Abstract
Exposure to high levels of environmental lead, or biomarker evidence of high body lead content, is associated with anaemia, developmental and neurological deficits in children, and increased mortality in adults. Adverse effects of lead still occur despite substantial reduction in environmental exposure. There is genetic variation between individuals in blood lead concentration but the polymorphisms contributing to this have not been defined. We measured blood or erythrocyte lead content, and carried out genome-wide association analysis, on population-based cohorts of adult volunteers from Australia and UK (N = 5433). Samples from Australia were collected in two studies, in 1993–1996 and 2002–2005 and from UK in 1991–1992. One locus, at ALAD on chromosome 9, showed consistent association with blood lead across countries and evidence for multiple independent allelic effects. The most significant single nucleotide polymorphism (SNP), rs1805313 (P = 3.91 × 10−14 for lead concentration in a meta-analysis of all data), is known to have effects on ALAD expression in blood cells but other SNPs affecting ALAD expression did not affect blood lead. Variants at 12 other loci, including ABO, showed suggestive associations (5 × 10−6 > P > 5 × 10−8). Identification of genetic polymorphisms affecting blood lead reinforces the view that genetic factors, as well as environmental ones, are important in determining blood lead levels. The ways in which ALAD variation affects lead uptake or distribution are still to be determined.
Introduction
Many biological and pathological effects are determined by a combination of exposure to an event or toxic substance and individual susceptibility. Harmful effects of toxic elements are usually traced to environmental exposure, but previous studies have shown evidence for genetic variation in humans for concentrations of arsenic, cadmium, lead and mercury in blood (1,2). Some loci which may affect concentrations of these elements in blood were identified using genetic linkage analysis (2).
The effects of environmental variation on lead, and of lead concentrations in blood or tissues, have been extensively studied, particularly because of its effects on intellectual development in infancy and childhood (3–6). Adult cognitive function may also be affected (7–9). There is evidence that higher lead values are associated with hypertension (10), peripheral vascular disease (11), increased adult mortality (12,13), reproductive impairment (14,15), renal impairment (16,17) and altered immune function (18,19). These adverse effects have prompted extensive measures to reduce lead exposure, mainly through removal of lead from paints and petrol (gasoline). Reduction in the lead content of petrol has produced changes in indices of body burden (20–23), but evidence of harm from lead exposure is still appearing. Successive rounds of the US National Health and Nutrition Examination Survey (NHANES), both before and after lead reduction measures, have shown associations between blood lead and both overall and cause-specific mortality (12,13,24–26). Results suggest a 40–60% increase in adjusted mortality with increasing lead concentration across the range encountered in the USA population, for both cardiovascular and cancer deaths and at both times of survey. Although blood lead has a half-life ∼30 days and is therefore said to be a short-term marker of exposure, it predicts long-term effects such as mortality.
A number of attempts have been made to test for effects of genetic polymorphisms in candidate genes on blood or bone lead values. To date the results are mixed, with allelic associations both reported and denied for variants in ALAD (aminolevulinate dehydratase) and VDR (1,25-dihydroxyvitamin D3 receptor) (27,28). Two meta-analyses of published data (29,30) suggest a small but statistically significant effect of rs1800435, a non-synonymous coding variant in ALAD (Lys59Asn, often described as ALAD 1/2), on blood lead. Because of the clinical association between iron deficiency and lead toxicity, an association with iron status, which may up- or down-regulate intestinal divalent cation transporters, and specifically with HFE (hemochromatosis) genotype has also been proposed (31,32).
A few studies have considered the general question of familial similarity of toxic element concentrations and whether any such similarity is due to shared genes or shared environment. A family-based study of blood lead (33,34) showed evidence for shared environmental effects in young siblings that diminished with age, and no spousal correlation, whereas a twin study (35) suggested additive genetic effects in women and shared environmental effects in men. We previously found significant heritability for blood lead, measured in erythrocytes (1), and have now extended the study of genetic causes of variation in lead concentrations using genome-wide association analysis on cohorts from Australia and UK.
Results
Blood lead distributions
The distribution of blood lead results varied by location and time. For the pregnant women in the ALSPAC study, with samples collected in 1991–1992, the mean was 0.177 µmol/l, SD 0.071 µmol/l (3.67 µg/dl, SD 1.47 µg/dl), N = 4285. For the first Australian study, with samples collected between 1993 and 1996, the mean estimated blood lead concentration (based on an assumed mean whole-blood haemoglobin concentration of 147 g/l) was 0.249 µmol/l, SD 0.129 µmol/l (5.16 µg/dl, SD 2.67 µg/dl), N = 2926. The second Australian study, with samples collected between 2001 and 2005, gave mean estimated blood lead 0.124 µmol/l, SD 0.083 µmol/l (2.57 µg/dl, SD 1.72 µg/dl), N = 1324. These statistics are derived from all participants with lead measurements, whether or not genotyping was available for the genome-wide association study (GWAS).
A small proportion of people had lead measurements from both the Australian projects, and for these the correlation between estimated blood lead results on the two occasions was r = 0.466, N = 78, P = 1.5 × 10−5. A paired test on these subjects showed that the mean decrease in blood lead across time was 0.148 ± 0.013 (standard error of mean) µmol/l, 3.06 ± 0.27 µg/dl (t = 11.76, P = 5.98 × 10−19).
Genetic associations
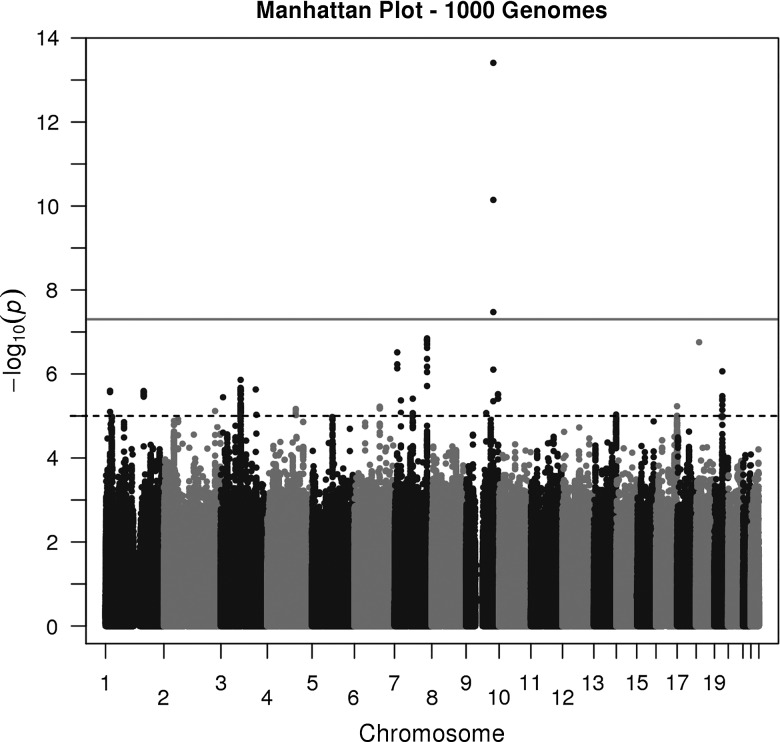
We present the allelic association results from Australian and UK cohorts as a Manhattan plot in Figure 1 and list the significant and suggestive loci in Table 1. Further details are given in Supplementary Material, Table S1 and Supplementary Material, Figures S1 and S2. Genome-wide significant (P < 5 × 10−8) results were found on chromosome 9, and the single nucleotide polymorphisms (SNP) showing association were located within or close to the ALAD gene. A regional plot for the ALAD region is shown in Figure 2. The most significant result in the meta-analysis (P = 3.91 × 10−14) was for rs1805313, which is intronic. This was also the most significant SNP in the ALSPAC data (P = 2.22 × 10−7), whereas in the QIMR data it was the second most significant (P = 1.65 × 10−9) and the most significant was rs8177800 (P = 2.76 × 10−12). The previously reported non-synonymous coding polymorphism (ALAD 1/2, rs1800435, Lys59Asn) did not show genome-wide significant association in our data (P = 0.0045).
Figure 1.
Manhattan plot for blood lead, showing results from meta-analysis of QIMR and ALSPAC data. The continuous and interrupted horizontal lines show the thresholds for genome-wide significant associations (P < 5 × 10−8) and for P < 5 × 10−5.
Table 1.
Genome-wide significant (P < 5.0 × 10−8) and suggestive (5.0 × 10−8 < P < 5.0 × 10−6) loci from meta-analysis of 1000 Genomes-imputed data, showing the SNP with smallest P-value from each locus.
| SNP | Chr | BP (Build 37) | A1 | A2 | Nearest gene | QIMR |
ALSPAC |
Meta-analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R2 | Freq A1 | Beta | SE | P | INFO | Freq A1 | Beta | SE | P | P | ||||||
| Significant | ||||||||||||||||
| rs1805313 | 9 | 116 151 191 | A | G | ALAD | 0.470 | 0.726 | 0.265 | 0.044 | 1.65 × 10−9 | 1 | 0.683 | 0.142 | 0.027 | 2.22 × 10−7 | 3.91 × 10−14 |
| Suggestive | ||||||||||||||||
| rs12136530 | 1 | 19 761 429 | A | G | CAPZB | 0.813 | 0.813 | 0.077 | 0.042 | 0.069 | 0.859 | 0.793 | 0.160 | 0.035 | 3.80 × 10−6 | 2.51 × 10−6 |
| rs2662776 | 1 | 163 165 029 | A | G | RGS5 | 0.993 | 0.475 | 0.116 | 0.028 | 2.67 × 10−5 | 0.999 | 0.467 | 0.066 | 0.026 | 0.011 | 2.54 × 10−6 |
| rs76153987 | 3 | 9 214 817 | T | C | SRGAP3 | 0.464 | 0.044 | −0.350 | 0.097 | 3.19 × 10−4 | 0.970 | 0.050 | −0.195 | 0.060 | 0.001 | 3.58 × 10−6 |
| rs9863067 | 3 | 84 147 443 | C | G | GBE1-CADM2 | 0.887 | 0.914 | 0.121 | 0.051 | 0.018 | 0.996 | 0.904 | 0.194 | 0.044 | 1.29 × 10−5 | 1.38 × 10−6 |
| rs79019069 | 3 | 148 481 023 | A | G | AGTR1/CPB1 | 0.645 | 0.974 | 0.408 | 0.112 | 2.76 × 10−4 | 0.889 | 0.974 | 0.276 | 0.087 | 0.001 | 2.33 × 10−6 |
| rs116864947 | 7 | 11 705 786 | T | C | THSD7A | 0.773 | 0.984 | −0.381 | 0.130 | 0.003 | 0.996 | 0.983 | −0.431 | 0.102 | 2.31 × 10−5 | 3.06 × 10−7 |
| rs6462018 | 7 | 27 519 118 | G | A | EVX1-HIBADH | 0.877 | 0.515 | −0.101 | 0.029 | 6.02 × 10−4 | 0.945 | 0.504 | −0.084 | 0.027 | 0.002 | 4.26 × 10−6 |
| rs798338 | 7 | 77 917 038 | A | C | MAGI2 | 0.965 | 0.359 | −0.072 | 0.029 | 0.013 | 0.992 | 0.354 | −0.111 | 0.027 | 5.27 × 10−5 | 3.88 × 10−6 |
| rs60580184 | 7 | 138 850 967 | A | G | TTC26 | 0.856 | 0.026 | 0.299 | 0.090 | 9.61 × 10−4 | 0.998 | 0.025 | 0.340 | 0.082 | 3.12 × 10−5 | 1.42 × 10−7 |
| rs550057 | 9 | 136 146 597 | T | C | ABO | 0.943 | 0.240 | −0.089 | 0.033 | 0.007 | 0.994 | 0.266 | −0.116 | 0.030 | 1.02 × 10−4 | 3.02 × 10−6 |
| rs144653651 | 18 | 12 909 504 | A | G | PTPN2-SEH1L | 0.386 | 0.013 | 0.124 | 0.221 | 0.574 | 0.990 | 0.075 | 0.265 | 0.050 | 1.53 × 10−7 | 1.75 × 10−7 |
| rs16968074 | 19 | 33 884 341 | A | G | PEPD | 0.911 | 0.711 | −0.110 | 0.032 | 5.50 × 10−4 | 0.975 | 0.686 | 0.101 | 0.028 | 4.08 × 10−4 | 8.64 × 10−7 |
A1 is the effect allele, R2 and INFO are measures of SNP imputation quality.
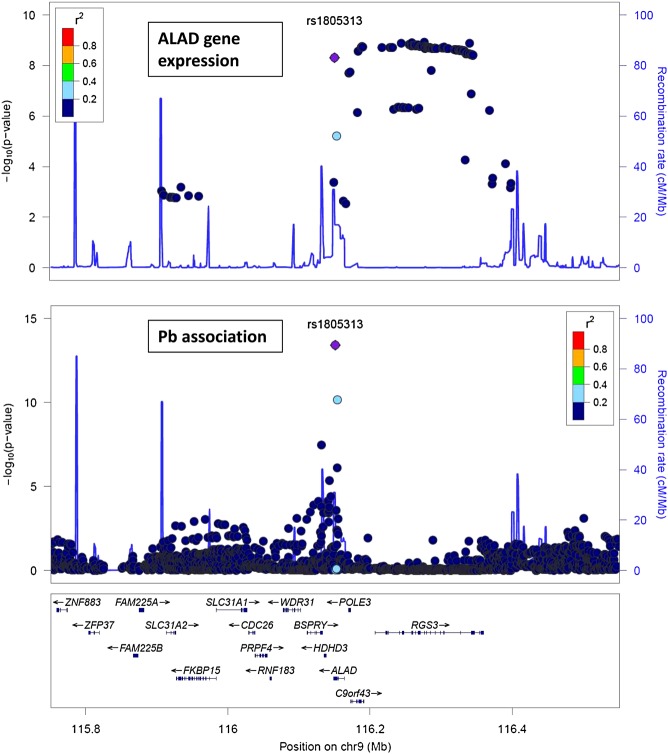
Figure 2.
Comparison of location of allelic effects on blood lead around the ALAD locus (bottom panel), and effects on ALAD expression in blood cells (top panel; data from 36 and http://genenetwork.nl/bloodeqtlbrowser/). Note the cluster of independent SNPs across RGS3 which affect ALAD expression but not blood lead. One hundred and twenty-eight out of the 129 SNPs plotted for ALAD expression are also in the plot for association with blood lead.
Conditional analysis of SNPs within 1.0 Mbp of rs1805313 (N = 3203 SNPs) revealed two SNPs with suggestive evidence of independent association; rs10121150 in the BSPRY gene and rs8177812 in the ALAD gene (Table 2, Supplementary Material, Fig. S3). rs10121150 at the BSPRY locus, which is not in strong linkage disequilibrium with rs1805313 (r2 = 0.020, D′ = 0.372), met genome-wide significance in the unadjusted analysis (Pmeta = 3.35 × 10−8), and this effect was only slightly attenuated after conditioning on rs1805313 (Pmeta = 5.06 × 10−7). In contrast, rs8177812 at the ALAD locus, which is in partial LD with rs1805313 (r2 = 0.406, D′ = 1), was non-significant in the unadjusted analysis (Pmeta = 0.935), but showed suggestive evidence for association after conditioning on rs1805313 (Pmeta = 2.27 × 10−6; Supplementary Material, Fig. S3). Results for the non-synonymous coding SNP rs1800435 were essentially unchanged after adjusting for effects of rs1805313 (P = 0.0038, compared with 0.0045 originally).
Table 2.
Significant and suggestive (P < 5.0 × 10−6) SNPs from meta-analysis of 1000 Genomes-imputed data, from conditional analysis.
| SNP | Chr | BP (Build 37) | A1 | A2 | Nearest gene | Conditioning | QIMR |
ALSPAC |
Meta-analysis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Freq A1 | Beta | SE | P | Freq A1 | Beta | SE | P | P | |||||||
| Conditioning on rs1805313 only | |||||||||||||||
| rs10121150 | 9 | 116 131 695 | A | G | BSPRY | Before | 0.797 | −0.122 | 0.036 | 6.57 × 10−4 | 0.786 | −0.143 | 0.033 | 1.12 × 10−5 | 3.35 × 10−8 |
| After | 0.797 | −0.105 | 0.035 | 0.003 | 0.786 | −0.133 | 0.032 | 4.48 × 10−5 | 5.06 × 10−7 | ||||||
| rs8177812 | 9 | 116 151 527 | A | G | ALAD | Before | 0.112 | 0.041 | 0.056 | 0.461 | 0.122 | −0.016 | 0.039 | 0.676 | 0.935 |
| After | 0.112 | 0.221 | 0.056 | 6.91 × 10−5 | 0.122 | 0.131 | 0.046 | 0.005 | 2.27 × 10−6 | ||||||
| Conditioning on rs10121150 and rs8177812 | |||||||||||||||
| rs1805313 | 9 | 116 151 191 | A | G | ALAD | Before | 0.726 | 0.265 | 0.044 | 1.65 × 10−9 | 0.683 | 0.142 | 0.027 | 2.22 × 10−7 | 3.91 × 10−14 |
| After | 0.726 | 0.274 | 0.044 | 4.13 × 10−10 | 0.683 | 0.183 | 0.032 | 1.95 × 10−8 | 1.63 × 10−16 | ||||||
| rs8177800 | 9 | 116 154 099 | C | T | ALAD | Before | 0.918 | 0.611 | 0.087 | 2.76 × 10−12 | 0.926 | 0.183 | 0.056 | 9.64 × 10−4 | 7.17 × 10−11 |
| After | 0.918 | 0.576 | 0.087 | 3.66 × 10−11 | 0.926 | 0.182 | 0.056 | 0.001 | 2.69 × 10−10 | ||||||
First, the allele count for the most significant SNP from the meta-analysis (rs1805313) was included as a covariate in association analysis, and two other SNPs (rs10121150 and rs8177812) then showed suggestive association with blood lead. Second, allele counts for rs10121150 and rs8177812 were included as covariates. A1 is the effect allele.
Several other loci contained SNPs which did not reach genome-wide significance but showed suggestive evidence of association (P < 5 × 10−6; Table 1, Supplementary Material, Fig. S2), including variants within the ABO gene. Gene-based analysis using VEGAS showed no additional significant genes. Estimation of the proportion of phenotypic variance associated with all typed SNPs, using GCTA on the unrelated ALSPAC subjects, gave an estimate of 37 ± 14% for the SNP-based heritability.
Gene expression
Multiple SNPs, including rs1805313, have effects on ALAD expression in unfractionated, non-transformed cells from whole blood (36). We compared allelic effects of SNPs on lead results with the reported effects on ALAD expression to check whether effects on gene expression give a plausible explanation for the effects on blood lead. A comparison of SNP associations with ALAD expression in whole blood and with blood lead concentration is shown in Figure 2.
Another SNP, rs818702, which is not in LD with rs1805313 (r2 = 0.003, D′ = 0.080) and has been reported to affect ALAD expression in liver (37) but not blood cells, shows only nominal association with blood lead in our data (P = 3.96 × 10−5).
Linkage and association
In view of our previous results (2) suggesting linkage of blood lead to a locus on chromosome 3, with SLC4A7 as a strong candidate gene, we examined the region of chromosome 3 between the microsatellite markers D3S3726 (at 19.5 Mbp) and D3S1300 (at 60.5 Mbp), with a particular emphasis on the region around the linkage peak at ∼28 Mbp. The strongest allelic association was for rs62255214, which is within 200 kb of SLC4A7, but this was not genome-wide significant (P = 3.14 × 10−5).
Discussion
After combining results from our UK and Australian studies, we found significant allelic associations for blood lead on chromosome 9 at ALAD. Three SNPs at this locus showed genome-wide significant associations. These were consistent between the Australian and UK datasets, but with some heterogeneity of effect sizes which may be due to differences in SNP imputation. The strongest effect on our combined data was for rs1805313, which is intronic, and we found evidence of independent effects for other SNPs in the region. Although the conditional analysis did not reveal additional SNPs that met the criteria for genome-wide significance, the region examined was limited and the two independent SNP associations with P < 5 × 10−6 at this locus are unlikely to arise by chance.
The mechanism behind these allelic associations is uncertain. Aminolevulinate dehydratase (E.C. 4.2.1.24) converts delta-aminolevulinic acid to porphobilinogen, an essential early step in the synthesis of porphyrins and haem. This enzyme has previously been associated with lead at a number of levels. Lead inhibits ALAD activity, and measurement of erythrocyte ALAD has been used as a biomarker of lead exposure (38). There have been many studies of the relationship between ALAD polymorphisms and blood or bone lead concentration (summarized in 29 and 30). Other papers suggest that ALAD polymorphisms modify the relationship between either lead exposure or blood lead, and various neurological or haematological aspects of lead toxicity. These previous studies have focussed on the non-synonymous coding SNP rs1800435, Lys59Asn (which they usually refer to as ALAD 1/2). A meta-analysis by Zhao et al. (30) suggested a small effect of ALAD 1/2 on blood lead, while Scinicariello et al. (29) suggested that the effect is only seen in lead-exposed subjects. However, although our data show highly significant association with blood lead at the ALAD locus, we find far less evidence of association for this non-synonymous coding SNP despite our large sample size and consistency of results across two populations (in Australia and UK).
SNP effects are often postulated to affect gene expression; data from blood cells or liver show associations between genotype and ALAD expression for the most significant SNP in our results (rs1805313), among others. One possible model is that these SNPs affect ALAD concentration in the erythrocytes; ALAD binds lead; so the SNPs affect erythrocyte lead concentration. However, the pattern of SNP associations with ALAD expression across the nearby region is inconsistent with this explanation, because a cluster of SNPs extending across RGS3 and not in linkage disequilibrium with rs1805313 show equally strong association with ALAD expression but not blood lead (see Fig. 2). One caveat is that the ALAD expression data are derived from studies on peripheral blood cells, and it is possible that effects on expression in the erythrocyte precursors may differ. Other explanations are possible, but the metabolic step catalysed by ALAD has no obvious connection with lead uptake or accumulation.
Although ALAD is the first gene to consider in relation to our SNP association findings, it is not the only candidate. Firstly, one of the SNPs which showed suggestive association with blood lead after conditioning on rs1805313, rs10121150, is in an intron of BSPRY, This gene is annotated by UniProt (http://www.uniprot.org/uniprot/Q5W0U4#section_comments) as coding for a zinc finger protein, with ‘zinc ion binding’ and ‘calcium ion transport’ functions. Other evidence (39) implies that BSPRY is involved in epithelial Ca2+ signalling on the grounds that it is co-expressed with other genes with this postulated function. Secondly, this SNP (rs10121150), and others in linkage disequilibrium with it, affect expression of SLC31A2, another transport gene which primarily transports copper (40) but might also affect other divalent cations including lead. However, other SNPs with stronger effects on SLC31A2 expression did not affect lead concentration.
It is important to consider whether variation at ALAD affects the whole-body burden of lead, or only the erythrocyte results. The ALAD result may mean that some people are more likely to suffer lead toxicity because the polymorphism leads to higher blood lead levels. Alternatively, these people could paradoxically be more resistant to lead toxicity (for a given blood lead level) because the lead is bound to ALAD and not available to damage neural or erythrocyte-precursor tissues. There are implications for variation in vulnerability to high blood (and maybe total body) lead from similar levels of exposure; and for interpretation of blood lead results and the relationship between blood lead and risk of toxicity. At present, and pending further data, we cannot be sure whether rs1805313 and other variants near ALAD affect whole-body lead burden and lead-related risk.
The suggestive loci need confirmation or refutation from future studies, but the association at ABO is interesting. This locus has previously shown allelic associations for low-density lipoprotein cholesterol and the cell adhesion molecule ICAM-1, coronary heart disease, venous thromboembolism and coagulation factors, thyroid disease and erythrocyte traits (http://www.genome.gov/gwastudies/, accessed 1 October 2014). The SNPs which reached suggestive significance for lead in this study include rs507666, which is associated with A1 blood group status (41). However, none of these findings provide obvious clues to the possible relationship between ABO and lead concentration.
The previous linkage finding on chromosome 3, centred on SLC4A7 (2), is not explained or supported by our allelic association results. There is no association in the relevant region with P < 10−5 so the current results certainly rule out a common-variant effect large enough to account for the previous linkage result. There could still be an effect from SNPs of large effect which are poorly tagged by the genotyped and imputed variants or the original linkage finding could represent a Type I error.
Our study has both limitations and strengths. It was conducted on adults not known to be occupationally exposed to lead, and we only measured blood (or erythrocyte) lead concentrations. Bone or total body lead content may be subject to different effects. As well as larger studies to detect additional loci, GWAS should be extended to include non-invasive assessment of bone lead. In view of the suggestions in published meta-analyses that allelic effects at ALAD may be greater in occupationally exposed people, and in children, effects of rs1805313 should be checked in these groups. Nevertheless this study, which we believe to be the first genome-wide assessment of SNP associations with blood lead, was based on two substantial datasets which showed consistent results. It emphasises the existence of genetic variation in the response to environmental lead sources, and shows highly significant and diverse effects at or near the ALAD locus.
Materials and Methods
Australia (Queensland Institute of Medical Research)
Our analysis is based on results for 2603 adults with phenotype and genotype data who participated in one or both of two studies run from the Queensland Institute of Medical Research (QIMR). Information on study participants is summarized in Supplementary Material, Table S2.
The first of these studies recruited twins born before 1964 who were enrolled in a volunteer registry (the Australian Twin Registry). Subjects and methods for this study are described in ref. (2). Briefly, participants completed a postal questionnaire in 1989 and a telephone interview in 1993–1994, and provided a blood sample in 1993–1996. We initially determined zygosity from responses to questions about physical similarity, but this has now been updated for those with data included in this paper using SNP-typing results. Participants gave written informed consent, and the studies were approved by the appropriate ethics committees. Blood was collected from 1134 men and 2241 women.
As blood samples had been fractionated to provide plasma, buffy coat for DNA extraction and erythrocytes, we used erythrocytes rather than whole blood for elemental analysis. Samples were stored at −80°C. Before analysis, the erythrocytes were thawed at room temperature and diluted 1:20 in ammonia/ethylenediaminetetraacetic acid solution containing rhodium as an internal standard. Lead concentrations were measured by inductively coupled-plasma mass spectrometry (ICP-MS) on a PerkinElmer Elan 5000 mass spectrometer (PerkinElmer Inc., Wellesley, MA, USA) or a Varian UltraMass (Varian Inc., Palo Alto, CA, USA). Haemoglobin concentration was then measured on the diluted samples using the cyanmethemoglobin method.
Analytical precision for lead measurement in the Australian study was calculated from results on high and low quality control (QC) materials that were analysed with each batch of samples. At a mean lead concentration of 0.14 μmol/l, the between-day standard deviation was 0.018 μmol/l (coefficient of variation 13.1%) and at 1.81 μmol/l it was 0.169 μmol/l (9.4%). For 117 subjects, erythrocyte lead was measured on two separate blood tubes taken on the same occasion, with good correlation between the replicates (r = 0.954).
Results were log-transformed and analysis batch, haemoglobin concentration in the thawed sample and analytical QC data were used as covariates in preliminary steps which generated standardized residuals for subsequent analysis, as previously described (1). Of the 2926 participants from this 1993–1996 Twin Study with measurements of lead concentration in erythrocytes, 1570 had genome-wide SNP genotyping data. Genotypes were determined using Illumina chips; methods, QC steps and imputation of untyped HapMap 2 SNPs were as previously described (42).
The second of the Australian studies which generated data on lead concentrations took place in 2001–2005 and was designed to characterize loci affecting alcohol or nicotine dependence through a genome-wide association approach (43). This was a twin-family design, in which relatives of participants in our earlier twin studies were also recruited. Data were again obtained through telephone interviews, and blood samples were obtained from 8396 people. Again, element concentrations were determined on erythrocyte fractions by ICP-MS, but with an Agilent 7500 system (Agilent Technologies Inc., Santa Clara, CA, USA). Covariate adjustment and generation of standardized residuals were carried out in the same way as for the previous study. A total of 1104 people from the nicotine-alcohol study had both phenotypic data and genome-wide SNP data.
After allowing for overlap of 71 genotyped people who had data from participation in both the earlier and later Australian studies, for whom the earlier result was used, there were 2603 valid sets of phenotype and genotype results.
Genotypes for untyped SNPs were imputed using haplotypes from the 1000 Genomes project (Europeans in the 1000G release on 4 August 2010), the observed genotypes were phased using the ‘MACH’ program followed by imputation using the ‘minimac’ program. Poorly imputed SNPs (with r2 < 0.3) were excluded from further analysis. Genome-wide allelic association analysis was carried out on the standardized residuals for erythrocyte lead, pre-adjusted for sex and age, using an additive model accounting for within-family relatedness in Merlin (44) (http://www.sph.umich.edu/csg/abecasis/Merlin/, accessed 7 June 2012).
UK (Avon Longitudinal Study of Parents and Children)
The Avon Longitudinal Study of Parents and Children (ALSPAC), also known as The Children of the Nineties Study, was designed to understand the ways in which the physical and social environments interact over time with genetic inheritance to affect health, behaviour and development in infancy, childhood and then into adulthood (45,46). Information on study participants is summarized in Supplementary Material, Table S2. The study area (formerly known as Avon) is an area bordering the Severn estuary, with a total population of 1 million, including Bristol, a major city of population 0.5 million, and surrounding areas including small towns, villages and farming communities. Eligible women were those who were pregnant, resident in the study area and had an expected date of delivery between 1 April 1991 and 31 December 1992. They were recruited as early in pregnancy as possible. Of all mothers who were interested in taking part, an estimated 80% of the eligible population were included and answered at least one questionnaire. Ethical approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees. Please note that the study website contains details of all the data that is available through a fully searchable data dictionary (http://www.bris.ac.uk/alspac/researchers/data-access/data-dictionary/).
Maternal blood samples were collected in acid washed vacutainers by midwives on the first occasion on which they saw the pregnant women. Samples were kept as whole blood in the original tubes stored at 4°C. Trace metal analysis was performed by the Centers for Disease Control, Atlanta, GA in 2009–2010 on a sub-sample of ALSPAC mothers, broadly representative of the whole cohort (47). Clotted whole blood was digested by adding concentrated nitric acid and heating in a microwave at a controlled temperature and time. Addition of rhenium prior to heating allowed the correction of results for any loss due to evaporation. The digestion matrix was diluted 1:9 by volume using internal standards (Ir and Te) at a constant concentration. Diluted liquid samples were introduced as an aerosol into the inductively coupled-plasma dynamic reaction cell mass spectrometry (ICP-DRC-MS) through a nebulizer and spray chamber carried by a flowing argon stream.
Analytical QC information could not be accessed for the ALSPAC study, but repeated measurement data were available. Blood lead was measured on some participants on two different occasions. Forty subjects had repeated measures up to 11 weeks apart (intraclass correlation 0.88); 24 of them had repeated measures up to 8 weeks apart (intraclass correlation 0.93); 13 of them had repeated measures up to 6 weeks apart (intraclass correlation 0.94). As the time interval decreases in UK data, the within-person correlation converges towards that found for replicated measurements on the same occasion for the Australian participants.
Ten thousand and fifteen women (mothers from the ALSPAC cohort) were genotyped using the Illumina 660 quad SNP chip which contains 557 124 SNP markers. Markers with minor allele frequency < 1%, SNPs with >5% missing genotypes and any markers that failed an exact test of Hardy–Weinberg equilibrium (P < 1 × 10−6) were excluded from further analyses. Samples were excluded if they displayed >5% missingness, had indeterminate X chromosome heterozygosity or extreme autosomal heterozygosity. Samples which might contribute to population stratification were identified by multidimensional scaling of genome-wide identity-by-state pairwise distances using the four HapMap2 populations as a reference, and then excluded. Cryptic relatedness was assessed using a Pi-hat of >0.125 which is expected to correspond to roughly 12.5% alleles shared IBD or a relatedness at the first cousin level. Related subjects that passed all other QC thresholds were retained during subsequent phasing and imputation, but then one from each pair of putatively related individuals were excluded from all genetic association analyses.
Nine thousand and forty-eight subjects and 526 688 SNPs passed QC filters. We combined mothers' genotypes with cleaned genome-wide SNP data from 9115 ALSPAC children (see 48 for details) and removed a further 321 subjects due to potential sample mismatches. This resulted in a dataset of 17 842 subjects including 6305 mother-offspring duos. We estimated haplotypes using ShapeIT (v2.r644) which utilises relatedness during phasing (49). Additional genotypes were imputed using IMPUTE version2 (50) to a 1000 Genomes reference panel that contained all available ethnicities with singleton and monomorphic sites removed (May 2011 1000 genomes release, and the phase release was December 2013 from the impute2 website (https://mathgen.stats.ox.ac.uk/impute/impute_v2.html#reference)). Eight thousand and one hundred ninety-six eligible mothers with available genotype data remained after exclusion of related subjects using the cryptic relatedness measures described above. Only imputed genotypes with minor allele frequencies ≥1%, INFO ≥0.4 and which were biallelic were considered for association (N = 8 269 388 SNPs). Of these 8196 mothers with cleaned genetic data, 2830 mothers also had phenotype data available.
Blood lead concentrations were log10 transformed to approximate normality. Batch was included as a random effect and standardized residuals were derived. We then performed genome-wide association analysis on these residuals using the software package SNPTEST (51).
Further analysis
Results from the Australian and UK data were compared and combined using fixed-effects inverse-variance meta-analysis in METAL (52) (http://www.sph.umich.edu/csg/abecasis/Metal/, accessed 6 June 2012). SNPs were excluded if the minor allele frequency was <1%, the INFO score was <0.4 (for ALSPAC) or the R2 score was <0.3 (for QIMR). Of note, 6 391 392 SNPs that met the inclusion criteria and were present in all studies were included in the meta-analysis. Each dataset was checked for acceptable Q–Q plots, with estimated λ values of 1.005 and 1.012 obtained for the Australian and UK results, respectively. Results were visualized using LocusZoom (53) (http://csg.sph.umich.edu/locuszoom/, accessed 6 June 2012) and R (54). Conditional analysis, in which the most significant SNP at each locus was included as a covariate in order to detect independent effects at such loci, was carried out on each of the datasets and results were combined by meta-analysis. Gene-based analysis with VEGAS (55) (http://gump.qimr.edu.au/VEGAS/, accessed 30 October 2012) was used to check whether any genes showed over-representation of nominally significant SNPs, as might occur if several variants in a gene, not in linkage disequilibrium with each other, affect the phenotype. Regions around SNPs which showed genome-wide significance were checked for expression QTLs using ‘Blood eQTL browser’ (http://genenetwork.nl/bloodeqtlbrowser/) and data on liver eQTLs from ref. (37).
Supplementary Material
Funding
QIMR Study: Sample collection and the recruitment and interviewing of participants were funded by grants AA007535, AA013320, AA013321, AA013326 and DA012854 from the US National Institutes of Health to A.C.H., N.G.M., P.A.F.M. and the late Richard Todd. Biomarker and trace element measurement was supported by AA014041 to J.B.W. G.W.M. is supported by the National Health and Medical Research Council of Australia Fellowship Scheme.
ALSPAC Study: The UK Medical Research Council, the Wellcome Trust (grant ref: 092731) and the University of Bristol currently provide core support for the Avon Longitudinal Study of Parents and Children. This work was partly conducted in the Medical Research Council Integrative Epidemiology Unit, a research unit supported by the Medical Research Council (MC_UU_12013 to G.D.S.). The study was supported by an Australian Research Council Future Fellowship (FT130101709 to D.M.E.) and a Medical Research Council Programme grant (MC_UU_12013/4 to D.M.E.). Genotyping in ALSPAC mothers was funded by a Wellcome Trust Grant to D.A.L. (WT088806) and her contribution to this study was supported by an MRC grant (MC_UU_12013/5).
Supplementary Material
Acknowledgements
QIMR Study: We thank the twins and other members of their families for their participation, and the staff of the Genetic Epidemiology and Molecular Epidemiology Units, QIMR Berghofer Medical Research Institute, for project management, subject recruitment and interviews, sample collection and sample processing. Trace element measurements were made using the facilities of the Department of Clinical Biochemistry, Royal Prince Alfred Hospital, Sydney, Australia.
ALSPAC Study: We are extremely grateful to all the individuals who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. We thank the Centre National de Genotypage for generating the ALSPAC GWA data. The assays of the maternal blood samples were carried out at the Centers for Disease Control and Prevention with funding from the US National Oceanic and Atmospheric Administration (NOAA), and we acknowledge the contribution of Robert Jones to this work.
This publication is the work of the authors and they will serve as guarantors for the contents of this paper.
Conflict of Interest statement. None declared.
References
- 1.Whitfield J.B., Dy V., McQuilty R., Zhu G., Montgomery G.W., Ferreira M.A., Duffy D.L., Neale M.C., Heijmans B.T., Heath A.C., et al. (2007) Evidence of genetic effects on blood lead concentration. Environ. Health Perspect., 115, 1224–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitfield J.B., Dy V., McQuilty R., Zhu G., Heath A.C., Montgomery G.W., Martin N.G. (2010) Genetic effects on toxic and essential elements in humans: arsenic, cadmium, copper, lead, mercury, selenium and zinc in erythrocytes. Environ. Health Perspect., 118, 776–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pocock S.J., Smith M., Baghurst P. (1994) Environmental lead and children's intelligence: a systematic review of the epidemiological evidence. BMJ, 309, 1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartz J. (1994) Low-level lead exposure and children’s IQ: a meta-analysis and search for a threshold. Environ. Res., 65, 42–55. [DOI] [PubMed] [Google Scholar]
- 5.Canfield R.L., Henderson C.R., Jr, Cory-Slechta D.A., Cox C., Jusko T.A., Lanphear B.P. (2003) Intellectual impairment in children with blood lead concentrations below 10 microg per deciliter. N. Engl. J. Med., 348, 1517–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Needleman H. (2004) Lead poisoning. Annu. Rev. Med., 55, 209–222. [DOI] [PubMed] [Google Scholar]
- 7.Weisskopf M.G., Wright R.O., Schwartz J., Spiro A., III, Sparrow D., Aro A., Hu H. (2004) Cumulative lead exposure and prospective change in cognition among elderly men: the VA Normative Aging Study. Am. J. Epidemiol., 160, 1184–1193. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz B.S., Hu H. (2007) Adult lead exposure: time for change. Environ. Health Perspect., 115, 451–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shih R.A., Hu H., Weisskopf M.G., Schwartz B.S. (2007) Cumulative lead dose and cognitive function in adults: a review of studies that measured both blood lead and bone lead. Environ. Health Perspect., 115, 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navas-Acien A., Guallar E., Silbergeld E.K., Rothenberg S.J. (2007) Lead exposure and cardiovascular disease—a systematic review. Environ. Health Perspect., 115, 472–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Navas-Acien A., Selvin E., Sharrett A.R., Calderon-Aranda E., Silbergeld E., Guallar E. (2004) Lead, cadmium, smoking, and increased risk of peripheral arterial disease. Circulation, 109, 3196–3201. [DOI] [PubMed] [Google Scholar]
- 12.Lustberg M., Silbergeld E. (2002) Blood lead levels and mortality. Arch. Intern. Med., 162, 2443–2449. [DOI] [PubMed] [Google Scholar]
- 13.Schober S.E., Mirel L.B., Graubard B.I., Brody D.J., Flegal K.M. (2006) Blood lead levels and death from all causes, cardiovascular disease, and cancer: results from the NHANES III mortality study. Environ. Health Perspect., 114, 1538–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hertz-Picciotto I. (2000) The evidence that lead increases the risk for spontaneous abortion. Am. J. Ind. Med., 38, 300–309. [DOI] [PubMed] [Google Scholar]
- 15.Sallmen M. (2001) Exposure to lead and male fertility. Int. J. Occup. Med. Environ. Health., 14, 219–222. [PubMed] [Google Scholar]
- 16.Staessen J.A., Lauwerys R.R., Buchet J.P., Bulpitt C.J., Rondia D., Vanrenterghem Y., Amery A. (1992) Impairment of renal function with increasing blood lead concentrations in the general population. The Cadmibel Study Group. N. Engl. J. Med., 327, 151–156. [DOI] [PubMed] [Google Scholar]
- 17.Lin J.L., Lin-Tan D.T., Hsu K.H., Yu C.C. (2003) Environmental lead exposure and progression of chronic renal diseases in patients without diabetes. N. Engl. J. Med., 348, 277–286. [DOI] [PubMed] [Google Scholar]
- 18.Dietert R.R., Lee J.E., Hussain I., Piepenbrink M. (2004) Developmental immunotoxicology of lead. Toxicol. Appl. Pharmacol., 198, 86–94. [DOI] [PubMed] [Google Scholar]
- 19.Dietert R.R., Piepenbrink M.S. (2006) Lead and immune function. Crit. Rev. Toxicol., 36, 359–385. [DOI] [PubMed] [Google Scholar]
- 20.Luo W., Zhang Y., Li H. (2003) Children's blood lead levels after the phasing out of leaded gasoline in Shantou, China. Arch. Environ. Health, 58, 184–187. [DOI] [PubMed] [Google Scholar]
- 21.Pirkle J.L., Brody D.J., Gunter E.W., Kramer R.A., Paschal D.C., Flegal K.M., Matte T.D. (1994) The decline in blood lead levels in the United States. The National Health and Nutrition Examination Surveys (NHANES). JAMA, 272, 284–291. [PubMed] [Google Scholar]
- 22.Schuhmacher M., Belles M., Rico A., Domingo J.L., Corbella J. (1996) Impact of reduction of lead in gasoline on the blood and hair lead levels in the population of Tarragona Province, Spain, 1990–1995. Sci. Total Environ., 184, 203–209. [DOI] [PubMed] [Google Scholar]
- 23.Wietlisbach V., Rickenbach M., Berode M., Guillemin M. (1995) Time trend and determinants of blood lead levels in a Swiss population over a transition period (1984–1993) from leaded to unleaded gasoline use. Environ. Res., 68, 82–90. [DOI] [PubMed] [Google Scholar]
- 24.Jemal A., Graubard B.I., Devesa S.S., Flegal K.M. (2002) The association of blood lead level and cancer mortality among whites in the United States. Environ. Health Perspect., 110, 325–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menke A., Muntner P., Batuman V., Silbergeld E.K., Guallar E. (2006) Blood lead below 0.48 micromol/L (10 microg/dL) and mortality among US adults. Circulation, 114, 1388–1394. [DOI] [PubMed] [Google Scholar]
- 26.Patel N., Adewoyin T., Chong N.V. (2008) Age-related macular degeneration: a perspective on genetic studies. Eye, 22, 768–776. [DOI] [PubMed] [Google Scholar]
- 27.Kelada S.N., Shelton E., Kaufmann R.B., Khoury M.J. (2001) Delta-aminolevulinic acid dehydratase genotype and lead toxicity: a HuGE review. Am. J. Epidemiol., 154, 1–13. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz B.S., Lee B.K., Lee G.S., Stewart W.F., Simon D., Kelsey K., Todd A.C. (2000) Associations of blood lead, dimercaptosuccinic acid-chelatable lead, and tibia lead with polymorphisms in the vitamin D receptor and [delta]-aminolevulinic acid dehydratase genes. Environ. Health Perspect., 108, 949–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scinicariello F., Murray H.E., Moffett D.B., Abadin H.G., Sexton M.J., Fowler B.A. (2007) Lead and delta-aminolevulinic acid dehydratase polymorphism: where does it lead? A meta-analysis. Environ. Health Perspect., 115, 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y., Wang L., Shen H.B., Wang Z.X., Wei Q.Y., Chen F. (2007) Association between delta-aminolevulinic acid dehydratase (ALAD) polymorphism and blood lead levels: a meta-regression analysis. J. Toxicol. Environ. Health A, 70, 1986–1994. [DOI] [PubMed] [Google Scholar]
- 31.Barton J.C., Patton M.A., Edwards C.Q., Griffen L.M., Kushner J.P., Meeks R.G., Leggett R.W. (1994) Blood lead concentrations in hereditary hemochromatosis. J. Lab. Clin. Med., 124, 193–198. [PubMed] [Google Scholar]
- 32.Wright R.O., Silverman E.K., Schwartz J., Tsaih S.W., Senter J., Sparrow D., Weiss S.T., Aro A., Hu H. (2004) Association between hemochromatosis genotype and lead exposure among elderly men: the normative aging study. Environ. Health Perspect., 112, 746–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hopper J.L., Balderas A., Mathews J.D. (1982) Analysis of variation in blood lead levels in Melbourne families. Med. J. Aust., 2, 573–576. [PubMed] [Google Scholar]
- 34.Hopper J.L., Mathews J.D. (1983) Extensions to multivariate normal models for pedigree analysis. II. Modeling the effect of shared environment in the analysis of variation in blood lead levels. Am. J. Epidemiol., 117, 344–355. [DOI] [PubMed] [Google Scholar]
- 35.Bjorkman L., Vahter M., Pedersen N.L. (2000) Both the environment and genes are important for concentrations of cadmium and lead in blood. Environ. Health Perspect., 108, 719–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westra H.J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E., et al. (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet., 45, 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schadt E.E., Molony C., Chudin E., Hao K., Yang X., Lum P.Y., Kasarskis A., Zhang B., Wang S., Suver C., et al. (2008) Mapping the genetic architecture of gene expression in human liver. PLoS Biol., 6, e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernard A., Lauwerys R. (1987) Metal-induced alterations of delta-aminolevulinic acid dehydratase. Ann. N. Y. Acad. Sci., 514, 41–47. [DOI] [PubMed] [Google Scholar]
- 39.Kohn K.W., Zeeberg B.M., Reinhold W.C., Pommier Y. (2014) Gene expression correlations in human cancer cell lines define molecular interaction networks for epithelial phenotype. PLoS ONE, 9, e99269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schweigel-Rontgen M. (2014) The families of zinc (SLC30 and SLC39) and copper (SLC31) transporters. Curr. Top. Membr., 73, 321–355. [DOI] [PubMed] [Google Scholar]
- 41.Pare G., Chasman D.I., Kellogg M., Zee R.Y., Rifai N., Badola S., Miletich J.P., Ridker P.M. (2008) Novel association of ABO histo-blood group antigen with soluble ICAM-1: results of a genome-wide association study of 6578 women. PLoS Genet., 4, e1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medland S.E., Nyholt D.R., Painter J.N., McEvoy B.P., McRae A.F., Zhu G., Gordon S.D., Ferreira M.A., Wright M.J., Henders A.K., et al. (2009) Common variants in the trichohyalin gene are associated with straight hair in Europeans. Am. J. Hum. Genet., 85, 750–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heath A.C., Whitfield J.B., Martin N.G., Pergadia M.L., Goate A.M., Lind P.A., McEvoy B.P., Schrage A.J., Grant J.D., Chou Y.L., et al. (2011) A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biol. Psychiatry, 70, 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet., 30, 97–101. [DOI] [PubMed] [Google Scholar]
- 45.Boyd A., Golding J., Macleod J., Lawlor D.A., Fraser A., Henderson J., Molloy L., Ness A., Ring S., Davey Smith G. (2013) Cohort profile: the ‘Children of the 90s’—the index offspring of the Avon Longitudinal Study of Parents and Children. Int. J. Epidemiol., 42, 111–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fraser A., Macdonald-Wallis C., Tilling K., Boyd A., Golding J., Davey Smith G., Henderson J., Macleod J., Molloy L., Ness A., et al. (2013) Cohort profile: the Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int J Epidemiol., 42, 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor C.M., Golding J., Hibbeln J., Emond A.M. (2013) Environmental factors predicting blood lead levels in pregnant women in the UK: the ALSPAC study. PLoS ONE, 8, e72371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fatemifar G., Hoggart C.J., Paternoster L., Kemp J.P., Prokopenko I., Horikoshi M., Wright V.J., Tobias J.H., Richmond S., Zhurov A.I., et al. (2013) Genome-wide association study of primary tooth eruption identifies pleiotropic loci associated with height and craniofacial distances. Hum. Mol. Genet., 22, 3807–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delaneau O., Marchini J., Zagury J.F. (2012) A linear complexity phasing method for thousands of genomes. Nat. Methods, 9, 179–181. [DOI] [PubMed] [Google Scholar]
- 50.Howie B.N., Donnelly P., Marchini J. (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet., 5, e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marchini J., Howie B. (2010) Genotype imputation for genome-wide association studies. Nat. Rev. Genet., 11, 499–511. [DOI] [PubMed] [Google Scholar]
- 52.Willer C.J., Li Y., Abecasis G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics, 26, 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ihaka R., Gentleman R. (1996) R: a language for data analysis and graphics. J. Comput. Graph. Stat., 5, 299–314. [Google Scholar]
- 55.Liu J.Z., McRae A.F., Nyholt D.R., Medland S.E., Wray N.R., Brown K.M., Hayward N.K., Montgomery G.W., Visscher P.M., Martin N.G., et al. (2010) A versatile gene-based test for genome-wide association studies. Am. J. Hum. Genet., 87, 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.