Abstract
In many animal cells, stimulation of cell surface receptors coupled to G proteins or tyrosine kinases mobilizes Ca2+ influx through store-operated Ca2+ release-activated Ca2+ (CRAC) channels. The ensuing Ca2+ entry regulates a wide variety of effector cell responses including transcription, motility, and proliferation. The physiological importance of CRAC channels for human health is underscored by studies indicating that mutations in CRAC channel genes produce a spectrum of devastating diseases including chronic inflammation, muscle weakness, and a severe combined immunodeficiency syndrome. Moreover, from a basic science perspective, CRAC channels exhibit a unique biophysical fingerprint characterized by exquisite Ca2+-selectivity, store-operated gating, and distinct pore properties and therefore serve as fascinating ion channels for understanding the biophysical mechanisms of ion permeation and gating. Studies in the last two decades have revealed the cellular and molecular choreography of the CRAC channel activation process, and it is now established that opening of CRAC channels is governed through direct interactions between the pore-forming Orai proteins, and the ER Ca2+ sensors, STIM1 and STIM2. In this review, we summarize the functional and structural mechanisms of CRAC channel regulation, focusing on recent advances in our understanding of the conformational and structural dynamics of CRAC channel gating.
Keywords: Calcium, CRAC channel, STIM1, Orai1, SOCE
Introduction
Calcium (Ca2+) is a multifunctional signaling ion that regulates a wide range of biological processes including neurotransmitter release, muscle contraction, and regulation of enzymes. Mobilization of cellular Ca2+ elevation in response to receptor stimulation commonly occurs through release of Ca2+ ions from intracellular Ca2+ stores or influx across the plasma membrane (PM) through Ca2+-permeable ion channels. Among these pathways, store-operated Ca2+ entry (SOCE), so named for its regulation by the free [Ca2+] of the endoplasmic reticulum (ER) Ca2+ stores, is a widespread Ca2+ entry mechanism in animal cells that delivers Ca2+ to refill ER stores and evoke cellular Ca2+ signals. These Ca2+ signals in turn regulate a wide range of effector functions including gene expression, proliferation, and cytokine release 1; 2. The best known and most extensively characterized store-operated channels are the Ca2+-release-activated-Ca2+ (CRAC) channels. Endowed with several distinguishing biophysical features including exquisite Ca2+ selectivity and a low unitary conductance 3, CRAC channels are assembled from two protein families: the Orai proteins, which form the ion channel pore, and the stromal interaction molecule (STIM) proteins, which function as ER calcium sensors and activators of the CRAC channel. The importance of CRAC channels for human health is underscored by a growing list of genetic studies that have revealed that patients deficient in STIM1 or Orai1 or who bear loss- or gain-of-function STIM1/Orai1 mutations suffer from serious health issues including immunodeficiency, autoimmunity, muscle defects, and bleeding disorders 4. In this review, we will present an overview of the structural underpinnings of CRAC channel ion conduction and activation mechanisms, and discuss molecules implicated in the regulation of these channels.
Store-operated Ca2+ entry
In the 1980s, a number of calcium imaging studies identified a link between inositol 1,4,5-triphosphate (IP3) production and increased Ca2+ influx through the plasma membrane although the exact mechanism that linked the two events was unclear 5; 6; 7; 8. In 1986, Putney proposed what was then an unconventional idea that IP3 mobilizes Ca2+ from intracellular stores which then activates plasma membrane Ca2+ influx pathway 9. Although a number years would pass before anyone understood the full details of this process, many studies in this period provided a rigorous biophysical description of CRAC channels and the mechanisms underlying regulation of SOCE. An important advance during this time was the electrophysiological identification of a small (a few pA), voltage-insensitive Ca2+ current which developed concurrently with a rise in intracellular calcium concentrations in response to intracellular dialysis of IP3 6; 7; 10; 11; 12. As described below, subsequent investigations provided the link between this unique Ca2+ current and store-depletion.
A central development that connected the plasma membrane Ca2+ influx pathway to the ER Ca2+ stores was the discovery of the plant lactone, thapsigargin (TG), which studies had shown was a selective inhibitor of the sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) localized in the ER membrane 13; 14; 15. Thapsigargin permitted experimental depletion of ER Ca2+ stores without receptor stimulation, in essence by-passing the generation of IP3 14; 15. With this tool, Putney and co-workers then showed that ER Ca2+ store release by TG triggered PM Ca2+ entry and that this process was the same as the Ca2+ influx caused by IP3 13. In a subsequent landmark study, Hoth and Penner 16; 17 coined the term ICRAC to describe the highly selective, inward-rectifying Ca2+ current across the PM resulting from depletion of ER stores in mast cells. Likewise, Zweifach and Lewis 18 identified a similar current activated by thapsigargin-mediated depletion of ER Ca2+ stores in T-cells. Further, these investigators found that the thapsigargin (TG) -activated current was identical to the Ca2+ current seen following T-cell stimulation, paving the way for understanding the physiological significance of CRAC channels for T-cell function.
Molecular components of the CRAC channel pathway
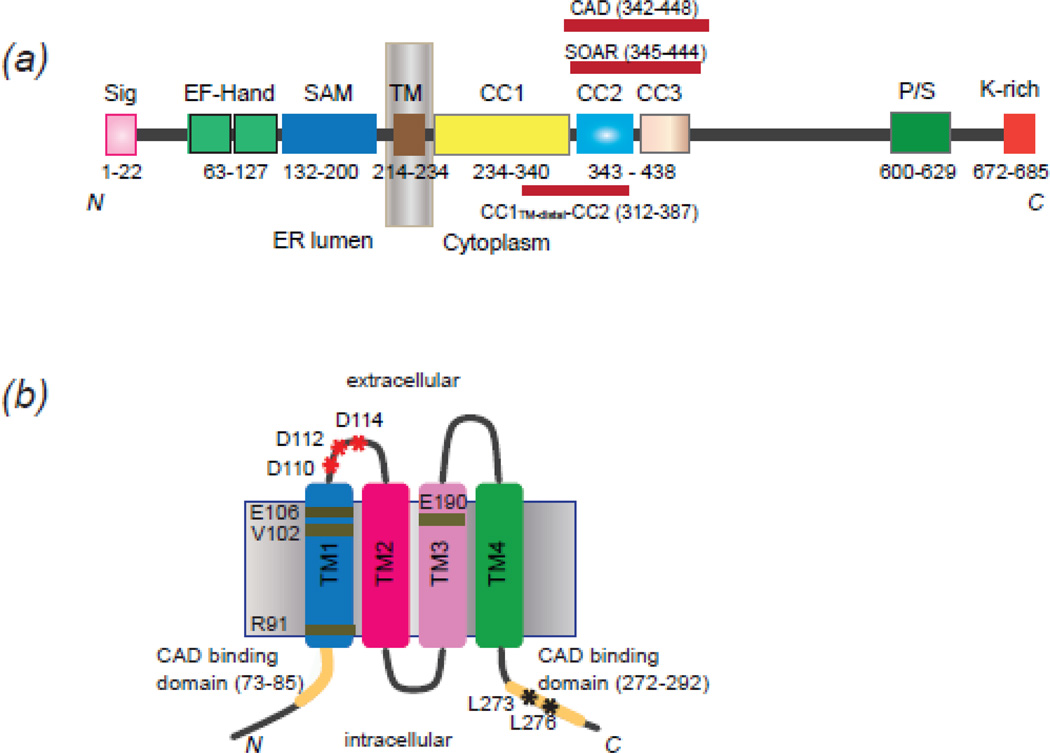
It would take another decade after the characterization of ICRAC for the molecules mediating this Ca2+influx to be identified. First, came the identification of STIM1 in 2005 using RNAi screens as a regulator of store-operated Ca2+ entry in Drosophila and HeLa cells 19; 20. STIM1 encodes a type I single pass membrane protein of 77-kDa, localized primarily in the ER membrane (Fig 1A). The ER luminal residing portion contains an EF-hand (EF), which imparts the protein with Ca2+ sensing functionality 19; 21, and a sterile alpha motif (SAM), which regulates STIM oligomerization 22. The cytosolic C-terminus is organized into several distinct modules, the most critical of which is a roughly 100 amino acid region, most commonly referred to as the CRAC Activation Domain (CAD)23; 24; 25; 26; 27, that is necessary and sufficient to activate ICRAC. A year later several studies utilizing genetic approaches, including linkage analysis of SCID patients 28 and RNAi screens in Drosophila 28; 29; 30, identified the PM protein Orai1 as essential component of ICRAC. Shortly after its discovery, a number of Orai1 mutants capable of altering the ion selectivity of CRAC current provided evidence that Orai1 was the PM Ca2+ influx channel 25; 31. Orai1 encodes the pore forming subunit of the CRAC channel and is comprised of four transmembrane domains (TM1–4) and cytosolic N- and C-termini, both of which harbor STIM1 binding sites (Fig 1B) 23; 24; 27; 32; 33.
Figure 1. Topology and functional domains of STIM1 and Orai1.
A) Topology of STIM1 and its functional domains. The domains include: signal peptide (Sig), EF-Hand (canonical and non-canonical EF-hand), sterile alpha motif (SAM), transmembrane domain (TM), coiled-coil domain (CC), CRAC Activation Domain (CAD), STIM Orai1-Activating Region (SOAR), Proline-Serine-rich domain (P/S), Lysine-rich domain (K-rich). The STIM1312–387 fragment discussed in the text is shown as a red bar below. B) Topology of Orai1. Each Orai1 monomer includes four transmembrane domains (TM1–4). Residues important for Orai1 function discussed in the text are marked, and CAD binding regions of N- and C- termini are highlighted in golden yellow.
Overview of CRAC channel activation steps
CRAC channels undergo a distinctive activation process in which STIM1 and Orai1 accumulate in distinct puncta in opposite membranes 34. When ER stores are full, the EF-hand motif of STIM1 is bound with Ca2+ and this causes STIM1 to be uniformly distributed in the ER membrane. However, upon store depletion, Ca2+ unbinding from the EF-hand triggers STIM1 oligomerization and migration of the oligomers to the ER-PM junctions, resulting in the appearance of distinct STIM1 puncta 21; 22; 33; 35. Orai1 also concomitantly redistributes within the PM to accumulate at sites opposite to STIM1 36. This juxtaposition allows for interactions of the two proteins to occur leading to channel activation 33; 35. While STIM1 and Orai1 appear to be both necessary and sufficient to reconstitute SOCE 27, recent findings indicate that the association of these proteins is facilitated and stabilized by several accessory proteins; these will be discussed further below.
Molecular attributes of the CRAC channel
Pore Architecture
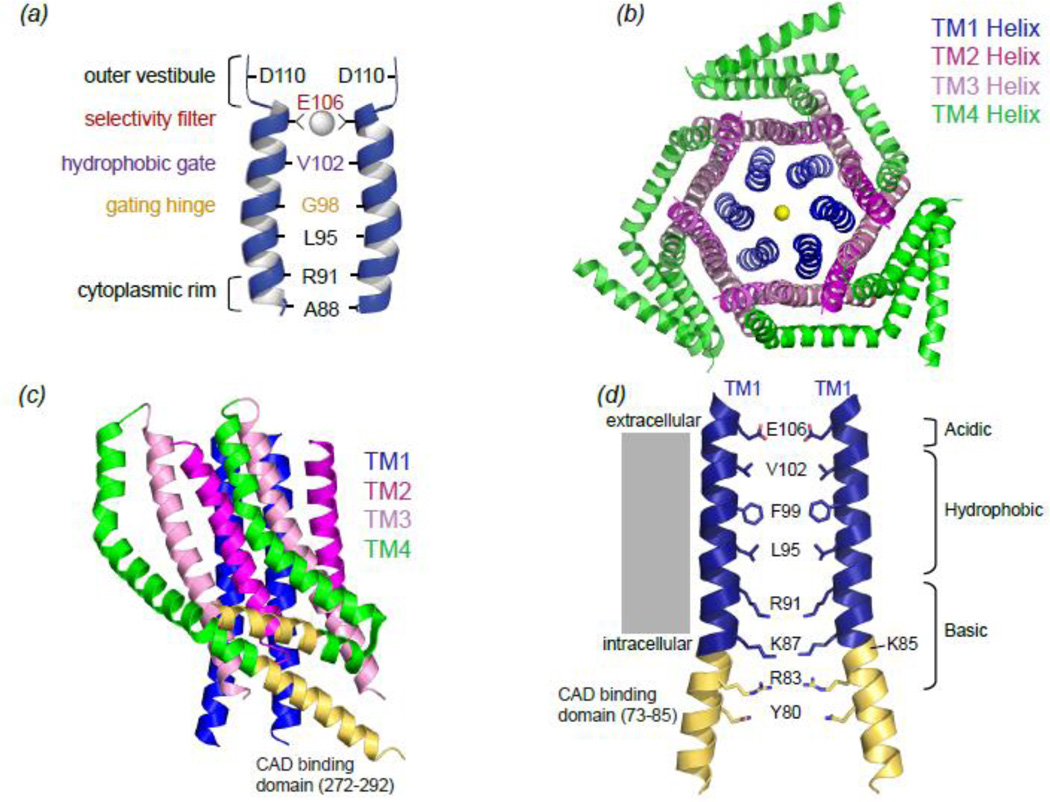
Following identification of STIM1 and Orai1 as two essential components of the CRAC channel, many efforts were directed at understanding the structural attributes of the CRAC channel pore and the basis of its high Ca2+ selectivity. In early studies, several acidic residues in TM1 (E106), TM3 (E190), and the TM1-TM2 (D110, D112, and D114) loop were identified whose mutation changed ion selectivity or permeation (Fig1B)25; 29; 31; 37. Subsequently, a study that mapped the accessibility of engineered cysteines to small, water-soluble thiol-reactive reagents to the open channel pore, found that the CRAC channel pore is flanked exclusively by residues of the TM1 segment (E106, V102, G98, L95, and R91), with the other segments (e.g. TM3 located) far from the central pore axis (Fig2A,Fig1B)38. A complementary study that examined the ability of engineered cysteines to form disulfide bonds with each other across the closed channel pore, came to similar conclusions39. Together, these studies indicated that the Orai1 channel is narrow across much of the ion conduction pathway with residue E106 forming a critical molecular determinant of the high Ca2+ selectivity of the CRAC channel (Fig.2A). Additionally, the close correspondence of TM1 residues in the two studies, which used active (open) and non-functional (closed) channels suggests that the conformational difference in the ion conduction pathway between open and closed states is not large.
Figure 2. Structural features of the Orai channel.
A) Schematic of pore-lining residues in the TM1 helix of Orai1 identified by cysteine scan studies and their postulated functional roles. B) A top-down view of the hexameric Drosophila Orai channel crystal structure (PDB ID code 4HKR) with a Ca2+ ion lodged in the ion conduction pathway. The TM1 helices (blue) of the six monomers form an inner concentric ring, the TM2 (magenta) and TM3 (light pink) helices form a second concentric ring layer, and TM4 helices comprise the outermost helical layer. C) Side-view of two adjacent Orai monomers highlighting the anti-parallel coiled-coil interaction between two Orai C-termini. The CAD binding domain in each Orai1 C-terminus (a.a. 272–292) is highlighted (golden yellow). D) Side view of the dOrai pore showing two of the six helices. The pore-lining residues of TM1 and the N-terminus extension helix are also shown to highlight the three distinct chemical environments of the of the ion conduction pathway. The N-terminal CAD binding domain (a.a. 73–85) is shown in yellow to highlight the overlap between this region and the cytoplasmic region of the pore. Amino acid numbering corresponds to human Orai1.
The crystal structure of the Drosophila Orai channel (a.a 132–341 with the C224S/C283T/P276R/P277R quadruple mutation to facilitate crystallization), has largely confirmed these findings (Fig2D) 40. This 3.35 Angstrom (Å) structure, widely presumed to be of the closed channel (STIM1-free) shows an Orai complex composed of six subunits, with the transmembrane domains arranged in concentric layers around the central aqueous pore formed by the TM1 helices (Fig. 2B). TM2 and TM3 form the second layer surrounding TM1, shielding it from the surrounding lipid bilayer and likely providing structural support. TM4 is the outermost and presumably the most lipid-exposed segment. The N- and C-termini were found to be extensions of the TM1 and TM4 helices, respectively. The hexameric stoichiometry was unexpected because several previous studies had indicated that CRAC channels are tetramers of Orai subunits41; 42; 43; 44, with one group even suggesting that resting state CRAC channels exists as dimers with STIM1 assembling the dimers into tetramers45; 46. However, many of these early studies reporting dimeric and tetrameric resting-state Orai utilized subunit counting or functional analysis of concatemers, which likely underestimated the channel stoichiometry. By contrast, the hexameric structure of Hou et al (2012) is supported by observations made using cross-linking and size exclusion chromatography coupled with light scattering 40 and that are in close agreement with the previously reported observations of Park et al (2009), who noted that detergent-solubilized, CAD-free Orai elutes as a multimer of ~290 kDa about six-eight times the size of the observed monomers24.
Given the relatively modest resolution of the crystal structure (3.35 Å), some uncertainty remains about the exact positioning of many side-chains. However, the close correspondence in the general pore architecture and identity of pore-lining residues between the cysteine scanning studies and the X-ray crystal structure indicate that the pore model is likely to be essentially correct. Notably, the identity of pore-lining residues determined by the McNally et al 38 SCAM study is, with the exception of G98, identical to that reported by the crystal structure (Figure2A,2D). The structure reveals a fascinating complexity in the chemical environment of the ion conduction pathway, exhibiting three discrete regions. The first extracellular third of the pore contains a negative electrostatic potential derived from the six TM1 E106 glutamates that form the selectivity filter. The middle region exhibits a hydrophobic environment with three turns of hydrophobic residues (V102, F99, and L95) that would be expected to make extensive van der Waals contacts with each other. The proposed hydrophobic gate38, V102, and the gating hinge 47, G98, discussed in further detail below, are located within this region. Finally, the lower third region formed by the cytoplasmic membrane proximal N-terminal region contains several basic residues (R91, K87, R83) and contains a small portion of the proposed N-terminal CAD binding site (residues 73–85) (Fig2D). These basic residues have been suggested to coordinate anions 40, which is highly unusual for a cation selective channel. It has been suggested that anion binding may be a vital part of the gating mechanism that keeps the pore closed in the resting state 40. However, whether binding of anions within the pore occurs under physiological conditions is unclear and remains to be determined.
STIM1 binding sites on Orai1
Structure-function and biochemical studies have shown that STIM1 interacts with two sites in Orai1, located in the intracellular C- and N-termini 24; 27. In the Orai1 C-terminus, the relevant interaction site is a helical domain that is well-resolved in the drosophila Orai structure (Fig.2C). Interestingly and contrary to early expectations, the C-termini of adjacent Orai monomers appear to be self-associated in an anti-parallel coiled-coil helical configuration mediated by interaction of Ile316 (hL273) with Leu319 (hL276) (Figure2C). Previous studies have shown that substitutions of these hydrophobic residues, L273S 23; 48 and L276D 33, diminish STIM1 binding and disrupt channel activation. Whether these residues and the anti-parallel coiled-coil motif in which they are located form a binding surface for STIM1 is not conclusively established. However, a recent NMR complex of a human Orai1 C-terminal fragment (a.a. 272–292) with a human STIM1 fragment (a.a. 312–387) (Fig 3D) indicates that these residues dissociate to interface with STIM149. Interestingly, a careful analysis of the Orai1 C-terminus sequence and that of the cytoplasmic region of STIM1 by Korzeniowski et al 50 revealed a striking similarity between the coiled-coil Orai1 C-terminus and an acidic coiled-coil segment of STIM1. This similarity led to initial proposals that the acidic sequence in STIM1 can compete with the Orai C-terminus for CAD/SOAR binding to keep CAD hidden in the resting state of STIM1. One prediction therefore is that STIM1 binds the Orai C-terminus via coiled-coil packing, a possibility supported by the recent NMR complex structure49.
Figure 3. Structural features of the STIM1 C-terminus.
A) Schematic of the human SOAR crystal structure (PDB ID code 3TEQ). The two monomers (gray and violet) form a parallel dimer, with interactions between the CC2 and CC3 of each monomer. B) Schematic of the C.elegans STIM crystal structure (PDB ID code 3TER) encompassing domains CC1-CC3. In C. elegans, the CC2 and CC3 domains and dimer arrangements are similar to the human SOAR structure (monomers shown in blue and pink). C) NMR structure of STIM1 encompassing amino acids 312–387 (PDB ID code 2MAJ). The two monomers in yellow and green are arranged in anti-parallel manner. CC2 (yellow) interacts with CC2’ (green) of the other monomer, likewise CC1 (yellow) associates with CC1’(green). This structure is displayed vertically on its long axis for easier comparison with the two crystal structures. D) Solution structure of the complex between STIM1312–387 and Orai1272–292 (PDB ID code 2MAK). The STIM1312–387 monomers (cyan and blue) are anti-parallel, and the two identical CC2:CC2’ interfaces form composite binding surfaces for the two Orai1 C272–292 (magenta) fragments. Homotypic interactions of CC1:CC1’ are also observed. The model figures were generated using PyMOL.
Much less is known about the second STIM1 binding site on the Orai N-terminus. Biochemical studies using peptide fragments of the Orai1 N-terminus have traced the binding site to residues between 73 and 85 in Orai124; 32. Functional studies indicate that this site is essential for CRAC channel gating 24 and also contributes to the overall interaction of Orai1 with STIM1 51; 52. Interestingly, as noted above, this region of Orai1 also flanks the ion conduction pathway (Fig. 2), raising the possibility that STIM1 interaction at this locus may underlie the gating of CRAC channels. However, the molecular details of how STIM1 interacts with this second site and what changes ensue in the CRAC channel from this binding are largely unknown. The overlap in the functions of the two sites for gating and STIM1-Orai1 interaction51; 52 suggests that rather than forming distinct sites with distinct functions, the C- and N-terminal STIM1 sites may assemble to form a single binding pocket for STIM1. One residue previously implicated in gating (K85 Orai1 or K157 in drosophila Orai) (Fig2D) 53 faces away from the pore and toward the cytosolic C-terminal binding site of Orai1, raising the possibility that a yet-to-be-defined motif in STIM1/CAD nestles into the external face of the Orai1 73–85 helix to induce pore opening.
Mechanism of CRAC channel gating
The binding of STIM1 induces conformational changes in the Orai1 protein, culminating in the removal of a barrier for ion conduction, or a gate, in the pore. Two plausible models have been proposed to describe the nature and location of the CRAC channel gate. In one model, the channel gate was localized through analysis of state-dependent differences in accessibility of bath applied MTS reagents in closed and open channels54. The results of this study revealed the presence of a significant barrier for ion conduction in the closed channel towards the extracellular region of the pore, above the probe site at G98. Mutational analysis uncovered V102, a residue located close to the external pore entrance (Fig.1B,Fig.2A,Fig.2D), as a strong candidate for the gate, with substitutions to more polar residues (Ala, Ser, Thr, or Cys) yielding constitutively open channels54. Moreover, substitution of the Val at this position to other amino acids suggested that V102 serves as a hydrophobic gate, with the side-chains of the six pore-facing Val residues forming a large desolvated barrier to cations. Consistent with this hypothesis, molecular dynamics simulations indicate that an Ala substitution (V102A) lowers the energetic barrier for water occupancy in the pore55, explaining the constitutive phenotype of the V102A Orai1 channel. Additionally, mutational analysis at G98 has lead to the suggestion that this pore-lining glycine may function as a hinge, a common role of glycine in other ion channels47. However, what is not clear yet in this model is the nature of the structural rearrangement that causes the gate to open to permit ion conduction.
An alternative viewpoint is that the CRAC channel gate is formed by constriction of the inner pore near the residue R9147. This model was based in part on findings showing that cysteine substitutions at R91 can be cross-linked across the pore47. In addition, the crystal structure study has suggested that cytoplasmic N-terminus containing a cluster of positive charges (Fig.2D) may coordinate negatively charged anions in the inner pore to occlude the pore40. Gating in this model is envisioned to be driven by widening of the aperture of the helices where the pore narrows. However, it should be noted that cross-linking of Cys residues engineered at pore-lining positions is seen at many locations along the length of the pore 39, and there is as yet no functional evidence for regulation of gating by intracellular anions. Regardless, the two models described above differ both on the location of the channel gate (outer vs inner pore) as well as on the gating mechanism (hydrophobic vs. steric/electrostatic gating). One possibility therefore, is that there are two gates, located at both ends of the pore. On the other hand, it is also possible that gating occurs across an extended length of the pore from energetic barriers spanning the permeation pathway in TM1. Although the differing ideas have limitations and underscore the need for additional studies to define the nature and location of the channel gates, the models provide a useful starting point to determine the structural rearrangements in the pore that underlie gating.
A fascinating aspect of CRAC channel gating uncovered by these studies, conspicuously seen in V102X mutant channels, is strong functional coupling of permeation and gating. The constitutively active V102C/A mutants are poorly selective for Ca2+ when expressed in the absence of STIM1 45. Remarkably, associating the mutant channels to STIM1 by co-expressing STIM1 and depleting stores converts these relatively non-selective channels into highly Ca2+-selective channels52; 54. In fact, this phenotype is not unique to the V102X mutants, for increasing the amount to STIM1 bound to the wild-type Orai1 channels also alters the current-voltage relationship, effectively increasing the Ca2+ selectivity of Orai154. The strong coupling of selectivity and gating is unusual among ion channels, and indicates that the traditional postulates on the separation of gating and permeation do not apply to CRAC channels. Moreover, this finding opens the possibility that Orai channels might function either as highly Ca2+-selective or non-selective channels depending on how they are activated, considerably expanding their range of functions to include Na+ influx that may contribute to membrane depolarization under some conditions. More work is needed to examine this possibility.
STIM dynamics and activation process
STIM is the ER Ca2+ sensor
The STIM proteins are composed of several functional domains with distinct molecular and functional features. The initial step in STIM1 activation is triggered by decrease in Ca2+ concentration in the ER lumen. The luminal portion of STIM1 is made up of a canonical EF-hand domain that pairs with an atypical non-Ca2+-binding EF-hand and an adjacent SAM domain (Fig. 1A). At rest, the luminal Ca2+ level is ~400–600 µM 35; 56, and Ca2+ is bound to EF-hand domain forming a stable complex with the SAM domain. Following ER store depletion, Ca2+ dissociates from the EF-hand and leads to destabilization of the EF-hand-SAM complex resulting in exposure of hydrophobic regions of the two domains. These exposed hydrophobic regions then self-associate to drive oligomerzation of STIM1 57; 58; 59; 60. This process is reversible and repletion of the ER store facilitates Ca2+ binding to the EF-hand domain and complex formation with SAM domain, leading to deactivation of Orai 61; 62.
Although very similar in structure, the two human homologs, STIM1 and STIM2, have notable functional differences. The Ca2+ dissociation constant (Kd) of STIM2 is ~400 µM or about 2-fold higher than that of STIM1 (~200 µM) 63. As a consequence, STIM2 is a more sensitive Ca2+ sensor, requiring smaller decreases in luminal ER Ca2+ concentrations for its activation 63; 64. Paradoxically, despite the higher sensitivity to ER Ca2+ store depletion, the EF-SAM domains of STIM2 exhibit slower unfolding and aggregation kinetics, resulting in slower and smaller levels of CRAC channel activation 59; 65. Thus, on balance, the overall lower efficacy of STIM2 activation may be a protective mechanism for cells to control runaway activation of CRAC channels and may be relevant for generating finely tuned Ca2+ signals such as oscillations.
Depletion of ER Ca2+ stores causes STIM oligomerization
A key event in the choreography of CRAC channel activation is the formation of higher order STIM1 oligomers triggered by ER Ca2+ store depletion. STIM appears to exist as a dimer at rest 46, with oligomerization likely forming higher order polymers of the elementary dimers. The available evidence indicates that both luminal as well as cytoplasmic regions of STIM1 are necessary for this process, with each complementing and strengthening interactions formed by the other. The molecular motif that controls the initial trigger for oligomerization appears to reside in the luminal EF-SAM domains. Ca2+ unbinding from the EF-hand caused by store depletion induces dimerization of the EF-SAM domains 58; 66. In turn, this is postulated to bring the CC1 domains in the trans-side of the membrane close to each other, triggering self-association of the cytoplasmic STIM1 regions and exposing regions in CC3 that ultimately form higher order STIM1 oligomers through interactions in this domain 67. Interestingly, artificially clustering the luminal EF-SAM domains without depleting ER Ca2+ stores is sufficient to trigger STIM1 oligomerization and activation of CRAC channels, 35, suggesting that STIM1 oligomerization is an intrinsic, self-organizing feature of STIM1, and the Ca2+ bound EF hands act as a brake to keep this process in check. The structural mechanisms of STIM1 oligomerization and organization of the oligomers remain largely unknown.
Translocation of STIM1 to the ER-PM junctions
Once STIM1 oligomerizes, the clustered STIM1 translocate to the ER-PM junctions and aggregate into puncta 19; 61; 68. The C-terminal, polybasic domain of STIM1 is critical for STIM1 accumulation at the plasma membrane and this is thought to involve interactions of the K-rich domain with negatively charged phospholipids 69; 70; 71. Interestingly, over-expressing Orai1 appears to diminish the requirement for the K-rich domain for puncta formation, suggesting that the K-rich region facilitates STIM1 accumulation at the ER-PM junctions, but is not needed for Orai1 interaction. Activation of CRAC channels, is however, significantly slowed in mutants lacking the K-rich domain, consistent with the notion that targeting of STIM1 to the ER-PM junctions enhances the local concentration of STIM1 near Orai1leading to faster and more efficient CRAC channel activation 24.
Cytoplasmic domains of STIM
CRAC activation domain – the minimal catalytic region of STIM1
As noted above, the cytoplasmic portion of STIM is composed of many functional domains that fulfill distinct roles in STIM function. A domain consisting of about 100 amino acids, encompassing CC2 and CC3 STIM1 region (Fig1A), was identified independently by several groups as the minimal unit capable of binding to and activating Orai channels. This domain, variously named CRAC Activation Domain (CAD), STIM1 Orai1-Activating Region (SOAR), or coiled-coil domain containing region b9 (CCb9) 24; 26; 72, is able to constitutively activate Orai channels when over-expressed as a soluble protein 24; 67; 73. The crystal structure of the CAD/SOAR domain, solved by Yang et al, shows that this domain assembles as a dimer. Within each monomer, the coiled-coil domains CC2 and CC3 form a hair-pin motif (Fig3A), and the monomers are parallel with respect to each other (gray and pink in Fig3A)49; 74. The prevailing model suggests that at rest this active site is hidden through interactions with other flanking domains of the STIM1 C-terminus, thereby preventing its exposure and ability to activate Orai channels. We will return to this idea below.
The inhibitory role of CC1 in STIM activation – different proposed modes of inhibition
Numerous studies have implied that the cytosolic region of STIM1 (c-STIM1) binds weaker to Orai1 than CAD and exhibits reduced activity 24; 26; 72. This observation raised the possibility that the domains flanking CAD reduce the activity of c-STIM1 through a mechanism wherein CAD exposure or activity is impeded. Because the initial trigger for STIM1 activation would presumably arise from the luminal, Ca2+ sensing side of STIM1, the absence of the luminal portion would likely cause c-STIM1 to be retained in a locked, inactivated state, reducing its ability to activate Orai1 channels.
Consistent with this possibility, subsequent studies identified CC1 as a vital part of this clamping mechanism. One influential early study 50 proposed that an acidic motif of STIM1 CC1 that resembles a previously identified acidic motif on the Orai1 C-terminus 75, functions as a decoy to keep the C-terminus folded in a quiescent state by interacting with a cluster of basic residues in CAD 75; 76. This model was supported by observations that Ala substitutions at the acidic residues in CC1 constitutively activated STIM1, whereas neutralizing the basic region of CAD prevented STIM1 from interacting with Orai150. From this model, it was inferred that a conformational change from the luminal side caused by ER Ca2+ store depletion releases CAD from the intramolecular CC1 clamp, in the trans-side of the ER membrane, priming CAD for interaction with Orai1 at its basic residues 50. A more detailed analysis of the mutations at the acidic and basic regions of CC1 and CC2 including charge reversal substitutions, however, indicated that the simple decoy model as originally proposed is unlikely to be correct. Specifically, charge reversal substitutions of the acidic residues that were predicted to disrupt the CC1-CC2 interaction had no effects on STIM1 function77. Further, both the crystal structure of the C. elegans CC1-CC3 domains as well as the solution NMR structure of STIM1 fragments show that the acidic motif in CC1 is located too far from the basic CC2 region to participate in direct interactions (Fig 3B, 3C). Nevertheless, this early study was influential in raising the idea of an intramolecular clamp involving interactions between CC1 and CAD that keeps STIM1 in a quiescent state.
FRET based studies have provided more direct support for this idea. In one approach, Muik and colleagues generated an intramolecular FRET sensor by attaching CFP and YFP probes to the ends of a cytoplasmic STIM1 fragment encompassing CC1-CC3. FRET measurements with this probe (which was named the Orai activating STIM fragment sensor or OASF sensor) suggested that at rest CC1 and CC3 are in close proximity to each other. However, activating mutations such as the gain-of-function L251S and L416S/L423S mutations resulted in a marked decrease in intramolecular FRET. Because these mutations did not change the dimeric/oligomeric status of OASF as assessed by SEC-MALS, the decrease in FRET was interpreted as arising due to extension of STIM1 CC1-CC3 78. These findings were supported and extended by Zhou et al 79. By mapping distances between specific regions of the STIM1 C-terminus using Tb3+-acceptor energy transfer, Zhou et al 79 found that introducing the gain of function L251S mutation within CC1 extended the STIM1 C-terminus. Interestingly, their results suggested that the CC1 domains of the two monomers are not self-associated in the quiescent state of the STIM1, but dimerize in the active state. Together, these studies have led to a model wherein unbinding of Ca2+ from the EF-SAM domain following ER Ca2+ store depletion results in the self-association of the CC1 domains in the trans-side of the ER membrane. This homodimerization of CC1 then releases CAD from CC1, enabling physical extension of STIM1 and exposure of the CAD and the distal polybasic segment, leading to Orai binding and CRAC channel activation (Fig5) 79. Important unresolved issues that await clarification include the nature of the CC1-CAD and Orai1-CAD binding interfaces, the molecular determinants of how CAD is buried in the quiescent STIM1 state, and the conformational changes in CAD itself that enable Orai channel activation.
Figure 5.
Schematics of the two proposed STIM1 activation models. A,B) Yang et al. STIM1 activation model based on the CAD/SOAR crystal structure. In the resting state, the Ca2+-bound luminal domain of STIM1 keeps the cytoplasmic regions in a folded, inactive state. Store depletion leads to exposure of the pre-formed CAD dimer, extending it towards the plasma membrane and allowing interactions with Orai1 at the apical ends of the R-shaped CAD monomers. C) STIM1 activation model based on the recent STIM1 NMR structure. In contrast to the crystal structure model, this model postulates that the STIM1-Orai1 interaction occurs along the length of CC2 and that the CAD dimer as hypothesized in the crystal structure undergoes internal rearrangements to accommodate the Orai1 peptides. Abbreviations used: EF-Hand (EFh), sterile alpha motif (SAM), coiled-coil domain (CC), Lysine-rich domain (K).
Other proposed C-terminal STIM1 activation inhibitory regions
In addition to CC1, there are additional autoinhibitory regions in the cytoplasmic part of STIM1 that affect CRAC channel and STIM1 function. Kawasaki et al demonstrated that a 31 amino acid region (aa445–475) immediately C-terminal to the CAD domain interferes with store-dependent and independent CRAC channel activation 72. Likewise, Muik et al found that a 11-residue region (aa 475–485) at the C-terminal end of CAD reduces CRAC channel activity 73. At present, how these autoinhibtory regions impede STIM mediated channel activation remains unknown and whether they affect CAD function or STIM1-Orai1 binding remains to be determined.
Overview of the available STIM C-terminus structures
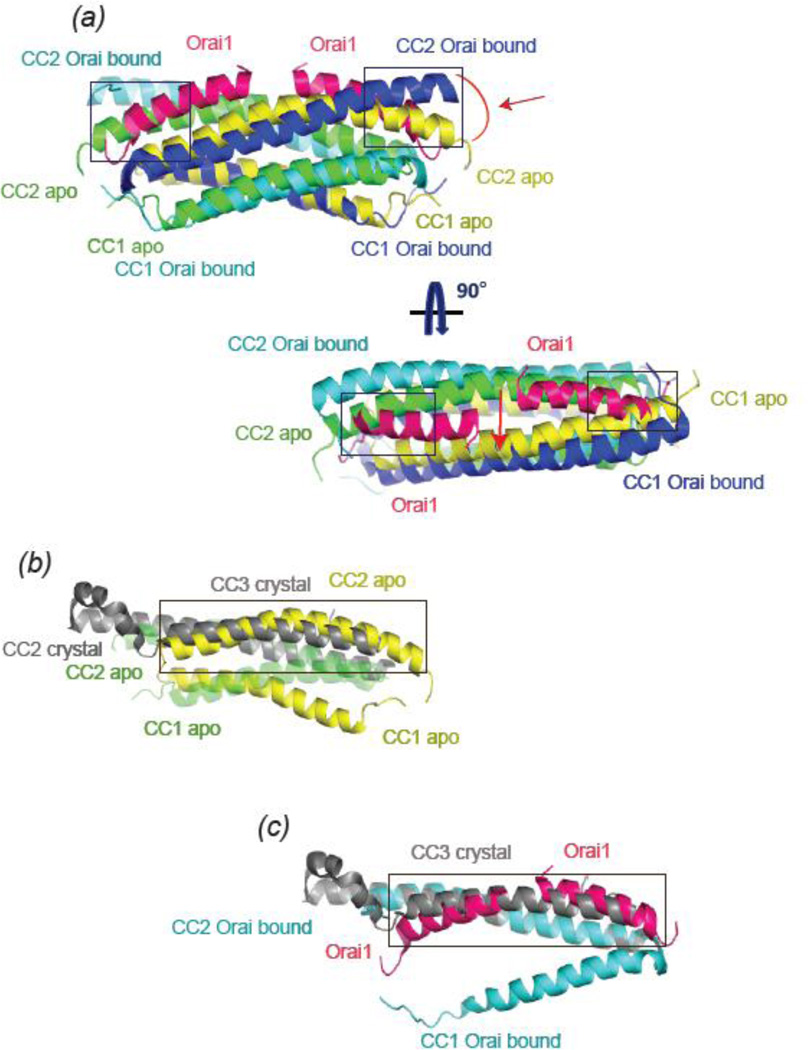
The recent atomic resolution structures provided new insights into self-association of the cytosolic parts of STIM1 and the potential conformational changes that underlie STIM1 activation 49; 74. The crystal structure of human SOAR shows CC2-CC3 helices forming a hairpin motif, and two monomers are parallel with CC2 engaging CC3’ of the other monomer and vice versa (Fig3A). The monomer SOAR structure is letter R shaped and made up of 4 α helices; each helix represents a stroke of the letter (Fig3A). Yang et al also reported a crystal structure of the longer cytoplasmic CC1-CC3 fragment of C. elegans STIM 74. The SOAR region of the protein is analogous to the human SOAR structure; however, CC1 is located at the base of the inverted tepee-shaped CAD dimer, far removed from the postulated active site of CAD, which is located at the top of the inverted tepee (Fig3B).
The NMR STIM1 structure, obtained from a STIM1 fragment (a.a. 312–387) that includes approximately one third of the C-terminal region of CC1 (CC1 TM-distal) and CC2, is also made up of helices in hairpin motif 49. However, the dimer arrangement of the CC2 domains is drastically different from the crystal structure. The two CC2 domains of the NMR structure are antiparallel to each other and participate in homotypic interactions. The structure also presents antiparallel CC1 TM-distal: CC1 TM-distal’ supercoiled dimer (Fig3C). Alignment of the two structures reveals that CC3 of the crystal structure and the nonaligned CC2’ of the NMR structure occupy an overlapping region (Fig4B). This overlap between the two domains implies the two structures must be in different conformations, and suggests that CC1 and/or CC3 segments have to rearrange their positions with respect to CC2 as a part of the STIM1 activation process.
Figure 4. Alignnment of the crystal and NMR STIM1 structures.
A) Alignment of NMR STIM1312–387 (yellow and green) dimer structure in the presence and absence of Orai1 C272–292 (magenta). The C-alpha atoms of chain A of apo-NMR structure (PDB 2MAJ) were structurally aligned with the C-alpha atoms of chain A or Orai complexed STIM structure (PDB 2MAK). Conformational changes are observed when Orai1 C-terminal peptide binds to the STIM1 dimer, where the two CC2 domains of the dimer are separated and C-terminal half of the CC2 moves away. These changes are highlighted with arrows. Orai1 peptide overlaps with CC2 domains of the STIM1 alone structure and these regions are boxed. Top view of the aligned structures is also shown, and the overlapping regions of the Orai1 fragments and the CC2 domain of the apo-STIM1are boxed. B) Superimposition of NMR apo STIM1312–387 (green and yellow) with SOAR crystal structure (grey). When the CC2 domain of the green monomer of the NMR structure is aligned with the CC2 of the SOAR structure, CC3 domain (grey) of SOAR clashes with CC2’ domain of the yellow NMR structure monomer, and the overlapping region is boxed. The green monomer is shown as a more transparent cartoon for clarity. In order to generate this figure, the C-alpha atoms of chain A or apo-NMR structure (PDB 2MAJ) were structurally aligned with the C-alpha atoms of chain A of the crystal SOAR structure (PDB 3TEQ). C) Structural alignment of CC2 domain (grey) of crystal SOAR structure with CC2 domain of STIM1312–387 (cyan) in complex with Orai1 C272–292 (magenta). Orai1 peptide (magenta) occupies the same space as CC3 domain of SOAR structure (boxed). Only one monomer (cyan) of the dimer complex structure is shown for clarity. The C-alpha atoms of chain A of Orai-complexed STIM1 structure (PDB 2MAK) were structurally aligned with the C-alpha atoms of chain A of the crystal SOAR structure (PDB 3TEQ). All the figures were aligned and made using PyMOL.
Comparison of the longer C.elegans STIM structure and the NMR structure shows an additional key distinction in the orientation and interactions of the CC1 domains. The NMR structure shows homotypic interactions in the CC1 distal domains. By contrast, the CC1 domains of the C.elegans crystal structure are far apart from each other (Fig. 3B,C). C.elegans STIM has a significantly shorter CC1 domain than human STIM1, and it is possible its domain arrangements and CC1 function may differ from its human homolog. However, based on these incongruent structures, it is difficult to discern how the CAD or STIM1 dimers are assembled, and how CC1 may function as an autoinhibitory clamp in inactive state. Both structures, however, challenge the idea that the CC1 acidic region interacts with the basic patch in CC2 via electrostatic interactions. The two regions are too far apart for association, raising the possibility that the observed gain-of-function (for the acidic amino acid mutations) or loss-of-function (for the mutations of the basic residues) phenotypes50 arise from disruption of other interactions or the secondary structure of the CAD domain.
Structural basis of STIM-Orai interaction
As noted above, previous studies indicate that CAD directly binds to N- and C- termini of Orai1 to open the CRAC channel, but the structural basis of this interaction is only now beginning to be elucidated. Early studies employing structure-function analysis postulated that a patch of acidic residues on the Orai1 C-terminus are located at the binding interface of STIM1, presumably interacting with a cluster of basic residues in STIM1 CC2 75; 76. The results of this analysis, however, were far from definitive and additional mutations of Orai1 acidic residues that elicited only modest phenotypes were not easily explained. Domain grafting studies in CAD between STIM1 and STIM2, which exhibit different Orai1 binding affinities as assessed by FRET, also implicated the STIM CC2 domain for Orai1 binding 80. The most direct evidence, however, has come from the recent NMR structure of STIM1 (a.a. 312–387) complexed with a small Orai1 fragment (human 272–291). This structure reveals a fascinating supercoiling interaction between the Orai1 CC domain and the STIM1 CC2 domain, with the binding interface composed of both hydrophobic and electrostatic interactions between CC2 of STIM1 and the Orai1 C-terminus272– 291 (Fig3D). The alignment of the NMR apo-STIM1 structure with the NMR Orai-bound STIM1 structure reveals a separation between the two CC2 and CC2’domains with the C-terminal half of CC2 moving away to make room for Orai binding (Fig 4A) 49. This implies that at least a partial disruption of the STIM1 dimer interaction has to occur to accommodate Orai1 C-terminal binding 49. Moreover, the structural alignment of the NMR complex structure and the apo-SOAR crystal structure shows that the Orai1 C-terminal fragment binds to CC2 at exactly the same position where CC3 in the SOAR crystal structure is located (Fig4C). It would seem therefore that CC3 has to dissociate from CC2 to permit CC2-Orai1 binding. Thus, once CAD is released from CC1 clamp following store depletion, it likely undergoes further conformational change to interact with Orai.49.
A noted above, the Orai1 C-terminus contains a significant stretch of negatively charged residues. In the solution NMR holo-structure, these negatively charges residues interact with complementary positively charged surface of STIM1 that include the lysines and arginines in the sequence at positions 365–366, 377, and 382–387. Interestingly, in the apo-STIM1312–387 NMR structure, the anticipated negatively charged decoy residues 50 of the STIM1 CC1 domain form a small negative patch on the opposite side of Orai1 C-terminal binding surface, but Orai1 binding brings the acidic stretch of residues closer to the identical amino acids of the other STIM1 monomer, creating a large continuous negatively charge surface 49. Therefore, the clustering of the acidic residues may be important for allosteric changes of STIM1instead of being an intramolecular clamp.
Alignment of the structures of STIM1 bound Orai1 C-terminal peptide and the corresponding crystal structure of the D. Melanogaster Orai C-terminal region provides additional interesting clues on the possible conformational changes that might occur in the Orai C-terminus following STIM1 binding 40; 49. The comparison suggests that the antiparallel C-terminal configuration of Orai is qualitatively similar between the two structures, and that there are no large-scale conformational changes in the Orai1 helices (e.g., straightening of each Orai C-terminus) upon STIM1 binding. However, the interhelix angles and orientation of the Orai1 C-termini in the NMR structure are different from that in the Orai crystal structure 49. Given the low sequence identity between the D.Melanogaster Orai and human Orai1, it is not certain whether drosophila Orai uses the same activation mechanism and if the observed structural discrepancies arise from different activation mechanisms utilized by the different species. Nevertheless, based on the comparison of the two structures, Stathopulos et al have suggested that STIM1 binding induces changes in the Orai1 C-termini, which are ultimately important for efficiently gating the channel49.
In summary, the available structures reveal at least three distinct conformations of the STIM1 CC1-CC3 segments. Moreover, the crystal and NMR apo structures, are not readily consistent with each other. Thus, a fundamental question that needs to be resolved is whether these distinct structures represent bona fide transition states of STIM1 at different steps of the STIM1 activation. Stathopulos et al, have proposed that the three models (two apo and one Orai1 bound) do represent conformations of STIM1 in three different states: inactive for the crystal structure, an intermediate apo NMR structure (discussed in section above), and active Orai-bound structure 49. An interplay of different domains was proposed with store depletion eliciting self-association of the membrane proximal CC1 region, leading to cytosolic domain reorganization, prompting the distal CC1 domains to self-associate (as seen in the apo NMR structure) (Fig3C and Fig5). Establishment of the CC1 dimer is presumed to release CC3 from CC2 and yields CC2:CC2’ interaction observed in the apo structure (Fig3C). The disengaged CC3 domains in the homodimer are then free to self-associate and assemble into higher order oligomers. This intermediate conformation of STIM1 is primed to bind to Orai1 at the ER-PM junction, and finally, Orai1 binding displaces the CC2:CC2’ interaction, as seen in the complex NMR structure (Fig3D and Fig5) 49.
Together, these structural studies give us significant insights but also reveal considerable limitations in the available literature. One major caveat is that all the available STIM structures are derived from small fragments. How these fragments are assembled in the context of the full length STIM1 remains unclear. Moreover, as noted above, the available structures contradict each other in fundamental ways that cannot be easily resolved unless one invokes the notions of transition states, but whether such transition states in fact exist in the context of full-length STIM1 is unknown. Additionally, many functional results are not easily reconcilable with the structural studies (for example the optimal STIM:Orai1 subunit stoichiometry from functional studies is 2:1 whereas the NMR study suggests a 1:1 binding ratio), raising questions about possible gaps in the structural information currently available. Finally, none of these studies elucidate the structural basis of CAD/SOAR interaction with the Orai1 N-terminus, although one residue in STIM1 (F394) has been suggested to be important for this interaction 80. Still, these limitations do not diminish the value of the structural studies, as they provide a firm framework and testable ideas for further study of the conformational dynamics of STIM1 activation.
Regulators of SOCE
It is now clear that STIM1 and Orai1 are essential for SOCE and can reconstitute Ca2+ influx even in a system of purified components in liposomes 27. However, as might be expected from a multiprotein signaling pathway involving aggregation of the two core components into complexes, the subcellular localization, efficiency of interaction, and activity of STIM1 and Orai1 is fine-tuned by numerous regulatory proteins. These regulators span a wide range of families including scaffolds, Ca2+ binding proteins, and chaperones that exhibit both transient as well as stable interactions with STIM1 and Orai1 to control the assembly and disassembly of STIM1-Orai1 interactions at the ER-PM junctions. Collectively, these proteins and interactions form the CRAC channel signalplex and optimize Ca2+ entry for the effector function on hand. Here, we briefly review some key members of this signalplex and the available evidence for what is known about how they regulate STIM1/Orai1 function. In nearly all of these examples, however, the molecular basis of how these proteins regulate CRAC channel function are unknown. Hence, these proteins likely represent an important focus of future investigations to understand the molecular/structural mechanisms of CRAC channel regulation.
Septin, a member of the conserved GTP binding scaffold protein was identified as a regulator of SOCE from a genome-wide RNA interference screen81. Consistent with its previously established role in organizing membrane domains in the yeast and around cilia, septin reportedly facilitates the organization of the membrane domains necessary for STIM1-Orai1 communication. Knockdown of septin is reported to alter the diffuse localization of Orai1 channels in the plasma membrane, suggesting that septins are necessary for proper organization of closed Orai1 channels in plasma membrane. In addition, septin is reported to enhance translocation of STIM1 to the ER-PM junction, thereby stabilizing Orai1 clusters through organization of lipid microdomains around the complex. These actions of septins are thought to enhance the efficacy of STIM1-Orai1 coupling and SOCE 81. Likewise, CRAC regulatory protein 2A (CRACR2A) is a cytoplasmic, EF-hand containing protein that was discovered in a large-scale affinity purification to bind to Orai1. The protein reportedly also binds directly to STIM1 and, together with the Orai1 N-terminus, is thought to form a ternary complex to stabilize the interaction between Orai1 and STIM1, facilitate clustering of STIM1 and Orai1 at the ER-PM junction, and enhance SOCE82. Srikanth et al have reported that the two EF-hands of CRACR2A regulate its interaction with STIM1 and Orai1 in Ca2+-dependent manner, with low [Ca2+]i favoring the association of CRACR2A with CRAC channel, whereas high [Ca2+]i leading to its dissociation from STIM1-Orai1. Moreover, the implicated CRACR2A binding site on Orai1 N-terminus partially overlaps with the CaM binding site 83, raising the possibility of potential competition between STIM1 and CRACR2A/B proteins for binding on Orai1 82. These hypotheses await further study. Junctate, a Ca2+ binding ER membrane protein that is widely associated with facilitating the formation of the triad/dyad junctions in muscle tissue, is also reported to be a STIM1 interacting protein important for coupling of ER-PM junctions84. EF-hand mutations in junctate facilitate STIM1 clustering without store depletion and recruit STIM1 to the ER-PM junctions without phosphinositide and Orai1 interaction, leading to activation of CRAC channels. Srikanth et al suggested that translocation of STIM1 clusters through the action of junctate permits STIM1 accumulation at the ER-PM junctions, ensuring efficient and timely assembly of CRAC channels 84.
Unlike the above molecules that facilitate SOCE, the protein SARAF (SOCE-associated regulatory factor) is reported to diminish SOCE by decoupling STIM-Orai interactions. Like STIM1, SARAF is an ER membrane protein with a similar topology, but its ER facing region does not appear to contain any known Ca2+ binding motifs. Analogous to STIM1, the C-terminal portion contains a serine-proline rich domain followed by basic residues. SARAF is reported to bind both STIM1 and STIM2 and translocates to ER-PM junctions in STIM dependent manner following store depletion. Its interaction with STIM1 is reported to facilitate Ca2+ dependent slow inactivation through a mechanism involving dissociation of STIM1 clusters, hence playing a role in negative regulation of SOCE 85. Likewise, Calmodulin has been implicated in feedback regulation of CRAC current, by serving as the Ca2+-sensor that transduces Ca2+ signal through the CRAC channel pore to initiate fast Ca2+-dependent inactivation (CDI) 83; 86. CDI is widely presumed to serve as a key mechanism to limit the amount of Ca2+ that enters the cell through the CRAC channel to rein in Ca2+-dependent cell damage. These effects are thought to be driven by Ca2+-calmodulin binding to the N-terminus of Orai1 in a region that overlaps with the STIM1 binding site. Although the exact mechanism remains unclear, one possibility is that the binding of Ca2+-CAM to the Orai1 N-terminus displaces STIM1 from its binding site at this location, thereby destabilizing activation gating and reducing CRAC channel activity. Interestingly, calmodulin was also shown to bind to STIM1 and STIM2 in a Ca2+-dependent manner and may exert completely unrelated mode of modulation via STIM 87. Golli, an alternative splice variant of the myelin basic protein is reported to negatively regulate SOCE 88. Golli is constitutively anchored to plasma membrane through myristolylation and mutation of the myristolylation site eliminated its ability to inhibit SOCE, implying membrane targeting of the protein is important for its inhibitory role 88. Golli directly binds to C-terminal domain of STIM1 and co-localizes with STIM1 and Orai1 in the puncta after thapsigargin treatment 89. Partner of STIM1 (POST) is a large protein with 10 predicted transmembrane-spanning segments and was identified in affinity purification of Orai1. The majority of the POST protein is thought to be located in the ER membrane but analogous to STIM1, a small fraction (5–10%) is reported to be also present in the PM. POST binds to STIM1, following store depletion and comigrate to the ER-PM junction. POST is not required for STIM1-Orai1 interaction and does not influence CRAC channel activation. Upon store depletion, POST-STIM1 complex binds to a variety of molecules, such as SERCA, PMCA, Na/K ATPase, nuclear transporters, exportins, and importins-β, and through these interactions, POST is speculated to assemble signaling complexes around the CRAC channel 90.
Future directions
In the past decade, we have witnessed impressive advances in our understanding of the mechanisms SOCE, including the identification of the STIM and Orai protein families, elucidation of the cellular and molecular choreography of the CRAC channel activation process, and illumination of the structures of the Orai and STIM1 proteins. Still, we only have limited insight into the details of how STIM1 binding to Orai1 opens the channel pore. Hence mechanistic studies that elucidate the conformational steps of the CRAC channel gating mechanism are a critical future direction. Moreover, there are many unresolved cell biological questions surrounding the activation of CRAC channels including the nature and components of the ER-plasma membrane junctions, the role of the many regulatory proteins implicated in SOCE, and how these proteins control the formation and maintenance of these specialized cellular structures. Also important is the molecular basis of drug action and the quest for developing CRAC channels based therapies for immune and other diseases. It is clear that looking forward, work on CRAC channels will be highly multidisciplinary involving the disciplines of physiology, structural biology, cell biology, genetics, as well as drug discovery. The collective application of these disciplines should illuminate many of the key gaps listed above and lead to even greater understanding of the operation and clinical relevance of these intriguing ion channels.
Table 1.
Regulators of SOCE
| Regulator name |
Cellular localization | Effect on SOCE | Binding partner(s)/possible mechanism |
References |
|---|---|---|---|---|
| Septin | Resides at the ER-PM junctions. Organizes membrane domains to facilitate STIM1-Orai1 interactions at the ER-PM junctions |
Enhances SOCE | STIM1:Orai1 complex |
[81] |
|
CRAC regulatory protein 2A (CRACR2A) |
Resides in the cytoplasm. Facilitates clustering of STIM1 and Orai1 at the ER-PM junctions. |
Enhances SOCE Regulates SOCE in [Ca2+]i dependent manner |
STIM1 Orai1 N-terminus |
[82] |
| Junctate | Resides in the ER membrane. Recruits STIM1 to the ER-PM junctions |
Ensures efficient and timely assembly of STIM1/Orai1 complexes at the ER-PM junctions. |
STIM1 | [84] |
|
SOCE- associated regulatory factor (SARAF) |
Resides in the ER membrane. Translocates to ER-PM junction in STIM dependent manner and facilitates dissolution of STIM1 clusters to turn off SOCE. |
Diminishes SOCE | STIM1 STIM2 |
[85] |
| Calmodulin | Resides in the cytoplasm. Site of action not established. |
Reduces SOCE through calcium- dependent inactivation. |
Orai STIM |
[83,86,87] |
| Golli | Anchored at the plasma membrane. Cellular function unknown. |
May inhibit SOCE, mechanism unknown. |
C-terminal domain of STIM1 |
[88,89] |
|
Partner of STIM1 (POST) |
Resides primarily in the ER membrane. Binds to STIM1 and comigrates to the ER-PM junction after store depletion. Organizes signaling molecules around the CRAC channel. |
No direct effect on SOCE but thought to organize signaling molecules around the CRAC channel. |
STIM1 | [90] |
Research Highlights.
Store-operated CRAC channels are critical for human immunity and regulate many cellular functions.
CRAC channels are encoded by the Orai proteins and activated by the ER Ca2+ sensors, the STIM proteins.
This review summarizes our current understanding of the structural mechanisms of CRAC channel regulation.
Acknowledgements
The authors would like to thank members of the laboratory for helpful discussions and Amit Jairaman for useful comments on the manuscript. The work described in this review was supported by NIH grant NS057499 and by an AHA postdoctoral fellowship to Ann Shim.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 2.Parekh AB, Penner R. Store depletion and calcium influx. Physiol Rev. 1997;77:901–930. doi: 10.1152/physrev.1997.77.4.901. [DOI] [PubMed] [Google Scholar]
- 3.Parekh AB, Putney JW., Jr Store-operated calcium channels. Physiol Rev. 2005;85:757–810. doi: 10.1152/physrev.00057.2003. [DOI] [PubMed] [Google Scholar]
- 4.Feske S. CRAC channelopathies. Pflugers Arch. 2010;460:417–435. doi: 10.1007/s00424-009-0777-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berridge MJ, Heslop JP, Irvine RF, Brown KD. Inositol trisphosphate formation and calcium mobilization in Swiss 3T3 cells in response to platelet-derived growth factor. Biochem J. 1984;222:195–201. doi: 10.1042/bj2220195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardner P, Alcover A, Kuno M, Moingeon P, Weyand CM, Goronzy J, Reinherz EL. Triggering of T-lymphocytes via either T3-Ti or T11 surface structures opens a voltage-insensitive plasma membrane calcium-permeable channel: requirement for interleukin-2 gene function. J Biol Chem. 1989;264:1068–1076. [PubMed] [Google Scholar]
- 7.Kuno M, Gardner P. Ion channels activated by inositol 1,4,5-trisphosphate in plasma membrane of human T-lymphocytes. Nature. 1987;326:301–304. doi: 10.1038/326301a0. [DOI] [PubMed] [Google Scholar]
- 8.von Tscharner V, Prod’hom B, Baggiolini M, Reuter H. Ion channels in human neutrophils activated by a rise in free cytosolic calcium concentration. Nature. 1986;324:369–372. doi: 10.1038/324369a0. [DOI] [PubMed] [Google Scholar]
- 9.Putney JW., Jr A model for receptor-regulated calcium entry. Cell Calcium. 1986;7:1–12. doi: 10.1016/0143-4160(86)90026-6. [DOI] [PubMed] [Google Scholar]
- 10.Lewis RS, Cahalan MD. Mitogen-induced oscillations of cytosolic Ca2+ and transmembrane Ca2+ current in human leukemic T cells. Cell Regul. 1989;1:99–112. doi: 10.1091/mbc.1.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matthews G, Neher E, Penner R. Second messenger-activated calcium influx in rat peritoneal mast cells. J Physiol. 1989;418:105–130. doi: 10.1113/jphysiol.1989.sp017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penner R, Matthews G, Neher E. Regulation of calcium influx by second messengers in rat mast cells. Nature. 1988;334:499–504. doi: 10.1038/334499a0. [DOI] [PubMed] [Google Scholar]
- 13.Takemura H, Hughes AR, Thastrup O, Putney JW., Jr Activation of calcium entry by the tumor promoter thapsigargin in parotid acinar cells. Evidence that an intracellular calcium pool and not an inositol phosphate regulates calcium fluxes at the plasma membrane. J Biol Chem. 1989;264:12266–12271. [PubMed] [Google Scholar]
- 14.Thastrup O, Cullen PJ, Drobak BK, Hanley MR, Dawson AP. Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2(+)-ATPase. Proc Natl Acad Sci U S A. 1990;87:2466–2470. doi: 10.1073/pnas.87.7.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thastrup O, Dawson AP, Scharff O, Foder B, Cullen PJ, Drobak BK, Bjerrum PJ, Christensen SB, Hanley MR. Thapsigargin, a novel molecular probe for studying intracellular calcium release and storage. 1989. Agents Actions. 1994;43:187–193. doi: 10.1007/BF01986687. [DOI] [PubMed] [Google Scholar]
- 16.Hoth M, Penner R. Calcium release-activated calcium current in rat mast cells. J Physiol. 1993;465:359–386. doi: 10.1113/jphysiol.1993.sp019681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoth M, Penner R. Depletion of intracellular calcium stores activates a calcium current in mast cells. Nature. 1992;355:353–356. doi: 10.1038/355353a0. [DOI] [PubMed] [Google Scholar]
- 18.Zweifach A, Lewis RS. Mitogen-regulated Ca2+ current of T lymphocytes is activated by depletion of intracellular Ca2+ stores. Proc Natl Acad Sci U S A. 1993;90:6295–6299. doi: 10.1073/pnas.90.13.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE, Jr, Meyer T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roos J, DiGregorio PJ, Yeromin AV, Ohlsen K, Lioudyno M, Zhang S, Safrina O, Kozak JA, Wagner SL, Cahalan MD, Velicelebi G, Stauderman KA. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang SL, Yu Y, Roos J, Kozak JA, Deerinck TJ, Ellisman MH, Stauderman KA, Cahalan MD. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature. 2005;437:902–905. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baba Y, Hayashi K, Fujii Y, Mizushima A, Watarai H, Wakamori M, Numaga T, Mori Y, Iino M, Hikida M, Kurosaki T. Coupling of STIM1 to store-operated Ca2+ entry through its constitutive and inducible movement in the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2006;103:16704–16709. doi: 10.1073/pnas.0608358103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muik M, Frischauf I, Derler I, Fahrner M, Bergsmann J, Eder P, Schindl R, Hesch C, Polzinger B, Fritsch R, Kahr H, Madl J, Gruber H, Groschner K, Romanin C. Dynamic coupling of the putative coiled-coil domain of ORAI1 with STIM1 mediates ORAI1 channel activation. J Biol Chem. 2008;283:8014–8022. doi: 10.1074/jbc.M708898200. [DOI] [PubMed] [Google Scholar]
- 24.Park CY, Hoover PJ, Mullins FM, Bachhawat P, Covington ED, Raunser S, Walz T, Garcia KC, Dolmetsch RE, Lewis RS. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 2009;136:876–890. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeromin AV, Zhang SL, Jiang W, Yu Y, Safrina O, Cahalan MD. Molecular identification of the CRAC channel by altered ion selectivity in a mutant of Orai. Nature. 2006;443:226–229. doi: 10.1038/nature05108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan JP, Zeng W, Dorwart MR, Choi YJ, Worley PF, Muallem S. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat Cell Biol. 2009;11:337–343. doi: 10.1038/ncb1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, Meraner P, Kwon HT, Machnes D, Oh-hora M, Zimmer J, Huang Y, Stura A, Rao A, Hogan PG. STIM1 gates the store-operated calcium channel ORAI1 in vitro. Nat Struct Mol Biol. 2010;17:112–116. doi: 10.1038/nsmb.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feske S, Gwack Y, Prakriya M, Srikanth S, Puppel SH, Tanasa B, Hogan PG, Lewis RS, Daly M, Rao A. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- 29.Vig M, Beck A, Billingsley JM, Lis A, Parvez S, Peinelt C, Koomoa DL, Soboloff J, Gill DL, Fleig A, Kinet JP, Penner R. CRACM1 multimers form the ion-selective pore of the CRAC channel. Curr Biol. 2006;16:2073–2079. doi: 10.1016/j.cub.2006.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang SL, Yeromin AV, Zhang XH, Yu Y, Safrina O, Penna A, Roos J, Stauderman KA, Cahalan MD. Genome-wide RNAi screen of Ca(2+) influx identifies genes that regulate Ca(2+) release-activated Ca(2+) channel activity. Proc Natl Acad Sci U S A. 2006;103:9357–9362. doi: 10.1073/pnas.0603161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prakriya M, Feske S, Gwack Y, Srikanth S, Rao A, Hogan PG. Orai1 is an essential pore subunit of the CRAC channel. Nature. 2006;443:230–233. doi: 10.1038/nature05122. [DOI] [PubMed] [Google Scholar]
- 32.Li Z, Lu J, Xu P, Xie X, Chen L, Xu T. Mapping the interacting domains of STIM1 and Orai1 in Ca2+ release-activated Ca2+ channel activation. J Biol Chem. 2007;282:29448–29456. doi: 10.1074/jbc.M703573200. [DOI] [PubMed] [Google Scholar]
- 33.Navarro-Borelly L, Somasundaram A, Yamashita M, Ren D, Miller RJ, Prakriya M. STIM1-Orai1 interactions and Orai1 conformational changes revealed by live-cell FRET microscopy. J Physiol. 2008;586:5383–5401. doi: 10.1113/jphysiol.2008.162503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis RS. The molecular choreography of a store-operated calcium channel. Nature. 2007;446:284–287. doi: 10.1038/nature05637. [DOI] [PubMed] [Google Scholar]
- 35.Luik RM, Wang B, Prakriya M, Wu MM, Lewis RS. Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature. 2008;454:538–542. doi: 10.1038/nature07065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu P, Lu J, Li Z, Yu X, Chen L, Xu T. Aggregation of STIM1 underneath the plasma membrane induces clustering of Orai1. Biochem Biophys Res Commun. 2006;350:969–976. doi: 10.1016/j.bbrc.2006.09.134. [DOI] [PubMed] [Google Scholar]
- 37.Yamashita M, Navarro-Borelly L, McNally BA, Prakriya M. Orai1 mutations alter ion permeation and Ca2+-dependent fast inactivation of CRAC channels: evidence for coupling of permeation and gating. J Gen Physiol. 2007;130:525–540. doi: 10.1085/jgp.200709872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McNally BA, Yamashita M, Engh A, Prakriya M. Structural determinants of ion permeation in CRAC channels. Proc Natl Acad Sci U S A. 2009;106:22516–22521. doi: 10.1073/pnas.0909574106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y, Ramachandran S, Oh-Hora M, Rao A, Hogan PG. Pore architecture of the ORAI1 store-operated calcium channel. Proc Natl Acad Sci U S A. 2010;107:4896–4901. doi: 10.1073/pnas.1001169107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou X, Pedi L, Diver MM, Long SB. Crystal structure of the calcium release-activated calcium channel Orai. Science. 2012;338:1308–1313. doi: 10.1126/science.1228757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji W, Xu P, Li Z, Lu J, Liu L, Zhan Y, Chen Y, Hille B, Xu T, Chen L. Functional stoichiometry of the unitary calcium-release-activated calcium channel. Proc Natl Acad Sci U S A. 2008;105:13668–13673. doi: 10.1073/pnas.0806499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madl J, Weghuber J, Fritsch R, Derler I, Fahrner M, Frischauf I, Lackner B, Romanin C, Schutz GJ. Resting state Orai1 diffuses as homotetramer in the plasma membrane of live mammalian cells. J Biol Chem. 2010;285:41135–41142. doi: 10.1074/jbc.M110.177881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maruyama Y, Ogura T, Mio K, Kato K, Kaneko T, Kiyonaka S, Mori Y, Sato C. Tetrameric Orai1 is a teardrop-shaped molecule with a long, tapered cytoplasmic domain. J Biol Chem. 2009;284:13676–13685. doi: 10.1074/jbc.M900812200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mignen O, Thompson JL, Shuttleworth TJ. Orai1 subunit stoichiometry of the mammalian CRAC channel pore. J Physiol. 2008;586:419–425. doi: 10.1113/jphysiol.2007.147249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Demuro A, Penna A, Safrina O, Yeromin AV, Amcheslavsky A, Cahalan MD, Parker I. Subunit stoichiometry of human Orai1 and Orai3 channels in closed and open states. Proc Natl Acad Sci U S A. 2011;108:17832–17837. doi: 10.1073/pnas.1114814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Penna A, Demuro A, Yeromin AV, Zhang SL, Safrina O, Parker I, Cahalan MD. The CRAC channel consists of a tetramer formed by Stim-induced dimerization of Orai dimers. Nature. 2008;456:116–120. doi: 10.1038/nature07338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang SL, Yeromin AV, Hu J, Amcheslavsky A, Zheng H, Cahalan MD. Mutations in Orai1 transmembrane segment 1 cause STIM1-independent activation of Orai1 channels at glycine 98 and channel closure at arginine 91. Proc Natl Acad Sci U S A. 2011;108:17838–17843. doi: 10.1073/pnas.1114821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frischauf I, Muik M, Derler I, Bergsmann J, Fahrner M, Schindl R, Groschner K, Romanin C. Molecular determinants of the coupling between STIM1 and Orai channels: differential activation of Orai1–3 channels by a STIM1 coiled-coil mutant. J Biol Chem. 2009;284:21696–21706. doi: 10.1074/jbc.M109.018408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stathopulos PB, Schindl R, Fahrner M, Zheng L, Gasmi-Seabrook GM, Muik M, Romanin C, Ikura M. STIM1/Orai1 coiled-coil interplay in the regulation of store-operated calcium entry. Nat Commun. 2013;4:2963. doi: 10.1038/ncomms3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Korzeniowski MK, Manjarres IM, Varnai P, Balla T. Activation of STIM1-Orai1 involves an intramolecular switching mechanism. Sci Signal. 2010;3:ra82. doi: 10.1126/scisignal.2001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McNally BA, Somasundaram A, Jairaman A, Yamashita M, Prakriya M. The C- and N-terminal STIM1 binding sites on Orai1 are required for both trapping and gating CRAC channels. J Physiol. 2013;591:2833–2850. doi: 10.1113/jphysiol.2012.250456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Derler I, Plenk P, Fahrner M, Muik M, Jardin I, Schindl R, Gruber HJ, Groschner K, Romanin C. The extended transmembrane Orai1 N-terminal (ETON) region combines binding interface and gate for Orai1 activation by STIM1. The Journal of biological chemistry. 2013;288:29025–29034. doi: 10.1074/jbc.M113.501510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lis A, Zierler S, Peinelt C, Fleig A, Penner R. A single lysine in the N-terminal region of store-operated channels is critical for STIM1-mediated gating. J Gen Physiol. 2010;136:673–686. doi: 10.1085/jgp.201010484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McNally BA, Somasundaram A, Yamashita M, Prakriya M. Gated regulation of CRAC channel ion selectivity by STIM1. Nature. 2012;482:241–245. doi: 10.1038/nature10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong H, Fiorin G, Carnevale V, Treptow W, Klein ML. Pore waters regulate ion permeation in a calcium release-activated calcium channel. Proc Natl Acad Sci U S A. 2013;110:17332–17337. doi: 10.1073/pnas.1316969110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen WW, Frieden M, Demaurex N. Local cytosolic Ca2+ elevations are required for stromal interaction molecule 1 (STIM1) de-oligomerization and termination of store-operated Ca2+ entry. J Biol Chem. 2011;286:36448–36459. doi: 10.1074/jbc.M111.269415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stathopulos PB, Li GY, Plevin MJ, Ames JB, Ikura M. Stored Ca2+ depletion-induced oligomerization of stromal interaction molecule 1 (STIM1) via the EF-SAM region: An initiation mechanism for capacitive Ca2+ entry. J Biol Chem. 2006;281:35855–35862. doi: 10.1074/jbc.M608247200. [DOI] [PubMed] [Google Scholar]
- 58.Stathopulos PB, Zheng L, Li GY, Plevin MJ, Ikura M. Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry. Cell. 2008;135:110–122. doi: 10.1016/j.cell.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 59.Zheng L, Stathopulos PB, Li GY, Ikura M. Biophysical characterization of the EF-hand and SAM domain containing Ca2+ sensory region of STIM1 and STIM2. Biochem Biophys Res Commun. 2008;369:240–246. doi: 10.1016/j.bbrc.2007.12.129. [DOI] [PubMed] [Google Scholar]
- 60.Zheng L, Stathopulos PB, Schindl R, Li GY, Romanin C, Ikura M. Auto-inhibitory role of the EF-SAM domain of STIM proteins in store-operated calcium entry. Proc Natl Acad Sci U S A. 2011;108:1337–1342. doi: 10.1073/pnas.1015125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liou J, Fivaz M, Inoue T, Meyer T. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc Natl Acad Sci U S A. 2007;104:9301–9306. doi: 10.1073/pnas.0702866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smyth JT, Dehaven WI, Bird GS, Putney JW., Jr Ca2+-store-dependent and -independent reversal of Stim1 localization and function. J Cell Sci. 2008;121:762–772. doi: 10.1242/jcs.023903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brandman O, Liou J, Park WS, Meyer T. STIM2 is a feedback regulator that stabilizes basal cytosolic and endoplasmic reticulum Ca2+ levels. Cell. 2007;131:1327–1339. doi: 10.1016/j.cell.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou Y, Mancarella S, Wang Y, Yue C, Ritchie M, Gill DL, Soboloff J. The short N-terminal domains of STIM1 and STIM2 control the activation kinetics of Orai1 channels. J Biol Chem. 2009;284:19164–19168. doi: 10.1074/jbc.C109.010900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stathopulos PB, Zheng L, Ikura M. Stromal interaction molecule (STIM) 1 and STIM2 calcium sensing regions exhibit distinct unfolding and oligomerization kinetics. J Biol Chem. 2009;284:728–732. doi: 10.1074/jbc.C800178200. [DOI] [PubMed] [Google Scholar]
- 66.Stathopulos PB, Li GY, Plevin MJ, Ames JB, Ikura M. Stored Ca2+ depletion-induced oligomerization of stromal interaction molecule 1 (STIM1) via the EF-SAM region: An initiation mechanism for capacitive Ca2+ entry. The Journal of biological chemistry. 2006;281:35855–35862. doi: 10.1074/jbc.M608247200. [DOI] [PubMed] [Google Scholar]
- 67.Covington ED, Wu MM, Lewis RS. Essential role for the CRAC activation domain in store-dependent oligomerization of STIM1. Mol Biol Cell. 2010;21:1897–1907. doi: 10.1091/mbc.E10-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu MM, Buchanan J, Luik RM, Lewis RS. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J Cell Biol. 2006;174:803–813. doi: 10.1083/jcb.200604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Korzeniowski MK, Popovic MA, Szentpetery Z, Varnai P, Stojilkovic SS, Balla T. Dependence of STIM1/Orai1-mediated calcium entry on plasma membrane phosphoinositides. The Journal of biological chemistry. 2009;284:21027–21035. doi: 10.1074/jbc.M109.012252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walsh CM, Chvanov M, Haynes LP, Petersen OH, Tepikin AV, Burgoyne RD. Role of phosphoinositides in STIM1 dynamics and store-operated calcium entry. Biochem J. 2010;425:159–168. doi: 10.1042/BJ20090884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Calloway N, Owens T, Corwith K, Rodgers W, Holowka D, Baird B. Stimulated association of STIM1 and Orai1 is regulated by the balance of PtdIns(4,5)P(2) between distinct membrane pools. J Cell Sci. 2011;124:2602–2610. doi: 10.1242/jcs.084178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kawasaki T, Lange I, Feske S. A minimal regulatory domain in the C terminus of STIM1 binds to and activates ORAI1 CRAC channels. Biochem Biophys Res Commun. 2009;385:49–54. doi: 10.1016/j.bbrc.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Muik M, Fahrner M, Derler I, Schindl R, Bergsmann J, Frischauf I, Groschner K, Romanin C. A Cytosolic Homomerization and a Modulatory Domain within STIM1 C Terminus Determine Coupling to ORAI1 Channels. J Biol Chem. 2009;284:8421–8426. doi: 10.1074/jbc.C800229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang X, Jin H, Cai X, Li S, Shen Y. Structural and mechanistic insights into the activation of Stromal interaction molecule 1 (STIM1) Proc Natl Acad Sci U S A. 2012;109:5657–5662. doi: 10.1073/pnas.1118947109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Calloway N, Vig M, Kinet JP, Holowka D, Baird B. Molecular clustering of STIM1 with Orai1/CRACM1 at the plasma membrane depends dynamically on depletion of Ca2+ stores and on electrostatic interactions. Mol Biol Cell. 2009;20:389–399. doi: 10.1091/mbc.E07-11-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Calloway N, Holowka D, Baird B. A basic sequence in STIM1 promotes Ca2+ influx by interacting with the C-terminal acidic coiled coil of Orai1. Biochemistry. 2010;49:1067–1071. doi: 10.1021/bi901936q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu F, Sun L, Hubrack S, Selvaraj S, Machaca K. Intramolecular shielding maintains the ER Ca2+ sensor STIM1 in an inactive conformation. J Cell Sci. 2013;126:2401–2410. doi: 10.1242/jcs.117200. [DOI] [PubMed] [Google Scholar]
- 78.Muik M, Fahrner M, Schindl R, Stathopulos P, Frischauf I, Derler I, Plenk P, Lackner B, Groschner K, Ikura M, Romanin C. STIM1 couples to ORAI1 via an intramolecular transition into an extended conformation. EMBO J. 2011;30:1678–1689. doi: 10.1038/emboj.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou Y, Srinivasan P, Razavi S, Seymour S, Meraner P, Gudlur A, Stathopulos PB, Ikura M, Rao A, Hogan PG. Initial activation of STIM1, the regulator of store-operated calcium entry. Nat Struct Mol Biol. 2013;20:973–981. doi: 10.1038/nsmb.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang X, Wang Y, Zhou Y, Hendron E, Mancarella S, Andrake MD, Rothberg BS, Soboloff J, Gill DL. Distinct Orai-coupling domains in STIM1 and STIM2 define the Orai-activating site. Nat Commun. 2014;5:3183. doi: 10.1038/ncomms4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sharma S, Quintana A, Findlay GM, Mettlen M, Baust B, Jain M, Nilsson R, Rao A, Hogan PG. An siRNA screen for NFAT activation identifies septins as coordinators of store-operated Ca2+ entry. Nature. 2013;499:238–242. doi: 10.1038/nature12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Srikanth S, Jung HJ, Kim KD, Souda P, Whitelegge J, Gwack Y. A novel EF-hand protein, CRACR2A, is a cytosolic Ca2+ sensor that stabilizes CRAC channels in T cells. Nat Cell Biol. 2010;12:436–446. doi: 10.1038/ncb2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mullins FM, Park CY, Dolmetsch RE, Lewis RS. STIM1 and calmodulin interact with Orai1 to induce Ca2+-dependent inactivation of CRAC channels. Proc Natl Acad Sci U S A. 2009;106:15495–15500. doi: 10.1073/pnas.0906781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Srikanth S, Jew M, Kim KD, Yee MK, Abramson J, Gwack Y. Junctate is a Ca2+-sensing structural component of Orai1 and stromal interaction molecule 1 (STIM1) Proc Natl Acad Sci U S A. 2012;109:8682–8687. doi: 10.1073/pnas.1200667109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Palty R, Raveh A, Kaminsky I, Meller R, Reuveny E. SARAF inactivates the store operated calcium entry machinery to prevent excess calcium refilling. Cell. 2012;149:425–438. doi: 10.1016/j.cell.2012.01.055. [DOI] [PubMed] [Google Scholar]
- 86.Litjens T, Harland ML, Roberts ML, Barritt GJ, Rychkov GY. Fast Ca2+-dependent inactivation of the store-operated Ca2+ current (ISOC) in liver cells: a role for calmodulin. J Physiol. 2004;558:85–97. doi: 10.1113/jphysiol.2004.065870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bauer MC, O’Connell D, Cahill DJ, Linse S. Calmodulin binding to the polybasic C-termini of STIM proteins involved in store-operated calcium entry. Biochemistry. 2008;47:6089–6091. doi: 10.1021/bi800496a. [DOI] [PubMed] [Google Scholar]
- 88.Feng JM, Hu YK, Xie LH, Colwell CS, Shao XM, Sun XP, Chen B, Tang H, Campagnoni AT. Golli protein negatively regulates store depletion-induced calcium influx in T cells. Immunity. 2006;24:717–727. doi: 10.1016/j.immuni.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 89.Walsh CM, Doherty MK, Tepikin AV, Burgoyne RD. Evidence for an interaction between Golli and STIM1 in store-operated calcium entry. Biochem J. 2010;430:453–460. doi: 10.1042/BJ20100650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Krapivinsky G, Krapivinsky L, Stotz SC, Manasian Y, Clapham DE. POST, partner of stromal interaction molecule 1 (STIM1), targets STIM1 to multiple transporters. Proc Natl Acad Sci U S A. 2011;108:19234–19239. doi: 10.1073/pnas.1117231108. [DOI] [PMC free article] [PubMed] [Google Scholar]