Abstract
Primary open angle glaucoma (POAG), a major cause of blindness worldwide, is a complex disease with a significant genetic contribution. We performed Exome Array (Illumina) analysis on 3504 POAG cases and 9746 controls with replication of the most significant findings in 9173 POAG cases and 26 780 controls across 18 collections of Asian, African and European descent. Apart from confirming strong evidence of association at CDKN2B-AS1 (rs2157719 [G], odds ratio [OR] = 0.71, P = 2.81 × 10−33), we observed one SNP showing significant association to POAG (CDC7–TGFBR3 rs1192415, ORG-allele = 1.13, Pmeta = 1.60 × 10−8). This particular SNP has previously been shown to be strongly associated with optic disc area and vertical cup-to-disc ratio, which are regarded as glaucoma-related quantitative traits. Our study now extends this by directly implicating it in POAG disease pathogenesis.
Introduction
Glaucoma is the leading cause of irreversible visual impairment and blindness, affecting >60 million people worldwide, and it is estimated that the number of affected individuals will reach 80 million in 2020 (1–3). Primary open angle glaucoma (POAG) is the most prevalent form of glaucoma in most populations and is characterized by progressive retinal ganglion cell (RGC) loss that causes characteristic structural changes of the optic nerve with associated with visual field loss in the face of an open drainage angle in the eye. POAG has a strong genetic component that has been well documented (4). Indeed, several susceptibility loci have been identified for POAG through the use of linkage and association studies (5). Genes known to contribute to glaucoma include myocillin (MYOC), optineurin (OPTN), TANK-binding kinase 1 (TBK1) and WD repeat domain 36 (WDR36) (6–12). However, mutations in these genes account for no >5–10% of all POAG cases in the general population (5).
It is likely that POAG, as a complex trait, results from the interactions of multiple genes and environmental factors (13–15). Genome-wide association studies (GWAS) have provided further insights into the genetic basis of POAG (16). The first GWAS on POAG was conducted on 1263 POAG cases and 34 877 controls from Iceland. Genome-wide significant association was detected at the CAV1–CAV2 locus on Chromosome 7q31 and subsequently was replicated in a multi-ethnic sample collection from Sweden, the UK, Australia, Hong Kong and China (17). This was rapidly followed by five other GWAS studies, which utilized either advanced or non-advanced POAG cases derived from populations of European or East Asian ancestries (18–22). These latter studies led to the discovery of nine additional genetic regions associated with POAG disease risk (TMCO1, CDKN2B-AS1, SIX1-SIX6, an intergenic region on chromosome 8q22, ABCA1, GAS7, AFAP1, GMDS and PMM2). Several of these genetic loci have been replicated in ethnically diverse populations, demonstrating them to be bona fide POAG associations with global implications (23–25).
Similar in concept and laboratory chemistry to the whole-genome genotyping chip design, the exome array approach evaluates putative functional coding variants selected from the exome sequences of >12 000 individuals (26). In addition, the exome array also contains >5000 common variant SNPs from GWAS arrays with a minor allele frequency (MAF) exceeding 5% which can serve as ancestry informative markers. Exome array genotyping allows us to specifically explore the possible contribution of potentially functional coding variants in POAG disease susceptibility.
Results
Common genetic variants in CDKN2B-AS1 and TGFBR3-CDC7 are associated with POAG
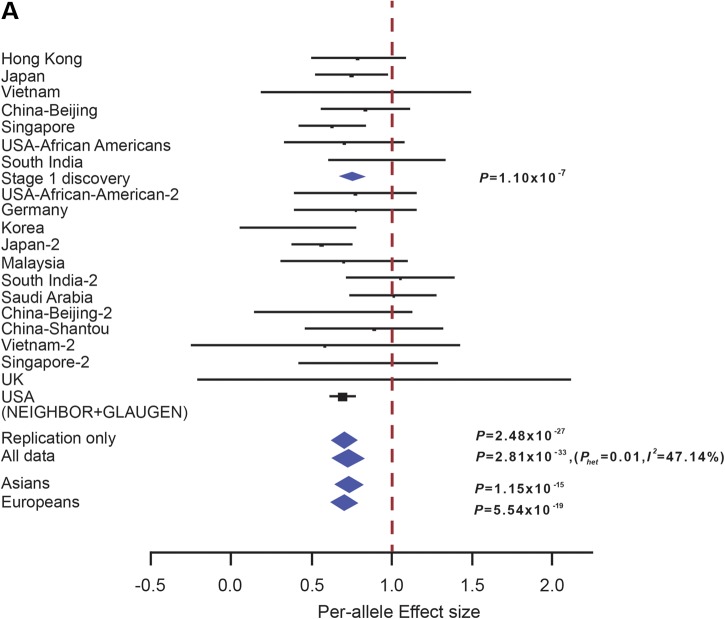
We conducted a two-stage Exome Chip discovery and replication on POAG cases and normal controls. For the discovery stage (Stage 1), genotyping was performed using the Illumina Infinium HumanExomeBeadChip (v1.0) on a total of 3822 POAG cases and 10 426 normal controls drawn from seven countries (Table 1). In addition to the ≈247 000 SNP markers present on the standard Illumina Exome array (26), we also included an extra 25 000 coding frame SNP markers obtained from exome sequencing of 2000 individuals of East Asian descent. Stringent quality control (QC) filters were applied to both SNPs and samples: per SNP call rate ≥99%, per-sample call rate ≥95%, non-monomorphic SNPs and non-significant deviation from Hardy–Weinberg equilibrium (HWE) P ≥ 10−6. Samples under suspicion of cross-contamination and biologically related samples were removed by verification of extreme heterozygosity and identical by descent/identical by state information, if applicable. We further performed principal component analysis (PCA) to verify that cases and controls were well-matched ancestrally (Supplementary Material, Fig. S1) (27–32). As a result, a total of 3504 POAG cases and 9746 controls passing QC filters were included for association analysis using unconditional logistic regression, with adjustments for the principal components (PCs). From these datasets, a total of 2206 POAG cases represent entirely new patient collections, which have not been previously reported (Supplementary Material, Table S1). Each study-specific point estimate was then summarized using fixed-effects meta-analysis (Nmeta-analysis = 7 collections). A quantile–quantile plot derived from the meta-analysis P-values showed no significant dispersion of test statistics from the expected distribution (λGC = 1.042; Supplementary Material, Fig. S2) suggesting that the association results were not confounded by cryptic population stratification. Using additive effect models, we observed experiment-wide significant (P = 0.05/272 000 SNPs = 1.84 × 10−7) evidence of association at CDKN2B-AS1 (rs2157719 [G], per-allele OR = 0.73, P = 1.10 × 10−7) (Supplementary Material, Fig. S3).
Table 1.
Sample collections of POAG cases and controls for Stages 1 (discovery) and 2 (replication)
| Collection | N cases | N controls | Ethnicity | Age of cases | Age of controlsa | Collection comment |
|---|---|---|---|---|---|---|
| Singapore | 850 | 2347 | Singaporean Chinese | 71.6 ± 10.1 | 58.88 ± 9.6 | New recruitment |
| Japan | 923 | 640 | Japanese | 65.3 ± 13.1 | 71.9 ± 5.8 | Previously GWAS in Nakano et al. (25) |
| USA-African-Americans | 590 | 636 | African American | 65.4 ± 12.3 | 54.8 ± 9.8 | New recruitment |
| China-Beijing | 587 | 461 | Northern Chinese | 58.5 ± 12.5 | Population-based controls | New recruitment |
| Hong Kong | 375 | 2962 | Southern Chinese | 62.3 ± 15.3 | Population-based controls | Previously described for replication in Thorleifsson et al. (17) |
| South India | 121 | 716 | Indian | 60.9 ± 12.0 | 51.0 ± 6.5 | New recruitment |
| Vietnam | 58 | 1984 | Vietnamese | 63 ± 7.1 | Population-based controls | New recruitment |
| Total discovery | 3504 | 9746 | 2206 cases are new discovery samples | |||
| Stage 2 replication | ||||||
| Singapore-2 | 520 | 5473 | Singaporean Chinese | 71.1 ± 10 | Population-based controls | New recruitment |
| Japan-2 | 935 | 996 | Japanese | 64.3 ± 14.0 | 57.5 ± 13.9 | N = 411 used for replication in Nakano et al. (25) |
| USA-Afican-American 2 | 497 | 304 | African-American | 69.1 ± 11.0 | 66.7 ± 13.1 | New recruitment |
| South India-2 | 453 | 2496 | Indian | 62.5 ± 9.9 | 58.9 ± 10.1 | New recruitment |
| Korea | 400 | 454 | Korean | 59.0 ± 11.8 | 40.3 ± 14.1 | New recruitment |
| Saudi Arabia | 236 | 655 | Middle Eastern | 60.8 ± 12.7 | 54.4 ± 11.7 | New recruitment |
| Malaysia | 132 | 2540 | Malay | 65.1 ± 8.2 | 58.7 ± 11.0 | New recruitment |
| China-Beijing 2 | 115 | 251 | Northern Chinese | 54.2 ± 12.4 | 71.53 ± 7.16 | New recruitment |
| UK | 336 | 6090 | European | 71.4 ± 10.8 | Population-based controls | New recruitment |
| China-Shantou | 247 | 289 | Southern Chinese | 52.9 ± 19.4 | 75.7 ± 6.1 | Previously described for replication in Thorleifsson et al. (17) |
| Germany | 56 | 142 | European | 67.9 ± 11.4 | 78.4 ± 8.9 | New recruitment |
| Vietnam-2 | 76 | 245 | Vietnamese | 52.4 ± 17.4 | 51.3 ± 17.8 | New recruitment |
| France | 80 | 75 | European | 75.6 ± 8.5 | 73.5 ± 8.3 | New recruitment |
| China-Shanghai, Chengdu 2 | 181 | 286 | Southern Chinese | 54.7 ± 16.5 | 84.7 ± 11.7 | Previously described in Chen et al. (19) |
| China-Shanghai, Chengdu | 608 | 1005 | Southern Chinese | 49.6 ± 17.0 | 62.9 ± 12.1 | Previously described in Chen et al. (19) |
| USA (NEIGHBOR) | 2170 | 2347 | European descendant | 66.4 | 68 | Previously described in Wiggs et al. (22) |
| USA (GLAUGEN) | 976 | 1140 | European descendant | 63.6 | 65.5 | Previously described in Wiggs et al. (22) |
| Australia (ANZRAG) | 1155 | 1992 | European descendant | 60.5 ± 14.3 | 55.6 ± 14.4 | Previously described in Gharahkhani et al. (20) |
| Total replication | 9173 | 26 780 | 3425 cases are new replication samples | |||
| Total all samples | 12 677 | 36 526 | 5631 cases are new in this report. | |||
aPopulation-based controls are ascertained from large-scale studies and do not have demographic data available. Based on many well-described examples, both by others and us, the frequency of POAG in the general population is uncommon (i.e. <5%). In this regard, the false-negative rate for POAG status in the population-based controls is likely to be low and thus the effect of loss of statistical power is negligible.
All SNP markers showing P < 0.0005 in the discovery stage were followed up in a validation stage (Stage 2) comprised of up to 9173 POAG cases and 26 780 controls. A total of 21 SNPs at 20 independent loci were brought forward for validation genotyping using Sequenom MassARRAY iPLEX or in silico look-ups if genome-wide genotyping data were available (see Supplementary Material, Table S2).
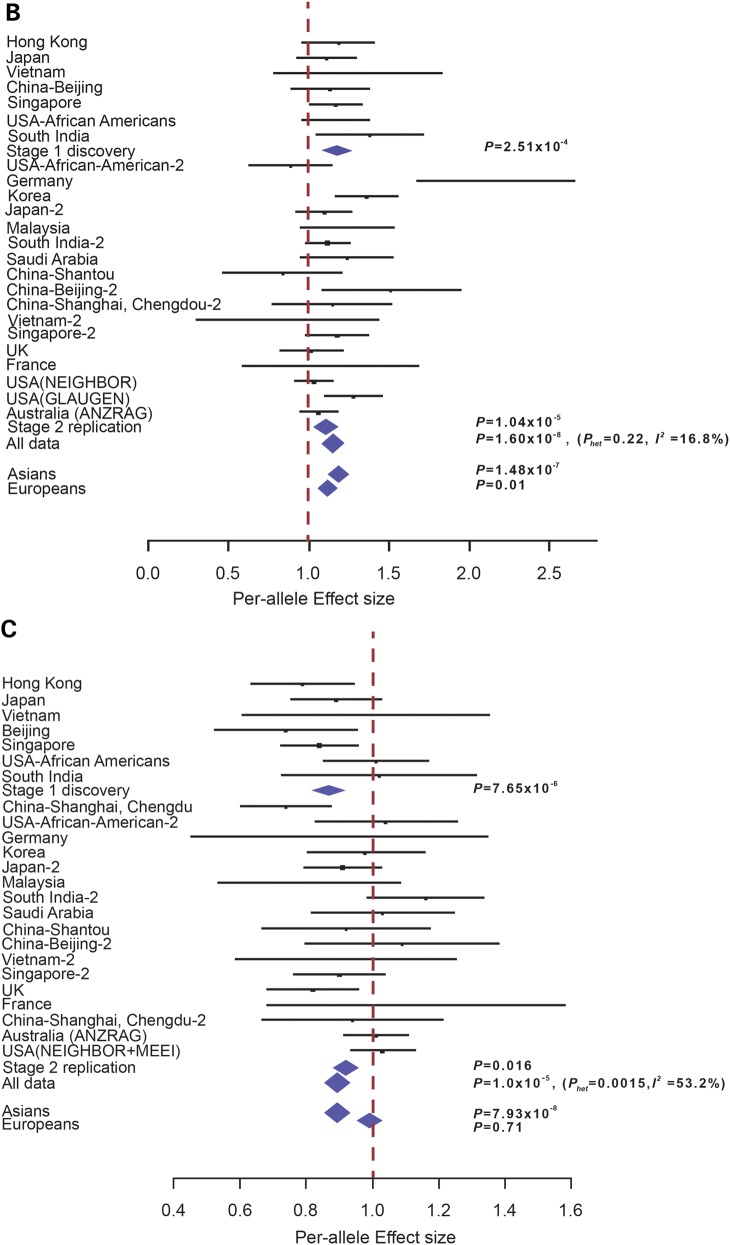
CDKN2B-AS1 rs2157719 once again showed significant evidence of association in the meta-analysis of all replication collections (OR = 0.70, P = 2.48 × 10−27) as well as meta-analysis of all samples tested (rs2157719 [G], OR = 0.71, Pmeta = 2.81 × 10−33) (Supplementary Material, Table S2 and Fig. S4). No heterogeneity of effect between Asians (OR = 0.72, P = 1.15 × 10−15) and Europeans (OR = 0.69, P = 5.54 × 10−19) were detected for this marker (Fig. 1A), consistent with the multiple previous reports describing association at this locus (18,22,25). Apart from CDKN2B-AS1 rs2157719, a second SNP marker (CDC7-TGFBR3 rs1192415) showed clear evidence of replication in Stage 2 (OR = 1.12, P = 1.04 × 10−5, Fig. 1B and Supplementary Material, Table S2). On meta-analysis with the exome-chip discovery findings, genome-wide significant evidence of association was observed for rs1192415 (OR = 1.13, Pmeta = 1.6 × 10−8) (Fig. 1B; Supplementary Material, Table S2). Although the risk allele ranged from between 11 and 34% across all ethnic groups studied for this marker, the association observed appeared to be uniform and consistent across most groups with little overall heterogeneity (I2 index< 20%, Supplementary Material, Table S3). For rs1192514, the association appeared stronger in Asians (OR = 1.17, P = 1.48 × 10−7) compared with Europeans (OR = 1.10, P = 0.01) (Fig. 1B), although the difference was not statistically significant (Phet = 0.17). One other marker (FNDC3B rs4894796) was nominally significant in the replication stage (OR = 0.95, P = 0.02) and remained suggestively associated with POAG in the overall meta-analysis (OR = 0.93, P = 1.40 × 10−5) (Fig. 1C; Supplementary Material, Table S2). However, the association appeared to be nearly entirely driven by the Asian POAG collections (OR = 0.89) compared with the Europeans (OR = 0.99, Phet between Asians and Europeans = 0.0042) (Fig. 1C). A recently reported GWAS on POAG showed strong association with common SNP markers mapping to AFAP1 (rs4478172) (20). Looking up on our exome dataset, we successfully genotyped rs7437940 (r2 = 0.18, D′ = 0.97 with rs4478172) which also mapped within AFAP1. We note significant association at this AFAP1 marker (rs7437940: Stage 1 P = 1.94 × 10−5, Stage 2 P = 0.08, P-value for meta-analysis = 4.25 × 10−6) (Supplementary Material, Table S2), which supports the previous report (20).
Figure 1.
Continued
Figure 1.
Forest plots showing evidence of association between SNPs: (A) CDKN2B-AS1 rs2157719, (B) CDC7/TGFBR3 rs1192415 and (C) FNDC3B rs4894796. The vertical line represents a per-allele odds ratio of 1.00. The oblongs represent point estimates (referring to the per-allele odds ratio), with the height of the oblongs inversely proportional to the standard error of the point estimates. Horizontal lines indicate the 95% confidence interval for each point estimate. Meta-analysis of Stages 1 and 2, OR, Pmeta and I2 was labeled on the right-hand side for corresponding analysis. For rs4894796 genotyping, see Supplementary Material, Information for sample collections.
Expression of POAG-associated genes in ocular tissues
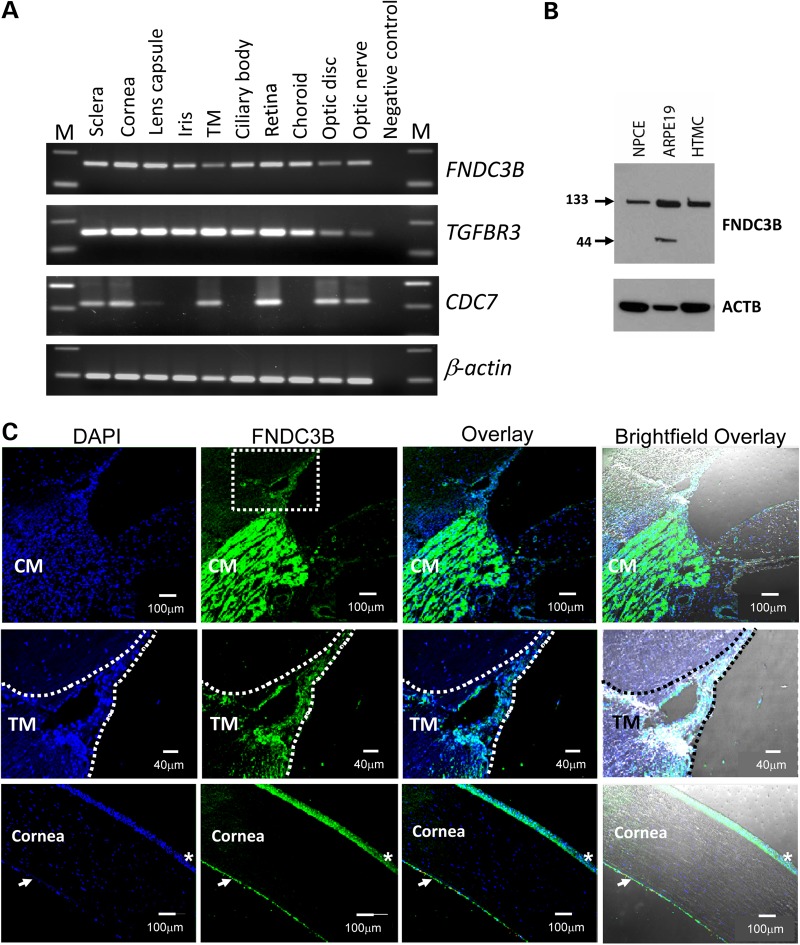
We examined the mRNA expression of CDC7, TGFBR3 and FNDC3B in multiple eye tissues. Expression of all three genes was observed in tissues relevant to POAG such as the trabecular meshwork, optic disc and nerve. In contrast to TGFBR3 and FNDC3B, which were expressed in all tested ocular tissues, CDC7 expression was absent in the iris, ciliary body and choroid (Fig. 2).
Figure 2.
Analysis of FNDC3B, TGFBR3 and CDC7 expression in ocular tissues. (A) The FNDC3B-specific 162 bp and TGFBR3-specific 152 bp amplification product was observed in all analyzed ocular tissues. CDC7-specific 242 bp product was observed in sclera, cornea, trabecular meshwork, retina, optic disc and optic nerve. The ubiquitously expressed gene, ACTB was used as the normalizing control. A no template sample acted as the negative control (NC) to ensure non-contamination of the RT–PCR reaction mix. The variable M denotes molecular-weight marker. (B) Immunoblot of whole cell lysates from NPCE, retinal pigment epithelial (ARPE19) and HTM cells, probed for FNDC3B and β-actin, as a loading control. Positions of the ∼133 and ∼44 kDa forms of FNDC3B are indicated. All ocular cells analyzed expressed the ∼133 kDa protein, while ARPE19 cells expressed a smaller ∼44 kDa isoform of FNDC3B. (C) Immunolocalization of FNDC3B in human eye tissues. Strong immunofluorescence labeling of FNDC3B (green) was seen in the ciliary muscle (CM) (top row). Scale bar: 100 µm. In the trabecular meshwork (TM, middle row), FNDC3B (green) labeling was relatively weaker. Scale bar: 40 µm. FNDC3B positive immunoreactivity was also observed in cornea epithelial (*) and cornea endothelial cells (white arrows) (bottom row). Nuclei were stained with DAPI (blue). Scale bar: 100 µm.
We also investigated the localization of FNDC3B protein in normal ocular tissues, focusing our attention on cornea and the tissues of the outflow pathway. FNDC3B could be immunolocalized to cells in the trabecular meshwork and all three layers of the cornea (Fig. 2). Immunolocalization of TGFBR3 and CDC7 could not be similarly investigated due to lack of availability of specific antibodies.
Analysis of rare variants from the exome-chip discovery collection
We next proceeded to conduct gene-based tests on mutational load to further investigate the role of low-frequency variants in POAG for all patient collections in the discovery stage. Gene-based tests are an alternative to single-marker tests for association, which are often underpowered to detect association with rare variants. To more directly address the impact of low frequency, non-synonymous genetic variants, we considered only the variants with MAF of <5% (33). As a result, we were able to assess a total of 7822 genes having at least two such variants using the sequence kernel association optimal test (34). We note two genes (MYO18A and SULF2) which showed nominal evidence of association on burden test (P = 6.26 × 10−7 and P = 5.78 × 10−5, respectively), but these findings are primarily driven by the small Vietnamese collection (N = 58 cases and N = 1984 controls). Removal of this dataset resulted in a marked reduction of the burden test association results (Supplementary Material, Table S4).
Discussion
The present study supports the association of POAG a locus that has been previously implicated with vertical cup-disc ratio, a glaucoma-associated endophenotype. This locus is defined by rs1192415, a common SNP marker mapping to the intergenic region between CDC7 and TGFBR3 (allele [G], OR = 1.13, Pmeta = 1.6 × 10−8). We were also able to strongly corroborate the established association of POAG with CDKN2B-AS1.CDC7-TGFBR3 rs1192415 has previously been reported to be strongly associated with overall optic disc area, which together with VCDR is a glaucoma-related quantitative trait (35,36). Examination of this locus in >4700 POAG cases and >90 000 controls of European descent in a recent study showed direction of effect consistent with our data (OR = 1.06, P = 0.12) (37). Indeed, in the European section of our study (N = 4773 cases and 11786 controls), we note a similar effect size (OR = 1.10, P = 0.01) in keeping with that shown by Springelkamp and others. The effect of this locus appeared to be much stronger in Asians (OR = 1.17, P = 1.48 × 10−7), leading to a genome-wide significant association upon meta-analysis of all collections. Each copy of the rs1192415 minor allele is associated with a relatively modest increase in POAG risk of 1.13 at exome-wide significance. The consistency of the effect across 21 of 24 POAG collections (Phet = 0.22, I2 index for heterogeneity = 16.9%), lend further credence to the observed association.
It is of note that the G allele of the rs1192415 is associated with increase in disc area and therefore a larger VCDR. This may indicate a lower threshold for diagnosing glaucoma in those individuals harboring the risk allele (38). However, as the diagnosis of POAG in this study was not based solely upon the presence of an increased cup–disc ratio but also compatible visual field loss, it is unlikely that selection bias has influenced the result obtained with rs1192415. We note that FNDC3B was one of 16 loci associated with central corneal thickness (sentinel SNP being rs4894535) in a meta-analysis conducted on >20 000 individuals of European and Asian descent (39). That study also showed marker rs4894535 to be associated with POAG in 2979 cases and 7399 controls of European descent (OR = 0.83, P = 5.6 × 10−4). We were able to confirm association of rs4894535 with POAG in 5810 cases and 13 175 controls with readily available DNA for genotyping across 13 collections in this study (OR = 0.91, P = 0.0018; Supplementary Material, Table S5), thus lending further support for this association.
It is noteworthy that the FNDC3B SNP we report here (rs4894796) was nominally associated with POAG in the overall meta-analyses (OR = 0.93, P = 1.4 × 10−5). In particular, the association at rs4894796 was observed to be particularly strong in Asians (OR = 0.89, P = 7.93 × 10−8) compared with Europeans (OR = 0.99, P = 0.71). Unsurprisingly, rs4894796 was found within a different linkage disequilibrium (LD) block than the SNP associated with central corneal thickness and POAG (rs4894535). The low LD between rs4894796 and rs4894535 suggests that the association signal at the latter SNP is not likely to be driven by rs4894796. An in-depth inspection of the LD patterns of this genetic region between European and East Asian populations, which currently appear visually identical according to available heat maps (http://www.hapmap.org), may be useful to better understand the degree of LD dissimilarity between populations and its bearing on the results obtained for FNDC3B. Of note, a further SNP (rs6445055) at FNDC3B showed genome-wide significant association for intraocular pressure (IOP) (P = 4.9 × 10−8), but only marginal association was seen for POAG per se (P = 0.03, OR = 0.92) in 4284 POAG cases and 95 560 controls of European descent (21). As the FNDC3B SNP associations with POAG are still suggestive rather than affirmative it is presumptive at this stage to speculate that it could be one of the links for IOP-dependent mechanisms of POAG.
This is one of the largest studies on the genetics of POAG, yet the power to detect genes with small effects was limited. One crucial reason for this apparent loss of power could be the scope of the genetic content which was used for interrogation (as exomic content only comprises <2% of the entire human genome, much useful data could be missed if the bulk of the true positive genetic associations for POAG lie in the non-coding regions of the genome). The fact that our discovery cohort was not mono-ethnic may also have reduced the power to detect ethnic specific variants of small effect sizes due to significant differences in allele frequencies between ethnic groups. The phenotypic heterogeneity and the lack of standardized clinical criteria across the cohorts may also have contributed to a loss in power. The level of IOP is commonly used to subdivide POAG into two subtypes: POAG with high IOP (>21 mmHg; named high-tension glaucoma, HTG) and NTG with normal IOP (<21 mmHg). In our cohorts, we had differing number of these subtypes with some cohorts having more NTG or HTG than others. In this study, we also did not subdivide POAG subjects into HTG and NTG to discover subtype-specific variants, focusing instead on identifying genetic variants for the overarching phenotype of POAG. However, when CDC7-TGFBR3 rs1192415 and FNDC3B rs4894796 were analyzed within NTG and HTG subgroups, they did not show differences in strength of association by subtype (data not shown).
All three loci we report in this study (CDKN2B-AS1, TGFBR3-CDC7 and FNDC3B) contain genes which may contribute to the regulation of transforming growth factor-β (TGF-β) signaling. TGF-β has been implicated previously in glaucomatous optic nerve damage and RGC death (40–42). The TGF-β family includes TGFβ1, TGFβ2 and TGFβ3, all of which bind to TGF-β receptor type-2 (TGFBR2). All TGF-β family members are dimeric polypeptide growth factors that inhibit the progression of cell cycle, which in turn may lead to terminal differentiation or apoptosis (40,43,44). TGF-β also modulates developmental and repair processes in several tissues. TGF-β signaling has been implicated in a wide variety of diseases including inflammation, autoimmune disorders, fibrosis, cancer, cataracts as well as glaucoma (40,43,44). The most strongly POAG-associated locus, CDKN2B-AS1, has been shown to regulate the transcription of cyclin-dependent kinase inhibitor 2A and 2B (CDKN2A and CDKN2B) (45), which inhibit cell proliferation via the TGF-β pathway by inducing G1-phase cell cycle arrest (44). Burdon et al. reported the up-regulation of CDKN2A and CDKN2B in response to elevated IOP (18). Collectively, these data suggest a link between the most well-recognized physiological risk factor of POAG and a downstream molecular response that may lead to RGC death. In this context, both CDC7 and TGFBR3 are also of interest and could have relevance to glaucomatous optic nerve damage and RGC death.
CDC7 encodes a cell division cycle protein with kinase activity that also interacts with CDKN2A. TGFBR3 is a TGF-β super family co-receptor, and is the most abundant of all TGF-β receptors (46). Through protein crystallography, murine TGFR-3 ZP domain (ZP-C) has been recently identified as a novel major TGF-β-binding site (47). It has been suggested that TGFBR3 may serve to enhance the binding of TGF-β ligands to TGF-β type II receptors by binding TGF-β and presenting it to TGFBR2, the receptor for all three TGF-β ligands. A linkage between FNDC3B, an oncogene and TGF-β signaling was also reported recently by Cai et al. Overexpression of FNDC3B was shown to induce epithelial-to-mesenchymal transition and activate several cancer pathways, including PI3-kinase/Akt, Rb1 and TGF-β signaling (48). FNDC3B also induced expression of all three TGF-β ligands and promoted TGFBR1 cell-surface localization (48). This connection of POAG-associated genes/loci with the TGF-β signaling pathway therefore lends further credence to the hypothesis of TGFB pathway involvement in glaucoma.
However, despite these attractive speculations on the genes vicinal to the associated SNPs, it is possible that these variants may also affect distant as yet unidentified target genes. Definitive evidence for the involvement of these genes in the pathogenesis of POAG awaits confirmation in other datasets, as well as the identification and characterization of functional variants.
Materials and Methods
Sample collections
Ethics statement
Ethics approval was obtained from the Centralized Institutional Review Board for the Singaporean patient sample and data collection and for the conduct of this study. All study protocols for patient and control sample collection were approved by the respective relevant Medical Ethics Committees of each participating site. All studies were conducted under the tenets of the Declaration of Helsinki with written informed consent obtained from all participants.
POAG and health controls subjects inclusion criteria
POAG cases in this study were defined by the following criteria: the presence of glaucomatous optic neuropathy (defined as loss of neuroretinal rim with a vertical cup : disc ratio of >0.7 or an inter-eye asymmetry of >0.2 and/or notching attributable to glaucoma) with compatible visual field loss, open angles on gonioscopy, and absence of secondary causes of glaucomatous optic neuropathy. POAG patients with a mean IOP without treatment that is consistently <21 mmHg on diurnal testing are classified as having NTG, whereas those with a mean IOP without treatment that is consistently >21 mmHg are classified as having HTG. Patients who were unable to give informed consent, or with secondary glaucoma due to trauma, uveitis, neovascularization, pseudoexfoliation, pigment dispersion, etc., were excluded from this study.
Controls in this study were recruited in a hospital-based or population-based manner. Hospital-based controls were all generally over the age of 40 years and confirmed to have no sign of glaucoma or other major eye diseases except for mild cataract and mild refractive errors (defined as |SE| < 3D) by an ophthalmic examination. These subjects at time of recruitment had IOP of <21 mmHg with open angles, healthy optic nerves, normal visual fields, and no family history of glaucoma. Population-based controls were ethnically matched healthy individuals over the age of 40 years, unless indicated otherwise (see Table 1; Supplementary Material for more details of the samples used in this study).
Genotyping and data QC
Study participants in the discovery stage were genotyped using the Illumina's Infinium HumanExomeBeadChip (Version 1.0) + Semi-Custom BeadChip (Illumina Inc.) that contains ∼250 000 SNPs of base content and an additional 25 000 East Asian-specific polymorphisms located on the coding frame. Stringent QC filters were applied after the laboratory work in genotyping completed. SNP markers that had missingness exceeding 5%, gross departure from HWE (P-value <1e−6) or were monomorphic were excluded from subsequent analysis. Likewise, individual samples with an overall call rate <95% were excluded. Samples were subjected to biological relationship verification by using the principle of variability in allele sharing according to the degree of relationship. Identity-by-state information was derived by PLINK. Those individuals who showed evidence of cryptic relatedness were removed before PC analysis was conducted. In addition, samples showing gender discrepancies between the clinical gender and genetically inferred gender were removed. A total of 1008 samples were excluded after rigorous application of QC filters (568 sample exclusions were due to PCA ancestral outliers, 246 were excluded due to per-sample call rate <95%, 168 were excluded due to first-degree familial relationships detected from the discovery stage exome-chip genotyping, and 26 were excluded due to suspicions of sample contamination). PC analysis was undertaken using EIGENSTRAT to account for spurious associations resulting from ancestral differences of individual SNPs (49). PCs showing significant effect on univariate analysis were used to correct for any underlying population substructure. We adjusted for the top three PCs (PC1–PC3) for Singapore, Hong Kong, Japan, USA-African Americans, China-Beijing and Vietnam. The Indian POAG collection was adjusted for the top 10 PCs (PC1–PC10), as there was more population substructure in this collection. After adjustment, we observed minimal evidence of genomic inflation (λGC = 1.042), thereby suggesting that this well-described method of controlling for population stratification was adequate in our study. Genotyping clouds for the key SNPs CDKN2B-AS1 rs2157719, CDC7-TGFBR3rs1192415 and FNDC3Brs4894796 were directly visualized (Supplementary Material, Fig. S5) to ensure good quality.
For Stage 2 (replication stage), genotyping was performed using the Sequenom MassArray platform (www.sequenom.com). Samples recruited at the latter stage of replication were genotyped for the key SNPs CDC7-TGFBR3rs1192415, and FNDC3Brs4894796 using pre-developed Taqman Assays (Applied Biosystems, Foster City, CA, USA; www.appliedbiosystems.com)
Statistical analysis
We contrasted the genotypes between POAG cases and healthy controls via single-SNP analysis using unconditional logistic regression fitted for genotype trend effects (1-degree-of-freedom score test). To do this, the PLINK software [version 1.07] (50) was used for modeling within a logistic regression framework, adjusting for age, gender and genetic ancestry (reflected by PCs). Manhattan and LD plots were created using Haploview [version 3.2] (51). Q–Q and regional association plots were created using the software R [www.r-project.org ] (52). Meta-analysis summarizing the results across all cohorts was performed using both fixed and random-effects modeling weighted in an inverse-variance manner (53). This method weighs each study according to effective sample size and cohort-specific MAF of the associated variants. To avoid an otherwise unacceptable number of false positive signals as an artifact of multiple testing, the threshold for exome-wide significance, P < 2 × 10−7, was considered to be statistically significant. Heterogeneity of the meta-analyses was calculated by measuring I2.
Expression analysis of genes
Expression of CDC7, TGFBR3 and FNDC3B was assessed by semi-quantitative reverse transcription–PCR (RT–PCR) using gene-specific primers (CDC7-forward 5′-TTTTCTCCCCAGCGTGACC-3′, CDC7-reverse 5′-GCAATTTTCTCTTCAGGTCCTAC-3′; TGFBR3-forward 5′-TCTCCTCAGTCCACATCCAC-3′, TGFBR3-reverse 5′-TGCTGATGAAAACTGGACCAC-3′; FNDC3B-forward 5′-AGCATCATCTTCCCCACACA-3′, FNDC3B-reverse 5′-AAGAAGGAGGGCTGTTGAGG-3′) on total RNA extracted from a variety of ocular tissues (cornea, sclera, retina and retinal pigment epithelium, iris, lens capsule and optic nerve) as described earlier (54). We used the ubiquitously expressed ACTB gene (forward 5′-CCAACCGCGAGAAGATGA-3′and reverse 5′-CCAGAGGCGTACAGGGATAG-3′) as amplification and normalizing control.
Western blotting
Cell lines obtained from American Type Culture Collection (Manassas, VA, USA) were the human retinal pigment epithelial cell line (APRE19), human Trabecular Meshwork cell line (HTM) was purchased from ScienCellResearch Laboratories (Carlsbad, CA, USA) and the human non-pigmented ciliary epithelial cell line (NPCE) is a kind gift from Prof. Miguel Coca-Prados from Yale School of Medicine. Cell lysates were obtained by lysing individual cell lines with lysis buffer (50 mm Tris–HCl, pH 8, 150 mm NaCl, 1.0% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 0.2 mm NaVO4, 10 mm NaF, 0.4 mm EDTA and 10% glycerol). SDS–PAGE resolved proteins were transferred to Hybond-C Extra nitrocellulose membranes (Amersham Life Science Inc., Arlington Heights, IL, USA). Membranes were blocked and blotted by 5% nonfat milk, 0.1% Tween 20 in Tris-buffered saline (20 mm Tris–HCl, pH 7.6, 150 mm NaCl) for 1 h before incubation with FNDC3Bfor 1 h (1 : 250) (Sigma-Aldrich Corp., St. Louis, MO, USA). Actin-horseradish peroxidase (HRP) (1 : 50 000) from Santa Cruz Biotechnology (Dallas, TX, USA). The bound primary antibodies were detected by horseradish peroxidase-conjugated secondary antibodies (GE Healthcare Biosciences, Pittsburgh, PA, USA), and visualized by Luminata Forte Western HRP substrate (Millipore, Bedford, MA, USA).
Immunofluorescence confocal microscopy
Immunofluorescence confocal microscopy was performed on antigen retrieved 4 µm paraffin sections. Blocking of tissue sections was performed with blocking buffer (5% nonfat milk, 5% FBS, 0.1% PBS-Tween; 1× pen/strep) for 1 h at RT. FNDC3B antibody (Sigma-Aldrich Corp.) was diluted (1 : 50) into blocking buffer and incubated overnight at 4°C. Secondary FITC (1 : 300)-labelled anti-rabbit antibody (Jackson Laboratories, Westgrove, PA, USA) was also diluted in blocking buffer and incubated at RT for 1 h followed by application of Vectashield with 4′,6-diamidino-2-phenyl-indole (DAPI) (Vector Laboratories, Burlingame, CA, USA). Coverslips were then used to overlay the sections and stored in the dark at 4°C until viewing with Olympus Fluoview 1000 confocal microscope (Olympus Optical Co. Ltd., Tokyo, Japan).
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
We thank all the study participants, the staff from all involved studies and sites who contributed to this study. The authors J.E.C., K.P.B., D.A.M., and others acknowledge the support of Ms Bronwyn Usher-Ridge in patient recruitment and data collection, and Dr Patrick Danoy and Dr Johanna Hadler for genotyping of the ANZRAG cohort. They also acknowledge David C. Whiteman and Graham Radford-Smith for providing access to the control samples used with the ANZRAG glaucoma cases. The authors D.M. thank Dr Philippe Gohier and Dr Ghislaine Jallet for patient recruitment and data collection and acknowledge the technical assistance provided by the Centre de Recherche Biologique (Dr Odile Blanchet) and Centre de Recherche Clinique (Pr Marc-Antoine Custeau), University Hospital, Angers, France. The Singapore study of E.N.V., T.A. and C.C.K was supported by a Biomedical Research Council (BMRC) grant in Singapore, Ref: BMRC 10/1/35/19/675. This research was also partly supported by a grant (NMRC/TCR/008-SERI/2013) from the Singapore National Research Foundation under its Translational and Clinical Research Flagship Programme and administered by the Singapore Ministry of Health's National Medical Research Council. We also acknowledge the following source of funding support for recruitment and genotyping of population-based cohorts SIMES and SCES: National Medical Research Council, Singapore (NMRC/TCR/002-SERI/2008, (R626/47/2008TCR), CSA R613/34/2008, NMRC 0796/2003, STaR/0003/2008), the National Research Foundation of Singapore, the Biomedical Research Council, Singapore (BMRC 09/1/35/ 19/616 and 08/1/35/19/550) and Genome Institute of Singapore (GIS/12-AR2105). The Singapore Tissue Network and the Genome Institute of Singapore, Agency for Science, Technology and Research, Singapore provided services. The research of M.A.H. was supported by NIH R01 EY023646, EY13315 (MAH) and by P30-EY005722. The Japanese sample collection from Kyoto was supported by the grants from the Collaborative Development of Innovative Seeds of Japan Science and Technology Agency (JST) to M.K. and K.T., from the Ministry of Health, Labor and Welfare of Japan to M.N., K.M., K.T. and S.K., and from Santen Pharmaceutical Co. Ltd. to S.K. and K.T. The collection of Japanese POAG cases by N.F. was supported by a grant provided by the Ministry of Health, Labor and Welfare of Japan. The glaucoma research of WNL is supported by National Natural Science Foundation of China project (81030016). The research of Y. Chen was supported by National Natural Science Foundation of China (81200723) and glaucoma research of Xinghuai Sun was supported by Special Scientific Research Project of Health Professions (201302015). This research project was supported by the National Natural Science Foundation of China (81170883 (Z.Y.) and 81430008 (Z.Y.)). The glaucoma research of C.P.P. is supported by the Health and Medical Research Fund (HMRF, Ref: 01122236 and 11120801), Hong Kong and the General Research Fund from the Research Grants Council (grant number 468810), Hong Kong. Support for recruitment of Australian & New Zealand Registry of Advanced Glaucoma (ANZRAG) was provided by the Royal Australian and New Zealand College of Ophthalmology (RANZCO) Eye Foundation. Genotyping was funded by the National Health and Medical Research Council of Australia (#535074 and #1023911). This work was also supported by funding from NHMRC #1031362 awarded to Jamie E. Craig, NHMRC #1037838 awarded to Alex W. Hewitt, NHMRC #1048037 awarded to Stuart L. Graham, NHMRC #1009844 awarded to Robert J. Casson and Ivan Goldberg, NHMRC #1031920 and an Alcon Research Institute grant awarded to David A. Mackey, an Allergan Unrestricted grant awarded to Andrew J. White, the BrightFocus Foundation and a Ramaciotti Establishment Grant. S.M. is supported by Australian Research Council (ARC) and Australian National Health & Medical Research Council (NHMRC) Fellowships. Controls for the ANZRAG discovery cohort were drawn from the Australian Cancer Study, the Study of Digestive Health, and from a study of inflammatory bowel diseases. The Australian Cancer Study was supported by the Queensland Cancer Fund and the National Health and Medical Research Council (NHMRC) of Australia (Program no. 199600, awarded to David C. Whiteman, Adele C. Green, Nicholas K. Hayward, Peter G. Parsons, David M. Purdie and Penelope M. Webb, and program number 552429, awarded to David C. Whiteman). The Study of Digestive Health was supported by grant number 5 RO1 CA 001833 from the National Cancer Institute (awarded to David C. Whiteman). The Barrett's and Esophageal Adenocarcinoma Genetic Susceptibility Study (BEAGESS) sponsored the genotyping of oesophageal cancer and Barrett's oesophagus cases, which were used as unscreened controls in the ANZRAG discovery cohort. BEAGESS was funded by grant R01 CA136725 from the National Cancer Institute Collection of Saudi POAG cases and controls was supported by the Glaucoma Research Chair at College of Medicine, King Saud University, Riyadh, Saudi Arabia. NEI Glaucoma Human Genetics Collaboration (NEIGHBOR): genotyping services for the NEIGHBOR study were provided by the CIDR and were supported by the NEI through grant HG005259-01 (J.L.W.). Additionally, CIDR is funded through a federal contract from the NIH to The Johns Hopkins University, contract number HHSN268200782096C. Collecting and processing samples for the NEIGHBOR data set was supported by the NEI through American Recovery and Reinvestment Act (ARRA) grants 3R01EY015872-05S1 (J.L.W.) and 3R01EY019126-02S1 (M.A.H.). Genotype imputation and meta-analysis were supported by EY022305 (J.L.W.). Funding for the collection of cases and controls was provided by the following NIH grants: EY015543 (R.R. Allingham); EY006827 (D. Gaasterland); HL73042, HL073389, EY13315, EY023646 (M.A.H.); CA87969, CA49449, UM1 CA167552 (J.H. Kang); EY009149 (P.R. Lichter); HG004608 (C. McCarty); EY008208 (F.A. Medeiros); EY015473 (L.R.P.); EY012118 (M. Pericak-Vance); EY015682 (A. Realini); EY011671, EY09580 (J.E. Richards); EY013178 (J.S. Schuman); RR015574, EY015872, EY010886, EY009847, EY014104 (J.L.W.); EY011008, EY144428, EY144448 and EY18660 (K. Zhang). J.L.W. and L.R.P. are also supported by the Harvard Glaucoma Center for Excellence and Research to Prevent Blindness. J.N.C.B. is supported by NIH T32 EY007157 (CWRU) and T32 EY21453-2 (VUMC). MEEI case-control sample: genotyping for the Massachusetts Eye and Ear Infirmary (MEEI) case-control sample was performed at the Broad Institute of MIT and Harvard with funding support from the NIH GEI (Gene Environment Initiative) (U01HG04424 and U01HG004728). The GENEVA Coordinating Center (U01HG004446) assisted with genotype cleaning. Imputation was supported by NIH EY022305 (JLW). Collection of cases and controls was supported by NIH EY015872 (JLW) and NIH P30 014104 (JLW). Funding to pay the Open Access publication charges for this article was provided by Singapore Eye Research Institute.
Conflict of Interest statement. None declared.
References
- 1.Quigley H.A. (1996) Number of people with glaucoma worldwide. Br. J. Ophthalmol., 80, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quigley H.A., Broman A.T. (2006) The number of people with glaucoma worldwide in 2010 and 2020. Br. J. Ophthalmol., 90, 262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tham Y.C., Li X., Wong T.Y., Quigley H.A., Aung T., Cheng C.Y. (2014) Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology, 121, 2081–2090. [DOI] [PubMed] [Google Scholar]
- 4.Allingham R.R., Liu Y., Rhee D.J. (2009) The genetics of primary open-angle glaucoma: a review. Exp. Eye Res., 88, 837–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan B.J., Wang D.Y., Lam D.S., Pang C.P. (2006) Gene mapping for primary open angle glaucoma. Clin. Biochem., 39, 249–258. [DOI] [PubMed] [Google Scholar]
- 6.Sheffield V.C., Stone E.M., Alward W.L., Drack A.V., Johnson A.T., Streb L.M., Nichols B.E. (1993) Genetic linkage of familial open angle glaucoma to chromosome 1q21-q31. Nat. Genet., 4, 47–50. [DOI] [PubMed] [Google Scholar]
- 7.Stone E.M., Fingert J.H., Alward W.L., Nguyen T.D., Polansky J.R., Sunden S.L., Nishimura D., Clark A.F., Nystuen A., Nichols B.E., et al. (1997) Identification of a gene that causes primary open angle glaucoma. Science (New York, NY), 275, 668–670. [DOI] [PubMed] [Google Scholar]
- 8.Rezaie T., Child A., Hitchings R., Brice G., Miller L., Coca-Prados M., Heon E., Krupin T., Ritch R., Kreutzer D., et al. (2002) Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science (New York, NY), 295, 1077–1079. [DOI] [PubMed] [Google Scholar]
- 9.Sarfarazi M., Child A., Stoilova D., Brice G., Desai T., Trifan O.C., Poinoosawmy D., Crick R.P. (1998) Localization of the fourth locus (GLC1E) for adult-onset primary open-angle glaucoma to the 10p15-p14 region. Am. J. Hum. Genet., 62, 641–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monemi S., Spaeth G., DaSilva A., Popinchalk S., Ilitchev E., Liebmann J., Ritch R., Heon E., Crick R.P., Child A., et al. (2005) Identification of a novel adult-onset primary open-angle glaucoma (POAG) gene on 5q22.1. Hum. Mol. Genet., 14, 725–733. [DOI] [PubMed] [Google Scholar]
- 11.Fingert J.H. (2011) Primary open-angle glaucoma genes. Eye (London, UK), 25, 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fingert J.H., Robin A.L., Stone J.L., Roos B.R., Davis L.K., Scheetz T.E., Bennett S.R., Wassink T.H., Kwon Y.H., Alward W.L., et al. (2011) Copy number variations on chromosome 12q14 in patients with normal tension glaucoma. Hum. Mol. Genet., 20, 2482–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan B.J., Leung Y.F., Wang N., Lam S.C., Liu Y., Tam O.S., Pang C.P. (2004) Genetic and environmental risk factors for primary open-angle glaucoma. Chin. Med. J., 117, 706–710. [PubMed] [Google Scholar]
- 14.Wiggs J.L., Damji K.F., Haines J.L., Pericak-Vance M.A., Allingham R.R. (1996) The distinction between juvenile and adult-onset primary open-angle glaucoma. Am. J. Hum. Genet., 58, 243–244. [PMC free article] [PubMed] [Google Scholar]
- 15.Kang J.H., Wiggs J.L., Rosner B.A., Hankinson S.E., Abdrabou W., Fan B.J., Haines J., Pasquale L.R. (2010) Endothelial nitric oxide synthase gene variants and primary open-angle glaucoma: interactions with sex and postmenopausal hormone use. Invest. Ophthalmol. Visual Sci., 51, 971–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirschhorn J.N., Daly M.J. (2005) Genome-wide association studies for common diseases and complex traits. Nat. Rev., 6, 95–108. [DOI] [PubMed] [Google Scholar]
- 17.Thorleifsson G., Walters G.B., Hewitt A.W., Masson G., Helgason A., DeWan A., Sigurdsson A., Jonasdottir A., Gudjonsson S.A., Magnusson K.P., et al. (2010) Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat. Genet., 42, 906–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burdon K.P., Macgregor S., Hewitt A.W., Sharma S., Chidlow G., Mills R.A., Danoy P., Casson R., Viswanathan A.C., Liu J.Z., et al. (2011) Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat. Genet., 43, 574–578. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y., Lin Y., Vithana E.N., Jia L., Zuo X., Wong T.Y., Chen L.J., Zhu X., Tam P.O., Gong B., et al. (2014) Common variants near ABCA1 and in PMM2 are associated with primary open-angle glaucoma. Nat. Genet., 46, 1115–1119. [DOI] [PubMed] [Google Scholar]
- 20.Gharahkhani P., Burdon K.P., Fogarty R., Sharma S., Hewitt A.W., Martin S., Law M.H., Cremin K., Bailey J.N., Loomis S.J., et al. (2014) Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat. Genet., 46, 1120–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hysi P.G., Cheng C.Y., Springelkamp H., Macgregor S., Bailey J.N., Wojciechowski R., Vitart V., Nag A., Hewitt A.W., Hohn R., et al. (2014) Genome-wide analysis of multi-ancestry cohorts identifies new loci influencing intraocular pressure and susceptibility to glaucoma. Nat. Genet., 46, 1126–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiggs J.L., Yaspan B.L., Hauser M.A., Kang J.H., Allingham R.R., Olson L.M., Abdrabou W., Fan B.J., Wang D.Y., Brodeur W., et al. (2012) Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet., 8, e1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan B.J., Wang D.Y., Pasquale L.R., Haines J.L., Wiggs J.L. (2011) Genetic variants associated with optic nerve vertical cup-to-disc ratio are risk factors for primary open angle glaucoma in a US Caucasian population. Invest. Ophthalmol. Visual Sci., 52, 1788–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao D., Jiao X., Liu X., Hennis A., Leske M.C., Nemesure B., Hejtmancik J.F. (2012) CDKN2B polymorphism is associated with primary open-angle glaucoma (POAG) in the Afro-Caribbean population of Barbados, West Indies. PLoS ONE, 7, e39278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakano M., Ikeda Y., Tokuda Y., Fuwa M., Omi N., Ueno M., Imai K., Adachi H., Kageyama M., Mori K., et al. (2012) Common variants in CDKN2B-AS1 associated with optic-nerve vulnerability of glaucoma identified by genome-wide association studies in Japanese. PLoS ONE, 7, e33389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huyghe J.R., Jackson A.U., Fogarty M.P., Buchkovich M.L., Stancakova A., Stringham H.M., Sim X., Yang L., Fuchsberger C., Cederberg H., et al. (2013) Exome array analysis identifies new loci and low-frequency variants influencing insulin processing and secretion. Nat. Genet., 45, 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.(ANZgene), Australia and New Zealand Multiple Sclerosis Genetics Consortium. (2009) Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat. Genet., 41, 824–828. [DOI] [PubMed] [Google Scholar]
- 28.Kooner J.S., Saleheen D., Sim X., Sehmi J., Zhang W., Frossard P., Been L.F., Chia K.S., Dimas A.S., Hassanali N., et al. (2011) Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat. Genet., 43, 984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reveille J.D., Sims A.M., Danoy P., Evans D.M., Leo P., Pointon J.J., Jin R., Zhou X., Bradbury L.A., Appleton L.H., et al. (2010) Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat. Genet., 42, 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoglinger G.U., Melhem N.M., Dickson D.W., Sleiman P.M., Wang L.S., Klei L., Rademakers R., de Silva R., Litvan I., Riley D.E., et al. (2011) Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat. Genet., 43, 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khor C.C., Davila S., Breunis W.B., Lee Y.C., Shimizu C., Wright V.J., Yeung R.S., Tan D.E., Sim K.S., Wang J.J., et al. (2011) Genome-wide association study identifies FCGR2A as a susceptibility locus for Kawasaki disease. Nat. Genet., 43, 1241–1246. [DOI] [PubMed] [Google Scholar]
- 32.Sawcer S., Hellenthal G., Pirinen M., Spencer C.C., Patsopoulos N.A., Moutsianas L., Dilthey A., Su Z., Freeman C., Hunt S.E., et al. (2011) Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature, 476, 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonnefond A., Skrobek B., Lobbens S., Eury E., Thuillier D., Cauchi S., Lantieri O., Balkau B., Riboli E., Marre M., et al. (2013) Association between large detectable clonal mosaicism and type 2 diabetes with vascular complications. Nat. Genet., 45, 1040–1043. [DOI] [PubMed] [Google Scholar]
- 34.Lee S., Wu M.C., Lin X. (2012) Optimal tests for rare variant effects in sequencing association studies. Biostatistics, 13, 762–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramdas W.D., van Koolwijk L.M., Ikram M.K., Jansonius N.M., de Jong P.T., Bergen A.A., Isaacs A., Amin N., Aulchenko Y.S., Wolfs R.C., et al. (2010) A genome-wide association study of optic disc parameters. PLoS Genet., 6, e1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khor C.C., Ramdas W.D., Vithana E.N., Cornes B.K., Sim X., Tay W.T., Saw S.M., Zheng Y., Lavanya R., Wu R., et al. (2011) Genome-wide association studies in Asians confirm the involvement of ATOH7 and TGFBR3, and further identify CARD10 as a novel locus influencing optic disc area. Hum. Mol. Genet., 20, 1864–1872. [DOI] [PubMed] [Google Scholar]
- 37.Springelkamp H., Höhn R., Mishra A., Hysi P.G., Khor C.C., Loomis S.J., Bailey J.N., Gibson J., Thorleifsson G., Janssen S.F., et al. (2015) Meta-analysis of genome-wide association studies identifies novel loci that influence cupping and the glaucomatous process. Nat. Commun., 5, 4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garway-Heath D.F., Ruben S.T., Viswanathan A., Hitchings R.A. (1998) Vertical cup/disc ratio in relation to optic disc size: its value in the assessment of the glaucoma suspect. Br. J. Ophthalmol., 82, 1118–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Y., Vitart V., Burdon K.P., Khor C.C., Bykhovskaya Y., Mirshahi A., Hewitt A.W., Koehn D., Hysi P.G., Ramdas W.D., et al. (2013) Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat. Genet., 45, 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuchshofer R. (2011) The pathogenic role of transforming growth factor-beta2 in glaucomatous damage to the optic nerve head. Exp. Eye Res., 93, 165–169. [DOI] [PubMed] [Google Scholar]
- 41.Pasquale L.R., Dorman-Pease M.E., Lutty G.A., Quigley H.A., Jampel H.D. (1993) Immunolocalization of TGF-beta 1, TGF-beta 2, and TGF-beta 3 in the anterior segment of the human eye. Invest. Ophthalmol. Visual Sci., 34, 23–30. [PubMed] [Google Scholar]
- 42.Pena J.D., Taylor A.W., Ricard C.S., Vidal I., Hernandez M.R. (1999) Transforming growth factor beta isoforms in human optic nerve heads. Br. J. Ophthalmol., 83, 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Massague J., Blain S.W., Lo R.S. (2000) TGFbeta signaling in growth control, cancer, and heritable disorders. Cell, 103, 295–309. [DOI] [PubMed] [Google Scholar]
- 44.Hannon G.J., Beach D. (1994) p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature, 371, 257–261. [DOI] [PubMed] [Google Scholar]
- 45.Pasmant E., Sabbagh A., Vidaud M., Bieche I. (2011) ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J., 25, 444–448. [DOI] [PubMed] [Google Scholar]
- 46.Blobe G.C., Liu X., Fang S.J., How T., Lodish H.F. (2001) A novel mechanism for regulating transforming growth factor beta (TGF-beta) signaling. Functional modulation of type III TGF-beta receptor expression through interaction with the PDZ domain protein, GIPC. J. Biol. Chem., 276, 39608–39617. [DOI] [PubMed] [Google Scholar]
- 47.Diestel U., Resch M., Meinhardt K., Weiler S., Hellmann T.V., Mueller T.D., Nickel J., Eichler J., Muller Y.A. (2013) Identification of a novel TGF-beta-binding site in the Zona Pellucida C-terminal (ZP-C) domain of TGF-beta-receptor-3 (TGFR-3). PLoS ONE, 8, e67214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai C., Rajaram M., Zhou X., Liu Q., Marchica J., Li J., Powers R.S. (2012) Activation of multiple cancer pathways and tumor maintenance function of the 3q amplified oncogene FNDC3B. Cell Cycle (Georgetown, Tex), 11, 1773–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet., 38, 904–909. [DOI] [PubMed] [Google Scholar]
- 50.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrett J.C., Fry B., Maller J., Daly M.J. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics (Oxford, England), 21, 263–265. [DOI] [PubMed] [Google Scholar]
- 52.Ihaka R., Gentleman R. (1996) R: a language for data analysis and graphics. J. Comput. Graph. Stat., 5, 299–314. [Google Scholar]
- 53.Nalls M.A., Plagnol V., Hernandez D.G., Sharma M., Sheerin U.M., Saad M., Simon-Sanchez J., Schulte C., Lesage S., Sveinbjornsdottir S., et al. (2011) Imputation of sequence variants for identification of genetic risks for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet, 377, 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vithana E.N., Aung T., Khor C.C., Cornes B.K., Tay W.T., Sim X., Lavanya R., Wu R., Zheng Y., Hibberd M.L., et al. (2011) Collagen-related genes influence the glaucoma risk factor, central corneal thickness. Hum. Mol. Genet., 20, 649–658. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.