Abstract
Because adhesion of leukocytes to endothelial cells is the first step of vascular-neuronal inflammation, inhibition of adhesion and recruitment of leukocytes to vascular endothelial cells will have a beneficial effect on neuroinflammatory diseases. In this study, we used the pRNA of bacteriophage phi29 DNA packaging motor to construct a novel RNA nanoparticle for specific targeting to transferrin receptor (TfR) on the murine brain-derived endothelial cells (bEND5) to deliver ICAM-1 siRNA. This RNA nanoparticle (FRS-NPs) contained a FB4 aptamer targeting to TfR and a siRNA moiety for silencing the intercellular adhesion molecule-1 (ICAM-1). Our data indicated that this RNA nanoparticle was delivered into murine brain-derived endothelial cells. Furthermore, the siRNA was released from the FRS-NPs in the cells and knocked down ICAM-1 expression in the TNF-α–stimulated cells and in the cells under oxygen-glucose deprivation/reoxygenation (OGD/R) condition. The functional end points of the study indicated that FRS-NPs significantly inhibited monocyte adhesion to the bEND5 cells induced by TNF-α and OGD/R. In conclusion, our approach using RNA nanotechnology for siRNA delivery could be potentially applied for inhibition of inflammation in ischemic stroke and other neuroinflammatory diseases, or diseases affecting endothelium of vasculature.
Keywords: blood–brain barrier, endothelial cell, inflammation, intercellular adhesion molecule-1, ischemia, RNA nanotechnology, siRNA, transferrin receptor, oxygen-glucose deprivation
Introduction
It has been proven that leukocytes play a key role in neuronal injury after cerebral ischemia reperfusion.1 These cells are rapidly recruited to the injury site after microvascular reperfusion1,2 and exert detrimental effects like increasing the blood–brain barrier (BBB) permeability3 and inducing cytotoxicity.4,5 These detrimental effects require adherence of leukocytes to the vascular endothelium first after vascular injury.1 Although other cellular adhesion molecules (CAMs), like vascular cell adhesion molecule (VCAM-1), and E-selectin, also play a role in adhesion and migration of inflammatory cells into the tissue,6 adherence of leukocytes to endothelial cells occurs primarily through interaction with ICAM-1 on the endothelial cells. ICAM-1, also known as CD54, plays an important role in cell-cell adhesion, extravasation, and inflammatory response.7 Normally, ICAM-1 is expressed at low levels on brain endothelium and perivascular astrocytes, but its expression rises dramatically in pathological conditions such as ischemic stroke,8 brain trauma, multiple sclerosis (MS),9 experimental autoimmune encephalomyelitis (EAE),10 Alzheimer's disease,11 and inflammatory conditions in vitro.12 ICAM-1 facilitates migration of leukocytes into the brain and initiates adhesion of microglia to neurons, which causes neuronal injury and death under disease conditions.13,14 Above pathophysiological changes initiate and exacerbate activation of macrophage and microglia throughout the central nerve system (CNS). These observations are implicated in the development of neuronal injury and death under neuroinflammation. Because adhesion of leukocytes to endothelial cells is the first step of vascular-neuronal inflammation, inhibition of adhesion and recruitment of leukocytes to vascular endothelial cells will have a beneficial effect on the outcome of neuroinflammatory diseases. A murine anti-human ICAM antibody (Enlimomab) was shown to reduce brain damage in the animal ischemic stroke model.15,16 Therapeutic effect of Enlimomab was shown as inhibition of neutrophil adhesion to and migration through the vascular endothelium by blocking ICAM-1 function.15 However, clinical trials of Enlimomab failed for treating stroke patients. The patients treated with Enlimomab were harmed compared to placebo controls.17 The failure of Enlimomab was due to excessive immune response induced by mouse IgG in treated patients.18 Although the clinical application of this anti-ICAM-1 antibody failed, targeting ICAM-1 is still a valuable therapeutic strategy for treating ischemic stroke and other neuroinflammatory diseases if the immunogenic problem of the drug is solved. The immunogenic problem of the drug can be solved by either humanized anti-ICAM-1 antibody or oligonucleotide RNA approach as we proposed in this study.
The discovery of RNA interference (RNAi) gives scientists a powerful tool to study gene functions and to manipulate gene expression.19,20 The exogenous small interfering RNAs (siRNAs) can knock-down genes of interest in the cells.19 RNAi could be an attractive alternative approach for inhibition of ICAM-1 function because of its unique characteristics including specificity in gene targeting, low immunogenicity, and simplicity of design and production. Although siRNA holds exciting promise for treatment of brain diseases, its delivery challenges have hampered the use of siRNA as a therapeutic agent. Such a progress cannot be made until the gap for siRNA delivery is bridged. The progress for such drugs like siRNA in brain disease therapy depends on finding solutions to the delivery problem, which occurs at the BBB. As a physiological barrier, the BBB prevents access of an estimated >98% of all drugs into the brain.21 Most current approaches used for brain delivery of macromolecular drugs are invasive, such as intra-cerebral injection causing brain tissue damage. This highlights the need for novel noninvasive approaches for brain delivery of macromolecular drugs. The receptor-mediated transport systems presented by brain endothelial cells at the BBB are excellent target for brain drug delivery. TfRs and insulin receptor are the most studied receptors for brain drug delivery. Neurotrophic factors,22 peptide hormones,23 and immunoliposomes encapsulating expression plasmids24 have been delivered into the brain by targeting the TfRs at therodent BBB. Neurotrophins25 and genes26 have been delivered into the CNS of the primate animals by targeting insulin receptors at the BBB. However, due to large sizes and protein contents, above formulations are either rapidly eliminated from body or trigger immune responses, and none of these approaches have been translated to clinical therapy now. In this study, we explored an aptamer-directed RNA complex as an alternative approach for brain drug delivery.
Aptamers are oligonucleotides that bind to target molecules with antibody-like high specificity and high affinity.27 Aptamers are selected from large random sequence poll by a process called “systematic evolution of ligands by exponential enrichment” (SELEX) to bind to various cellular targets.28,29 In addition to their exquisite molecular recognition properties, aptamers offer other advantages over antibodies, including being easily engineered in a test tube by chemical synthesis, and the fact that they elicit little or no detectable immune response when injected into animals.30 Because of their high specificity and affinity, aptamers can be developed to target extracellular targets such as receptors for drug delivery. Aptamers can be tethered to, and deliver, a variety of secondary reagents specifically to targeted cells so that nontargeted cells are not exposed to secondary reagents, thus unwanted side-effects can be reduced or eliminated. FB4, a RNA aptamer (shown in Figure 1a), specifically binds to the extracellular domain of mouse TfR. FB4 has been successfully used to deliver a lysosomal enzyme into deficient cells to correct the defective glycosaminoglycan degradation in these cells.31 In this study, we have synthesized the novel RNA chimeras (FRS-NP) which specifically targets TfR on the brain endothelial cells by the TfR binding sequence of FB4 for delivery of siRNA with a goal to inhibit monocyte adhesion under inflammatory and ischemia/reperfusion conditions in vitro.
Figure 1.
Formation of FRS-NPs. (a) The structure of FB4 RNA aptamer: The box is the outlining the truncated form of FB4, which binds to TfR-ECD.31 (b) Phi29 pRNA (Ab') sequence and secondary structure. The right-and left-hand loops are circled in orange and green, respectively. The double-stranded helical domain on the 5′/3′ ends is framed, and the domain for dimer formation is shaded. The curved line points to the two interacting loops. (c) Design of chimeric FB4-pRNA. (d) The formation of pRNA-siRNA/FB4-pRNA dimer (FRS-NPs).
Results
Nomenclature, construction, and synthesis of RNA chimeras
A 117-nucleotide motor RNA (pRNA) of bacterial virus phi29 was used as a delivery vector.32,33,34 It contains two functional domains: the double-stranded helical domain at 5′/3′ end and the intermolecular binding domain. These two domains fold independently, and change of the primary sequences of helical region doesn't affect pRNA structure and folding as long as the two strands are paired.34 Therefore, the helical region at the 5′/3′ end of pRNA can be replaced by siRNA or other RNA sequences without affecting the formation of RNA multimers mediated by base-pairing of upper and lower loops in the intermolecular binding domain.32 The nomenclature of pRNA subunits is shown in Figure 1b. The upper-case and lower-case letters are used to represent the right- and left-hand loops of pRNA, respectively. Matched letters indicate complementarity, whereas different letters indicate noncomplementary loops. For example, pRNA (Ab') contains right-hand loop A (5′ G45G46A47C48) circled in orange and left-hand loop b' (3′ U85G84C83G82) circled in green, which can pair with the left-hand loop a' (3′ C85C84U83G82) and right-hand loop B (5′ A45C46G47C48), respectively, of pRNA (Ba'). To construct the FB4-pRNAand pRNA-siRNA (ICAM-1) chimeras, the 5′/3′ end of pRNA helical domain (between 23G and 97C) was replaced by the truncated part of FB4 (Figure 1c) or the siRNA. The generation of FRS-NP was achieved by mixing equal molar amounts of FB4-pRNA (Ab') and pRNA-siRNA (ICAM-1) (Ba') through interaction of loop Aa' and Bb' respectively (Figure 1d). To confirm that the RNA chimeric complexes retained their correct folding and capability for intermolecular interaction, the RNA chimeras were analyzed on a native gel. As expected, the native gel showed that Ab' and Ba' pRNA chimeras efficiently formed pRNA-siRNA/FB4-pRNA heterodimers, FRS-NPs, as indicated by mobility of the bands. Furthermore, the 108 nt Ab' FB4-pRNA monomers and 216 nt FRS-NP ran slightly faster than the 117 nt pRNA monomers and 220 nt pRNA dimers respectively (Figure 2a).
Figure 2.
Synthesis and characterization of FRS-NPs. (a) The formation of dimers and monomers were demonstrated by native-PAGE gel. Native polyacrylamide gel showed monomer and dimer of the pRNA chimeras exhibiting different migration rates. The images on the right side are Cy3 images of monomers and dimers. The images on the left side are EB staining images of monomer and dimers. (b) dsRNA release by Dicer or bEND5 cell lysates processing. The 32P-labeled chimeric RNA complexes were incubated with purified recombinant Dicer or lysate of bEND5 cells for 30 minutes, and then separated on denaturant PAGE/urea gel. 22 nt siRNA and RNA Marker were loaded as reference to estimate the size of RNA.
Processing of FRS-NPs into siRNA by Dicer or lysate of bEND5 cells
Exogenous ICAM-1 siRNA in the FRS-NPs was expected to retained biological functions in cells. To determine whether siRNA sequence can be released to induce knockdown effect, FRS-NPs were labeled with 32P for evaluation of siRNA release. For dicer processing experiments, 5′-[γ32P]-RNA complex were treated with recombinant purified Dicer, which cleaves double-stranded RNA (dsRNA) into short double-stranded siRNA fragments (22 base pairs). As expected, the short dsRNA (about 24 base pairs) was released after incubation with Dicer solution (Figure 2b). In the lysate processing experiments, there was a dsRNA with about 30 base pairs released after incubation with lysate of bEND5 cells (Figure 2b). The above results indicated that the short dsRNA was released from FRS-NPs, and therefore had the potential to silence target gene expression after delivered into the cells.
Targeted delivery of pRNA chimeras into brain-derived endothelial cells and acute brain slices
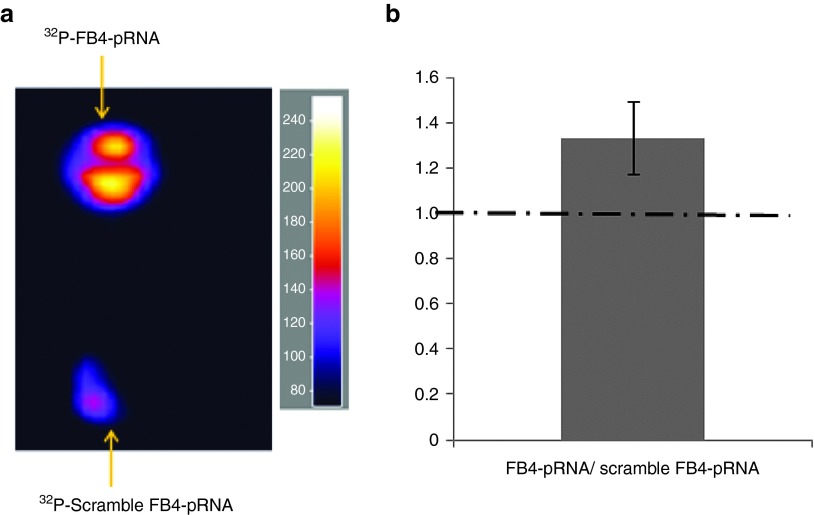
From the Dicer or cell lysate processing experiments, we know that dsRNA can be released from the FRS-NPs. The next question is whether the FRS-NPs can be delivered into the target cells? To answer this question, we have performed uptake studies using bEND5 cells and acute mouse coronal brain sections. From the confocal data, we could identify the localization of synthesized monomers and FRS-NPs in brain-derived endothelial cells. There were clearly binding and internalization of FB4-pRNA monomers and FRS-NPs by bEND5 cells. Moreover, the binding and uptake mechanism appears to be specific, because parallel experiments, performed under identical incubation and imaging conditions, with the untargeted controls, which had been identically labeled, did not result in any detectable binding or internalization (Figure 3a). The further experiments have been performed by radioactively labeling RNA complexes to investigate the mechanism of cellular uptake of FRS-NPs. As shown in Figure 3c, cellular uptake of FRS-NPs in bEND5 cells was significantly higher than that in TfR negative bEND5 cells or that of scramble-a RNA complexes in bEND5 cells (P < 0.05, Figure 3c). The Figure 3b is the representative image of TfR knockdown by siRNA in bEND5 cells. Furthermore, the uptake ratio of FB4-pRNA to scramble FB4-pRNA in acute mouse brain section was 1.32 ± 0.16 (mean ± CI), which was more than 1(Figure 4, P < 0.05). This indicates that uptake of FB4-pRNA monomer by acute mouse brain slices is significantly higher than that of scramble FB4-pRNA, which contains scramble FB4 aptamer binding sequence. These results have demonstrated the delivery specificity of FRS-NPs into brain endothelial cells by FB4 RNA aptamer.
Figure 3.
Delivery of FRS-NPs to bEND5 cells.(a) Binding and internalization of FB4-pRNA monomer and FRS-NPs. Cy3-pRNA monomer and Cy3-pRNA dimer are the comparable controls ofCy3-FB4-pRNA monomer and dimer respectively. (b) Representative western blotting image of TfR expression after siRNA silencing in bEND5 cells. (c) Cellular uptake of FRS-NPs measured by 32P radioactive labeled RNA complexes. Mean ± SD, n = 4, P < 0.05.
Figure 4.
Uptake of FB4-pRNA monomer by acute mouse brain sections measured by autoradiography. (a) Uptake images. (b) Statistical analysis: n = 10, mean ± CI, 95% confidence level, P < 0.05.
FRS-NPs inhibited ICAM-1 expression in TNF-α–stimulated bEND5 cells
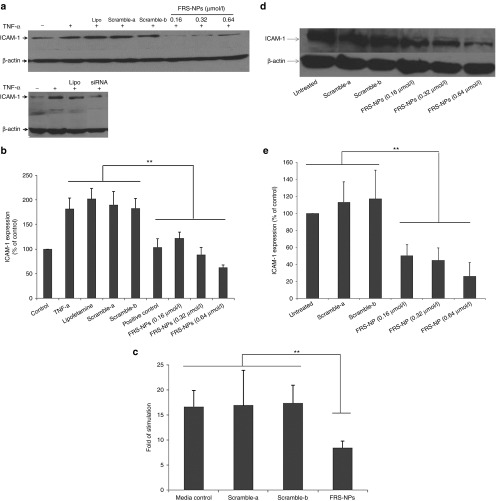
We demonstrated that FRS-NPs could be delivered into bEND5 cells and acute brain sections. Here, we further investigated effect of FRS-NPs on ICAM-1 expression in inflammatory model in vitro using bEND5 cells. As shown in Figure 5a,b, the expression of ICAM-1 was increased to 182 ± 22% of control after stimulated by TNF-α. However, FRS-NPs reversed this increase in ICAM-1 expression induced by TNF-α in a dose-dependent manner. The ICAM-1 levels were 122 ± 12, 82 ± 14, and 63 ± 5% of untreated control in treated groups at concentrations of 0.16,0.32, and 0.64 µmol/l of FRS-NPs respectively. The FRS-NPs with scramble FB4 (scramble-a) and scramble ICAM-1 siRNA (scramble-b) did not have silencing effect, in which ICAM-1 expression were190 ± 26 and 183 ± 19% of control respectively. Cells transfected with ICAM-1 siRNA (Santa Cruz) by lipofetamine-2000 was used as positive control, in which ICAM-1 expression was 104 ± 17% of untreated control, and lipofetamine alone did not affect ICAM-1 level which was 202 ± 21% of untreated control. Furthermore, mRNA level of ICAM-1 was also detected by RT-PCR. Compared with un-stimulated control, the mRNA level of ICAM-1 was increased to 16.6-folds after TNF-α challenge, while it was reduced to 8.4-folds after FRS-NPs incubation (Figure 5c, P < 0.01). Scramble-a and scramble-b RNA complexes did not have any effect on mRNA levels of ICAM-1, which were 16.9- and 17.3-folds of control respectively. These results indicated that this FRS-NP was able to significantly decrease the ICAM-1 expression in this inflammatory model induced by TNF-α. However, it is possible that the reduction of ICAM-1 expression may be due to inhibition of TNF-induced stimulation by FRS-NPs rather than inhibition of ICAM-1 directly by FRS-NPs. Therefore, we further investigate the effect of FRS-NPs on the basal level of ICAM-1 in bEND5 cells. The protein level of ICAM-1 were 50 ± 13, 45 ± 14, and 26 ± 16% of control in 0.16, 0.32, and 0.64 µmol/l of FRS-NPs treatment respectively (Figure 5d,e, P < 0.05).
Figure 5.
Effect of FRS-NPs on expression of ICAM-1 in TNF-α–induced inflammatory condition in bEND5 cells and on basal level of ICAM-1 in bEND5 cells. (a) The representative image of western blotting in ICAM-1 expression in inflammatory model in vitro. (b) Effect of FRS-NPs on expression of ICAM-1 in inflammatory model in vitro. Mean ± SD, n = 6, P < 0.05. (c) The mRNA level of ICAM-1 measured by RT-PCR. The positive control is the cells treated with 0.64 µmol/l siRNA in the present of lipofectamine2000. Mean ± SD, n = 4, P < 0.01. (d) The representative image of western blotting in ICAM-1 expression in normal bEND5 cells. (e) The effect of FRS-NPs on the basal level of ICAM-1 in bEND5 cells. Mean ± SD, n = 4, P < 0.05.
FRS-NPs inhibited ICAM-1 expression in OGD/R condition in vitro
The ICAM-1 protein level was increased to threefolds of the untreated control after OGD/R (Figure 6). The ICAM-1 level was 313 ± 26 of untreated control in OGD/R group, while the levels were reduced to 235 ± 25, 126 ± 14, and 61 ± 11% of untreated control in FRS-NPs treatment groups at concentrations of 0.16, 0.32, and 0.64 µmol/l respectively (Figure 6, P < 0.01). The FRS-NPs with scramble-a and scramble-b did not have silencing effect on ICAM-1 expressions, which were 340 ± 24 and 305 ± 12% of control respectively. In the positive control group, ICAM-1 expression was 220 ± 8% of untreated control and lipofetamine alone did not affect ICAM-1 level, which was 332 ± 16% of untreated control (Figure 6). These data showed that FRS-NPs inhibited the increase of ICAM-1 level induced by “hypoxia/reoxygenation” and restored the ICAM-1 expression back to its normal level and even less.
Figure 6.
Effect of FRS-NPs on expression of ICAM-1 in OGD/R in bEND5 cells. (a) Representative image of western blotting. (b) Statistic analysis of ICAM-1 expression in western blotting. Mean ± SD, n = 4–8, P < 0.01. The positive control is the cells treated with 0.64 µmol/l siRNA in the present of lipofectamine2000.
FRS-NPs inhibit monocyte adhesion to bEND5 cells in inflammatory and OGD/R conditions
Under certain disease conditions like ischemic stroke and neuroinflammation, adhesion of monocyte to endothelium is enhanced, and ICAM-1 plays a primary role in mediating monocyte adhesion to endothelial cells. In order to determine whether the FRS-NPs affect the monocyte adhesion to the brain endothelial cells, we performed U937 adhesion assay. When endothelial cells were stimulated by TNF-α, the adhesion of U937 cells to bEND5 cells was increased to 19.5 ± 1.1% compared to nonactivated endothelial cells with 8.8 ± 0.6% adherent monocytes. FRS-NPs at concentrations of 0.16, 0.32, and 0.64 µmol/l reduced the number of adherent U937 cells to 6.6 ± 1.6, 8.6 ± 1.5, and 6.5 ± 0.7% respectively. To exclude any nonspecific inhibition in adhesion, the experiments were done with scramble FRS-NPs, which did not result in significant decrease in adhesion to U937 cell with 21.9 ± 1.8 and 17.5 ± 3.4% in scramble-a and scramble-b group respectively (Figure 7a, P < 0.01). Furthermore, FRS-NPs also significantly inhibited adhesion of monocyte to endothelial cells under OGD/R condition. The adhesion of monocyte was 14.4 ± 1.3, 12.6 ± 1.7, and 8.5 ± 2.7% at treatment of 0.16, 0.32, and 0.64 µmol/l FRS-NPs respectively. The scramble-a and scramble-b did not have significant effect on adhesion of U937 cell to bEND5 cells with 18.9 ± 1.9 and 17.7 ± 0.4% respectively (Figure 7b, P < 0.01).
Figure 7.

Effect of FRS-NPs on monocytes adhesion to bEND5 cells.(a) Effect of FRS-NPs on adhesion of U937 cell to bEND5 cells in inflammatory condition. (b) Effect of FRS-NPs on adhesion of U937 cell to bEND5 cells in OGD/R condition. Mean ± SD, n = 3, P < 0.01.
Discussion
In this study, we constructed a completely RNA-based chimera using a RNA aptamer and a package RNA of bacteriophage phi29 to deliver the anti-ICAM-1 siRNA into brain-derived endothelial cells with a goal to inhibit inflammation. The FRS-NP has a targeting moiety for RNA complex transport, the TfR binding sequence of FB4 aptamer, and an RNA-silencing moiety, the siRNA, which is recognized and processed by Dicer in a manner similar to the processing of microRNAs.35 The FB4 aptamer binds to the cell surface receptor, TfR, on brain endothelial cells mediating uptake of FRS-NP chimera, and delivers therapeutic siRNA targeting ICAM-1 overexpressed in most neuroinflammatory diseases including cerebral ischemia/reperfusion. The pRNAs in the FRS-NP chimera were used as vectors to carry anti-ICAM-1 siRNA and FB4. The pRNA is derived from the phi29 DNA-packaging RNA, and has been proved of the potential being safe, noninfectious/nonpathogenic and resistant to degradation.36 The pRNA can be manipulated to produce chimeric RNAs that form dimer, trimer, hexamer through interaction between right- and left-hand loops, which gives a bottom-up approach for drug delivery.33,36 Fusion of the pRNA with a variety of foreign moieties did not impede the formation of dimer, trimer or interfere with moiety function.33,36 The ability of pRNA chimeras to form dimers makes them extremely useful for targeting delivery of siRNA into targeted cells.32,34 The chimeric pRNAs with various foreign moieties have been successfully delivered into a number of cells.32,34,36
Construction of FRS-NP was confirmed by native gel analysis, which indicated the RNA chimeric complexes retained their correct folding and capability for intermolecular interaction. To have knockdown effect on ICAM-1 expression, the double-stranded siRNA duplexes must be released from FRS-NPs after entering cells. Our experiments showed that the double-stranded siRNA duplexes were released after processed by Dicer or incubated with lysate of brain-derived endothelial cells. The size of the double-stranded siRNA released from FRS-NPs in lysate of targeted cells is larger than 22 nt siRNA (Figure 2b), and the double-stranded siRNA with released in cell lysate has more potential in silencing the ICAM-1 gene compared to the 22 nt siRNA positive control (Figures 5b and 6b). This result is consistent with recent reports indicating that the 25–30 nt double-stranded siRNA duplexes are more potent than 21-mer siRNAs.37,38 The observed increased potency in silencing gene expression using longer siRNA is thought to be resulted from providing dicer with a substrate instead of siRNA, which improves the rate or efficiency of entry of the siRNA into RISC as facilitated by dicer.39 The other explanation is that the siRNA of pRNA-siRNA may be more stable than the siRNA itself within cells.40
TfRs, the delivery targets of FRS-NP, are highly expressed on microvascular endothelial cells of the BBB, glia and neurons in the CNS.41 The bEND5 cells we used in the study have been proved to highly express TfR.42 Furthermore, TfRs are up-regulated in some brain disease like ischemic stroke.43 The relative high expression and up-regulation of TfRs at the BBB give more selectivity for delivery of siRNA to the brain under disease conditions. Ligand targeting function of the FRS-NP was evaluated by confocal microscopy, radioactive approach and by uptake study in acute brain slices. When brain endothelial cells or acute brain sections were incubated with these FRS-NPs or pRNA-FB4 without transfection reagents, there were significant binding/uptake of RNA complexes, while there showed no binding/uptake of RNAs without FB4 or with scrambled FB4 (Figures 3a and 4). Furthermore, cellular uptake of FRS-NPs was significantly higher than that in TfR negative bEND5 cells or uptake of scramble-a in normal bEND5 cells (Figure 3c). This indicates that the transport of FRS-NPs is TfR dependent, the FRS-NPs-TfR recognition, rather than the molecular conformation. The pharmacological and functional end points were evaluated in the study to demonstrate feasibility of our novel approach in therapy of neuro-vascular inflammation. In absence of transfection agent, the FRS-NPs knocked down ICAM-1 expression in bEND5 cells under inflammatory condition (TNF-α stimulation) (Figure 5a–c). In addition, we found that FRS-NPs knocked down the basal level of ICAM-1 in normal bEND5 cells in the absence of TNF (Figure 5d,e). Therefore, the effect of FRS-NPs in TNF-α stimulation condition is ICAM-1 specific. Similarly, FRS-NPs also inhibited ICAM-1 expression in OGD/R condition (Figure 6). The functional study indicated that FRS-NPs significantly inhibited monocyte adhesion to the bEND5 cells induced by TNF-α and OGD/R condition (Figure 7). This finding is significant because FRS-NPs could be potentially applied for treatment of ischemic stroke and other neuroinflammatory diseases, or diseases affecting endothelium of vasculature.
In contrast to most reported antibody delivery methods, the approach used in this study contains only RNA (RNA aptamer, pRNA, and siRNA). The chimeric RNAs would be expected to produce less immune response than protein-mediated delivery approaches.30 In addition, RNAs can easily be synthesized in large quantities at relatively low cost and are feasible to a variety of chemical modifications that improve both stability and pharmacokinetics in vivo. Furthermore, the smaller size of this RNA chimera compared with that of antibodies has better tissue penetration for delivery.44,45 Importantly, the approach described in this study provides preliminary information for cell type–specific siRNA delivery, which is an ideal therapeutic application of siRNA. In this proof-of-concept study, we have targeted ICAM-1 gene specifically with an RNA complex in brain-derived endothelial cells expressing the cell-surface receptor, TfR. We found that the FRS-NPs inhibited ICAM-1 gene expression, which resulted in decrease of monocyte adhesion to the brain-derived endothelial cells in culture (Figure 7). The approach could be utilized for delivery of drugs to the BBB or across BBB into the brain. Further studies will be needed to evaluate the stability and pharmacokinetics of the FRS-NPs in vivo. Of course, for application of FRS-NP in vivo, the RNA chimera has to be modified to improve its stability and pharmacokinetics, and such modifications would need to be tested to determine whether they interfere with cellular uptake and processing by Dicer.
Materials and methods
Construction of pRNA-siRNA, FB4-pRNA chimera and FRS-NP. The synthesis of RNA has been described previously.46 The DNA primers for pRNA-siRNA and FB4-pRNA chimeras were purchased from Integrated DNA Technologies. The linear DNA sequences were used as templates to generate PCR DNA fragments with primer pairs with Taq DNA polymerase (Thermo Scientific). FB4-pRNA represents a pRNA chimera (pRNA Ab') that harbors a TfR binding sequence of FB4; Full sequence of FB4-pRNA is 5′-GUUGAUUGCGUGUCAAUCAUGGCGGACGGAUUGCGGCCGUUGUCUGUGGCGUCCGUUCGUCAUGUGUAUGUUGGGGAUUAGGACGCUGAUUGAGUUCAGCCCACAUAC-3′ (Figure 1c). The bolded sequence is the binding sequence of FB4. The GGAC is the sequence of loop A, and the GCGU is the sequence of loop b'. pRNA-siRNA represents pRNA chimera (pRNA Ba') that harbors the sequences of ICAM-1 siRNA: 5′-AGCCCAACUCUUCUUGAUGUAUUGUCAUGUGUAUGUUGGGGAUUAACGCCUGAUUGAGUUCGACCCACAUACUUUGUUGAUUGUCCGUCAAUCAUGGCUUUACAUCAAGAAGAGUUGGGCUUU-3′. The bolded sequence is the sequences of ICAM-1 siRNA. GUCC is the sequence of loop a', and ACGC is the sequence of loop B. The loop A interacts with loop a', and loop b' interacts with loop B to form RNA chimera (FRS-NP) by binding of complementary sequence of loops. The control RNA chimera contains either the scramble sequence of FB4 (scramble-a: UGCGUGGUGUGGAUGGGGGACGCUCCGUCCUCUUC) or the scramble sequence of ICAM-1 siRNA (scramble-b: GAUUCACGCUUACUUCACU). Transcription products were purified by 8% PAGE/urea gel and eluted with elution buffer (0.5 mol/l sodium acetate, 0.1 mmol/l EDTA and 0.1% SDS). RNAs were precipitated in ethanol and resuspended in nuclease-free water. The generation of FRS-NPs were achieved by mixing equal molar amounts of FB4-pRNA (A-b') and pRNA-siRNA (ICAM-1) (Ba') through interaction of A-a' and B-b' respectively (Figure 1d). 5 mmol/l of magnesium chloride was included in all buffers to maintain the intermolecular interaction and folding of pRNA.32,34 Fluorescent RNA chimeras were generated using the Cy3 labeling kit (Applied Biosystems/Ambion, Austin, TX) in accordance with the manufacturer's instructions. To verify the formation of FRS-NPs, all RNAs were electrophoresed on 8% native-PAGE gel.
Cell culture, inflammation model in vitro, and OGD/R condition
Cell culture : The bEND5 cells derived from mouse brain and immortalized with polyoma middle T oncogene was gifted by Dr Ulrich Bickel from Texas Tech University. bEND5 cells were grown in DMEM media (Mediatech Cellgro, Herndon, VA) supplemented with 10% (v/v) FBS (fetal bovine serum) (Atlanta Bio, Lawrenceville, GA), 1 mmol/l sodium pyruvate, 4 mmol/l L-glutamine, 1% (v/v) nonessential amino acids, 1% (v/v) 100 IU/ml penicillin 100 mg/ml streptomycin (all from ATCC, Manassas, VA). The cell lines were maintained at 37 °C, 5% CO2 and 95% relative humidity. U937 cells (a monocyte cell line from ATCC) were cultured in RPMI 1640 medium supplemented with 2 mmol/l L-glutamine (Mediatech Cellgro, Manassas, VA), 10% heat-inactivated FBS and 100 IU/ml penicillin/streptomycin.
The inflammatory model in vitro. : The in vitro inflammatory model experiments were performed by adding 50 µg/ml of TNF-α to stimulate bEND5 cells overnight. Briefly, the bEND5 cells were incubated with FRS-NPs in serum free medium with 5 mmol/l Mg2+ for 1 hour; the treated cells were then cultured in normal complete medium. At 32 hours after FRS-NPs treatment, TNF-α was added and incubated overnight. The cell samples were obtained at 48 hours after FRS-NPs treatment.
The OGD/R condition : This condition was used to mimic ischemia/reperfusion in vitro. Briefly, after adding FRS-NPs or scramble RNSs into the media, hypoxia was immediately induced by placing cells in a sealed chamber (Billups-Rothenberg, Del Mar, CA) at 37 °C, which has been flushed with 95% N2/5% CO2 gas. The concentration of oxygen in the atmosphere was maintained at 0% oxygen and the PO2 in the medium was below 25 mmHg. Aglycemia was induced by using RPMI 1640 medium (without L-glucose) (Hyclone, Logan, UT). After 16 hours OGD, the cells were back to normal growth condition for 24 hours.
The bEND5 cells under different conditions were treated with various doses of RNA chimeras or the same dose of the vehicle scramble RNAs and evaluated by RT-PCR and western blotting to determine expression of ICAM-1, and by monocyte adhesion assay to investigate the effect of FRS-NPs on inflammation at functional level. The β-Actin was used as the protein loading control marker. The following antibodies and the respective dilutions were used in western blotting procedure. 1: 2,000 (dilution) β-Actin antibody (Cell Signaling Technology, Danvers, MA), 1:1,000 ICAM-1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Chimeric pRNA/ siRNA processing pRNA/siRNA was labeled with 32P-labeled ATP (PerkinElmer) using T4 Polynucleotide Kinase (Fermentas) according to the manufacturer's instructions. Then dsRNAs were processed by the purified recombinant Dicer (Genlantis, San Diego, CA) or bEND5 cell lysates. Processed RNAs were electrophoresed on 8% PAGE/urea gel and then visualized by phosphorimager.
Cellular uptake of FB4-pRNA chimera and FRS-NPs
Confocal imaging: Confocal imaging was performed using a Zeiss LSM 510 laser scanning confocal microscope to observe cellular uptake of RNAs. The same settings for laser power and photomultiplier gains were applied for imaging samples stained with specific antibodies. FB4-pRNA monomer, FRS-NPs and their corresponding controls were labeled with Cy3 using the Mirus Label IT Cy3 labeling kit (Mirusbio) according to the manufacturer's instructions. Briefly, bEND5 cells were incubated with Cy3-labeled RNAs at the concentration of 500 nmol/l in DMEM medium for 2 hours at 37 °C. After washing the cells were fixed with 4% paraformaldehyde for 5 minutes, and then counter-stained by two kinds of fluorescence dye to locate the cell membrane and cell nucleus: Alexa Fluor 488 phalloidin (probe for F-actin) and ToPro3 iodide (monomeric cyanine nucleic acid stains).
Cellular uptake of radioactively labeled FRS-NPs: FRS-NPs or scramble-a RNA complexes were labeled with 32P-labeled ATP (PerkinElmer) using T4 Polynucleotide Kinase (Fermentas) according to manufacture protocol. The TfR negative bEND5 cells were generated by transfecting CD71 siRNA with lipofectamine RNAiMAX (Life Technology, Grand Island, NY) according to manufacturer's protocol. Both TfR negative and normal bEND5 cells were incubated with 32P-labeled FRS-NPs or scramble-a control at the concentration of 0.25 μmol/l (~0.2 μCi) for 40 minutes at 37 °C, then the cells were washed three times with phosphate-buffered saline and lysed with RIPA buffer. Finally, the radioactivity was counted by LSC 6500-multiplepurpose scintillation counter (Beckman Counter).
Adhesion assay. U937 cells (a monocyte cell line) were labeled with 5 mg/ml BCECF-AM (2′,7′-bis(2-carboxyethyl)-5(6)-carboxyfluorescein acetoxymethyl ester, Sigma) for 30 minutes at 37 °C, and then the cells were washed and resuspended in serum-free media. For FRS-NPs treatment in TNF-α–stimulated bEND5 cells, bEND5 cells were cultured and incubated with FRS-NPs in a 24-well plate for 1 hour. At 32 hours after FRS-NPs treatment, TNF-α (50 ng/ml) was added and incubated for 16 hours, then the cells were cocultured with BCECF-AM-labeled U937 cells (106 cells/well) for 30 minutes at 37 °C. For FRS-NPs treatment in OGD/R bEND5 cells, the bEND5 cells were treated with FRS-NPs for 1 hour then subjected to OGD/R condition as described early, and then cells were cocultured with BCECF-AM-labeled U937 cells (106 cells/well) for 30 minutes at 37 °C. Nonadhering U937 cells were removed and cells were washed with phosphate-buffered saline, then cells were lysed in the buffer containing 0.1% Triton X-100 in 0.1 mol/l Tris–HCl (pH7.4). Fluorescence (F) was measured with a microplate fluorescence reader using excitation at 492 nm and emission at 535 nm. The monocyte adhesion was calculated as: Adhesion (%) = 100 × Fsample/Ftotal, (Ftotal: fluorescence intensity of 106 cells).
Autoradiography. The FB4-pRNA monomers were labeled with 32P-labeled ATP (PerkinElmer) using T4 Polynucleotide Kinase (Fermentas). Snap-frozen mouse brain specimens were obtained, and frozen coronal brain sections (20 μm) were prepared on slides with a cryostat at −20 °C. The slides were warmed to room temperature and briefly air-dried, and were incubated with the buffer (0.01 mol/l Na2HPO4, 0.15 mol/l NaCl, pH 7.4, 0.1% bovine serum albumin) containing one of the following solutions: (i) 0.16 µmol/l 32P-FB4-pRNA; (ii) 0.16 µmol/l 32P-scramble FB4-pRNA. After 2 hours of incubation at room temperature, the slides were washed with the buffer and air-dried, followed by exposed to phosphor screen in a cassette overnight and then scanned by phosphorimager.
Statistical analysis. Statistical analysis was performed by analysis of variance, followed by posttests (Dunett's test for comparison of experimental groups versus control conditions). P < 0.05 indicates statistical significance.
Acknowledgments
The research was supported by 2BGIA9090000 (J.K.H.) from the American Heart Association and CA151648 (P.G.). P.G. is a cofounder of Kylin Therapeutics, Inc., and Biomotor and RNA Nanotechnology Development Corp, Ltd. The authors declare no conflict of interest. We would like to thank Dr Peixuan Guo for his support to this work.
References
- Jin R, Yang G, Li G. Inflammatory mechanisms in ischemic stroke: role of inflammatory cells. J Leukoc Biol. 2010;87:779–789. doi: 10.1189/jlb.1109766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grønberg NV, Johansen FF, Kristiansen U, Hasseldam H. Leukocyte infiltration in experimental stroke. J Neuroinflammation. 2013;10:115. doi: 10.1186/1742-2094-10-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry VH, Anthony DC, Bolton SJ, Brown HC. The blood-brain barrier and the inflammatory response. Mol Med Today. 1997;3:335–341. doi: 10.1016/S1357-4310(97)01077-0. [DOI] [PubMed] [Google Scholar]
- Benakis C, Hirt L, Du Pasquier RA. Inflammation and stroke. Kardiovaskulare Medizin. 2008;12:143–150. [Google Scholar]
- Jordan J, Segura T, Brea D, Galindo MF, Castillo J. Inflammation as therapeutic objective in stroke. Curr Pharm Des. 2008;14:3549–3564. doi: 10.2174/138161208786848766. [DOI] [PubMed] [Google Scholar]
- Supanc V, Biloglav Z, Kes VB, Demarin V. Role of cell adhesion molecules in acute ischemic stroke. Ann Saudi Med. 2011;31:365–370. doi: 10.4103/0256-4947.83217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard AK, Rothlein R. Intercellular adhesion molecule-1 (ICAM-1) expression and cell signaling cascades. Free Radic Biol Med. 2000;28:1379–1386. doi: 10.1016/s0891-5849(00)00223-9. [DOI] [PubMed] [Google Scholar]
- Vemuganti R, Dempsey RJ, Bowen KK. Inhibition of intercellular adhesion molecule-1 protein expression by antisense oligonucleotides is neuroprotective after transient middle cerebral artery occlusion in rat. Stroke. 2004;35:179–184. doi: 10.1161/01.STR.0000106479.53235.3E. [DOI] [PubMed] [Google Scholar]
- Sobel RA, Mitchell ME, Fondren G. Intercellular adhesion molecule-1 (ICAM-1) in cellular immune reactions in the human central nervous system. Am J Pathol. 1990;136:1309–1316. [PMC free article] [PubMed] [Google Scholar]
- Bullard DC, Hu X, Schoeb TR, Collins RG, Beaudet AL, Barnum SR. Intercellular adhesion molecule-1 expression is required on multiple cell types for the development of experimental autoimmune encephalomyelitis. J Immunol. 2007;178:851–857. doi: 10.4049/jimmunol.178.2.851. [DOI] [PubMed] [Google Scholar]
- Frohman EM, Frohman TC, Gupta S, de Fougerolles A, van den Noort S. Expression of intercellular adhesion molecule 1 (ICAM-1) in Alzheimer's disease. J Neurol Sci. 1991;106:105–111. doi: 10.1016/0022-510x(91)90202-i. [DOI] [PubMed] [Google Scholar]
- Fabry Z, Waldschmidt MM, Hendrickson D, Keiner J, Love-Homan L, Takei F, et al. Adhesion molecules on murine brain microvascular endothelial cells: expression and regulation of ICAM-1 and Lgp 55. J Neuroimmunol. 1992;36:1–11. doi: 10.1016/0165-5728(92)90026-h. [DOI] [PubMed] [Google Scholar]
- Blann AD, Ridker PM, Lip GY. Inflammation, cell adhesion molecules, and stroke: tools in pathophysiology and epidemiology. Stroke. 2002;33:2141–2143. doi: 10.1161/01.str.0000029008.00497.d3. [DOI] [PubMed] [Google Scholar]
- Ishikawa M, Cooper D, Russell J, Salter JW, Zhang JH, Nanda A, et al. Molecular determinants of the prothrombogenic and inflammatory phenotype assumed by the postischemic cerebral microcirculation. Stroke. 2003;34:1777–1782. doi: 10.1161/01.STR.0000074921.17767.F2. [DOI] [PubMed] [Google Scholar]
- Bowes MP, Rothlein R, Fagan SC, Zivin JA. Monoclonal antibodies preventing leukocyte activation reduce experimental neurologic injury and enhance efficacy of thrombolytic therapy. Neurology. 1995;45:815–819. doi: 10.1212/wnl.45.4.815. [DOI] [PubMed] [Google Scholar]
- Zhang RL, Chopp M, Li Y, Zaloga C, Jiang N, Jones ML, et al. Anti-ICAM-1 antibody reduces ischemic cell damage after transient middle cerebral artery occlusion in the rat. Neurology. 1994;44:1747–1751. doi: 10.1212/wnl.44.9.1747. [DOI] [PubMed] [Google Scholar]
- Enlimomab Acute Stroke Trial Investigators Use of anti-ICAM-1 therapy in ischemic stroke: results of the Enlimomab Acute Stroke Trial. Neurology. 2001;57:1428–1434. doi: 10.1212/wnl.57.8.1428. [DOI] [PubMed] [Google Scholar]
- Furuya K, Takeda H, Azhar S, McCarron RM, Chen Y, Ruetzler CA, et al. Examination of several potential mechanisms for the negative outcome in a clinical stroke trial of enlimomab, a murine anti-human intercellular adhesion molecule-1 antibody: a bedside-to-bench study. Stroke. 2001;32:2665–2674. doi: 10.1161/hs3211.098535. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Karagiannis TC, El-Osta A. RNA interference and potential therapeutic applications of short interfering RNAs. Cancer Gene Ther. 2005;12:787–795. doi: 10.1038/sj.cgt.7700857. [DOI] [PubMed] [Google Scholar]
- Pardridge WM. Drug transport across the blood-brain barrier. J Cereb Blood Flow Metab. 2012;32:1959–1972. doi: 10.1038/jcbfm.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pardridge WM. Conjugation of brain-derived neurotrophic factor to a blood-brain barrier drug targeting system enables neuroprotection in regional brain ischemia following intravenous injection of the neurotrophin. Brain Res. 2001;889:49–56. doi: 10.1016/s0006-8993(00)03108-5. [DOI] [PubMed] [Google Scholar]
- Bickel U, Yoshikawa T, Landaw EM, Faull KF, Pardridge WM. Pharmacologic effects in vivo in brain by vector-mediated peptide drug delivery. Proc Natl Acad Sci USA. 1993;90:2618–2622. doi: 10.1073/pnas.90.7.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YF, Boado RJ, Pardridge WM. Absence of toxicity of chronic weekly intravenous gene therapy with pegylated immunoliposomes. Pharm Res. 2003;20:1779–1785. doi: 10.1023/b:pham.0000003375.13655.f9. [DOI] [PubMed] [Google Scholar]
- Boado RJ, Zhang Y, Zhang Y, Pardridge WM. Genetic engineering, expression, and activity of a fusion protein of a human neurotrophin and a molecular Trojan horse for delivery across the human blood-brain barrier. Biotechnol Bioeng. 2007;97:1376–1386. doi: 10.1002/bit.21369. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Schlachetzki F, Pardridge WM. Global non-viral gene transfer to the primate brain following intravenous administration. Mol Ther. 2003;7:11–18. doi: 10.1016/s1525-0016(02)00018-7. [DOI] [PubMed] [Google Scholar]
- Gu F, Zhang L, Teply BA, Mann N, Wang A, Radovic-Moreno AF, et al. Precise engineering of targeted nanoparticles by using self-assembled biointegrated block copolymers. Proc Natl Acad Sci USA. 2008;105:2586–2591. doi: 10.1073/pnas.0711714105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Madaio MP, Hodder S, Schwartz RS, Stollar BD. Responsiveness of autoimmune and normal mice to nucleic acid antigens. J Immunol. 1984;132:872–876. [PubMed] [Google Scholar]
- Chen CH, Dellamaggiore KR, Ouellette CP, Sedano CD, Lizadjohry M, Chernis GA, et al. Aptamer-based endocytosis of a lysosomal enzyme. Proc Natl Acad Sci USA. 2008;105:15908–15913. doi: 10.1073/pnas.0808360105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Huang F, Guo P. Construction of folate-conjugated pRNA of bacteriophage phi29 DNA packaging motor for delivery of chimeric siRNA to nasopharyngeal carcinoma cells. Gene Ther. 2006;13:814–820. doi: 10.1038/sj.gt.3302716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Shu D, Simon MN, Guo P. Gene cloning, purification, and stoichiometry quantification of phi29 anti-receptor gp12 with potential use as special ligand for gene delivery. Gene. 2003;315:145–152. doi: 10.1016/s0378-1119(03)00729-7. [DOI] [PubMed] [Google Scholar]
- Guo S, Tschammer N, Mohammed S, Guo P. Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Hum Gene Ther. 2005;16:1097–1109. doi: 10.1089/hum.2005.16.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- Khaled A, Guo S, Li F, Guo P. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano Lett. 2005;5:1797–1808. doi: 10.1021/nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarzguioui M, Rossi JJ, Kim D. Approaches for chemically synthesized siRNA and vector-mediated RNAi. FEBS Lett. 2005;579:5974–5981. doi: 10.1016/j.febslet.2005.08.070. [DOI] [PubMed] [Google Scholar]
- Rose SD, Kim DH, Amarzguioui M, Heidel JD, Collingwood MA, Davis ME, et al. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005;33:4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarzguioui M, Lundberg P, Cantin E, Hagstrom J, Behlke MA, Rossi JJ. Rational design and in vitro and in vivo delivery of Dicer substrate siRNA. Nat Protoc. 2006;1:508–517. doi: 10.1038/nprot.2006.72. [DOI] [PubMed] [Google Scholar]
- Tarapore P, Shu Y, Guo P, Ho SM. Application of phi29 motor pRNA for targeted therapeutic delivery of siRNA silencing metallothionein-IIA and survivin in ovarian cancers. Mol Ther. 2011;19:386–394. doi: 10.1038/mt.2010.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moos T. Immunohistochemical localization of intraneuronal transferrin receptor immunoreactivity in the adult mouse central nervous system. J Comp Neurol. 1996;375:675–692. doi: 10.1002/(SICI)1096-9861(19961125)375:4<675::AID-CNE8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Bhattacharya R, Osburg B, Fischer D, Bickel U. Targeted delivery of complexes of biotin-PEG-polyethylenimine and NF-kappaB decoys to brain-derived endothelial cells in vitro. Pharm Res. 2008;25:605–615. doi: 10.1007/s11095-007-9389-y. [DOI] [PubMed] [Google Scholar]
- Omori N, Maruyama K, Jin G, Li F, Wang SJ, Hamakawa Y, et al. Targeting of post-ischemic cerebral endothelium in rat by liposomes bearing polyethylene glycol-coupled transferrin. Neurol Res. 2003;25:275–279. doi: 10.1179/016164103101201508. [DOI] [PubMed] [Google Scholar]
- Pestourie C, Tavitian B, Duconge F. Aptamers against extracellular targets for in vivo applications. Biochimie. 2005;87:921–930. doi: 10.1016/j.biochi.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: an emerging class of therapeutics. Annu Rev Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- Zhang C, Lee CS, Guo P. The proximate 5' and 3' ends of the 120-base viral RNA (pRNA) are crucial for the packaging of bacteriophage phi 29 DNA. Virology. 1994;201:77–85. doi: 10.1006/viro.1994.1267. [DOI] [PubMed] [Google Scholar]