Abstract
Retinitis pigmentosa (RP), the most common form of inherited retinal degeneration, is clinically and genetically heterogeneous and can appear as syndromic or non-syndromic. Mucopolysaccharidosis type IIIC (MPS IIIC) is a lethal disorder, caused by mutations in the heparan-alpha-glucosaminide N-acetyltransferase (HGSNAT) gene and characterized by progressive neurological deterioration, with retinal degeneration as a prominent feature. We identified HGSNAT mutations in six patients with non-syndromic RP. Whole exome sequencing (WES) in an Ashkenazi Jewish Israeli RP patient revealed a novel homozygous HGSNAT variant, c.370A>T, which leads to partial skipping of exon 3. Screening of 66 Ashkenazi RP index cases revealed an additional family with two siblings homozygous for c.370A>T. WES in three Dutch siblings with RP revealed a complex HGSNAT variant, c.[398G>C; 1843G>A] on one allele, and c.1843G>A on the other allele. HGSNAT activity levels in blood leukocytes of patients were reduced compared with healthy controls, but usually higher than those in MPS IIIC patients. All patients were diagnosed with non-syndromic RP and did not exhibit neurological deterioration, or any phenotypic features consistent with MPS IIIC. Furthermore, four of the patients were over 60 years old, exceeding by far the life expectancy of MPS IIIC patients. HGSNAT is highly expressed in the mouse retina, and we hypothesize that the retina requires higher HGSNAT activity to maintain proper function, compared with other tissues associated with MPS IIIC, such as the brain. This report broadens the spectrum of phenotypes associated with HGSNAT mutations and highlights the critical function of HGSNAT in the human retina.
Introduction
Inherited retinal dystrophies (IRD) are a heterogeneous group of diseases, which cause visual loss owing to the progressive loss of photoreceptors in the retina. The different forms of IRD lie along a spectrum with differential cone and rod photoreceptor involvement. The most common form of IRD is retinitis pigmentosa (RP), with a worldwide prevalence of ∼1 in 4000. In RP, the disease process initially affects the rod photoreceptors to a more severe degree than the cones, and thus, the first clinical symptom is usually night blindness, followed by visual field loss. Ophthalmologic findings also include characteristic pigmentation of the midperipheral retina, attenuation of retinal arterioles and optic disk pallor (1). In RP patients, the phenotype can be limited to the eye (non-syndromic), or it can appear as part of a syndrome. For instance, patients with Usher syndrome have a combination of RP and hearing loss (2).
Besides being clinically heterogeneous, RP is also genetically heterogeneous. To date, >65 genes have been implicated in the etiology of non-syndromic RP, and >30 syndromic forms have been described [Retnet-Retinal Information Network; Online Mendelian Inheritance in Man (OMIM)]. Interestingly, certain genes are associated with both syndromic and non-syndromic forms of RP. For example, mutations of USH2A cause either type 2 Usher syndrome (RP and hearing loss) or non-syndromic RP (3); mutations of the BBS1, BBS2, ARL6 and BBS8 genes cause either Bardet-Biedl syndrome (RP, obesity, mental retardation, polydactyl, renal and genital abnormalities) or non-syndromic RP (4–7).
The mucopolysaccharidoses (MPS) are inherited lysosomal storage diseases characterized by an abnormal accumulation of incompletely degraded mucopolysaccharides (glycosaminoglycans, GAGs) in multiple tissues and organs. Eleven known specific lysosomal enzyme deficiencies can be distinguished by their biochemical, clinical and genetic features (MPS I-MPS IX) (8). MPS IIIC, also known as Sanfilippo syndrome type C, is a rare autosomal recessive disorder caused by the deficiency of the lysosomal membrane enzyme, heparan-alpha-glucosaminide N-acetyltransferase (HGSNAT), which catalyzes transmembrane acetylation of the terminal glucosamine residues of heparan sulfate prior to their hydrolysis by α-N-acetylglucosaminidase (9,10). Most MPS IIIC patients manifest symptoms during childhood, with progressive and severe neurological deterioration causing hyperactivity, sleep disorders and loss of speech, accompanied by behavioral abnormalities, neuropsychiatric problems, mental retardation, hearing loss and relatively minor visceral manifestations, such as hepatomegaly, mild dwarfism with joint stiffness, coarse faces and hypertrichosis (11,12). Moderate-to-severe retinopathy with associated electroretinographic (ERG) abnormalities is a prominent feature of MPS IIIC (13). Most MPS IIIC patients die in early adolescence. In this study, we report mutations in the HGSNAT gene in six patients with non-syndromic RP.
Results
Identification of HGSNAT mutations in non-syndromic RP patients
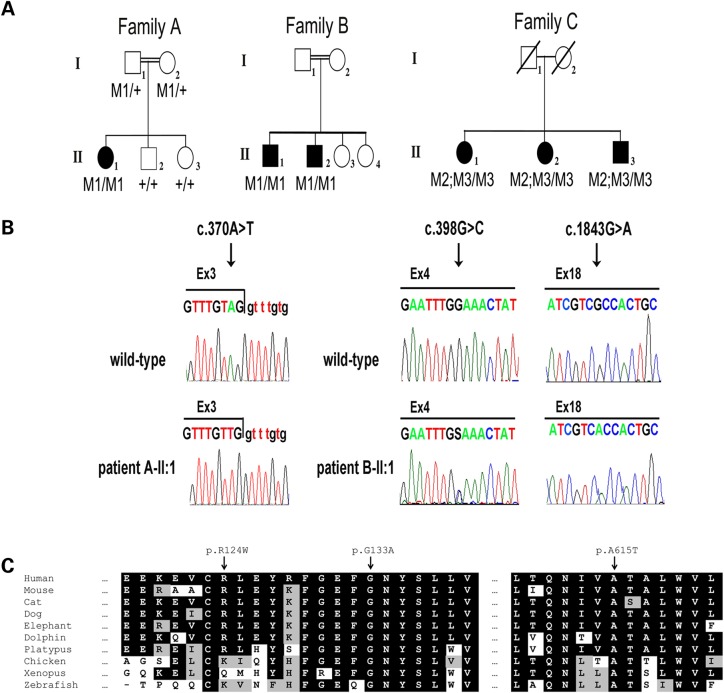
In a consanguineous Israeli family of Ashkenazi Jewish (AJ) descent (Family A, Fig. 1A), one of three siblings was diagnosed with RP at the age of 34 years (Patient A-II:1). This patient tested negative for several common founder mutations underlying RP in the AJ population, including the Alu insertion in the MAK gene, the p.K42E mutation of DHDDS and the c.1355_136delCA mutation of FAM161A (14–16). To identify the genetic cause of disease in this patient, WES analysis was performed. WES data were analyzed based on the hypothesis of autosomal recessive inheritance. WES resulted in 175 009 variants. A total of 4139 of these were rare variants (allele frequency <1% in 1000 genomes database, the Exome Variant Server and dbSNP), and 351 were either found inside exons and resulted in an amino acid change or were up to three bases from an intron–exon junction. Out of these, 30 were found in a homozygous state in the patient, and of these, six were completely novel and were either loss-of-function variants or predicted to be pathogenic. Additional manual review removed four variants owing to high probability of error (see Materials and Methods). The remaining two variants were located in the HGSNAT (GeneBank accession number NM_152419.2) and KAT6A (GeneBank accession number NM_001099412.1) genes. As HGSNAT mutations cause a syndrome with retinal degeneration, whereas KAT6A mutations are associated with leukemia, we concluded that the HGSNAT c.370A>T (p.R124W) variant was most likely the causative mutation (Supplementary material, Table S1). Segregation analysis via Sanger sequencing showed that the c.370A>T variant co-segregated with disease in the family (Fig. 1A and B).
Figure 1.
HGSNAT mutations identified in three families with non-syndromic RP. (A) Pedigrees of three families with HGSNAT mutations. Filled symbols represent affected individuals, whereas clear symbols represent unaffected individuals. A double line represents a consanguineous marriage. Genotypes of family members are indicated below them. M1: c.370A>T; M2: c.398G>C; M3: c.1843G>A. (B) Mutant and wild-type nucleotide sequence traces of the three mutations. Left: the boundary between HGSNAT exon and intron 3 in a non-carrier individual (wild-type) and in Patient A-II:1. Middle and right: nucleotide sequence traces of HGSNAT exons 4 and 18, respectively, in a non-carrier individual (wild-type) and in Patient C-II:1. (C) Multiple sequence alignments of the regions spanning the R124, G133 and A615 amino acids of the HGSNAT protein in various organisms. Conserved amino acids are indicated by a black background. Similar amino acids are indicated by a gray background.
Screening for the c.370A>T allele in a panel of 66 AJ non-syndromic RP patients who were previously found negative for known RP-causing AJ founder mutations was performed. In one of these patients (Patient B-II:1), the c.370A>T allele was also detected in a homozygous state. In contrast, c.370A>T was not detected in 211 AJ control individuals. The difference in c.370A>T allele frequencies between AJ patients and healthy control groups (4/134 versus 0/422) was statistically significant (P = 0.003 using the Fisher's exact test). Patient B-II:1 belonged to a consanguineous Israeli AJ family, and his affected sibling (Patient B-II:2) was also found to be homozygous for c.370A>T (Family B, Fig. 1A).
In an additional non-consanguineous Dutch family (Family C, Fig. 1A), three siblings were diagnosed with RP at an age of >45 years (Patients C-II:1, C-II:2 and C-II:3). Linkage analysis did not render any regions with known IRD disease genes. To identify the genetic cause of disease in this family, WES analysis was performed. WES data were analyzed based on the hypothesis of autosomal recessive inheritance. In Patients C-II:1, C-II:2 and C-II:3 of Family C, 9814 variants were shared in the exome data of all three siblings. Of these, 53 were rare variants (allele frequency <1% in dbSNP and the exomes of 5036 primarily Dutch individuals) and 28 were either found inside exons and resulted in an amino acid change or were up to 3 bases from an intron–exon junction. The only homozygous variant detected (c.1843G>A; p.A615T) was present in the HGSNAT gene, which also contained a heterozygous variant (c.398G>C; p.G133A). In addition, two heterozygous variants were detected in the ATP8B4 gene (GeneBank accession number NM_024837). As HGSNAT is expressed in the retina and HGSNAT mutations cause a syndrome with retinal degeneration, whereas this is not the case for ATP8B4, we concluded that the HGSNAT c.398G>C and c.1843G>A variants were the most plausible causative mutations (Supplementary material, Table S1). Sanger sequencing and copy number analysis (excluding the presence of a heterozygous deletion in HGSNAT) via qPCR confirmed that the three affected siblings of family C carried both the c.[398G>C;1843G>A] and c.1843G>A alleles (Fig. 1A and B). Segregation analysis could not be performed, because the parents are deceased.
Evaluation of the variants detected in HGSNAT
The HGSNAT c.370A>T and c.398G>C variants are both novel, as they were not found in 2184 alleles of the 1000 genomes database, 13 006 alleles of the Exome Variant Server, dbSNP and 10 572 alleles of exomes of 250 Israeli and 5036 primarily Dutch individuals.
The c.370A>T nucleotide substitution was predicted to result in the exchange of the moderately conserved, positively charged arginine into an uncharged tryptophan at position 124 of HGSNAT (p.R124W) (Fig. 1C). This sequence change was predicted to be pathogenic by both CADD (17) and PolyPhen-2 (18) but was predicted to be non-pathogenic by MutationTaster (19) (Supplementary material, Table S1). As c.370A>T is located in the penultimate nucleotide of exon 3 (Fig. 1B), it could also affect splicing. The wild-type (wt) HGSNAT intron 3 donor splice site is relatively weak, and the c.370A>T variant is predicted to further weaken it (Table 1). RT–PCR on leukocyte RNA of Patients A-II:1, B-II:1 and wt controls detected transcripts harboring exon 3 in both patients and controls, whereas transcripts that lacked exon 3 were only detected in patients (Fig. 2C). We therefore concluded that the c.370A>T variant leads to partial skipping of exon 3. Skipping of exon 3 is expected to cause a frameshift and generate a transcript which encodes for a truncated protein (p.Cys79Valfs*20) that lacks all transmembrane domains and the C-terminus, and therefore, it is likely non-functional. Moreover, given the nature of this mutation and the effects of the nonsense-mediated mRNA decay mechanism, this truncated protein may not be generated at all.
Table 1.
Analysis of HGSNAT intron 3 donor splice-site by splice-site prediction tools
| Prediction tool | Score |
|
|---|---|---|
| TAGgtttgt (wt) | TTGgtttgt (mutant) | |
| Analyzer Splice Tool | 75.81 | 67.27 |
| Human Splicing Finder Version 3.0 | 84.19 | 79.44 |
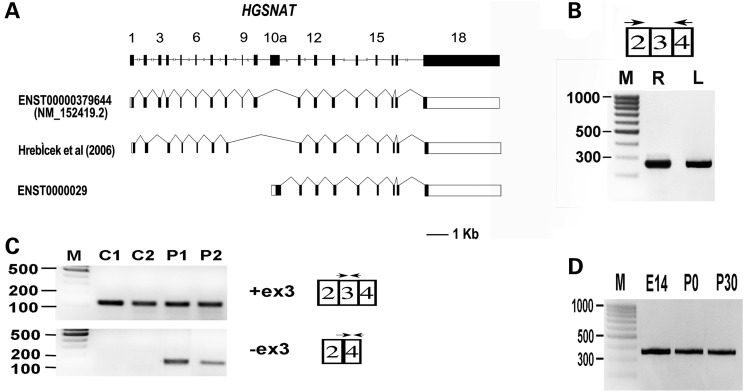
Figure 2.
Analysis of HGSNAT expression and splicing. (A) HGSNAT gene and splice-variants. Shown is a schematic representation of the human HGSNAT gene (drawn to scale), the main coding transcript (NM_152419.2) and two reported splice-variants lacking exon 3. In the splice-variants illustrations, filled boxes represent coding exons, whereas open boxes represent non-coding exons. (B) RT–PCR analysis of HGSNAT expression in human wt retina (R) and leukocytes (L). M: size marker (fragments sizes in base pairs are indicated to the left). HGSNAT expression was tested using primers located in exons 2 and 4. The obtained PCR product (280 bp) includes exons 2, 3 and 4. (C) RT–PCR analysis of exon 3 splicing in patient and control leukocytes. Transcripts harboring exon 3 were specifically amplified with forward and reverse primers located within exon 3 (115-bp product). Transcripts in which exon 3 was skipped were specifically amplified with a forward primer located on the border between exons 2 and 4, and a reverse primer located within exon 4 (130-bp product). C1, C2: wt controls. P1, P2: Patients A-II:1 and B-II:1. M: size marker. (D) RT–PCR analysis of Hgsnat expression in the mouse eye at different developmental time points: embryonic day 14 (E14), postnatal days 0 and 30 (P0, P30). The analysis indicates Hgsnat expression (360-bp product) at all time points tested. M: size marker.
The c.398G>C nucleotide substitution was predicted to result in the exchange of a highly conserved glycine into an alanine at position 133 of HGSNAT (p.G133A) (Fig. 1C). This substitution, located within the N-terminus, is predicted to be pathogenic by CADD, MutationTaster and PolyPhen-2.
The c.1843G>A is a rare variant, with a mean allele frequency of 0.59% in the European American population of the Exome Variant Server. A similar frequency, 0.56%, was detected in an exome variant database of 5036 primarily Dutch individuals. This variant is predicted to result in the exchange of the highly conserved alanine into a threonine at position 615 of HGSNAT (p.A615T) (Fig. 1C). The p.A615T variant has been reported twice in MPSIIIC patients, both times homozygously and in combination with another homozygous variant, p.W403C (10,20). It was shown that p.A615T leads to moderately reduced HGSNAT activity (50–70% of wt) (21,22).
Decreased HGSNAT activity in RP patients with HGSNAT mutations
The HGSNAT activity in blood leukocytes, measured in Patients A-II:1, C-II:1, C-II:2 and C-II:3, was decreased compared with healthy controls: 2.5, 3.6, 4.6 and 3.3 nmol/18 h/mg in the patients, respectively, versus 6.7–23.5 nmol/18 h/mg protein in healthy individuals. The HGSNAT activities in Patients C-II:1, C-II:2 and C-II:3 with non-syndromic RP are slightly higher than those measured in MPS IIIC patients (0.7–3.0 nmol/18 h/mg protein; nine patients of seven families), whereas the HGSNAT activity of Patient A-II:1 is within the upper range obtained in MPS IIIC patients. In Patient A-II:1, total urine GAGs were also determined and these were elevated (6.6 mg/mmol creatinine compared with controls: <3 mg/mmol creatinine).
To exclude the possibility that, in addition to retinal dystrophy, these patients might have mild manifestations of HGSNAT deficiency, such as coarse facial features, hypertrichosis, contractures, organomegaly, hearing impairment, behavioral and sleeping problems, recurrent infections, diarrhea, epilepsy or late onset of mental deterioration, a physical exam was performed. Patients A-II:1 and B-II:1 underwent a comprehensive physical exam for extra-ocular features, but no significant findings were observed, except for mild hearing impairment diagnosed in Patient A-II:1 at the age of 59 years. A general physical exam of Patients C-II:1 and C-II:2 also did not reveal any additional symptoms.
Ophthalmological findings
All five patients were diagnosed with non-syndromic RP, and in all patients, initial symptoms were night blindness and/or visual field loss. However, between patients, there were differences in clinical symptoms and in age of onset (Table 2).
Table 2.
Clinical characteristics of patients with non-syndromic RP owing to HGSNAT mutations
| Patient number (gender) | Age of onset | Initial symptoms | Age at diagnostic exam | Visual acuity |
Fundus | Color vision | Visual field | Fundus autofluorescence | OCT | ffERG |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OD | OS | Scot. | Phot. | |||||||||
| A-II:1 (F) | Childhood | Night blindness | 34 y | 20/30 | 20/200 | NR | NR | |||||
| 60 y | FC | FC | Extensive atrophic changes at the posterior pole and peripheral retina with bone spicule pigmentation at the midperiphery | |||||||||
| B-II:1 (M) | Childhood | Night blindness | 29 y | 20/40 | 20/40 | Peripapillary atrophy, foveal cystic edema, small crystals in the macula and mid periphery, pronounced choroidal vasculature | Ring scotoma 10o–20o (BE) | Hyperautofluorescent cystic foveal spaces, perifoveal inner hyperautofluorescent and outer hypoautofluorescent rings, hypoautofluorescent spots in the posterior pole | Thinning of the ellipsoid zone, foveal cystic edema and diffuse atrophy of the PR-RPE and thinning of the neurosensory retina in the posterior pole | MR | MR | |
| B-II:2 (M) | 18 years | Night blindness | 30 y | 20/30 | 20/40 | Normal optic discs, small crystals in the macula and inferior retina, pronounced choroidal vasculature in both eyes, OS: a few hyperpigmented bone spicules in the superior midperiphery |
Peripheral constriction (BE) | Perifoveal hyperautofluorescent ring surrounded by hypoautofluorescence and then diffuse hyperautofluorescence in the posterior pole, as well as hypoautofluorescent spots in the posterior pole and periphery | OD: thinning of the ellipsoid zone, diffuse atrophy of the PR-RPE and thinning of the neurosensory retina in the posterior pole and segments of epiretinal membrane. OS: more diffuse atrophy involving the fovea as well |
SR | SR | |
| C-II:1 (F) | 52 y | Night blindness, visual field loss, decreased visual acuity, photopsia, decreased contrast sensitivity | 52 y | 20/20 | 20/20 | Mild arteriolar narrowing | Reduced red color vision | Pericentral reduced sensitivity (Goldmann) | MR | MR | ||
| 66 y | 20/60 | 20/40 | Attenuated veins, central atrophic lesions with hyperpigmentation, midperipheral atrophy with bone spicules | Ring scotoma and central reduced sensitivity (Goldmann) | SR | SR | ||||||
| C-II:2 (F) | 47 y | Night blindness, visual field loss, blurred vision | 47 y | 20/20 | 20/600 | OD: paracentral atrophic RPE lesion, OS: RPE changes, attenuated veins | Normal | Ring scotoma with central reduced sensitivity OS>OD (Goldmann) | MR | MR | ||
| 66 y | 20/25 | 20/400 | OD: attenuated veins, waxy disc pallor, atrophic RPE lesion superotemporal to the fovea. OS: attenuated veins, in macula hard exudates and hyperpigmentation, loss of RPE in (mid)periphery |
OD: central reduced sensitivity and ring scotoma. OS: Central scotoma, and paracentral ring scotoma (Goldmann) |
OD: geographic atrophy superotemporal to the fovea. Remaining macula hyperautofluorescence. Hypoautofluorescence midperiphery and RPE loss. OS: macula and midperiphery hypoautofluorescence and RPE loss. Hyperautofluorescence surrounding macula |
OD: loss of RPE layer, apart from fovea. OS: loss of RPE layer, hard exudates parafoveal |
||||||
| C-II:3 (M) | 50 y | Visual field loss, reduced color vision | 50 y | 20/15 | 20/20 | No abnormalities | Reduced red color vision | Arcuate shaped scotoma in Bjerrum area, OS>OD (HFA-30) | N | N | ||
| 55 y | 20/15 | 20/20 | Waxy disc pallor, in periphery some pigmentation | Ring scotoma OS>OD (HFA-30) | ||||||||
F: female, M: male, y: years, OD: right eye, OS: left eye, BE: both eyes, HFA: humphrey field analyzer, OCT: optical coherence tomography, ffERG: full-field electroretinogram, Scot: scotopic, Phot: photopic., NR: non-recordable, N: normal (equal to or above the lower 5% of the range for a normal population: photopic ≥78 uV, scotopic ≥263 uV), MR: moderately reduced (1–5% of normal range: photopic ≥69 uV and <78 uV, scotopic ≥195 uV and <263 uV), SR: severely reduced (<1% of normal range: photopic <69 uV, scotopic <195 uV)
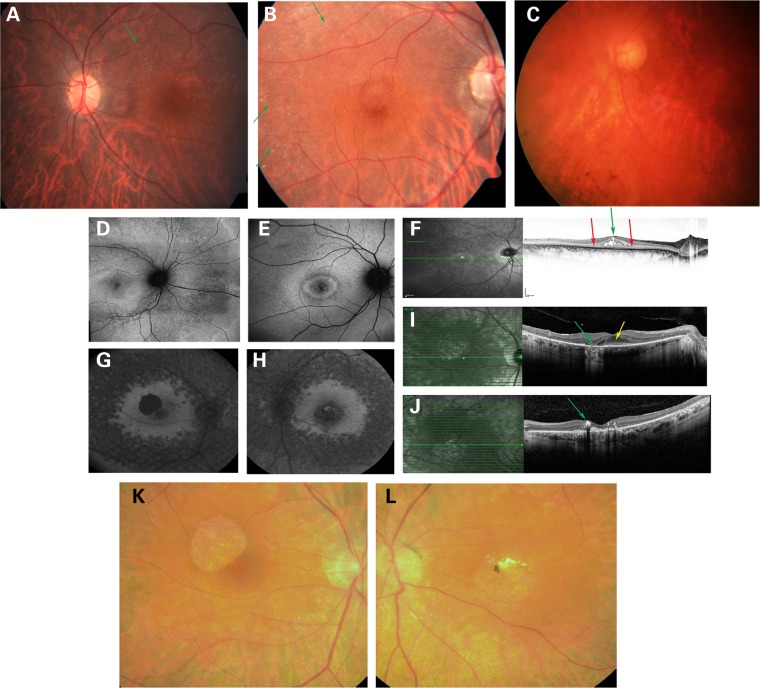
Patient A-II:1 had night blindness since childhood. She was diagnosed with RP at the age of 34 years. At this age, full-field ERG (ffERG) responses were non-detectable. At the age of 60 years, her visual acuity was severely compromised in both eyes. The fundus appearance revealed severe and extensive atrophic changes of the retina and the retinal pigmented epithelium (RPE) as well as choroidal sclerosis, particularly at the posterior pole, accompanied by bone-spicule pigmentation at the midperiphery, with sparse and narrow retinal blood vessels (Fig. 3C and Table 2).
Figure 3.
Fundus photographs, FAF and optical coherence tomography (OCT) of affected individuals with RP owing to HGSNAT mutations. (A) Fundus photograph of left eye in Patient B-II:2, showing small crystals in the posterior pole and midperiphery (green arrow) and pronounced choroidal vasculature. (B) Fundus photograph of right eye in Patient B-II:1, showing peripapillary atrophy, foveal cystic edema, small crystals in the macula and mid periphery (green arrows), pronounced choroidal vasculature, with normal retinal blood vessels and no bone spicules. (C) Fundus photograph of left eye in Patient A-II:1, showing severe and extensive atrophic changes of the retina and the RPE and choroidal sclerosis, mostly at the posterior pole, accompanied by bone spicule pigmentation at the midperiphery, with sparse and narrow retinal blood vessels. (D) FAF of right eye in Patient B-II:2, showing perifoveal hyperautofluorescent ring surrounded by hypoautofluorescence and then diffuse hyperautofluorescence in the posterior pole, as well as hypoautofluorescent spots in the posterior pole and periphery. (E) FAF of right eye in Patient B-II:1, showing hyperautofluorescent cystic foveal spaces, perifoveal inner hyperautofluorescent and outer hypoautofluorescent rings and hypoautofluorescent spots in the posterior pole. (F) OCT of right eye in Patient B-II:1, showing thinning of the ellipsoid zone (red arrows), the distal photoreceptors and neurosensory retina, with foveal cystic edema (green arrow). (G) FAF of right eye in Patient C-II:2, showing geographic atrophy superotemporal to the fovea, remaining macula hyperautofluorescence, hypoautofluorescence in the midperiphery and RPE loss. (H) FAF of left eye in Patient C-II:2, showing macula and midperiphery hypoautofluorescence and RPE loss, with hyperautofluorescence surrounding the macula. (I) OCT of right eye in Patient C-II:2, showing loss of the RPE layer (green arrow), apart for the fovea (yellow arrow). (J) OCT of left eye in Patient C-II:2, showing loss of the RPE layer, and parafoveal hard exudates (green arrow). (K) Fundus photograph of right eye in Patient C-II:2, showing attenuated veins, waxy disc pallor and an atrophic RPE lesion superotemporal to the fovea. (L) Fundus photograph of left eye in Patient C-II:2, showing attenuated veins, hard exudates and hyperpigmentation in the macula and loss of RPE in the midperiphery.
Patient B-II:1, with the same homozygous mutation as Patient A-II:1, had night blindness since childhood. At the age of 29 years, his best-corrected visual acuity (BCVA) was 20/40 on both eyes. Funduscopy revealed peripapillary atrophy, foveal cystic edema, small crystals in the macula and midperiphery, pronounced choroidal vasculature, with normal retinal blood vessels and no bone spicule-like pigmentation. Visual fields (VFs) showed ring scotomas in both eyes (Fig. 3B, E and F and Table 2). His brother, Patient B-II:2, noticed night blindness at the age of 18 years. At the age of 30 years, his BCVA was 20/30 and 20/40 in the right and left eye, respectively. Funduscopy revealed normal optic discs, small crystals in the macula and inferior retina, pronounced choroidal vasculature in both eyes and a few hyperpigmented bone spicule-like spots in the superior midperiphery of the left eye. VFs were evident for peripheral constriction in both eyes (Fig. 3A and D and Table 2). ffERG for both Patients B-II:1 and B-II:2 showed severe generalized rod-cone dysfunction, with an electronegative pattern (reduced b/a ratio).
In Patients C-II:1, C-II:2 and C-II:3 of Family C, the first clinical signs, e.g. night blindness and/or VF loss, occurred in the fifth to sixth decade of life. Detailed ophthalmologic analysis determined that all three siblings were affected by RP. The RP in these siblings is, however, atypical owing to the fact that two siblings have reduced red color vision and the late onset of disease (Fig. 3G–L and Table 2).
Analysis of HGSNAT expression
RT–PCR analysis was performed on RNA derived from normal human retina and leukocytes with primers located in HGSNAT exons 2 and 4. Both tissues yielded the expected product, harboring exons 2, 3 and 4 (Fig. 2B). However, while this product was obtained from human retina following 35 cycles of standard PCR, obtaining a product from leukocyte RNA required an additional set of amplification. This indicated that HGSNAT expression levels in the retina are much higher than those in blood leukocytes.
In the literature and in the public databases, there is evidence for HGSNAT transcripts lacking exon 3. Hrebicek et al detected a transcript in which exons 3, 9 and 10 were skipped in several human tissues (10), and the Ensembl genome browser contains a transcript with an alternative first exon (exon 10a, located downstream of exon 10) fused to exons 11–18 (ENST00000297798) (Fig. 2A). The functional significance of these shorter transcripts is unclear. Our analysis showed no evidence for a retinal transcript in which exon 3 was skipped (Fig. 2B). In addition, the transcript with an alternative first exon (exon 10a) was also not detected in retina and leukocytes, even following an additional set of PCR amplification (data not shown). We therefore conclude that these alternative transcripts are not expressed in retina.
To examine the ocular expression of Hgsnat at different developmental time points, we performed RT–PCR analysis of total RNA from mouse eye. Hgsnat was found to be expressed in the developing mouse eye as early as embryonic day 14 (E14), with equal high expression levels after birth [postnatal day 0 (P0) and postnatal day 30 (P30)] (Fig. 2D).
Discussion
In this study, we detected mutations in the HGSNAT gene in six patients with apparent non-syndromic RP. Three patients of two unrelated Israeli AJ families were homozygous for the c.370A>T (p.R124W) mutation, which causes partial skipping of exon 3 during splicing (p.Cys79Valfs*20), and three patients of one Dutch family were heterozygous for the c.398G>C (p.G133A) and homozygous for the c.1843G>A (p.A615T) mutation.
HGSNAT mutations have previously been associated with the severe and lethal disorder MPS IIIC (9,10). The main symptoms in MPS IIIC patients result from progressive degeneration of the central nervous system, and retinal degeneration is a prominent feature (11,13). Interestingly, two sisters with an attenuated course of the disease have been reported. One of them had borderline intelligence since infancy, whereas the other had normal development. They developed major symptoms at the third decade of life, including adult-onset dementia and RP. One of them died at 48 years; the other one was still alive at the age of 48 (23,24). The six patients with HGSNAT mutations in this study were diagnosed with retinal degeneration at the third to fifth decade of life. The ERG responses of Patients B-II:1, B-II:2, C-II:1 and C-II:2 showed a generalized rod-cone dysfunction with an electronegative pattern, which is a common finding in patients with MPS and other types of lysosomal storage diseases (25–27). However, all six patients in this study showed no signs of mental deterioration, behavioral or sleeping abnormalities (including Patients A-II:1, C-II:1, C-II:2 and C-II:3, who are currently in their sixties and seventies). Moreover, they have no other visceral, skeletal or facial signs associated with MPS IIIC, except for late-onset mild hearing impairment in Patient A-II:1, which might be age related. We therefore conclude that their disease is non-syndromic.
The HGSNAT mutations of Patients C-II:1, C-II:2 and C-II:3 lead to amino acid changes (p.G133A and p.A615T), whereas the c.370A>T mutation detected in Patients A-II:1, B-II:1 and B-II:2 was shown to result in partial skipping of HGSNAT exon 3 (p.Cys79Valfs*20) in blood leukocytes, as well as an amino acid change (p.R124W). We hypothesize that in the retina exon 3 is skipped as well, although this cannot be confirmed experimentally. As all patients in this study manifest no additional extra-ocular symptoms, other tissues that are usually affected by HGSNAT deficiency (especially the brain) must have sufficient levels of enzymatic activity. We therefore hypothesize that the retina requires higher levels of HGSNAT activity compared with other tissues. The same phenomenon has been reported for other genes, such as USH2A (3), CEP290 (28) and BBS1 (5), which are associated with both syndromic and non-syndromic forms of IRD. In these cases, severe (null) mutations are usually associated with syndromic IRD, whereas milder (hypomorphic) mutations or combinations of mutations are usually associated with non-syndromic IRD. The genotypes detected in HGSNAT in the non-syndromic RP patients (homozygous c.370A>T, and compound heterozygous c.1843G>A and c.[398G>C; 1843G>A]) have never been detected in patients with MPS IIIC. Furthermore, the HGSNAT activity in all but one non-syndromic RP patient was higher compared with MPS IIIC patients, indicating that indeed the HGSNAT mutations in the non-syndromic RP patients are milder compared with the mutations in MPS IIIC patients. Interestingly, Patient A-II:1, who had the lowest HGSNAT activity level, also had an earlier age of onset of RP and a more severe retinal dystrophy compared with Patients C-II:1, C-II:2 and C-II:3, with higher HGSNAT activity. Interestingly, in Family C, patients were initially diagnosed at the ages of 47–52 years, and at these ages, their central vision was still very good. This is an uncommon finding, as most RP patients are diagnosed between the first and the third decades of life and are legally blind by the age of 40 years (29).
There are marked differences between the retinal phenotypes observed in Patients A-II:1, B-II:1 and B-II:2, who are homozygous for the same HGSNAT mutation. These differences suggest the involvement of additional genetic and/or environmental modifying effects on the retinal phenotype caused by HGSNAT mutations. Patient A-II:1 has classic RP, characterized by extensive atrophic changes, bone spicule pigmentation, sparse and attenuated retinal blood vessels. Patient B-II:1 has macular edema, but no bone spicules, which are evident in Patient B-II:2. In addition, Patients B-II:1 and B-II:2 have prominent crystals in the retina, which are not evident in Patient A-II:1. Given the low rate of HGSNAT activity found in patients' lymphocytes, it would be interesting to use electron microscopy to test whether crystals are present in lymphocytes as well.
One of the HGSNAT alleles of the affected siblings of family C has two variants p.[G133A;A615T]. Most likely, there is a cumulative effect of both variants on HGSNAT activity, as was also shown for the p.[W403C;A615T] allele (21). Therefore, we believe that the combination of both HGSNAT variants on one allele, in conjunction with the p.A615T variant in trans, is responsible for RP in Family C. Because the p.A615T variant gives a maximal HGSNAT activity reduction of 50% (21,22) and given the frequency of the p.A615T allele in the healthy population (0.59%), it is unlikely that the homozygous presence of this variant alone can cause retinal dystrophy. It is still possible that this allele acts as a modifier in the retinal dystrophy of patients caused by other genes. It would be interesting to analyze patients with retinal dystrophy who carry mutations in other IRD-associated genes in addition to the p.A615T variant in HGSNAT, to determine whether this variant has an additional effect on the phenotype.
HGSNAT is already highly expressed in the developing mouse eye at E14 and to a same extent in the fully developed mouse eye. This indicates that the function of HGSNAT, transmembrane acetylation of the terminal glucosamine residues of heparan sulfate prior to their hydrolysis, is an important process during eye development, and also in the fully developed eye. This could be related to the high turnover of proteins in the retina. A similar ‘activity threshold’ model was recently proposed for two other genes encoding lysosomal proteins, i.e. CLN3 (30) and MFSD8 (31). Both were previously implicated in severe neurological syndromes (neuronal ceroid lipofuscinosis or Batten disease) and recently found to be mutated in non-syndromic IRDs. In particular for MFSD8, a clear-cut genotype–phenotype correlation was identified (31), strongly suggesting that an optimal function of the lysosome is essential for photoreceptor survival. Metabolic analysis on blood of IRD patients could therefore potentially be used to detect novel lysosomal gene defects and/or modifier alleles.
In summary, we report six patients with non-syndromic RP owing to HGSNAT mutations. This report broadens the spectrum of phenotypes associated with HGSNAT mutations and highlights the critical function of HGSNAT in the human retina.
Materials and Methods
Subjects and clinical evaluation
The study was approved by local Ethics Committees at the ascertaining institutes (Rambam Medical Center, Hadassah-Hebrew University Medical Center and Radboud University Medical Center). Informed consent was obtained from all participants. AJ control DNA samples were obtained from the National Laboratory for the Genetics of the Israeli Population at Tel Aviv University.
The standard ophthalmic examination included the measurement of BCVA using Snellen visual acuity charts and ophthalmoscopy. Fundus photography was performed by using Topcon TRC50IX (Topcon Corporation, Tokyo, Japan). The visual field testing was performed by Goldman perimetry or a Humphrey perimeter (Carl Zeiss Meditec, Jena, Germany) using central 30-2 threshold test. Cross-sectional images were obtained using spectral-domain optical coherence tomography (SD-OCT; Heidelberg Engineering, Heidelberg, Germany). Fundus autofluorescence (FAF) images were acquired using a confocal scanning laser ophthalmoscope (cSLO, Spectralis, Heidelberg Engineering). ffERG was conducted according to International Society for Clinical Electrophysiology of Vision guidelines (32), using the Espion E3 Electroretinography system (Diagnosys, Lowell, MA, USA) for Israeli patients, and the RETI-port system (Roland Consults, Stasche & Finger GmbH, Brandenburg an der Havel, Germany) for Dutch patients.
Assessment of HGSNAT activity
HGSNAT activity in blood leukocytes was measured using a fluorimetric assay with 4-methylumbelliferyl-beta-d-glucosaminide as a substrate (Moscerdam Substrates, Oestgeest, the Netherlands) (33). Total GAGs in urine were measured by spectrophotometric analysis using creatinine levels as a standard (34). Metabolic assays were performed at the Enzyme Laboratory, Department of Genetics and Metabolic Diseases, Hadassah-Hebrew University Medical Center for Israeli patients, and at the Translational Metabolic Laboratory, Radboud University Medical Center, for Dutch patients.
DNA analyses
Genomic DNA was extracted from leukocytes according to standard procedures (35). WES of Patient A-II:1 was performed at Otogenetics corporation (Norcross, GA, USA) using Roche NimbleGen V2 (44.1 Mbp) paired-end sample preparation kit and Illumina HiSeq2000 at a 30× coverage. Sequence reads were aligned to the reference human genome (GRCh37/hg19), and variants were called as previously described (36). WES of Patients C-II:1, C-II:2 and C-II:3 was performed using Agilent's Sure Select All Human Exome version 2 kit (50 Mb; Agilent Technologies, Santa Clara, CA, USA), followed by sequencing on a SOLiD4 sequencing platform (Life Technologies, Carlsbad, CA, USA). Sequence reads were aligned to the reference human genome (GRCh37/hg19) using Lifescope v2.1 software (Life Technologies), followed by variant calling on the aligned sequence via a customized pipeline (37).
HGSNAT exons were amplified from genomic DNA using the primers listed in Supplementary material, Table S2. Sanger sequencing of PCR products was used to verify the mutations and screen additional patients and controls.
In Patients C-II:1, C-II:2 and C-II:3, copy number variant analysis of HGSNAT exons 3 and 18 was performed on genomic DNA via qPCR using GoTaq qPCR Master Mix (Promega Corporation, Madison, WI, USA) to determine whether a heterozygous deletion was present in HGSNAT. The KEL and ZDHHC5 genes were used as standards (primers sequences in Supplementary material, Table S2).
RNA analyses
Total RNA was isolated from human blood lymphocytes and retina, and from mouse retinas using Tri reagent (Sigma–Aldrich, St Louis, MO, USA) and treated with RQ1 RNase-free DNase (Promega Corporation). Reverse transcription was performed with 1 µg of total RNA in a 20 µl of reaction volume using 200U of M-MLV Reverse Transcriptase and 100 ng of random primers (Stratagene, La Jolla, CA, USA). PCR was performed in a 25 µl reaction volume in the presence of 5× Readymix (LAROVA GmbH, Teltow, Germany) and 10 pmol of each forward and reverse primers (Supplementary material, Table S2).
Animal use
C57BL/6 mice were obtained from Harlan Laboratories, Inc. (Jerusalem, Israel). Animal care guidelines comparable to those published by the Institute for Laboratory Animal Research (Guide for the Care and Use of Laboratory Animals) were followed, and the research was approved by the committee for the supervision of animal experiments, Technion-Israel Institute of Technology.
Web-based tools and databases
Retnet-Retinal Information Network (http://www.sph.uth.tmc.edu/Retnet/)
OMIM (http://www.ncbi.nlm.nih.gov/omim)
The 1000 genomes database (http://www.1000genomes.org/)
EVS NHLBI Exome Sequencing Project (ESP) Exome Variant Server (http://evs.gs.washington.edu/EVS/)
dbSNP (http://www.ncbi.nlm.nih.gov/SNP/)
PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/)
Mutation Taster (http://www.mutationtaster.org/)
Analyzer Splice Tool (http://ibis.tau.ac.il/ssat/SpliceSiteFrame.htm)
Human Splicing Finder Version 3.0 (http://www.umd.be/HSF3/)
Ensembl genome browser (http://www.ensembl.org/index.html)
CADD (http://cadd.gs.washington.edu/)
Ocular Tissue Database (https://genome.uiowa.edu/otdb/)
Human Retinal Transcriptome (http://oculargenomics.meei.harvard.edu/index.php/ret-trans)
Supplementary Material
Funding
This work was supported by research grants from the Foundation Fighting Blindness (FFB) (BR-GE-0214-0639-TECH to T.B., D.S., R.L., H.N. and N.S., BR-GE-0510-0489-RAD to A.I.dH., C-GE-0811-0545-RAD01 to F.P.M.C.), Prof. Dr HJ Flieringa Foundation SWOO, the Rotterdam Eye Hospital (to F.P.M.C. and A.I.dH.), the Gelderse Blinden Stichting (to F.P.M.C.), the MD Fonds, the Janivo Stichting and the Stichting A.F. Deutman Researchfonds Oogheelkunde (to C.B.H.) and the Yedidut Research Fund (to E.B.). Funding to pay the Open Access publication charges for this article was provided by The Rappaport Family Institute for Research in the Medical Sciences.
Supplementary Material
Acknowledgements
We are grateful to the patients for their participation in this study.
Conflict of Interest statement. None declared.
References
- 1.Ayuso C., Millan J.M. (2010) Retinitis pigmentosa and allied conditions today: a paradigm of translational research. Genome Med., 2, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathur P., Yang J. (2015) Usher syndrome: hearing loss, retinal degeneration and associated abnormalities. Biochim. Biophys. Acta, 1852, 406–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rivolta C., Sweklo E.A., Berson E.L., Dryja T.P. (2000) Missense mutation in the USH2A gene: association with recessive retinitis pigmentosa without hearing loss. Am. J. Hum. Genet., 66, 1975–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aldahmesh M.A., Safieh L.A., Alkuraya H., Al-Rajhi A., Shamseldin H., Hashem M., Alzahrani F., Khan A.O., Alqahtani F., Rahbeeni Z., et al. (2009) Molecular characterization of retinitis pigmentosa in Saudi Arabia. Mol. Vis., 15, 2464–2469. [PMC free article] [PubMed] [Google Scholar]
- 5.Estrada-Cuzcano A., Koenekoop R.K., Senechal A., De Baere E.B., de Ravel T., Banfi S., Kohl S., Ayuso C., Sharon D., Hoyng C.B., et al. (2012) BBS1 mutations in a wide spectrum of phenotypes ranging from nonsyndromic retinitis pigmentosa to Bardet-Biedl syndrome. Arch. Ophthalmol., 130, 1425–1432. [DOI] [PubMed] [Google Scholar]
- 6.Riazuddin S.A., Iqbal M., Wang Y., Masuda T., Chen Y., Bowne S., Sullivan L.S., Waseem N.H., Bhattacharya S., Daiger S.P., et al. (2010) A splice-site mutation in a retina-specific exon of BBS8 causes nonsyndromic retinitis pigmentosa. Am. J. Hum. Genet., 86, 805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shevach E., Ali M., Mizrahi-Meissonnier L., McKibbin M., El-Asrag M., Watson C.M., Inglehearn C.F., Ben-Yosef T., Blumenfeld A., Jalas C., et al. (2015) Association between missense mutations in the BBS2 gene and nonsyndromic retinitis pigmentosa. JAMA Ophthalmol, 133, 312–318. [DOI] [PubMed] [Google Scholar]
- 8.Muenzer J. (2011) Overview of the mucopolysaccharidoses. Rheumatology (Oxford), 50(Suppl 5), v4–12. [DOI] [PubMed] [Google Scholar]
- 9.Fan X., Zhang H., Zhang S., Bagshaw R.D., Tropak M.B., Callahan J.W., Mahuran D.J. (2006) Identification of the gene encoding the enzyme deficient in mucopolysaccharidosis IIIC (Sanfilippo disease type C). Am. J. Hum. Genet., 79, 738–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hrebicek M., Mrazova L., Seyrantepe V., Durand S., Roslin N.M., Noskova L., Hartmannova H., Ivanek R., Cizkova A., Poupetova H., et al. (2006) Mutations in TMEM76* cause mucopolysaccharidosis IIIC (Sanfilippo C syndrome). Am. J. Hum. Genet., 79, 807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartsocas C., Grobe H., van de Kamp J.J., von Figura K., Kresse H., Klein U., Giesberts M.A. (1979) Sanfilippo type C disease: clinical findings in four patients with a new variant of mucopolysaccharidosis III. Eur. J. Pediatr., 130, 251–258. [DOI] [PubMed] [Google Scholar]
- 12.Valstar M.J., Ruijter G.J., van Diggelen O.P., Poorthuis B.J., Wijburg F.A. (2008) Sanfilippo syndrome: a mini-review. J. Inherit. Metab. Dis., 31, 240–252. [DOI] [PubMed] [Google Scholar]
- 13.Ashworth J.L., Biswas S., Wraith E., Lloyd I.C. (2006) Mucopolysaccharidoses and the eye. Surv. Ophthalmol., 51, 1–17. [DOI] [PubMed] [Google Scholar]
- 14.Bandah-Rozenfeld D., Mizrahi-Meissonnier L., Farhy C., Obolensky A., Chowers I., Pe'er J., Merin S., Ben-Yosef T., Ashery-Padan R., Banin E., et al. (2010) Homozygosity mapping reveals null mutations in FAM161A as a cause of autosomal-recessive retinitis pigmentosa. Am. J. Hum. Genet., 87, 382–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tucker B.A., Scheetz T.E., Mullins R.F., DeLuca A.P., Hoffmann J.M., Johnston R.M., Jacobson S.G., Sheffield V.C., Stone E.M. (2011) Exome sequencing and analysis of induced pluripotent stem cells identify the cilia-related gene male germ cell-associated kinase (MAK) as a cause of retinitis pigmentosa. Proc. Natl Acad. Sci. USA, 108, E569–E576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zelinger L., Banin E., Obolensky A., Mizrahi-Meissonnier L., Beryozkin A., Bandah-Rozenfeld D., Frenkel S., Ben-Yosef T., Merin S., Schwartz S.B., et al. (2011) A missense mutation in DHDDS, encoding dehydrodolichyl diphosphate synthase, is associated with autosomal-recessive retinitis pigmentosa in Ashkenazi Jews. Am. J. Hum. Genet., 88, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet., 46, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. (2010) A method and server for predicting damaging missense mutations. Nat. Methods, 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. (2014) MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods, 11, 361–362. [DOI] [PubMed] [Google Scholar]
- 20.Feldhammer M., Durand S., Mrazova L., Boucher R.M., Laframboise R., Steinfeld R., Wraith J.E., Michelakakis H., van Diggelen O.P., Hrebicek M., et al. (2009) Sanfilippo syndrome type C: mutation spectrum in the heparan sulfate acetyl-CoA: alpha-glucosaminide N-acetyltransferase (HGSNAT) gene. Hum. Mutat., 30, 918–925. [DOI] [PubMed] [Google Scholar]
- 21.Fedele A.O., Hopwood J.J. (2010) Functional analysis of the HGSNAT gene in patients with mucopolysaccharidosis IIIC (Sanfilippo C Syndrome). Hum. Mutat., 31, E1574–E1586. [DOI] [PubMed] [Google Scholar]
- 22.Feldhammer M., Durand S., Pshezhetsky A.V. (2009) Protein misfolding as an underlying molecular defect in mucopolysaccharidosis III type C. PLoS One, 4, e7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berger-Plantinga E.G., Vanneste J.A., Groener J.E., van Schooneveld M.J. (2004) Adult-onset dementia and retinitis pigmentosa due to mucopolysaccharidosis III-C in two sisters. J. Neurol., 251, 479–481. [DOI] [PubMed] [Google Scholar]
- 24.Ruijter G.J., Valstar M.J., van de Kamp J.M., van der Helm R.M., Durand S., van Diggelen O.P., Wevers R.A., Poorthuis B.J., Pshezhetsky A.V., Wijburg F.A. (2008) Clinical and genetic spectrum of Sanfilippo type C (MPS IIIC) disease in The Netherlands. Mol. Genet. Metab., 93, 104–111. [DOI] [PubMed] [Google Scholar]
- 25.Pradhan S.M., Atchaneeyasakul L.O., Appukuttan B., Mixon R.N., McFarland T.J., Billingslea A.M., Wilson D.J., Stout J.T., Weleber R.G. (2002) Electronegative electroretinogram in mucolipidosis IV. Arch. Ophthalmol., 120, 45–50. [DOI] [PubMed] [Google Scholar]
- 26.Tzetzi D., Hamilton R., Robinson P.H., Dutton G.N. (2007) Negative ERGs in mucopolysaccharidoses (MPS) Hurler-Scheie (I-H/S) and Hurler (I-H)-syndromes. Doc. Ophthalmol., 114, 153–158. [DOI] [PubMed] [Google Scholar]
- 27.Weleber R.G., Gupta N., Trzupek K.M., Wepner M.S., Kurz D.E., Milam A.H. (2004) Electroretinographic and clinicopathologic correlations of retinal dysfunction in infantile neuronal ceroid lipofuscinosis (infantile Batten disease). Mol. Genet. Metab., 83, 128–137. [DOI] [PubMed] [Google Scholar]
- 28.den Hollander A.I., Koenekoop R.K., Yzer S., Lopez I., Arends M.L., Voesenek K.E., Zonneveld M.N., Strom T.M., Meitinger T., Brunner H.G., et al. (2006) Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am. J. Hum. Genet., 79, 556–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartong D.T., Berson E.L., Dryja T.P. (2006) Retinitis pigmentosa. Lancet, 368, 1795–1809. [DOI] [PubMed] [Google Scholar]
- 30.Wang F., Wang H., Tuan H.F., Nguyen D.H., Sun V., Keser V., Bowne S.J., Sullivan L.S., Luo H., Zhao L., et al. (2014) Next generation sequencing-based molecular diagnosis of retinitis pigmentosa: identification of a novel genotype-phenotype correlation and clinical refinements. Hum. Genet., 133, 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roosing S., van den Born L.I., Sangermano R., Banfi S., Koenekoop R.K., Zonneveld-Vrieling M.N., Klaver C.C., van Lith-Verhoeven J.J., Cremers F.P., den Hollander A.I., et al. (2015) Mutations in MFSD8, encoding a lysosomal membrane protein, are associated with nonsyndromic autosomal recessive macular dystrophy. Ophthalmology, 122, 170–179. [DOI] [PubMed] [Google Scholar]
- 32.Hood D.C., Bach M., Brigell M., Keating D., Kondo M., Lyons J.S., Marmor M.F., McCulloch D.L., Palmowski-Wolfe A.M. and International Society For Clinical Electrophysiology of, V. (2012) ISCEV Standard for Clinical Multifocal Electroretinography (mfERG) (2011 Edition). Doc Ophthalmol, 124, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voznyi Ya V., Karpova E.A., Dudukina T.V., Tsvetkova I.V., Boer A.M., Janse H.C., van Diggelen O.P. (1993) A fluorimetric enzyme assay for the diagnosis of Sanfilippo disease C (MPS III C). J. Inherit. Metab. Dis., 16, 465–472. [DOI] [PubMed] [Google Scholar]
- 34.de Jong J.G., Wevers R.A., Liebrand-van Sambeek R. (1992) Measuring urinary glycosaminoglycans in the presence of protein: an improved screening procedure for mucopolysaccharidoses based on dimethylmethylene blue. Clin. Chem., 38, 803–807. [PubMed] [Google Scholar]
- 35.Grimberg J., Nawoschik S., Belluscio L., McKee R., Turck A., Eisenberg A. (1989) A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucl. Acids Res., 17, 8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eytan O., Morice-Picard F., Sarig O., Ezzedine K., Isakov O., Li Q., Ishida-Yamamoto A., Shomron N., Goldsmith T., Fuchs-Telem D., et al. (2013) Cole disease results from mutations in ENPP1. Am. J. Hum. Genet., 93, 752–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Ligt J., Willemsen M.H., van Bon B.W., Kleefstra T., Yntema H.G., Kroes T., Vulto-van Silfhout A.T., Koolen D.A., de Vries P., Gilissen C., et al. (2012) Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med., 367, 1921–1929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.