Abstract
The respiratory syncytial virus (RSV) fusion (F) glycoprotein prefusion conformation is the target of most RSV-neutralizing activity in human sera, but its metastability has hindered characterization. To overcome this obstacle, we identified prefusion-specific antibodies which were substantially more potent than the prophylactic antibody palivizumab. The co-crystal structure for one of these antibodies, D25, in complex with the F glycoprotein revealed that D25 locks F in its prefusion state by binding to a quaternary epitope at the trimer apex. Electron microscopy showed two other antibodies, AM22 and 5C4, also bind to the newly identified site of vulnerability, which we named antigenic site Ø. These studies should enable design of improved vaccine antigens and guide new approaches for passive prevention of RSV-induced disease.
Respiratory syncytial virus (RSV) infects nearly all children by 3 years of age (1), and is a leading cause of infant hospitalization and childhood wheezing (2, 3). Globally, RSV accounts for 6.7% of deaths among infants 1 month to 1 year old—more than any other single pathogen except malaria (4). The only intervention is passive administration of the licensed monoclonal antibody palivizumab (Synagis®), which recognizes the RSV fusion (F) glycoprotein (5, 6) and reduces incidence of severe disease (7). Clinical evidence that RSV F-specific antibodies can protect against disease has prompted a search for better antibodies (8–10) and a concerted effort to develop an effective vaccine (11).
RSV F is a type I fusion protein (12) that rearranges from a metastable prefusion conformation to a highly stable postfusion structure. Three previously described antigenic sites (I, II, and IV) associated with neutralizing activity (13–15) exist on the postfusion form of F (16, 17). Absorption of human sera with postfusion F, however, fails to remove the majority of F-specific neutralizing activity, suggesting there are neutralizing antigenic sites unique to the prefusion form (18). Thus, determining the prefusion RSV F structure and identifying antibodies that bind prefusion-specific antigenic sites have become converging priorities for developing new antibodies and vaccines to prevent RSV infection.
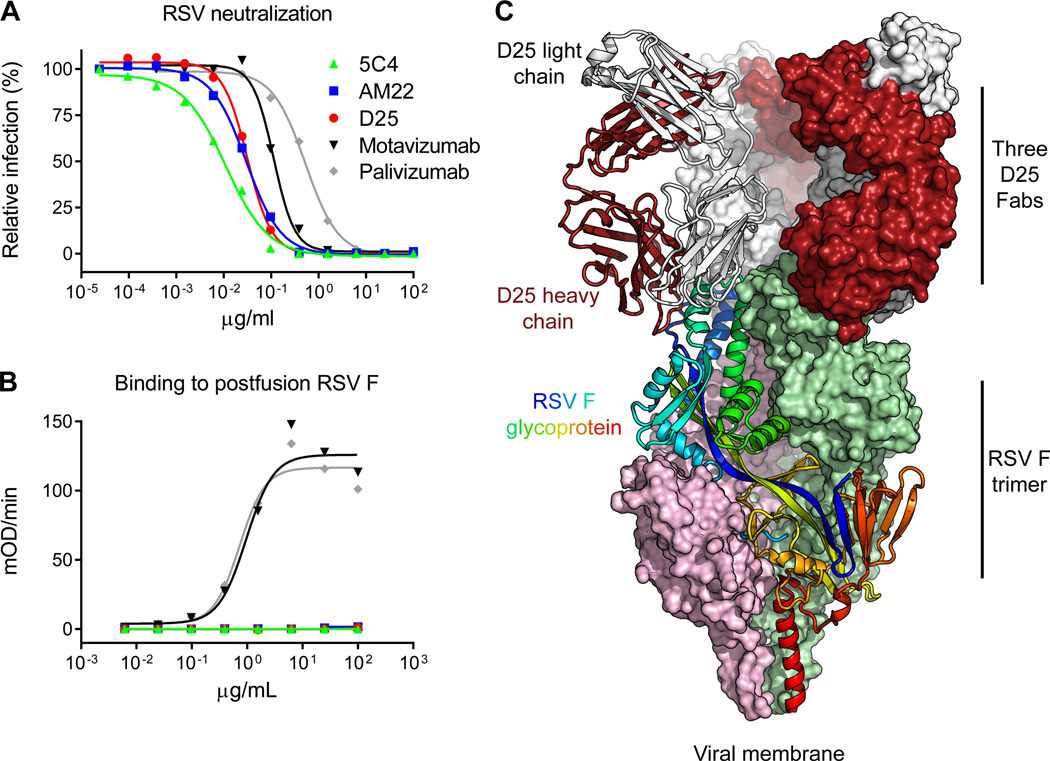
From mice immunized with gene-based vectors expressing the F protein (19), we isolated an antibody, 5C4, that was 50-fold more potent than palivizumab (Fig. 1A) and did not bind to a soluble form of RSV F stabilized in the postfusion conformation (16) (Fig. 1B). We determined that 5C4 shared these properties with two recently isolated human antibodies, D25 and AM22 (10, 20, 21) (Fig. 1A, B), and we hypothesized that these antibodies recognized the metastable prefusion conformation (22).
Figure 1. RSV neutralization, F glycoprotein recognition, and crystal structure of human antibody D25 in complex with the prefusion RSV F trimer.
The prefusion conformation of RSV F is metastable, and when expressed in a soluble form readily adopts the postfusion state; a number of potent antibodies, including D25, bind to a newly revealed antigenic site at the top of the prefusion F glycoprotein. (A) RSV neutralization by antibodies. Palivizumab is the FDA-approved prophylactic antibody that prevents severe RSV disease. (B) ELISA measuring antibody binding to postfusion F glycoprotein. For (A) and (B), data are representative of multiple independent experiments. (C) D25-RSV F trimer crystal structure in ribbon and molecular surface representations. One protomer of the F glycoprotein trimer is shown as ribbons and colored as a rainbow from blue to red, N-terminus of F2 to C-terminus of F1, respectively. Molecular surfaces are shown for the other two F protomers, colored pink and green. The D25 Fab bound to the F protomer shown in ribbons is also displayed in ribbon representation, with heavy chain colored red and light chain colored grey. The other D25 Fabs are colored the same, but shown in surface representation.
We focused our structural efforts on the human antibodies by first screening their binding to a panel of RSV F glycoprotein variants (23). We observed D25 and AM22 antibody binding to a construct, RSV F(+) Fd, comprising residues 1-513 fused to a C-terminal fibritin trimerization domain (24). However, we failed to form complexes by mixing purified RSV F(+) Fd with purified D25 or AM22, suggesting F was triggered during purification (25). To capture F in its prefusion state, RSV F(+) Fd was expressed as a complex with D25. Optimal expression was obtained from cotransfection of DNA encoding D25 Fab with DNA encoding RSV F(+) Fd (fig. S1). X-ray diffraction data to 3.6 Å resolution were obtained on crystals of this complex, and the structure was solved by molecular replacement using the unbound D25 Fab structure (table S1) and portions of the postfusion RSV F structure (16, 17) as search models. The structure was refined to Rcryst/Rfree of 21.3/26.7% (Fig. 1C) (table S1).
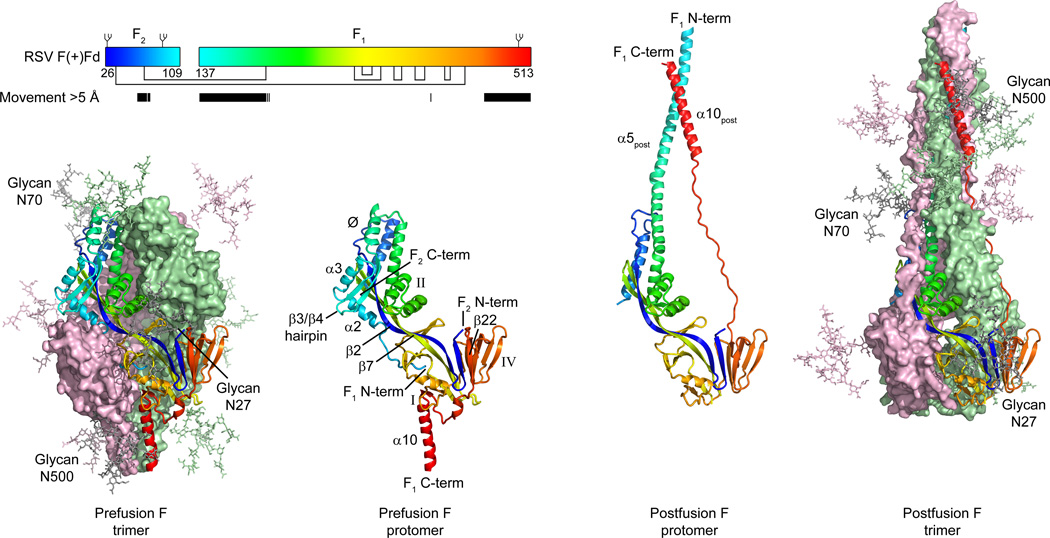
The D25-bound RSV F structure resembled the prefusion structure of the related parainfluenza virus 5 (PIV5) F glycoprotein (26, 27), indicating that D25-binding stabilizes RSV F in the prefusion conformation (fig. S2). Comparison with the postfusion RSV F glycoprotein structure (16, 17) revealed that the majority of secondary and tertiary structure was preserved in both pre- and postfusion states, with 215 residues showing less than 2 Å Cα deviation between the two structures (Fig. 2). In contrast, regions at the N- and C-termini of the F1 subunit showed dramatic conformational changes. The fusion peptide, located at the N-terminus of F1, and five secondary structure elements (α2, α3, α4 and the β3/β4 hairpin) rearrange and fuse with the α5-helix to form a single extended postfusion helix (α5post) of over 100 Å in length (fig. S3). At the C-terminus of F1, the sole parallel strand (β22) unravels, allowing the prefusion α10-helix to move towards the α5post-helix. Similar rearrangements were observed in the comparison of prefusion PIV5 and postfusion parainfluenza virus 3 (PIV3) F glycoprotein structures (27), indicating that F glycoproteins from both the Paramyxovirinae (PIV5/PIV3) and Pneumovirinae (RSV) subfamilies undergo similar conformational rearrangements to facilitate membrane fusion.
Figure 2. Structural rearrangement of RSV F.
To mediate virus-cell entry, the RSV F glycoprotein transitions from a metastable prefusion conformation to a stable postfusion conformation. Outer images display prefusion (left) and postfusion (right) trimeric structures, colored the same as in Fig. 1C. A complex glycan, shown as sticks, is modeled at each of the three N-linked glycosylation sites found in the mature protein. Inner images display a single RSV F protomer in ribbon representation, colored as a rainbow from blue to red, N-terminus of F2 to C-terminus of F1, respectively. Select secondary structure elements are labeled (correspondence with amino acid sequence in pre- and postfusion conformations is shown in fig. S3). Inset: schematic of the mature RSV F protein in the RSV F(+) Fd construct. The rainbow coloring of the boxes representing the F2 and F1 subunits matches that in the structures. Glycans are shown as branches on top of the boxes, and disulfide bonds are shown as black lines under the boxes. Amino acids that move more than 5 Å in the pre- and post-fusion conformations are indicated by black bars.
Despite overall similarities between the PIV5 and RSV F prefusion structures, there are distinct differences important for function and antigenicity. In the prefusion RSV F structure, the helix-loop-helix (α6 and α7) motif that constitutes antigenic site II is located further away from the 3-fold trimer axis than is the homologous region in PIV5 F (fig. S4). This repositioning exposes the face of the helix-loop-helix bound by palivizumab and motavizumab, allowing these antibodies to bind prefusion RSV F without first requiring a conformational rearrangement to occur as originally suggested based on modeling using the PIV5 F structure (28).
Another difference is that the RSV F fusion peptide is buried in the center of the trimer cavity, with its N-terminus located more than 40 Å away from the last visible F2 residue (Fig. 2). In contrast, the PIV5 F fusion peptide lies in a surface groove between subunits, is partially exposed to solvent (27), and undergoes minimal movement after protease cleavage (26). This suggests that in RSV F, a substantial structural rearrangement of the fusion peptide occurs after the F0 precursor is cleaved by the furin-like host protease to produce F1 and F2 subunits (29). In comparison to other structures of cleaved, type I fusion proteins (26, 30, 31), the location of the RSV F fusion peptide is most similar to that of influenza hemagglutinin (31) (fig. S5), which is surprising given that hemagglutinin is triggered by acidic pH in the endosome, whereas RSV F triggering is pH-independent and thought to occur at the cell surface (32).
In the RSV F prefusion trimer, the C-terminal helices (α10) form an inverted pyramid (fig S4). In contrast, the C-terminal helices in the PIV5 F trimer form a coiled-coil, which was stabilized by a coiled-coil GCN4-trimerization motif (27). The addition of this motif was sufficient to stabilize PIV5 and human metapneumovirus (hMPV) F proteins in the prefusion state (27, 33). The inverted pyramid conformation observed in the RSV F structure may explain why RSV F could not be expressed with the GCN4 motif, but could be expressed with the fibritin trimerization domain (24). The addition of the fibritin domain was not, however, sufficient to stabilize RSV F in the prefusion state, suggesting that it is not an optimal substitute for the native transmembrane domains that normally stabilize F in the viral membrane (34). The binding of antibody D25 was thus required to stabilize the prefusion trimer.
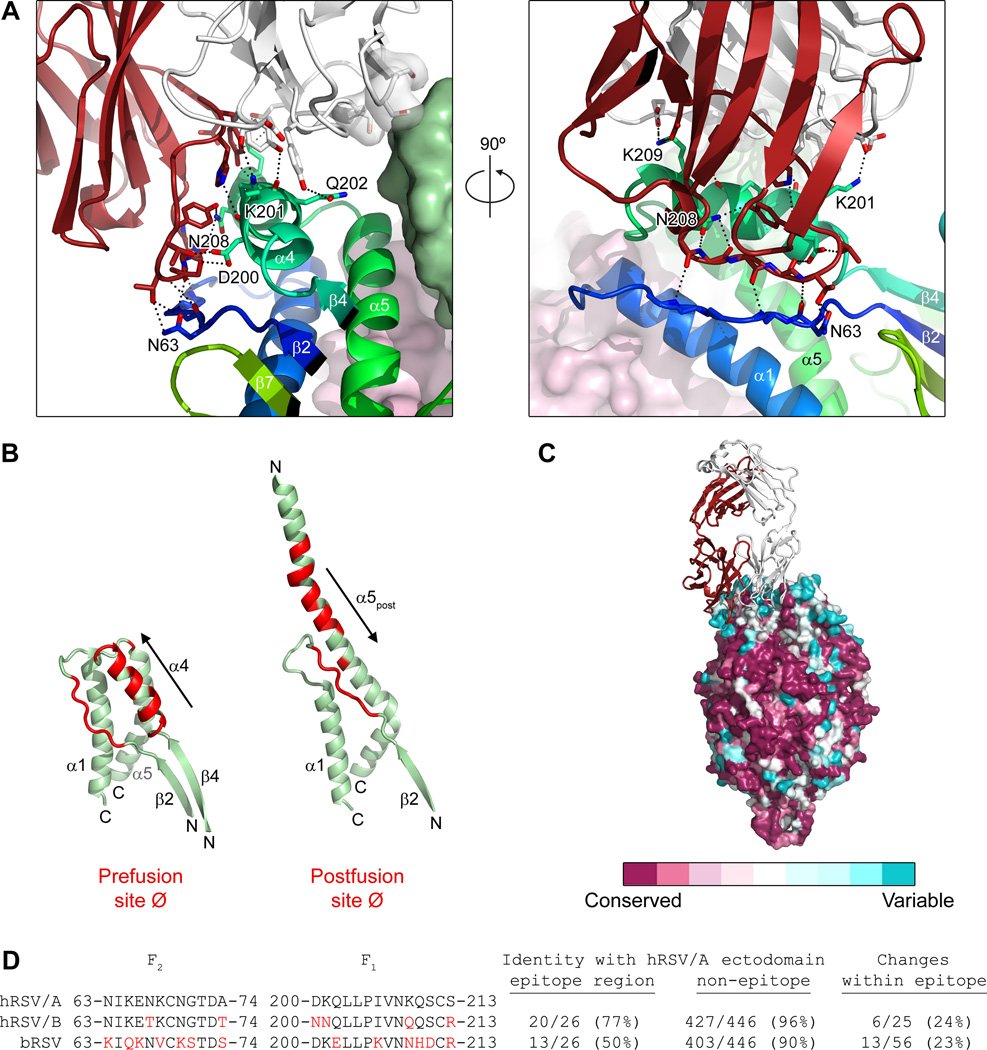
D25 recognizes a quaternary epitope at the membrane-distal apex of the RSV F glycoprotein (Fig. 1C). The D25-heavy chain interacts with one protomer (involving 638 Å2 of interactive-surface area on F) and the D25-light chain binds to the same protomer (373 Å2) and a neighboring protomer (112 Å2) (Fig. 3A). D25 contacts RSV F with 5 of its 6 complementarity-determining regions (CDR), with the heavy chain 3rd CDR interacting with the α4-helix (F1 residues 196-210) and forming intermolecular hydrogen bonds with F2 residues 63, 65, 66 and 68 in the loop between strand β2 and helix α1. Whereas the secondary structure of the D25 epitope remains mostly unchanged in pre- and postfusion conformations, the tertiary structure changes substantially, with helix α4 pivoting ~180° relative to strand β2 (Fig. 3B). This explains the failure of D25 to bind postfusion F and suggests D25 neutralizes RSV by fixing F in its prefusion conformation.
Figure 3. RSV F interface with D25.
Antibody D25 binds a quaternary epitope spanning two protomers at the apex of the prefusion F trimer. (A) Close-up of the interface between D25 and RSV F. Side chains of F residues interacting with D25 are labeled and shown as sticks. Oxygen atoms are colored red and nitrogen atoms are colored blue. Hydrogen bonds are depicted as dotted lines. The two images are related by a 90° rotation about the vertical axis. (B) Position and conformation of the D25 epitope on the prefusion and postfusion F molecules. RSV F residues at the D25 interface are colored red; polarity of α4 and α5post indicated with arrows, with fragment N- and C-termini indicated. (C) Surface representation of RSV F colored according to sequence conservation. Coloring was generated using the ConSurf server (40) using 178 F glycoprotein sequences from human RSV subtype A and B, as well as bovine RSV. (D) Sequence conservation of F residues in regions recognized by D25. Amino acids in human RSV subtype B (hRSV/B) or in bovine RSV (bRSV) that differ from hRSV/A are colored red. Ectodomain is defined as F residues 26-109 and 137-524.
D25 binds the least conserved region on the F glycoprotein (Fig. 3C). Although F proteins from human RSV A and B subtypes are highly related in sequence (447/472 or 94.7% of the amino acids comprising the mature F2/F1 ectodomain are identical), six naturally observed sequence variations are located in the region bound by D25 (Fig. 3D). Similarly, of the 56 amino acids in bovine RSV F that are not identical to the mature ectodomain of human RSV F subtype A, 13 are found in this same region (Fig 3D). Thus, the D25 epitope, at the apex of the prefusion RSV F structure, may be under immune pressure and serve as a determinant of subtype-specific immunity (35, 36).
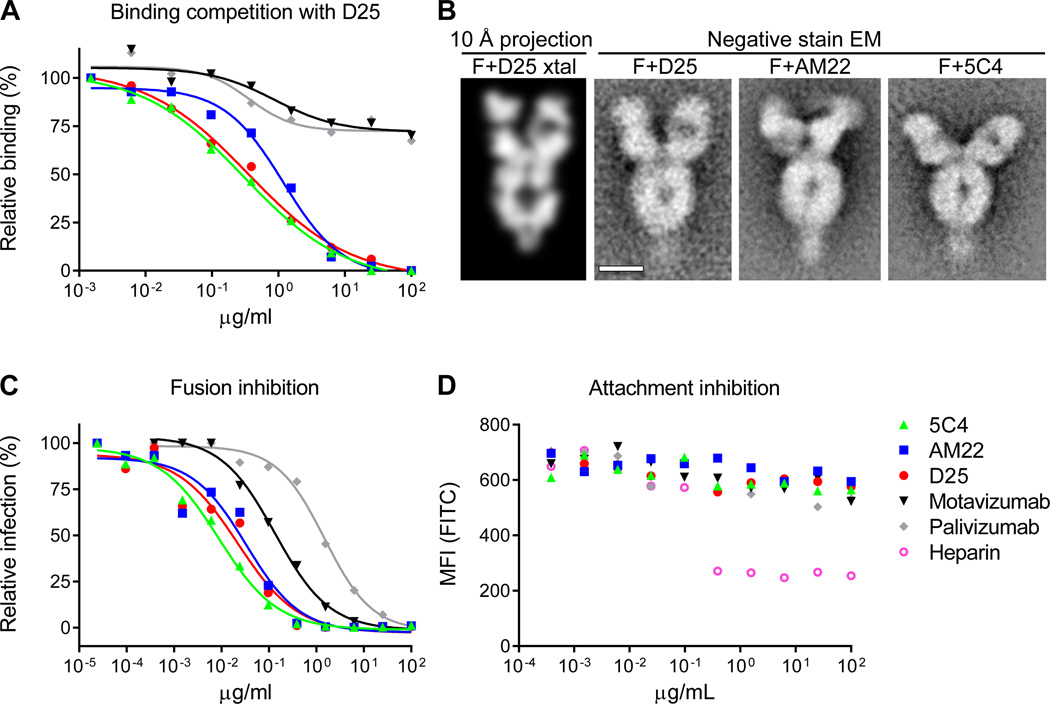
To investigate whether AM22 and 5C4 bind the D25 epitope, competition binding and electron microscopy (EM) experiments were performed. We determined that D25 binding to RSV-infected cells was competed by AM22 and 5C4 (Fig. 4A). In addition, negative-stain EM images of Fab D25-RSV F complexes resembled the EM images of Fab AM22-RSV F and Fab 5C4-RSV F complexes (Fig. 4B). Together, these results suggest antibodies D25, AM22 and 5C4 recognize the same or a highly related epitope, which we named “antigenic site Ø”. In addition to their extraordinary potency and prefusion-specificity (Fig. 1A), all three antibodies strongly inhibited fusion when added post-attachment (Fig. 4C), and all three were unable to block cell-surface attachment (Fig. 4D), suggesting that the putative RSV F receptor binds to a region on F not blocked by these antibodies. AM22 and D25 antibodies also neutralized similarly in both Fab and immunoglobulin contexts (fig. S6), indicating that avidity was not required for potent neutralization, in contrast to what was previously observed for some influenza-virus antibodies (37). Thus, antigenic site Ø-specific antibodies are distinct from neutralizing antibodies to other known antigenic sites on RSV F because of their exclusive recognition of the prefusion F structure and extremely high potency (fig. S7).
Figure 4. Antigenic site Ø.
Highly effective RSV-neutralizing antibodies target a site at the membrane-distal apex of the prefusion F trimer. (A) The ability of antibodies to block D25 binding to RSV-infected cells was measured as a function of antibody concentration. (B) Analysis of RSV F-Fab complexes by negative stain electron microscopy: Reprojection of a 12 Å slice through the crystal structure of RSV F + D25 Fab filtered to 10 Å resolution (left). A slice was used to emphasize visibility of the F glycoprotein cavity. Aligned average of 263 particles of RSV F + D25 Fab (middle - left). Aligned average of 550 particles of RSV F + AM22 Fab (middle - right). Aligned average of 171 particles of RSV F + 5C4 Fab (right). Scale bar is 50 Å. (C) Fusion inhibition and (D) attachment inhibition activity for antibodies targeting antigenic site Ø and F-specific antibodies targeting other antigenic sites. For the attachment-inhibition assay, heparin was used as a positive control. Data in panels (A) (C) and (D) are representative of multiple independent experiments.
Antigenic site Ø, located at the apex of the prefusion F trimer, should be readily accessible even on the crowded virion surface, which may explain the observation that most neutralizing activity in human sera induced by natural RSV infection is directed against the prefusion form of RSV F (18). That the three antibodies described in this paper recognize antigenic site Ø at different angles (Fig. 4B) and share low sequence homology suggests there may be many other antibodies within the repertoire capable of recognizing this prominent epitope. The high potency of antibodies against antigenic site Ø suggests they could be developed for passive prophylaxis of RSV-induced disease in neonates. Importantly, the immunogen-elicited 5C4 antibody provides proof-of-principle that antibodies against antigenic site Ø can be induced by gene-based immunization presenting the native form of F. Vaccine antigens stabilized in the prefusion conformation, perhaps facilitated by linking mobile and immobile portions of F with disulfide bonds, may further improve elicitation of antibodies to antigenic site Ø. Definition of the D25-RSV F structure thus provides the basis for new approaches to prevent RSV-induced disease.
Supplementary Material
Acknowledgments
The authors would like to thank members of the Structural Biology Section, Structural Bioinformatics Core Section, and Viral Pathogenesis Laboratory at the Vaccine Research Center for helpful comments, and the staff at SER-CAT (Southeast Regional Collaborative Access Team) for help with X-ray diffraction data collection. The data presented in this manuscript are tabulated in the main paper and the supplementary materials. Atomic coordinates and structure factors of the reported crystal structures have been deposited in the Protein Data Bank under accession codes 4JHA and 4JHW. Support for this work was provided by the Intramural Research Program (National Institute of Allergy and Infectious Diseases). Use of insertion device 22 (SER-CAT) at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract W-31-109-Eng-38.
Footnotes
References and Notes
- 1.Glezen W, Taber L, Frank A, Kasel J. Risk of primary infection and reinfection with respiratory syncytial virus. Am. J. Dis. Child. 1986;140:543. doi: 10.1001/archpedi.1986.02140200053026. [DOI] [PubMed] [Google Scholar]
- 2.Shay DK, et al. Bronchiolitis-Associated Hospitalizations Among US Children, 1980–1996. JAMA. 1999;282:1440. doi: 10.1001/jama.282.15.1440. [DOI] [PubMed] [Google Scholar]
- 3.Hall CB, et al. The Burden of Respiratory Syncytial Virus Infection in Young Children. N. Engl. J. Med. 2009;360:588. doi: 10.1056/NEJMoa0804877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lozano R, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2013;380:2095. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson S, et al. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J. Infect. Dis. 1997;176:1215. doi: 10.1086/514115. [DOI] [PubMed] [Google Scholar]
- 6.Beeler JA, van Wyke Coelingh K. Neutralization epitopes of the F glycoprotein of respiratory syncytial virus: effect of mutation upon fusion function. J. Virol. 1989;63:2941. doi: 10.1128/jvi.63.7.2941-2950.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The IMpact-RSV Study Group. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics. 1998;102:531. [PubMed] [Google Scholar]
- 8.Collarini EJ, et al. Potent High-Affinity Antibodies for Treatment and Prophylaxis of Respiratory Syncytial Virus Derived from B Cells of Infected Patients. J. Immunol. 2009;183:6338. doi: 10.4049/jimmunol.0901373. [DOI] [PubMed] [Google Scholar]
- 9.Wu H, et al. Development of motavizumab, an ultra-potent antibody for the prevention of respiratory syncytial virus infection in the upper and lower respiratory tract. J. Mol. Biol. 2007;368:652. doi: 10.1016/j.jmb.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 10.Kwakkenbos MJ, et al. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat. Med. 2010;16:123. doi: 10.1038/nm.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham BS. Biological challenges and technological opportunities for respiratory syncytial virus vaccine development. Immunol. Rev. 2011;239:149. doi: 10.1111/j.1600-065X.2010.00972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh EE, Hruska J. Monoclonal antibodies to respiratory syncytial virus proteins: identification of the fusion protein. J. Virol. 1983;47:171. doi: 10.1128/jvi.47.1.171-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arbiza J, et al. Characterization of two antigenic sites recognized by neutralizing monoclonal antibodies directed against the fusion glycoprotein of human respiratory syncytial virus. J. Gen. Virol. 1992;73:2225. doi: 10.1099/0022-1317-73-9-2225. [DOI] [PubMed] [Google Scholar]
- 14.Lopez JA, et al. Antigenic structure of human respiratory syncytial virus fusion glycoprotein. J. Virol. 1998;72:6922. doi: 10.1128/jvi.72.8.6922-6928.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.López JA, Peñas C, García-Barreno B, Melero JA, Portela A. Location of a highly conserved neutralizing epitope in the F glycoprotein of human respiratory syncytial virus. J. Virol. 1990;64:927. doi: 10.1128/jvi.64.2.927-930.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLellan JS, Yang Y, Graham BS, Kwong PD. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J. Virol. 2011;85:7788. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swanson KA, et al. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9619. doi: 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magro M, et al. Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc. Natl. Acad. Sci. U.S.A. 2012;109:3089. doi: 10.1073/pnas.1115941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Information on materials and methods is available on Science Online.
- 20.Spits H, Beaumont T. RSV-specific binding molecules and means for producing them. Patent Application 12/600,950. 2010
- 21.Beaumont T, Bakker AQ, Yasuda E. RSV specific binding molecule. Patent Application 12/898,325. 2012
- 22.Approximately 100-fold greater potency of AM22 and D25 relative to palivizumab are observed by Beaumont and colleagues in a plaque assay; in our flow-based neutralization assay, we observed a 15-fold improvement over palivizumab and a 3-fold improvement over motavizumab.
- 23.Pancera M, et al. N332-Directed Broadly Neutralizing Antibodies Use Diverse Modes of HIV-1 Recognition: Inferences from Heavy-Light Chain Complementation of Function. PLoS ONE. 2013;8:e55701. doi: 10.1371/journal.pone.0055701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank S, et al. Stabilization of short collagen-like triple helices by protein engineering. J. Mol. Biol. 2001;308:1081. doi: 10.1006/jmbi.2001.4644. [DOI] [PubMed] [Google Scholar]
- 25.Chaiwatpongsakorn S, Epand RF, Collins PL, Epand RM, Peeples ME. Soluble respiratory syncytial virus fusion protein in the fully cleaved, pretriggered state is triggered by exposure to low-molarity buffer. J. Virol. 2011;85:3968. doi: 10.1128/JVI.01813-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welch BD, et al. Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16672. doi: 10.1073/pnas.1213802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLellan JS, et al. Structural basis of respiratory syncytial virus neutralization by motavizumab. Nat. Struct. Mol. Biol. 2010;17:248. doi: 10.1038/nsmb.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.RSV F has the shortest fusion peptide of all paramyxoviruses, which may allow it to move from an initially exposed position to the central cavity of the trimer.
- 30.Lee JE, et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 32.The fusion peptide is connected to the surface-exposed α2 and α3 helices through a cylindrical opening between the protomers that is roughly 10 Å in diameter; this opening may be used as an exit path for the fusion peptide during triggering.
- 33.Wen X, et al. Structure of the human metapneumovirus fusion protein with neutralizing antibody identifies a pneumovirus antigenic site. Nat. Struct. Mol. Biol. 2012;19:461. doi: 10.1038/nsmb.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.“Breathing” of the F1 C-terminus could potentially trigger the F protein because the region near residue 461 is in contact with the fusion peptide of a neighboring protomer. During the transition to the postfusion state, F1 residues 461 to 513 move at least 8 Å, which could destabilize the fusion peptide and lead to triggering.
- 35.Chambers P, Pringle CR, Easton AJ. Sequence analysis of the gene encoding the fusion glycoprotein of pneumonia virus of mice suggests possible conserved secondary structure elements in paramyxovirus fusion glycoproteins. J. Gen. Virol. 1992;73:1717. doi: 10.1099/0022-1317-73-7-1717. [DOI] [PubMed] [Google Scholar]
- 36.Based on sequence analysis, a loop region in F glycoproteins was hypothesized to exist within the Paramyxoviridae family that might be under immune pressure (35). It has been demonstrated that binding of RSV sub-group specific monoclonal antibodies can be affected by site-directed mutations between F1 residues 200 and 216 (38), and that a peptide comprising F1 residues 205–225 could elicit neutralizing activity in rabbits, although a specific epitope was not defined (39).
- 37.Ekiert DC, et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489:526. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Connor AL, Bevitt DJ, Toms GL. Comparison of human respiratory syncytial virus A2 and 8/60 fusion glycoprotein gene sequences and mapping of sub-group specific antibody epitopes. J. Med. Virol. 2001;63:168. [PubMed] [Google Scholar]
- 39.Corvaisier C, Bourgeois C, Pothier P. Cross-reactive and group-specific immune responses to a neutralizing epitope of the human respiratory syncytial virus fusion protein. Arch. Virol. 1997;142:1073. doi: 10.1007/s007050050143. [DOI] [PubMed] [Google Scholar]
- 40.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucl. Acids Res. 2010;38:W529. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graham BS, Perkins MD, Wright PF, Karzon DT. Primary respiratory syncytial virus infection in mice. J. Med. Virol. 1988;26:153. doi: 10.1002/jmv.1890260207. [DOI] [PubMed] [Google Scholar]
- 42.Hallak LK, Collins PL, Knudson W, Peeples ME. Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology. 2000;271:264. doi: 10.1006/viro.2000.0293. [DOI] [PubMed] [Google Scholar]
- 43.Harlow E, Lane DP, et al. Antibodies: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 44.Chen M, et al. A flow cytometry-based assay to assess RSV-specific neutralizing antibody is reproducible, efficient and accurate. J Immunol Methods. 2010;362:180. doi: 10.1016/j.jim.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McLellan JS, et al. Design and characterization of epitope-scaffold immunogens that present the motavizumab epitope from respiratory syncytial virus. J. Mol. Biol. 2011;409:853. doi: 10.1016/j.jmb.2011.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otwinowski Z, Minor W. Methods Enzymol. Vol. 276. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 47.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen L, et al. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326:1123. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62:1243. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 53.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13419. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Majeed S, et al. Enhancing protein crystallization through precipitant synergy. Structure. 2003;11:1061. doi: 10.1016/s0969-2126(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 55.Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J. Struct. Biol. 2007;157:3. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 56.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 57.McLellan JS, et al. Structure of a major antigenic site on the respiratory syncytial virus fusion glycoprotein in complex with neutralizing antibody 101F. J. Virol. 2010;84:12236. doi: 10.1128/JVI.01579-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Magro M, Andreu D, Gomez-Puertas P, Melero JA, Palomo C. Neutralization of human respiratory syncytial virus infectivity by antibodies and low-molecular-weight compounds targeted against the fusion glycoprotein. J. Virol. 2010;84:7970. doi: 10.1128/JVI.00447-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.