene–environment study (COGS) (18).
Further details of the OCAC and CIMBA study populations as well as the genotyping, quality control and statistical analyses have been described elsewhere (18,55,56). All subjects from OCAC and CIMBA included in this analysis were of European descent. All recruiting centers in CIMBA and OCAC required participants to give written informed consent and collected data and blood samples under ethically approved protocols.
Integrating regulatory and germline genetic data
Annotation of SNPs was carried out using bedtools (57) with data sets of peak calls described in this paper for FAIRE, H3K4me1 and H3K27Ac of each of IOE4, IOE11, FT33, FT246 and EEC16. We used the snp138 track of the UCSC genome browser to annotate missense mutations in protein coding regions, and miRcode (58) to interrogate potential microRNA binding sites in untranslated sequences. Motif disruptions and enrichment calculations were determined according to the previously published algorithm (45), using position weight matrices as defined in Factorbook (59), Hocomoco (60) and Homer (53).
Analyses of enrichment and hierarchical clustering in cell-type specific enhancers
Enrichment of SNPs in enhancers of different cell types and within risk regions was calculated using the hypergeometric distribution using base functions in R. To assess relationships among the SNP/regulatory overlaps in different cell types, a matrix of 1 and 0 s denoting membership of each SNP in each cell type was constructed and a distance matrix calculated on Euclidean distance. The matrix was then further analyzed using the pvclust (61) package in R, which calculates the multi-scale bootstrapping P-value [method of Shimodaira (62)] on each putative cluster within the tree, as reported in Figure 2C.
Supplementary Material
Funding
This study was supported by the Genetic Associations and Mechanisms in Oncology (GAME-ON): a NCI Cancer Post-GWAS Initiative (U19-CA148112), a Program Project Development Grant from the Ovarian Cancer Research Fund (to S.A.G. and A.M.) and a pilot award from the Clinical and Translational Science Institute at USC to K.L. and S.A.G. K.L. is supported by a K99/R00 grant from the National Cancer Institute (Grant number 1K99CA184415-01). The in vitro aspects of this project were performed within the Norris Cancer Centre at USC, which is supported in part by award number P30CA014089 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Supplementary Material
Ackowledgements
We thank Charles Nicolet and the Data Production Facility at the USC Epigenome Center for library construction and high throughput sequencing. We would also like to acknowledge other members of the Ovarian Cancer Association Consortium and the Collaborative Oncological Gene-environment Study (COGS) (Jonathan Tyrer, Hoda Anton-Culver, Natalia Antonenkova, Helen Baker, Elisa V. Bandera, Yukie Bean, Matthias W. Beckmann, Andrew Berchuck, Maria Bisogna, Line Bjorge, Natalia Bogdanova, Louise A. Brinton, Angela Brooks-Wilson, Fiona Bruinsma, Ralf Butzow, Ian G. Campbell, Karen Carty, Jenny Chang-Claude, Ann Chen, Zhihua Chen, Linda S. Cook, Daniel W. Cramer, Julie M. Cunningham, Cezary Cybulski, Agnieszka Dansonka-Mieszkowska, Joe Dennis, Ed Dicks, Jennifer A. Doherty, Thilo Dörk, Andreas du Bois, Matthias Dürst, Diana Eccles, Douglas F. Easton, Robert P. Edwards, Ursula Eilber, Arif B. Ekici, Peter A. Fasching, Brooke L. Fridley, Yu-Tang Gao, Aleksandra Gentry-Maharaj, Graham G. Giles, Rosalind Glasspool, Ellen L. Goode, Marc T. Goodman, Jacek Grownwald, Patricia Harrington, Philipp Harter, Hanis Nazihah Hasmad, Alexander Hein, Florian Heitz, Michelle A.T. Hildebrandt, Peter Hillemanns, Estrid Hogdall, Claus Hogdall, Satoyo Hosono, Edwin S. Iversen, Anna Jakubowska ,Paul James, Allan Jensen, Bu-Tian Ji, Beth Y Karlan, Susanne Kruger Kjaer, Linda E. Kelemen, Melissa Kellar, Joseph L. Kelley, Lambertus A. Kiemeney, Camilla Krakstad, Jolanta Kupryjanczyk, Diether Lambrechts, Sandrina Lambrechts, Nhu D. Le, Shashi Lele, Arto Leminen, Jenny Lester, Douglas A. Levine, Dong Liang, Jolanta Lissowska, Karen Lu, Jan Lubinski, Lene Lundvall, Leon F.A.G. Massuger, Keitaro Matsuo, Valerie McGuire, John R. McLaughlin, Ian McNeish, Usha Menon, Francesmary Modugno, Kirsten B. Moysich, Steven A. Narod, Lotte Nedergaard, Roberta B. Ness, Mat Adenan Noor Azmi, Kunle Odunsi, Sara H. Olson, Irene Orlow, Sandra Orsulic, Rachel Palmieri Weber, Celeste L. Pearce, Tanja Pejovic, Liisa M. Pelttari, Jennifer Permuth-Wey, Catherine M. Phelan, Malcolm C. Pike, Elizabeth M. Poole, Harvey A. Risch, Barry Rosen, Mary Anne Rossing, Joseph H. Rothstein, Anja Rudolph, Ingo B. Runnebaum, Iwona K. Rzepecka, Helga B. Salvesen, Joellen M. Schildkraut, Ira Schwaab, Thomas A. Sellers, Xiao-Ou Shu, Yurii B Shvetsov, Nadeem Siddiqui, Weiva Sieh, Honglin Song, Melissa C. Southey, Lara Sucheston, Ingvild L. Tangen, Soo-Hwang Teo, Kathryn L. Terry, Pamela J. Thompson, Agnieszka Timorek, Ya-Yu Tsai, Shelley S. Tworoger, Jonathan Tyrer, Anne M. van Altena, Els Van Nieuwenhuysen, Ignace Vergote, Robert A. Vierkant, Shan Wang-Gohrke, Christine Walsh, Nicolas Wentzensen, Alice S. Whittemore, Kristine G. Wicklund, Lynne R. Wilkens, Yin-Ling Woo, Xifeng Wu, Anna H. Wu, Hannah Yang, Wei Zheng, Argyrios Ziogas); and The Consortium of Investigators of Modifiers of BRCA1/2 CIMBA.
Conflict of Interest statement. None declared.
Contributor Information
Collaborators: The Ovarian Cancer Association Consortium, The Consortium of Investigators of Modifiers of BRCA1/2, Jonathan Tyrer, Hoda Anton-Culver, Natalia Antonenkova, Helen Baker, Elisa V. Bandera, Yukie Bean, Matthias W. Beckmann, Andrew Berchuck, Maria Bisogna, Line Bjorge, Natalia Bogdanova, Louise A. Brinton, Angela Brooks-Wilson, Fiona Bruinsma, Ralf Butzow, Ian G. Campbell, Karen Carty, Jenny Chang-Claude, Ann Chen, Zhihua Chen, Linda S. Cook, Daniel W. Cramer, Julie M. Cunningham, Cezary Cybulski, Agnieszka Dansonka-Mieszkowska, Joe Dennis, Ed Dicks, Jennifer A. Doherty, Thilo Dörk, Andreas du Bois, Matthias Dürst, Diana Eccles, Douglas F. Easton, Robert P. Edwards, Ursula Eilber, Arif B. Ekici, Peter A. Fasching, Brooke L. Fridley, Yu-Tang Gao, Aleksandra Gentry-Maharaj, Graham G. Giles, Rosalind Glasspool, Ellen L. Goode, Marc T. Goodman, Jacek Grownwald, Patricia Harrington, Philipp Harter, Hanis Nazihah Hasmad, Alexander Hein, Florian Heitz, Michelle A.T. Hildebrandt, Peter Hillemanns, Estrid Hogdall, Claus Hogdall, Satoyo Hosono, Edwin S. Iversen, Anna Jakubowska, Paul James, Allan Jensen, Bu-Tian Ji, Beth Y Karlan, Susanne Kruger Kjaer, Linda E. Kelemen, Melissa Kellar, Joseph L. Kelley, Lambertus A. Kiemeney, Camilla Krakstad, Jolanta Kupryjanczyk, Diether Lambrechts, Sandrina Lambrechts, Nhu D. Le, Shashi Lele, Arto Leminen, Jenny Lester, Douglas A. Levine, Dong Liang, Jolanta Lissowska, Karen Lu, Jan Lubinski, Lene Lundvall, Leon F.A.G. Massuger, Keitaro Matsuo, Valerie McGuire, John R. McLaughlin, Ian McNeish, Usha Menon, Francesmary Modugno, Kirsten B. Moysich, Steven A. Narod, Lotte Nedergaard, Roberta B. Ness, Mat Adenan Noor Azmi, Kunle Odunsi, Sara H. Olson, Irene Orlow, Sandra Orsulic, Rachel Palmieri Weber, Celeste L. Pearce, Tanja Pejovic, Liisa M. Pelttari, Jennifer Permuth-Wey, Catherine M. Phelan, Malcolm C. Pike, Elizabeth M. Poole, Harvey A. Risch, Barry Rosen, Mary Anne Rossing, Joseph H. Rothstein, Anja Rudolph, Ingo B. Runnebaum, Iwona K. Rzepecka, Helga B. Salvesen, Joellen M. Schildkraut, Ira Schwaab, Thomas A. Sellers, Xiao-Ou Shu, Yurii B Shvetsov, Nadeem Siddiqui, Weiva Sieh, Honglin Song, Melissa C. Southey, Lara Sucheston, Ingvild L. Tangen, Soo-Hwang Teo, Kathryn L. Terry, Pamela J. Thompson, Agnieszka Timorek, Ya-Yu Tsai, Shelley S. Tworoger, Jonathan Tyrer, Anne M. van Altena, Els Van Nieuwenhuysen, Ignace Vergote, Robert A. Vierkant, Shan Wang-Gohrke, Christine Walsh, Nicolas Wentzensen, Alice S. Whittemore, Kristine G. Wicklund, Lynne R. Wilkens, Yin-Ling Woo, Xifeng Wu, Anna H. Wu, Hannah Yang, Wei Zheng, and Argyrios Ziogas
References
- 1.Parker S.C., Stitzel M.L., Taylor D.L., Orozco J.M., Erdos M.R., Akiyama J.A., van Bueren K.L., Chines P.S., Narisu N., Black B.L., et al. (2013) Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl Acad. Sci. USA, 110, 17921–17926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J., et al. (2012) Systematic localization of common disease-associated variation in regulatory DNA. Science, 337, 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Network C.G.A.R. (2011) Integrated genomic analyses of ovarian carcinoma. Nature, 474, 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed A.A., Etemadmoghadam D., Temple J., Lynch A.G., Riad M., Sharma R., Stewart C., Fereday S., Caldas C., Defazio A., et al. (2010) Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J. Pathol., 221, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiegand K.C., Shah S.P., Al-Agha O.M., Zhao Y., Tse K., Zeng T., Senz J., McConechy M.K., Anglesio M.S., Kalloger S.E., et al. (2010) ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med., 363, 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahman M., Nakayama K., Rahman M.T., Nakayama N., Ishikawa M., Katagiri A., Iida K., Nakayama S., Otsuki Y., Shih I.M., et al. (2012) Clinicopathologic and biological analysis of PIK3CA mutation in ovarian clear cell carcinoma. Hum. Pathol., 43, 2197–2206. [DOI] [PubMed] [Google Scholar]
- 7.Auersperg N. (2013) Ovarian surface epithelium as a source of ovarian cancers: unwarranted speculation or evidence-based hypothesis? Gynecol. Oncol., 130, 246–251. [DOI] [PubMed] [Google Scholar]
- 8.Leeper K., Garcia R., Swisher E., Goff B., Greer B., Paley P. (2002) Pathologic findings in prophylactic oophorectomy specimens in high-risk women. Gynecol. Oncol., 87, 52–56. [DOI] [PubMed] [Google Scholar]
- 9.Crum C.P., Herfs M., Ning G., Bijron J.G., Howitt B.E., Jimenez C.A., Hanamornroongruang S., McKeon F.D., Xian W. (2013) Through the glass darkly: intraepithelial neoplasia, top-down differentiation, and the road to ovarian cancer. J. Pathol., 231, 402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sainz de la Cuesta R., Eichhorn J.H., Rice L.W., Fuller A.F., Nikrui N., Goff B.A. (1996) Histologic transformation of benign endometriosis to early epithelial ovarian cancer. Gynecol. Oncol., 60, 238–244. [DOI] [PubMed] [Google Scholar]
- 11.Foster K.A., Harrington P., Kerr J., Russell P., DiCioccio R.A., Scott I.V., Jacobs I., Chenevix-Trench G., Ponder B.A., Gayther S.A. (1996) Somatic and germline mutations of the BRCA2 gene in sporadic ovarian cancer. Cancer Res., 56, 3622–3625. [PubMed] [Google Scholar]
- 12.Futreal P.A., Liu Q., Shattuck-Eidens D., Cochran C., Harshman K., Tavtigian S., Bennett L.M., Haugen-Strano A., Swensen J., Miki Y. (1994) BRCA1 mutations in primary breast and ovarian carcinomas. Science, 266, 120–122. [DOI] [PubMed] [Google Scholar]
- 13.Chui M.H., Ryan P., Radigan J., Ferguson S.E., Pollett A., Aronson M., Semotiuk K., Holter S., Sy K., Kwon J.S., et al. (2014) The histomorphology of Lynch syndrome-associated ovarian carcinomas: toward a subtype-specific screening strategy. Am. J. Surg. Pathol., 38, 1173–1181. [DOI] [PubMed] [Google Scholar]
- 14.Song H., Ramus S.J., Tyrer J., Bolton K.L., Gentry-Maharaj A., Wozniak E., Anton-Culver H., Chang-Claude J., Cramer D.W., DiCioccio R., et al. (2009) A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat. Genet., 41, 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolton K.L., Tyrer J., Song H., Ramus S.J., Notaridou M., Jones C., Sher T., Gentry-Maharaj A., Wozniak E., Tsai Y.Y., et al. (2010) Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat. Genet., 42, 880–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goode E.L., Chenevix-Trench G., Song H., Ramus S.J., Notaridou M., Lawrenson K., Widschwendter M., Vierkant R.A., Larson M.C., Kjaer S.K., et al. (2010) A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat. Genet., 42, 874–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bojesen S.E., Pooley K.A., Johnatty S.E., Beesley J., Michailidou K., Tyrer J.P., Edwards S.L., Pickett H.A., Shen H.C., Smart C.E., et al. (2013) Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat. Genet., 45, 371–384, 384e371–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pharoah P.D., Tsai Y.Y., Ramus S.J., Phelan C.M., Goode E.L., Lawrenson K., Buckley M., Fridley B.L., Tyrer J.P., Shen H., et al. (2013) GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat. Genet., 45, 362–370, 370e361–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen H., Fridley B.L., Song H., Lawrenson K., Cunningham J.M., Ramus S.J., Cicek M.S., Tyrer J., Stram D., Larson M.C., et al. (2013) Epigenetic analysis leads to identification of HNF1B as a subtype-specific susceptibility gene for ovarian cancer. Nat. Commun., 4, 1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen K., Ma H., Li L., Zang R., Wang C., Song F., Shi T., Yu D., Yang M., Xue W., et al. (2014) Genome-wide association study identifies new susceptibility loci for epithelial ovarian cancer in Han Chinese women. Nat. Commun., 5, 4682. [DOI] [PubMed] [Google Scholar]
- 21.Kuchenbaecker K.B., Ramus S.J., Tyrer J., Lee A., Shen H.C., Beesley J., Lawrenson K., McGuffog L., Healey S., Lee J.M., et al. (2015) Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat. Genet., 47, 164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrenson K., Sproul D., Grun B., Notaridou M., Benjamin E., Jacobs I.J., Dafou D., Sims A.H., Gayther S.A. (2011) Modelling genetic and clinical heterogeneity in epithelial ovarian cancers. Carcinogenesis, 32, 1540–1549. [DOI] [PubMed] [Google Scholar]
- 23.Lawrenson K., Grun B., Benjamin E., Jacobs I.J., Dafou D., Gayther S.A. (2010) Senescent fibroblasts promote neoplastic transformation of partially transformed ovarian epithelial cells in a three-dimensional model of early stage ovarian cancer. Neoplasia, 12, 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karst A.M., Levanon K., Drapkin R. (2011) Modeling high-grade serous ovarian carcinogenesis from the fallopian tube. Proc. Natl Acad. Sci. USA, 108, 7547–7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karst A.M., Drapkin R. (2012) Primary culture and immortalization of human fallopian tube secretory epithelial cells. Nat. Protoc., 7, 1755–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brueggmann D., Templeman C., Starzinski-Powitz A., Rao N.P., Gayther S.A., Lawrenson K. (2014) Novel three-dimensional in vitro models of ovarian endometriosis. J. Ovarian Res., 7, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhie S.K., Hazelett D.J., Coetzee S.G., Yan C., Noushmehr H., Coetzee G.A. (2014) Nucleosome positioning and histone modifications define relationships between regulatory elements and nearby gene expression in breast epithelial cells. BMC Genomics, 15, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., Consortium G.P. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature, 491, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurman R.J., Shih I.M. (2010) The origin and pathogenesis of epithelial ovarian cancer: a proposed unifying theory. Am. J. Surg. Pathol., 34, 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearce C.L., Templeman C., Rossing M.A., Lee A., Near A.M., Webb P.M., Nagle C.M., Doherty J.A., Cushing-Haugen K.L., Wicklund K.G., et al. (2012) Association between endometriosis and risk of histological subtypes of ovarian cancer: a pooled analysis of case-control studies. Lancet Oncol., 13, 385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dzatic-Smiljkovic O., Vasiljevic M., Djukic M., Vugdelic R., Vugdelic J. (2011) Frequency of ovarian endometriosis in epithelial ovarian cancer patients. Clin. Exp. Obstet. Gynecol., 38, 394–398. [PubMed] [Google Scholar]
- 32.Yamamoto S., Tsuda H., Takano M., Tamai S., Matsubara O. (2012) PIK3CA mutations and loss of ARID1A protein expression are early events in the development of cystic ovarian clear cell adenocarcinoma. Virchows Arch., 460, 77–87. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto S., Tsuda H., Takano M., Iwaya K., Tamai S., Matsubara O. (2011) PIK3CA mutation is an early event in the development of endometriosis-associated ovarian clear cell adenocarcinoma. J. Pathol., 225, 189–194. [DOI] [PubMed] [Google Scholar]
- 34.Bernstein B.E., Birney E., Dunham I., Green E.D., Gunter C., Snyder M., Consortium E.P. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunham I., Kundaje A., Aldred S.F., Collins P.J., Davis C.A., Doyle F., Epstein C.B., Frietze S., Harrow J., Kaul R., et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reade C.J., McVey R.M., Tone A.A., Finlayson S.J., McAlpine J.N., Fung-Kee-Fung M., Ferguson S.E. (2014) The fallopian tube as the origin of high grade serous ovarian cancer: review of a paradigm shift. J. Obstet. Gynaecol. Can., 36, 133–140. [DOI] [PubMed] [Google Scholar]
- 37.Lee Y., Miron A., Drapkin R., Nucci M.R., Medeiros F., Saleemuddin A., Garber J., Birch C., Mou H., Gordon R.W., et al. (2007) A candidate precursor to serous carcinoma that originates in the distal fallopian tube. J. Pathol., 211, 26–35. [DOI] [PubMed] [Google Scholar]
- 38.Dubeau L., Drapkin R. (2013) Coming into focus: the nonovarian origins of ovarian cancer. Ann. Oncol., 24(Suppl. 8), viii28–viii35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medeiros F., Muto M.G., Lee Y., Elvin J.A., Callahan M.J., Feltmate C., Garber J.E., Cramer D.W., Crum C.P. (2006) The tubal fimbria is a preferred site for early adenocarcinoma in women with familial ovarian cancer syndrome. Am. J. Surg. Pathol., 30, 230–236. [DOI] [PubMed] [Google Scholar]
- 40.Perets R., Wyant G.A., Muto K.W., Bijron J.G., Poole B.B., Chin K.T., Chen J.Y., Ohman A.W., Stepule C.D., Kwak S., et al. (2013) Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in brca;tp53;pten models. Cancer Cell, 24, 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherman-Baust C.A., Kuhn E., Valle B.L., Shih I.M., Kurman R.J., Wang T.L., Amano T., Ko M.S., Miyoshi I., Araki Y., et al. (2014) A genetically engineered ovarian cancer mouse model based on fallopian tube transformation mimics human high-grade serous carcinoma development. J. Pathol., 233, 228–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Howell V.M. (2014) Genetically engineered mouse models for epithelial ovarian cancer: are we there yet? Semin. Cell Dev. Biol., 27, 106–117. [DOI] [PubMed] [Google Scholar]
- 43.Antoniou A.C., Wang X., Fredericksen Z.S., McGuffog L., Tarrell R., Sinilnikova O.M., Healey S., Morrison J., Kartsonaki C., Lesnick T., et al. (2010) A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat. Genet., 42, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turnbull C., Ahmed S., Morrison J., Pernet D., Renwick A., Maranian M., Seal S., Ghoussaini M., Hines S., Healey C.S., et al. (2010) Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet., 42, 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hazelett D.J., Rhie S.K., Gaddis M., Yan C., Lakeland D.L., Coetzee S.G., Henderson B.E., Noushmehr H., Cozen W., Kote-Jarai Z., et al. (2014) Comprehensive functional annotation of 77 prostate cancer risk loci. PLoS Genet., 10, e1004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petersen G.M., Amundadottir L., Fuchs C.S., Kraft P., Stolzenberg-Solomon R.Z., Jacobs K.B., Arslan A.A., Bueno-de-Mesquita H.B., Gallinger S., Gross M., et al. (2010) A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet., 42, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turnbull C., Rapley E.A., Seal S., Pernet D., Renwick A., Hughes D., Ricketts M., Linger R., Nsengimana J., Deloukas P., et al. (2010) Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat. Genet., 42, 604–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stacey S.N., Sulem P., Masson G., Gudjonsson S.A., Thorleifsson G., Jakobsdottir M., Sigurdsson A., Gudbjartsson D.F., Sigurgeirsson B., Benediktsdottir K.R., et al. (2009) New common variants affecting susceptibility to basal cell carcinoma. Nat. Genet., 41, 909–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Q., Seo J.H., Stranger B., McKenna A., Pe'er I., Laframboise T., Brown M., Tyekucheva S., Freedman M.L. (2013) Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell, 152, 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li N.F., Wilbanks G., Balkwill F., Jacobs I.J., Dafou D., Gayther S.A. (2004) A modified medium that significantly improves the growth of human normal ovarian surface epithelial (OSE) cells in vitro. Lab. Invest., 84, 923–931. [DOI] [PubMed] [Google Scholar]
- 51.Li H., Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R., et al. (2012) Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature, 481, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell, 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Permuth-Wey J., Kim D., Tsai Y.Y., Lin H.Y., Chen Y.A., Barnholtz-Sloan J., Birrer M.J., Bloom G., Chanock S.J., Chen Z., et al. (2011) LIN28B polymorphisms influence susceptibility to epithelial ovarian cancer. Cancer Res., 71, 3896–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gaudet M.M., Kuchenbaecker K.B., Vijai J., Klein R.J., Kirchhoff T., McGuffog L., Barrowdale D., Dunning A.M., Lee A., Dennis J., et al. (2013) Identification of a BRCA2-specific modifier locus at 6p24 related to breast cancer risk. PLoS Genet., 9, e1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Couch F.J., Wang X., McGuffog L., Lee A., Olswold C., Kuchenbaecker K.B., Soucy P., Fredericksen Z., Barrowdale D., Dennis J., et al. (2013) Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet., 9, e1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quinlan A.R., Hall I.M. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jeggari A., Marks D.S., Larsson E. (2012) miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics, 28, 2062–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J., Zhuang J., Iyer S., Lin X., Whitfield T.W., Greven M.C., Pierce B.G., Dong X., Kundaje A., Cheng Y., et al. (2012) Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res., 22, 1798–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kulakovskiy I.V., Medvedeva Y.A., Schaefer U., Kasianov A.S., Vorontsov I.E., Bajic V.B., Makeev V.J. (2013) HOCOMOCO: a comprehensive collection of human transcription factor binding sites models. Nucleic Acids Res., 41, D195–D202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suzuki R., Shimodaira H. (2006) Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics, 22, 1540–1542. [DOI] [PubMed] [Google Scholar]
- 62.Shimodaira H. (2002) An approximately unbiased test of phylogenetic tree selection. Syst. Biol., 51, 492–508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.