Abstract
In the perinatal setting, chromosome imbalances cause a wide range of clinically significant disorders and increase risk for other particular phenotypes. As technologies have improved to detect increasingly smaller deletions and duplications, collectively referred to as copy number variants (CNVs), we are learning the significant role that these types of genomic variants play in human disease and their relatively high frequency in ~1% of all pregnancies. In this overview, we will highlight key aspects of CNV detection and interpretation used during the course of clinical care in the prenatal and neonatal periods. Since CNVs are one of the most frequent causes of a broad spectrum of human disorders, early diagnosis and accurate interpretation is important to implement timely interventions and targeted clinical management.
Keywords: copy number variant, CNV, chromosomal microarray, non-invasive prenatal testing, genomic databases, aneuploidy, prenatal, neonatal
INTRODUCTION
In the perinatal setting, chromosome abnormalities span a wide range of genomic imbalance, from polyploidy (the presence of three (triploidy) or four (tetraploidy) copies of every chromosome), to whole chromosome aneuploidy (typically involving only a single chromosome), to submicroscopic deletions and duplications that can only be detected by DNA-based copy number methods, such as Fluorescence In Situ Hybridization (FISH) or chromosomal microarray (CMA). As technologies have improved to detect smaller and smaller copy number variants (CNVs) across the genome, we are learning the very high frequency and important role that this type of genomic variation plays in human health and development.
CNVs have been identified as a common cause of a number of human diseases, many of which present in the neonatal period and/or early childhood. These include neurodevelopmental disorders (such as autism, intellectual disability and epilepsy), congenital heart defects, and other congenital anomalies.1-3 Not all CNVs, however, are disease-causing: some CNVs have been identified in apparently normal individuals.4,5 Whether a CNV is disease-causing or not depends on many factors, such as gene content (e.g., a CNV that is gene-rich is more likely to cause a phenotype than one containing few or no genes).6 Therefore, understanding the corresponding phenotypic effects of particular CNVs is becoming increasingly important in clinical medicine so we can define which CNVs cause a clinical phenotype versus those that are part of normal variation.
In this overview, we will highlight key aspects of copy number detection during the prenatal and neonatal periods. Many infants presenting to neonatology services for a possible genetic diagnosis may have had prenatal testing; it is important to understand which test was performed to interpret the results and know if additional genetic testing is warranted. If, on the other hand, prenatal testing was not done, then decisions will need to be made about which genetic test(s) are most appropriate to order. To make informed test ordering decisions, it is important for neonatologists and other providers to understand the limitations and benefits of the various laboratory technologies. Therefore, we will compare methods for CNV detection. We will also explore some of the more common CNVs associated with disease and how interpretation of CNVs is accomplished through the use of various resources, including online genomic databases. Given that CNVs are now appreciated as one of the most frequent causes of a broad spectrum of human disorders, early diagnosis and accurate interpretation is important to implement timely interventions and targeted clinical management.
METHODS FOR THE DETECTION OF COPY NUMBER VARIANTS
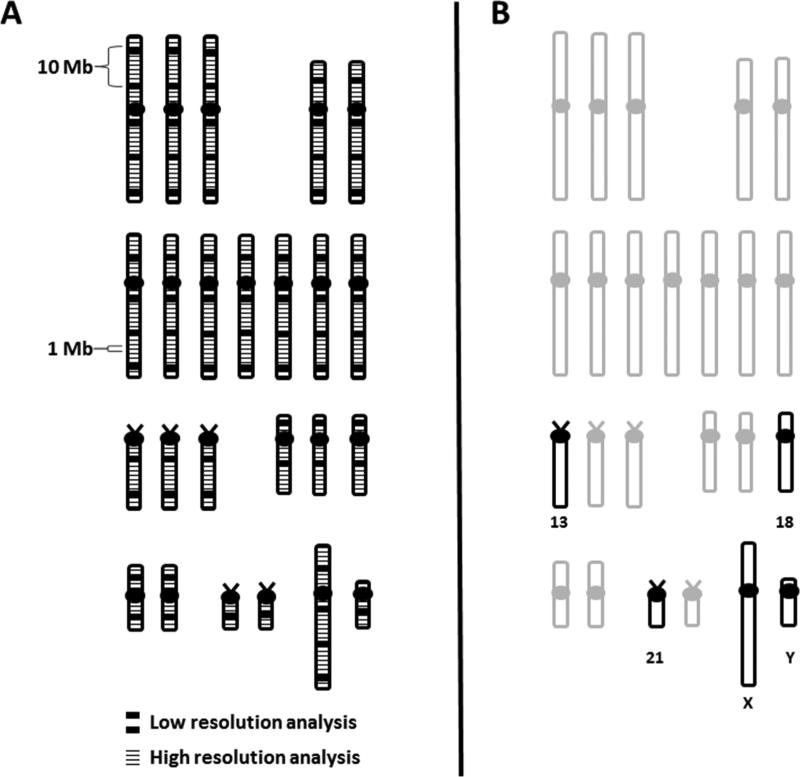
Various methods have been developed over the years for the detection of chromosomal deletions, duplications and rearrangements. As shown in Figure 1, some of these methods allow genome-wide analyses, where the entire chromosome complement is being interrogated, while others are targeted analyses and only examine specific regions of the genome. In addition, methods can differ in their level of resolution (i.e., how small of an imbalance can be detected) and the type of sample that can be analyzed. Table 1 summarizes the more commonly used cytogenetic methods for the detection of chromosome abnormalities and compares the benefits and limitations of each.
Figure 1.
Comparison of genome-wide versus targeted analyses for CNV detection using schematic diagrams of a human karyotype. A) Genome-wide analysis by G-banding or CMA. The thick black lines correspond to the lower resolution obtained from traditional G-banding analysis, while the thin black lines correspond to the higher resolution from newer techniques, like CMA. An example is depicted in the diagram, showing that CMA can detect an imbalance of 1 Megabase (Mb) in size that would be missed by G-banding. G-banding on the hand, could only detect larger imbalances, such as the 10 Mb abnormality shown. B) Targeted analysis. The only chromosomes being analyzed by targeted analysis are shown in black. The grey chromosomes would not be analyzed by targeted tests.
Table 1.
Benefits and limitations of tests used to detect copy number variation.
| Test Type | Test Evaluates | Test Benefits | Test Limitations |
|---|---|---|---|
| G-banded chromosome analysis | Genome-wide large scale deletions, duplications and structural rearrangementsa; aneuploidy | Can detect balanced rearrangements | Cannot detect small imbalances (<3-5 Mb); May miss low-level mosaic cases |
| Chromosomal microarray analysis | Genome-wide deletions and duplications; aneuploidy | Can detect small deletions and duplications not detected by G-banded karyotype; can be done using DNA isolated from uncultured cells | Cannot detect balanced rearrangements (e.g., translocations or inversions); Does not give information about the mechanism of an imbalance; May miss low-level mosaic cases |
| Chromosomal microarray analysis, with SNPs | Genone-wide deletions and duplications; aneuploidy | Able to detect some uniparental disomies; can detect regions of absence of heterozygosity (AOH) and consanguinity | Cannot detect balanced rearrangements (e.g., translocations or inversions); Does not give information about the mechanism of an imbalance; May miss low-level mosaic cases |
| Interphase FISH for aneuploidy | Aneuploidy of specific chromosomes (13, 18, 21, X, Y) | Performed on uncultured cells | Limited to certain regions of the genome |
| Non-invasive prenatal screening (NIPS) | Aneuploidy of specific chromosomes (13, 18, 21, X, Y) | Performed using a blood sample from the mother | Limited to certain regions of the genome |
Structural rearrangements can include translocations, inversions, supernumerary chromosomes, etc.
Of the techniques listed in Table 1, G-banded chromosome analysis and CMA are the only ones that are considered genome-wide analyses, where the entirety of each chromosome is being analyzed. However, the resolution of CMA far exceeds that of G-banding; genomic imbalances that could only be approximated by G-banding analysis can now be measured with precision by CMA based on the ability to link the probes contained on a microarray with the underlying DNA sequence coordinates. For these reasons, and others detailed later, CMA has become the first-tier test for clinical cytogenetic testing in the pediatric setting.
Most genome-wide microarrays used for clinical CMA now also include single nucleotide polymorphism (SNP) probes in addition to probes used for copy number detection. The addition of SNP probes offers several advantages. For example, SNP probes allow the detection of triploidy and some cases of tetraploidy.7 These abnormalities are usually not detectable by copy number analyses alone, but are important to identify in the prenatal setting since both are common causes of fetal loss. In addition, genomic regions with an absence of heterozygosity (AOH) may be detected. AOH can suggest the presence of uniparental disomy (UPD), where homologous chromosomes are both inherited from the same parent, instead of one from each parent; UPD for certain chromosomes has been associated with genetic disorders, such as Prader-Willi syndrome when both chromosomes 15 are inherited from the mother in about 20-25% of cases. AOH can also distinguish genomic regions that are identical by descent, which could increase risk for an autosomal recessive disorder if a deleterious mutation is present. The use of SNP arrays for these indications is not a diagnostic test, however both of these findings could prompt targeted diagnostic testing for UPD or sequencing of a specific autosomal recessive gene based on the patient's clinical phenotype.8
In contrast to genome-wide methods, targeted methods for the detection of cytogenetic aberrations are used to examine specific regions of the genome, such as aneuploidy for a single chromosome or deletion/duplication of a region associated with a known genetic syndrome. With the adoption of CMA, most targeted tests for microdeletion or microduplication syndromes are not used anymore, since many of these syndromes lack distinctive phenotypic findings and CMA can test for multiple regions in one assay instead of testing for one disorder at a time.1
Targeted tests are still predominantly used for aneuploidy testing of the chromosomes most frequently involved in human disorders, including 13, 18, 21, X and Y, particularly in the prenatal setting or when a trisomy is suspected in a neonate based on clinical features. Table 1 compares two targeted methods for aneuploidy detection: FISH and non-invasive prenatal screening (NIPS, discussed below in more detail).
PRENATAL DIAGNOSIS OF COPY NUMBER VARIANTS
As mentioned in the Introduction, infants presenting to neonatology services for a possible genetic diagnosis may or may not have had prenatal testing. It is important for providers to understand these laboratory tests and the results to accurately determine if any additional genetic testing is necessary. For example, if the mother had an amniocentesis with chromosome analysis during pregnancy and that test was normal, has a chromosome abnormality been ruled out or should other genetic testing be pursued?
Amniocentesis was first shown to be a safe and accurate method for prenatal diagnosis of genetic anomalies in the early 1970s.9 Since that time, approaches to prenatal screening and diagnosis for chromosomal aberrations have quickly evolved based on new technologies and emerging practices. When considering results from prenatal testing, it is important to understand the difference between diagnostic and screening tests, and between genome-wide versus targeted testing, since different levels of information are obtained.
Diagnostic tests provide an accurate representation of the fetal chromosome complement; currently, all prenatal diagnostic tests require an invasive procedure, such as amniocentesis or chorionic villi sampling, to obtain a sample directly from the fetus or placenta. Screening tests, on the other hand, have risk for false-positive and false-negative results, since the sample is not being directly obtained from the fetus. Some commonly used non-invasive screening tests for aneuploidy, which are performed on a blood sample from the mother of the fetus, include maternal serum screening and NIPS.
G-banded chromosome analysis has historically been the gold-standard for detecting genome-wide prenatal chromosome abnormalities. However, several large studies have now compared the diagnostic yield of G-banding to genome-wide CMA for prenatal diagnosis and shown that a significant proportion of clinically relevant chromosome aberrations are missed by G-banding alone.10,11 Callaway et al. (2013) recently carried out a systematic review of the literature, including more than 12,000 prenatal cases that had CMA after a normal karyotype. This analysis revealed clinically significant CNVs in 2.4% of cases, with the highest yield in cases ascertained for an abnormal ultrasound (6.5%). However, even cases referred due to increased maternal age or for other reasons, such as abnormal serum screening or parental anxiety, had significant yields of 1.0% and 1.1%, respectively. Despite these data, in the prenatal setting, array is still not considered standard of care for all pregnancies. The most recent recommendations from the American Congress of Obstetricians and Gynecologists (ACOG), published in 2013, allow CMA to replace a G-banded karyotype when ultrasound anomalies are detected and invasive testing is being pursued. Either a karyotype or CMA can be used in patients undergoing invasive testing with a structurally normal fetus.12 Therefore, if only a G-banded karyotype is performed prenatally and is normal, CMA should be ordered in a neonate presenting with features suggestive of a chromosomal disorder. Box 1 lists some of the more common clinical features that should prompt consideration of a chromosomal disorder; the presence of more than one of these findings in a patient raises the suspicion for a genetic etiology proportionately.
A rapidly evolving field in prenatal diagnosis is non-invasive prenatal screening (NIPS), also referred to as non-invasive prenatal testing (NIPT). Even though a recent study showed no increased risk for fetal loss due to invasive procedures13, the misperception that any invasive test carries some increased risk for fetal loss still exists. In addition, some women do not want invasive testing, independent of the risk for fetal loss. These two issues have been the main driving factors for technological developments for non-invasive screening methods and the uptake of non-invasive testing by patients. In fact, with the advent of NIPS in 2011, the number of amniocentesis and CVS procedures has significantly decreased, as demonstrated by data from several maternal-fetal medicine centers.14-17 NIPS is based on the detection of cell-free fetal DNA in maternal plasma using next-generation sequencing or other methods for fetal DNA assessment.14,18 At this time, NIPS is mainly used for the targeted detection of common aneuploidies (13, 18, 21, X and Y; shown in Figure 1B). However, the technology is already being refined to detect common microdeletion/duplication syndromes as well as genome-wide CNVs.19 Since NIPS is currently only a targeted screening test, a complete evaluation of the fetal genome is not obtained and clinically significant chromosome abnormalities could be missed. In addition, several cases of discordant results between NIPS and diagnostic cytogenetic testing have been reported (Pan 2013; Wang 2014). Therefore, in the context of a neonate presenting with features suggestive of a chromosomal disorder, if the only result from prenatal genetic testing is a normal NIPS test, additional genetic testing is warranted.
COPY NUMBER DETECTION IN THE NEONATAL PERIOD
Although some chromosome abnormalities may be suspected and tested for in the prenatal period due to ultrasound abnormalities or other clinical indications, most are not suspected until birth when dysmorphic features, congenital malformations or other anomalies are observed. Early studies of the frequency of chromosome abnormalities in newborns estimated the rate to be ~4% from chromosome analysis.20,21 Aneuploidies of chromosomes 21, X and Y were the most common abnormalities detected, with trisomy of chromosomes 13 or 18, unbalanced rearrangements and supernumerary chromosomes occurring less frequently.
With the advent of CMA it was hypothesized that the contribution of chromosomal imbalances in neonates was being underestimated. Indeed this hypothesis was proved to be true by a large study of 638 neonates with various birth defects who were referred for clinical CMA.22 Clinically significant imbalances were detected in 17.1% of patients, the majority of which would not have been identified by G-banding analysis. While there were various reasons for referral for CMA testing among the samples with abnormal findings, the highest diagnostic yield was observed in the author-defined category “possible chromosome abnormality +/− other birth defect” (66.7%). Other high yield clinical indications were “ambiguous genitalia +/− other birth defect” (33.3%), “dysmorphic features with multiple congenital anomalies +/− other birth defect” (24.6%) and “congenital heart disease +/− other birth defect” (21.8%). Overall, 2.5% of abnormal cases had whole chromosome aneuploidies while 12.7% had deletions or duplications
Importantly, at the time of this study, high-resolution genome-wide CMA analysis had not yet been developed for clinical testing. The arrays used in this study were targeted arrays (containing coverage over known clinically relevant regions of the genome, such as microdeletion/duplication syndromes, telomeres and centromeres) with only low resolution coverage across the rest of the genome, corresponding to approximately one targeted region per chromosome band at the 650 band level of resolution (~5-10 Megabases). Even with coverage at a lower resolution than employed in currently available clinical arrays, this study still identified abnormalities in a significant number (17.1%) of neonates. With the higher resolution arrays currently being used, this diagnostic yield is predicted to be even greater, demonstrating the importance of CMA in the clinical care of neonates.
Since congenital heart defects (CHDs) are one of the most frequent birth defects, and also a common indication for cytogenetic testing in the neonatal period, many studies have focused on the contribution of CNVs to isolated CHDs and CHDs with other associated defects. A recent review including data from 20 studies examined the diagnostic yield of CMA in CHDs.3 Clinically relevant CNVs were reported in 3-25% of patients with CHDs plus other associated defects, with many of these studies in the 17-20% range. Even in cases with isolated CHDs, the diagnostic yield was still significant with 3-10% of cases having a clinically relevant CNV. Thus most CHDs, whether observed in the context of additional phenotypic findings or as isolated defects, warrant consideration of CMA to detect pathogenic CNVs.
The most common submicroscopic CNV associated with CHDs is a deletion of 22q11.2. This CNV is estimated to occur in 1 in 2,000 to 1 in 4,000 livebirths. In addition to CHDs, the most common being conotruncal defects, individuals with a 22q11.2 deletion can show various clinical features including palatal abnormalities, hypocalcemia, immune deficiency and a range of neurodevelopmental disorders.23 In ~10% of cases, this deletion is inherited from an affected parent who usually has a more mild presentation than the proband; therefore, parental testing to determine inheritance is important for recurrence risk estimates and familial genetic counseling.
More broadly, the implementation of high-resolution genome-wide CMA for other common postnatal indications, such as developmental delay, intellectual disability, autism spectrum disorder or multiple congenital anomalies, has also demonstrated a diagnostic yield that far surpasses that of G-banding. A systematic literature review of 33 CMA studies of these patient populations estimated that ~15-20% have a clinically relevant CNV, compared to a yield of only ~3% from G-banding (the 3% estimate excluded Down syndrome and other recognizable chromosomal syndromes).1 These data ultimately resulted in CMA being recommended as the first-tier clinical test for individuals with developmental disorders or congenital anomalies by several groups, including the American College of Medical Genetics and Genomics (ACMG).1,24
CLINICAL INTERPRETATION OF COPY NUMBER VARIANTS
The use of CMA has obvious advantages over previous cytogenetic methods for diagnostic yield. Another invaluable benefit of CMA as a diagnostic test is the ability to immediately link the genomic coordinates from the DNA probes contained on the array to the human genome sequence to evaluate size, gene content and other elements that make up the architecture of the human genome. With the wide range of copy number variation present in the human genome, we are still learning which variation from individuals is causative of disease and which has a benign or negligible impact. Cataloging both benign and pathogenic regions of the genome is imperative to aid in the clinical interpretation of CNVs.
Recurrent and Non-recurrent CNVs
The collection of CNVs from across the genome has allowed us to compare overlapping CNVs to determine underlying mechanisms and the resulting phenotypic effects. Although it has been estimated that the majority (~75%) of CNVs occur at non-recurrent sites across the genome, ~25% of CNVs are mediated by non-allelic homologous recombination between flanking sequences of shared DNA sequence homology (commonly referred to as segmental duplications or genomic hotspots) and make up a class of CNVs termed “recurrent CNVs”. 25,26 Because these CNVs, which contain identical unique genomic regions of imbalance across patients, recur due to their underlying mechanism, they are frequently encountered during CMA analysis.
Table 2 lists some of the more frequently observed recurrent CNVs that are encountered during clinical CMA testing. Some of these CNVs (e.g., Prader-Willi/Angelman syndromes and 22q11.2 deletion syndrome) have been described for some time now because they were associated with a specific syndrome and detected either through high-resolution G-banding or FISH analyses. Other CNVs, with more variable phenotypes (e.g., deletions and duplications of 1q21.1 and 16p11.2), have only emerged recently due to our ability to detect smaller imbalances across the genome via CMA. Targeted research studies comparing the phenotype of individuals with many of these recurrent CNVs are now underway to better define the deleterious impact of each CNV.27,28
Table 2.
Frequently observed recurrent CNVs identified among clinical populations referred for CMA testinga.
| CNVb | Copy Number | Syndrome | Size | Genomic Coordinates (hg19) |
|---|---|---|---|---|
|
Highly Penetrant Phenotype
| ||||
| 7q11.23 (ELN) | deletion | Williams | 1.4 Mbc | chr7:72744455-74142513 |
| 8p23.1 (SOX7, CLDN23) | deletion | 3.6 Mb | chr8:8119296-11765719 | |
| 8p23.1 (SOX7, CLDN23) | duplication | 3.6 Mb | chr8:8119296-11765719 | |
| 15q11.2q13 BP2-3d (UBE3A) | deletion | Prader-Willi or Angelman | 4.8 Mb | chr15:23758391-28557186 |
| 17p11.2 (RAI1) | deletion | Smith-Magenis | 3.5 Mb | chr17:16757112-20219651 |
| 17p11.2 (RAI1) | duplication | Potocki-Lupski | 3.5 Mb | chr17:16757112-20219651 |
| 17q21.31 (MAPT, KANSL1) | deletion | Koolen-de Vries | ||
| 22q11.2 (TBX1, HIRA) | deletion | DiGeorge/Velo-cardio-facial | 2.9 Mb | chr22:18661726-21561514 |
|
Variable Clinical Phenotype | ||||
| 1q21.1 (GJA5) | deletion | 0.8 Mb | chr1:146577487-147394506 | |
| 1q21.1 (GJA5) | duplication | 0.8 Mb | chr1:146577487-147394506 | |
| 7q11.23 (ELN) | duplication | 1.4 Mb | chr7:72744455-74142513 | |
| 15q11.2q13 BP2-3c (UBE3A) | duplication | 4.8 Mb | chr15:23758391-28557186 | |
| 15q13.3 BP4-5 (KLF13, CHRNA7) | deletion | 1.3 Mb | chr15:31137105-32445408 | |
| 15q13.3 BP4-5 (KLF13, CHRNA7) | duplication | 1.3 Mb | chr15:31137105-32445408 | |
| 16p11.2 (TBX6) | deletion | 0.6 Mb | chr16:29649997-30199855 | |
| 16p11.2 (TBX6) | duplication | 0.6 Mb | chr16:29649997-30199855 | |
| 16p11.2 distal (SH2B1) | deletion | |||
| 16p11.2 distal (SH2B1) | duplication | |||
| 16p12.1 (CDR2, EEF2K) | deletion | |||
| 16p13.11 (MYH11) | deletion | |||
| 17q12 (HNF1B) | deletion | Renal cysts and diabetes | ||
| 17q12 (HNF1B) | duplication | |||
| 22q11.2 (TBX1, HIRA) | duplication | 2.9 Mb | chr22:18661726-21561514 | |
The list is divided into CNVs that have a highly penetrant phenotype and those with more variable phenotypic presentations. Within each category, the CNVs are listed in chromosomal order. The CNV list was compiled from multiple sources, but should not be considered exhaustive: DECIPHER syndrome list (https://decipher.sangeer.ac.uk/syndromes#overview), ClinGen pathogenic list (http://www.ncbi.nlm.nih.gov/dbvar, study ID nstd45) and additional references25,36-39
genes in CNV region included as landmarks for genomic location and are not necessarily known to be causative of phenotype
Mb, Megabase
BP, breakpoint
Interpretation Guidelines
The technical definition of a CNV is “a segment of DNA that is ≥ 1 kilobase (kb) in size that differs in copy number compared with a representative reference genome”.29 However, most CNVs that are less than ~400 kb in size are observed frequently in cohorts of apparently normal control individuals, and are therefore not believed to have appreciable effects on human health and/or development.30 Because of this, for the purposes of detecting CNVs as part of clinical testing, several organizations have recommended a resolution of ≥ 400 kb across the genome to avoid detection of these common, benign CNVs.1,31 It should be noted that some array designs used by clinical laboratories contain higher resolution coverage (~20-50 kb) over known disease causing genes in order to detect single gene deletions or duplications.
It is important to note that the term CNV must be qualified with additional information in order to understand the clinical relevance of the finding: 1) a CNV must be designated as a deletion or duplication, and 2) a CNV should have a defined category of clinical significance. As outlined in Table 3, the ACMG has defined five categories for interpreting the clinical significance of CNVs and examples of each are listed. CNVs included on clinical reports should be classified into one of these categories so that clinicians can review the laboratory findings and correlate with their patient's clinical phenotype. It is not uncommon (~10% of cases) that a CNV could be reported as “uncertain clinical significance” based on limited information that the laboratory had at the time of testing, but when a clinician reviews the CNV detected and pairs it with more detailed phenotypic data from the patient, a more definitive interpretation of “pathogenic” can often be made. This example highlights the critical need for coordinated communication between clinical laboratories and clinicians for accurate interpretation of genomic testing.
Table 3.
ACMG recommended categories for defining clinical significance of a CNV
| CATEGORY | DEFINITION | EXAMPLE |
|---|---|---|
| Pathogenic | CNV is documented as clinically significant consistently and in multiple publications and/or case databases | 22q11.2 deletion syndrome (DiGeorge/velocardiofacial syndrome) |
| Uncertain (no subclassification) | Clinical significance not know at time of reporting but CNV meets the laboratory reporting criteria; expected that CNVs in this category will shift toward pathogenic or benign over time | 500 kb deletion of chromosome 3 that contains 5 genes but the inheritance of the deletion is not known and there is nothing known about the 5 genes in the deleted interval |
| Sub-classifications: | Some evidence to increase the likelihood that the CNV is pathogenic | 500 kb de novo deletion of chromosome 3 that contains 5 genes and there is a single case report in the literature that has similar breakpoints and shares a distinct phenotype |
| Uncertain: Likely Pathogenic | ||
| Uncertain: Likely Benign | Some evidence to increase the likelihood that the CNV is benign | 500 kb deletion of chromosome 3 that does not contain any genes but is not found in any control databases |
| Benign | CNV is documented as benign consistently and in multiple publications and/or control databases or represents a common polymorphism (present in >1% of the population) | 2q37.3 telomere polymorphism (~200 kb in size and well-documented as benign in multiple studies) |
Adapted from Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, et al. Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee. American college of medical genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med 2011; 13(7):680-685; with permission.
When a CNV is identified, a number of characteristics of the genomic region that is either deleted or duplicated need to be considered in interpreting its significance. The bulleted list below documents some of the basic questions to investigate:
-
1)
Is the CNV included in databases of normal variation? If so, the CNV is considered a benign variant. If not, then the potential pathogenicity needs to be evaluated.
-
2)
Does the CNV contain a region of the genome known to cause a genomic syndrome when deleted or duplicated (e.g., a recurrent CNV region associated with a particular phenotype)? If so, then the CNV would be consistent with causing the syndrome that corresponds to either a deletion or duplication of that region, depending on the CNV finding.
-
3)
Does the CNV contain a gene that is known to cause a syndrome as a result of haploinsufficiency (deletion) or triplosensitivity (duplication)? If so, then the CNV that encompasses the entire gene would be consistent with causing the syndrome that corresponds to either a deletion or duplication of that gene, depending on the CNV finding.
-
4)
If the CNV does not overlap a known region or gene, what is the gene content and size of the CNV? In general, a larger imbalance with high gene content is more likely to be considered pathogenic.
-
5)
Is the CNV de novo? In general, a de novo CNV is more likely to be pathogenic than one inherited from a parent with an apparently normal phenotype.
-
6)
Is the CNV inherited? If a CNV is inherited, then it is important to evaluate the phenotype of the parent carrying the same CNV. The parent could be affected with the same clinical phenotype as the proband. Alternatively, the parent could have more subtle phenotypic effects than the proband caused by variable expressivity of the CNV. The CNV could also be a benign variant if it is observed frequently in the general population.
Genomic Resources for CNV Curation
Even though new genomic discoveries are made and published every day, the interplay between genomic variants and their impact on various systems involved in human development and function is still much of a mystery. Since many genomic variants are rare, community efforts are needed to assist in deciphering the clinical significance of genomic variants in an evidence-based manner. Towards the goal of curating genome-wide CNVs, multiple online genome resources, as detailed in de Leeuw et al. (2012)32, have been garnered from large-scale data sharing and are now publically available.
Table 4 lists some of the online resources for CNVs and their corresponding phenotypes that are most commonly used for interpreting clinical significance. The table includes three different types of tools that can be used to aid in CNV interpretation: 1) genome browsers, where one can enter the genomic coordinates of a particular CNV and use the browser to view its genomic content; 2) databases of CNVs submitted from case and control cohorts, which can be used to compare individual cases to other previously observed CNVs; 3) catalogs of phenotypic information collected from the literature or written by experts in the field that provide overviews of well-described syndromes or gene/disease associations. All of these resources are dynamic and evolving at a rate that largely depends on the discovery, data submission and curation efforts of researchers, clinical laboratories, clinicians and others.
Table 4.
Online, publically available genome resources with CNV and phenotype data.
| Database | Web Address | Case CNV Data | Control CNV Data | Phenotype Data |
|---|---|---|---|---|
| GENOME BROWSERS | ||||
| dbVara | http://www.ncbi.nlm.nih.gov/dbvar/ | X* | X* | X* |
| Ensembl | http://www.ensembl.org | X* | X* | X* |
| UCSC Genome Browser | https://genome.ucsc.edu/ | X* | X* | X* |
| DATABASES OF CNVS SUBMITTED FROM CASE AND CONTROL COHORTS | ||||
| ClinVarb | http://www.ncbi.nlm.nih.gov/clinvar/ | X | X | |
| DECIPHERc | https://decipher.sanger.ac.uk/ | X | X | |
| DGVd | http://dgv.tcag.ca/dgv/app/home | X | ||
| ECARUCAe | http://ecaruca.net | X | X | |
| CATALOGS OF PHENOTYPIC INFORMATION | ||||
| GeneReviews | http://www.genetests.org/resources/genereviews.php | X | ||
| MedGen | http://www.ncbi.nlm.nih.gov/medgen | X | ||
| OMIMf | http://www.omim.org/ | X | ||
| Orphanet | http://www.orpha.net/ | X | ||
displays data in browser format the is derived from several of the CNV and phenotype resources listed below
dbVar, Database of genomic structural variation
Main submission portal for data from the groups previously known as the International Standards for Cytogenomic Arrays (ISCA) and the International Consortium for Clinical Genomics (ICCG), which are now integrated into the Clinical Genome Resource (ClinGen)
DECIPHER, Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources
DGV, Database of Genomic Variation
European Cytogeneticists Association Register of Unbalanced Chromosome Aberrations
Online Mendelian Inheritance in Man
CONCLUSION AND FUTURE DIRECTIONS
As discussed above, CNVs provide a genetic etiology for a wide range of disorders diagnosed in the prenatal and neonatal periods. There are a growing number of examples where knowing a genetic etiology leads to genome-directed clinical care and improved medical management. For example, in neonates with 22q11.2 deletions, not only is it important to assess for all of the congenital anomalies associated with this CNV, but it is also important to monitor neonatal calcium levels. A recent study showed that neonatal seizures and neonatal hypocalcemia were predictors of a more severe level of intellectual disability. The authors thus concluded that early monitoring of calcium levels prior to seizure onset might improve outcomes in these patients by preventing damage to neurons caused by seizures.33
The continuing evolution of genomic technologies for the detection of CNVs and aneuploidy in the perinatal setting will allow earlier diagnosis of these conditions in fetuses and neonates. Indeed, next-generation whole-exome (WES) and whole-genome sequencing (WGS) methods are already being used to detect CNVs in postnatal samples and the feasibility of using WGS for non-invasive sequencing of a human fetus by analyzing parental blood samples was recently reported.34,35 As the decreasing costs of WES and WGS make broader adoption possible, one can envision an era of genomic medicine where it is feasible to routinely carry out these genome-wide methods for variant detection on neonatal or ultimately prenatal samples collected non-invasively. Through our increasing understanding of the interplay between genomic variants and health, we have the potential to realize the full benefits of personalized genomic medicine resulting in earlier interventions and improved outcomes.
KEY POINTS.
-
1)
Copy number variants (CNVs) are a common cause of a wide-range of human disorders, accounting for ~15% of neurodevelopmental disorders, cardiac abnormalities and other congenital anomalies.
-
2)
Various methods are available to detect CNVs, including those that can identify CNVs across the entire genome and those that only target specific regions of the genome (e.g., the common aneuploidies involving chromosomes 13, 18, 21, X and Y).
-
3)
Accurate clinical interpretation of CNVs requires incorporation of genotype plus phenotype information.
-
4)
Identifying a genetic etiology for a patient's phenotype can help to define targeted interventions and clinical management.
Box 1.
Selected clinical features that suggest the presence of a chromosomal disorder.
| Congenital anomalies |
| Examples: structural abnormalities of the heart, renal system, skeletal system, and/or brain |
| Dysmorphic features |
| Hypotonia |
| Intrauterine growth retardation |
| Failure to thrive |
| Microcephaly |
| Seizures |
| Ambiguous genitalia |
ACKNOWLEDGEMENTS
The authors would like to thank Erin Rooney Riggs, MS, for critically reviewing this article and Thomas Challman, MD, and Eli Williams, PhD, for useful discussions about selected information presented. This work was supported in part by NIH grants RO1 MH074090 and U41 HG006834 (both to CLM and DHL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement: David H. Ledbetter is a consultant to Natera, Inc.
REFERENCES
- 1*.Miller DT, Adam MP, Aradhya S, et al. Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86(5):749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mefford HC, Yendle SC, Hsu C, et al. Rare copy number variants are an important cause of epileptic encephalopathies. Ann Neurol. 2011;70(6):974–985. doi: 10.1002/ana.22645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3*.Lander J, Ware SM. Copy number variation in congenital heart defects. Curr Genet Med Rep. 2014;2:168–178. [Google Scholar]
- 4.Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36(9):949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 5.Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305(5683):525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 6.Coe BP, Girirajan S, Eichler EE. A genetic model for neurodevelopmental disease. Curr Opin Neurobiol. 2012;22(5):829–836. doi: 10.1016/j.conb.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lathi RB, Massie JA, Loring M, et al. Informatics enhanced SNP microarray analysis of 30 miscarriage samples compared to routine cytogenetics. PLoS One. 2012;7(3):e31282. doi: 10.1371/journal.pone.0031282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.South ST, Lee C, Lamb AN, Higgins AW, Kearney HM. Working Group for the American College of Medical Genetics and Genomics Laboratory Quality Assurance Committee. ACMG standards and guidelines for constitutional cytogenomic microarray analysis, including postnatal and prenatal applications: Revision 2013. Genet Med. 2013;15(11):901–909. doi: 10.1038/gim.2013.129. [DOI] [PubMed] [Google Scholar]
- 9.Nadler HL, Gerbie AB. Role of amniocentesis in the intrauterine detection of genetic disorders. N Engl J Med. 1970;282(11):596–599. doi: 10.1056/NEJM197003122821105. [DOI] [PubMed] [Google Scholar]
- 10.Wapner RJ, Martin CL, Levy B, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med. 2012;367(23):2175–2184. doi: 10.1056/NEJMoa1203382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11*.Callaway JL, Shaffer LG, Chitty LS, Rosenfeld JA, Crolla JA. The clinical utility of microarray technologies applied to prenatal cytogenetics in the presence of a normal conventional karyotype: A review of the literature. Prenat Diagn. 2013;33(12):1119–1123. doi: 10.1002/pd.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The American College of Obstetricians and Gynecologists Committee on Genetics and Society for Maternal-Fetal Medicine The use of chromosomal microarray analysis in prenatal diagnosis. ACOG committee opinion no. 581. Obstet Gynecol. 2013;122:1374–1377. doi: 10.1097/01.AOG.0000438962.16108.d1. [DOI] [PubMed] [Google Scholar]
- 13*.Akolekar R, Beta J, Picciarelli G, Ogilvie C, D'Antonio F. Procedure-related risk of miscarriage following amniocentesis and chorionic villus sampling: A systematic review and meta-analysis. Ultrasound Obstet Gynecol. 2014 Jul 17; doi: 10.1002/uog.14636. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 14*.Lo JO, Cori DF, Norton ME, Caughey AB. Noninvasive prenatal testing. Obstet Gynecol Surv. 2014;69(2):89–99. doi: 10.1097/OGX.0000000000000029. [DOI] [PubMed] [Google Scholar]
- 15.Beamon CJ, Hardisty EE, Harris SC, Vora NL. A single center's experience with noninvasive prenatal testing. Genet Med. 2014 Mar 27; doi: 10.1038/gim.2014.20. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 16.Louis-Jacques A, Burans C, Robinson S, Schofield E, Smulian J, Rochon M. Effect of commercial cell-free fetal DNA tests for aneuploidy screening on rates of invasive testing. Obstet Gynecol. 2014;123(Suppl 1):67S. [Google Scholar]
- 17.Pettit KE, Hull AD, Korty L, Jones MC, Pretorius DH. The utilization of circulating cell-free fetal DNA testing and decrease in invasive diagnostic procedures: An institutional experience. J Perinatol. 2014 May 29; doi: 10.1038/jp.2014.102. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 18.Bianchi DW, Platt LD, Goldberg JD, et al. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012;119(5):890–901. doi: 10.1097/AOG.0b013e31824fb482. [DOI] [PubMed] [Google Scholar]
- 19.Yu SC, Jiang P, Choy KW, et al. Noninvasive prenatal molecular karyotyping from maternal plasma. PLoS One. 2013;8(4):e60968. doi: 10.1371/journal.pone.0060968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hook EB. Chromosome abnormalities: Prevalence, risks and recurrence. In: Brock DJH, Rodeck CH, Ferguson-Smith MA, editors. Prenatal diagnosis and screening. Churchill Livingstone; Edinburgh: 1992. p. 351. [Google Scholar]
- 21.Jacobs PA, Browne C, Gregson N, Joyce C, White H. Estimates of the frequency of chromosome abnormalities detectable in unselected newborns using moderate levels of banding. J Med Genet. 1992;29(2):103–108. doi: 10.1136/jmg.29.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu XY, Phung MT, Shaw CA, et al. Genomic imbalances in neonates with birth defects: High detection rates by using chromosomal microarray analysis. Pediatrics. 2008;122(6):1310–1318. doi: 10.1542/peds.2008-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.McDonald-McGinn DM, Emanuel BS, Zackai EH. 22q11.2 deletion syndrome. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews [internet] University of Washington; 2013. [PubMed] [Google Scholar]
- 24.Manning M, Hudgins L. Professional Practice and Guidelines C. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med. 2010;12(11):742–745. doi: 10.1097/GIM.0b013e3181f8baad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaminsky EB, Kaul V, Paschall J, et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet Med. 2011;13(9):777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharp AJ, Locke DP, McGrath SD, et al. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet. 2005;77(1):78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanson E, Bernier R, Porche K, et al. The cognitive and behavioral phenotype of the 16p11.2 deletion in a clinically ascertained population. Biol Psych. 2014 Jun 16; doi: 10.1016/j.biopsych.2014.04.021. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28*.Bassett AS, McDonald-McGinn DM, Devriendt K, et al. Practical guidelines for managing patients with 22q11.2 deletion syndrome. J Pediatr. 2011;159(2):332–339. doi: 10.1016/j.jpeds.2011.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST. Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee. American college of medical genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011;13(7):680–685. doi: 10.1097/GIM.0b013e3182217a3a. [DOI] [PubMed] [Google Scholar]
- 30.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444(7118):444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kearney HM, South ST, Wolff DJ, et al. American college of medical genetics recommendations for the design and performance expectations for clinical genomic copy number microarrays intended for use in the postnatal setting for detection of constitutional abnormalities. Genet Med. 2011;13(7):676–679. doi: 10.1097/GIM.0b013e31822272ac. [DOI] [PubMed] [Google Scholar]
- 32.de Leeuw N, Dijkhuizen T, Hehir-Kwa JY, et al. Diagnostic interpretation of array data using public databases and internet sources. Hum Mutat. 2012 doi: 10.1002/humu.22049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheung EN, George SR, Andrade DM, Chow EW, Silversides CK, Bassett AS. Neonatal hypocalcemia, neonatal seizures, and intellectual disability in 22q11.2 deletion syndrome. Genet Med. 2014;16(1):40–44. doi: 10.1038/gim.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34*.Zhao M, Wang Q, Wang Q, Jia P, Zhao Z. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: Features and perspectives. BMC Bioinformatics. 2013;14(Suppl 11):S1. doi: 10.1186/1471-2105-14-S11-S1. Epub Sep 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitzman JO, Snyder MW, Ventura M, et al. Noninvasive whole-genome sequencing of a human fetus. Sci Transl Med. 2012;4(137):137ra76. doi: 10.1126/scitranslmed.3004323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cooper GM, Coe BP, Girirajan S, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43(9):838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreno-De-Luca D, Sanders SJ, Willsey AJ, et al. Using large clinical data sets to infer pathogenicity for rare copy number variants in autism cohorts. Mol Psych. 2013;18(10):1090–1095. doi: 10.1038/mp.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenfeld JA, Coe BP, Eichler EE, Cuckle H, Shaffer LG. Estimates of penetrance for recurrent pathogenic copy-number variations. Genet Med. 2013;15(6):478–481. doi: 10.1038/gim.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39*.Ledbetter DH, Riggs ER, Martin CL. Ginsburg GS, Willard HF, editors. Clinical applications of whole-genome chromosomal microarray analysis. Genomic and personalized medicine. (2nd ed.) 2013;1:133. [Google Scholar]