Abstract
Canine atopic dermatitis (AD) is clinically similar to human AD, implicating it as a useful model of human eosinophilic allergic disease. To identify cutaneous gene transcription changes in relatively early inflammation of canine AD, microarrays were used to monitor transcription in normal skin (n = 13) and in acute lesional AD (ALAD) and nearby visibly nonlesional AD (NLAD) skin (n = 13) from dogs. Scanning the putative abnormally transcribed genes, several potentially relevant genes, some abnormally transcribed in both NLAD and ALAD (e.g. IL6, NFAM1, MSRA, and SYK), were observed. Comparison for abnormally transcribed genes common to two related human diseases, human AD and asthmatic chronic rhinosinusitis with nasal polyps (aCRSwNP), further identified genes or gene sets likely relevant to eosinophilic allergic inflammation. These included: (1) genes associated with alternatively activated monocyte-derived cells, including members of the monocyte chemotactic protein (MCP) gene cluster, (2) members of the IL1 family gene cluster, (3) eosinophil-associated seven transmembrane receptor EMR1 and EMR3 genes, (4) interferon-inducible genes, and (5) keratin genes associated with hair and nail formation. Overall, numerous abnormally transcribed genes were observed only in canine AD; however, many others are common to related human eosinophilic allergic diseases and represent therapeutic targets testable in dogs with AD.
Keywords: Allergy, Atopic dermatitis, Asthma, Canine, Dog, Eczema, Eosinophil, Gene expression, High-density oligonucleotide arrays, Microarray, Rhinitis, Sinusitis
1. Introduction
Both human and canine atopic dermatitis (AD) are prevalent, and severe AD is particularly debilitating. Moreover, naturally occurring canine AD is clinically most similar to human AD – typically early onset, common environmental exposures and IgE reactivity (e.g. against house dust mite), Th2-predominant inflammation with ancillary eosinophil infiltration (i.e. “Th2 eosinophilic” inflammation), and complicating Staphylococcus and Malassezia infections. Some common molecular changes have also been reported in canine and human AD. For example, reduced ceramide (Reiter et al., 2009; Shimada et al., 2009; Yoon et al., 2011), increased CCR4, IL4, and S100A8 (Maeda et al., 2002; Nuttall et al., 2002; van Damme et al., 2009), and altered antimicrobial peptide levels (van Damme et al., 2009) have been shown in canine as well as human AD. Th2 eosinophilic inflammation at other epithelial surfaces also occurs in human allergic diseases such as asthma and eosinophilic esophagitis. The similarities between canine AD and related human eosinophilic allergic diseases indicate that canine AD should be a useful model to better understand and develop treatments for both dogs and humans.
Identifying common molecular networks between canine AD and human eosinophilic allergic diseases would provide the opportunity to test the importance of such networks in associated inflammation. A popular screening approach capable of identifying disease-associated molecular networks in an unbiased manner involves analysis of gene transcription using high-density oligonucleotide arrays, often referred to as microarrays. However, only one article using microarray technology to analyze gene transcription in canine AD skin has been reported, and this article presented limited data (Merryman-Simpson et al., 2008). Because identifying and controlling events relatively early in the inflammatory process is appealing, both acute lesional and nearby visibly nonlesional AD skin from dogs were analyzed here and compared to healthy skin from dogs using microarrays. Subsequent comparisons of these data to human AD (Olsson et al., 2006; Plager et al., 2007) and asthmatic chronic rhinosinusitis with nasal polyps (aCRSwNP) (Plager et al., 2010) data were performed to identify molecular networks common to canine AD and human eosinophilic allergic disease.
2. Materials and methods
2.1. Canine subjects and skin specimens
Thirteen dogs with AD diagnosed based on a characteristic history, clinical signs (Favrot et al., 2010), and exclusion of other pruritic skin disorders as deemed necessary were recruited. In addition, 13 normal dogs with no sign of atopic or other disease were enrolled. The study was approved by the University of Minnesota Institutional Animal Care and Use Committee (protocol number 0601A79512; University of Minnesota Assurance of Compliance number A3456). Canine Atopic Dermatitis Extent and Severity Index-2 (CADESI-2) scores or pruritus intensity using a visual analog scale from 0 to 10 were assessed to estimate overall AD severity (Table S1). Skin biopsies (8 mm) from dogs with AD were collected from sites of acute lesional skin and nearby visibly nonlesional skin (about 10 cm apart) that had not been topically treated with glucocorticoids or other immunosuppressants for at least three weeks before specimen collection. Oral immunosuppressants were also avoided for at least four weeks before skin biopsy collection, and other oral medications were minimized, and if used, are listed in Table S1. Immediately after collection, each skin biopsy was placed in room temperature RNAlater (Ambion, Austin, TX) for 2 min with mixing, transported on ice, and then a small portion of the biopsy was removed for formalin fixation. The larger portion of skin in RNAlater was held at 4 °C overnight and stored at −20 °C until total RNA isolation.
2.2. Total RNA isolation and high-density oligonucleotide microarray analysis
After removal from RNAlater, each skin specimen was frozen in liquid nitrogen, placed in ice-cold TRIzol (Invitrogen, Carlsbad, CA), and immediately shattered and homogenized with a Polytron homogenizer (Brinkmann Instruments, Westbury, NY). Total RNA was isolated using the TRIzol Plus RNA Purification System (Invitrogen) according to the manufacturer’s protocol. Purified total RNA was supplied to the Mayo CTSA Microarray Core facility, assessed for integrity using the Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA), and used for Affymetrix Canine 2 Genome (May 2005 version) high-density oligonucleotide microarray analyses according to Affymetrix-established protocols. To compensate for potential variability among separate microarray analyses, individual, non-pooled total RNA samples were isolated and analyzed in batches containing different specimen types (i.e. Normal, NLAD, and ALAD). Hybridized microarrays were scanned in the Affymetrix GeneArray Scanner. The resulting image files were analyzed to generate files with the x, y coordinates and average fluorescence intensities of each microarray cell. The 39 files with a .CEL extension (deposited at the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO)) were analyzed using Affymetrix Microarray Suite to quantify and scale the fluorescence data. Data analyses to identify differential transcription used GeneSpring GX 9.0 software (Agilent). Molecular network analyses of the identified differentially transcribed genes used Ingenuity Pathways Analysis software (Ingenuity Systems, Mountain View, CA).
To identify abnormal gene transcription common to canine AD and two related human eosinophilic allergic diseases, AD and aCRSwNP, a comparison based on identical Gene Symbols using Microsoft Office Access software was performed. Briefly, AD microarray data sets from Olsson et al. (2006) (GSE6012 in NCBI GEO; lesional AD skin, n = 10, and normal skin, n = 10) and from our group (GSE5667 in NCBI GEO; minimally lesional AD skin, n = 6, and nearby visibly nonlesional AD skin, n = 6, and normal skin, n = 5) (Plager et al., 2007) were imported separately into GeneSpring GX 9.0. After an initial low stringency comparison between AD and normal samples within each data set (fold change (Fc) ≥ 1.4 and unpaired nonparametric Mann–Whitney U test, p ≤ 0.22), subsequent analyses for differentially transcribed genes between AD and normal samples at Fc ≥ 2.0 and corrected p-value ≤0.1 (unpaired nonparametric test with Benjamini–Hochberg False Discovery Rate correction) were performed. A union set (i.e. a combined list) from the two AD to normal skin comparisons was generated. A similar list of differentially transcribed genes from the comparison of aCRSwNP to normal/rhinitis nasosinus tissue (Fc ≥ 2.0 and corrected p-value ≤0.05) was also used (Plager et al., 2010). Finally, a union set of the differentially transcribed genes identified here by comparing ALAD and Normal, NLAD and Normal, or ALAD and NLAD skin from dogs was generated (Fc ≥ 2.0 and corrected p-value ≤0.1), and this union set list from canine AD skin was compared to the Gene Symbol lists of differentially transcribed genes from human AD or aCRSwNP.
3. Results and discussion
Eosinophilic allergic diseases are prevalent, and severe disease can be debilitating. Unfortunately, highly representative animal models useful for testing potential therapeutic approaches to control or reverse such disease are lacking. Canine AD may be an exception in that it is clinically similar to human AD, and there may be important commonalities to other human eosinophilic allergic diseases as well. With this in mind and with the goal of identifying relatively early inflammatory changes, acute lesional (ALAD) and nearby visibly nonlesional (NLAD) AD skin (n = 13) and healthy skin (Normal; n = 13) from dogs (Table S1) was collected for microarray analysis. High-quality total RNA was isolated from each skin biopsy and analyzed individually using Affymetrix Canine 2 Genome microarrays. The resulting microarray mRNA transcript abundance data were grouped as ALAD, NLAD, or Normal. ALAD and Normal data (Table S2) and NLAD and Normal data (Table S3) were compared using an unpaired Mann–Whitney (nonparametric) analysis and ALAD and NLAD data (Table S4) were compared using a paired Mann–Whitney analysis with GeneSpring GX 9.0 software for differential transcription at ≥2.0 Fc and Benjamini–Hochberg False Discovery Rate-corrected p-value ≤0.1.
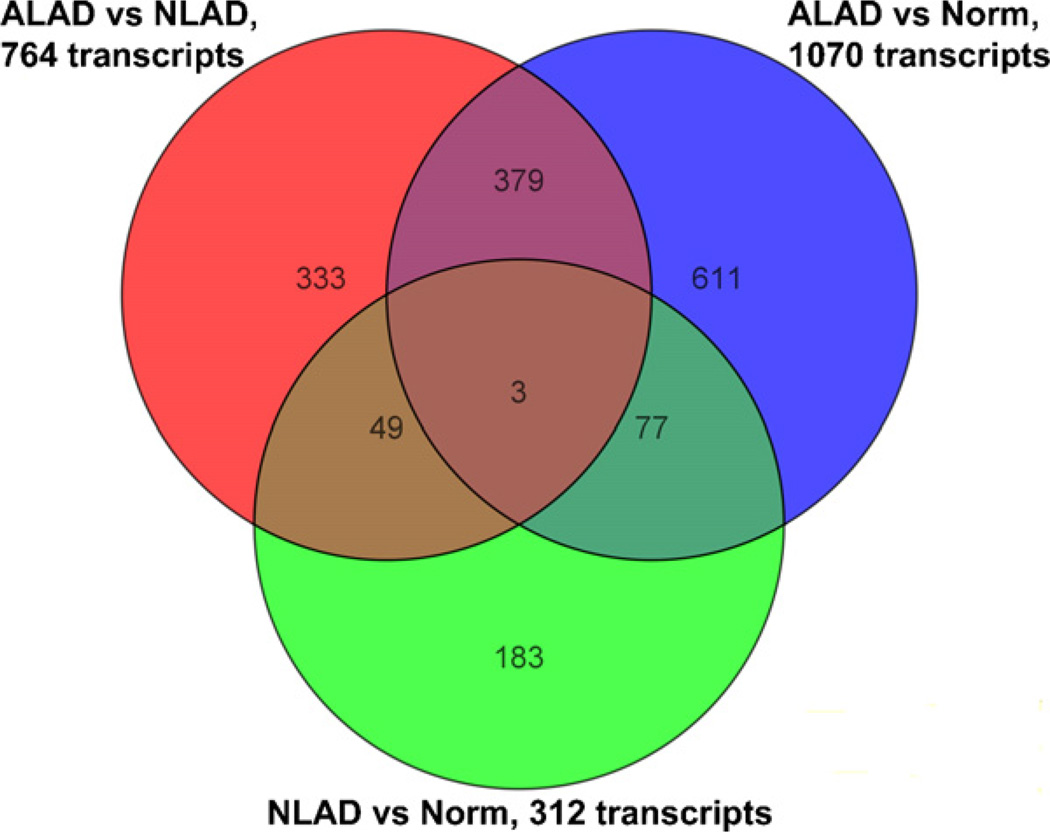
While use of a Benjamini–Hochberg False Discovery Rate-corrected p-value ≤0.1 is a relatively low stringency test, our focus on differential transcription at ≥2.0 Fc, and often in both NLAD and ALAD skin or in more than one Th2 eosinophilic disease, should help limit false positive identifications. Moreover, somewhat lower stringency analyses were chosen here to help identify interrelated genes that might not otherwise be tied together or genes that might only be differentially transcribed in a major subset of canine AD subjects. Even so, the data are also provided in an Excel file format allowing easy identification of differential transcription at any p-value less than 0.1 by applying the data “Sort” function to the “Corrected p-value” column of Tables S2–S4. Sorting in this way, it is seen that 77%, 32%, and 79% of the putative differentially expressed transcripts in Tables S2 (ALAD to Normal), S3 (NLAD to Normal), and S4 (ALAD to NLAD), respectively, were identified at corrected p-values <0.05. Thus, a substantial majority of differentially expressed transcripts in ALAD were identified at corrected p-value <0.05, and selected functional groupings of the corresponding abnormally transcribed genes in ALAD are shown in Table 1. The 39 raw microarray data files are available at NCBI GEO for re-analysis using any other criteria of choice as well (see Section 2). The total number and overlap of the differentially expressed transcripts identified here from comparison of ALAD or NLAD to Normal and ALAD to NLAD at ≥2.0 Fc and corrected p-value ≤0.1 are shown in Fig. 1.
Table 1.
Potential functional groupings of selected abnormally transcribed genes in ALAD skin from dogs.
| Functional grouping | Gene symbol(s) of abnormally transcribed genes (+, up- and −, down-regulated in ALAD) |
Comments |
|---|---|---|
| “Alternatively activated” (IL4- or IL13-induced) | ARG1(+)a, CCL2(+), CCL7(+), CCL8(+)b, CCL17(+), CHI3L1(+), CLEC4G(+), SAMSN1(HACS1)(+), TFEC(+) | a Produced by mouse alternatively activated macrophages (Martinez et al., 2009); b CCL2, CCL7, and CCL8 are members of the MCP gene cluster |
| Anti-apoptotic | BCL2A1(+), BIRC3(+) | |
| Cell adhesion | CADM1(−), ITGB2(+), ITGB6(+), SELE(+), SELL(+), SELP(+) | |
| Other chemokinesc, ILsc, and their receptors | CCL19(+), CCL28(+),CCR1(+), CCR3(+), CXCL10(+), IL6(+)d, IL8(+), IL20(+)d, IL26(+), IL1RAPL1(+), IL12RB2(−), IL13RA2(+), IL18BP(+) | c Also see “alternatively activated” and “IL1 family” sections in this table; d IL6 and IL20 transcription up in ALAD but down in NLAD |
| Collagens | COL7A1(−), COL10A1(+), COL11A2(−), COL13A(−), COL29A1(COL6A5)(+)e | e Sequence polymorphisms in the human ortholog are linked to AD |
| Complement-associated | C3(+), C5AR1(+), CFP(+) | |
| GPCRs (eosinophil-associated) | CCR3(+), EMR1(+), EMR3(+) | |
| GPRCs and associated signaling | ARHGEF12(+), ARHGEF4(+), GALR1(+), GPR65(+), GPR98(+),GPR112(−), GPR143(−), GPRC5D(−), GPSM3(+), PROKR2(−) | |
| IL1 familyf | IL1B(+), IL1F8(+), IL1F10(−) | f Also see Table 2 of this article |
| Innate immunity receptors and NLRs | CLEC4G(+), CLEC4M(+), MSR1(+), MST1R(+), NLRC5/P1/P3(+), NLRP9(−) | |
| Interferon-induced | GBP1(+), IFI44(+), IFITM2(+), ISG15(+), ISG20(+), MNDA(+), MX1(+), MX2(+), OAS1(+), OAS2(+) | Increased transcription of PILRA and TNFRSF14, receptors for HSV1 entry into human cells, was also seen in dog ALAD skin |
| Keratins | KRT2(+), KRT5/26/27/28/31/32/33B/35/37/39/72/81/82/85(−) | Several of the under-transcribed KRT genes and PADI3 have human orthologs associated with hair, nail, or skin barrier formation |
| LILRs | LILRB2(+), LILRB3(+) | These molecules bind MHC class I and transduce a negative signal that inhibits immune responses |
| Lipid-associated | FABP9(−), FADS1(−), LPCAT1(+), PLA2G2E(−), PLA2G4D(+), PLA2G4E(+), PLA2G4F(+), PLA2G6(−), PLCB4(−), PTGDS(−) | |
| L-tryptophan metabolism | INDO(IDO1)(+), INDOL1(IDO2)(−), KMO(+) | |
| Lymphocyte antigens | LY75(−), LY9(+), LY86(+), LY96(+) | |
| Mast cell-associated | CPA3(+), FCER1A(+)g, FCER1G(+)g, LOC448801(Mastin)(+), SRGN(+), SYK(+)h | g FCER1A and G also expressed by cutaneous dendritic cells; h SYK transcription also up in NLAD |
| Proteases and their inhibitors | ADAMTS20(−), ELA1(+), KLK2(−), KLK4(−), KLK8(+), KLK13(+), MMP1(+), MMP7(+), MMP9(−/+)i, MMP13(+), NLN(−), PI16(−), SERPINA3(+), SERPINA7(−), SERPINB4(+), SERPINB13(−), SERPINE1(+), SPINK4(+), TIMP1(+), TMPRSS3(−) | i One transcript variant of MMP9 is over- and one is under-transcribed in ALAD |
| S100s | S100A3(−), S100A8(+), S100A9(+), S100P(+) | |
| Solute carriersj | Numerous “SLC” genes putatively up- and down-regulated (e.g. SLC11A1(+) and SLC12A8(−)) | j Multiple SLC genes also appear to be differentially transcribed in NLAD |
| Transcription factors and zinc finger proteins | AFF3(+), BNC2(−), FOXF1(−), GLI1(−), GRHL3(−), HOXB3(−), HOXC12(+), HOXC13(−)k, KLF7(+), MKX(−), NFAM1(+)l, NFE2(+), NR0B2(−), PDX1(−), SPI1(PU.1)(+), TAL1(+), TBX1(−), TFEC(+), numerous “zinc finger” genes putatively up- and down-regulated (e.g. ZBTB22(−), ZC3H12A(+)m, ZFP106(−), ZNF208(+)) | k May play a role in hair or nail development; l a membrane receptor that activates the calcineurin/NFAT pathway. NFAM1, IKZF3 and MTA3, transcription is also up in NLAD; m CCL2-induced |
Abbreviations: AD, atopic dermatitis; ALAD, acute lesional AD; GPRCs, G-protein coupled receptors; HSV1, herpes simplex virus-1; LILRs, leukocyte Ig-like receptors; MCP, monocyte chemotactic protein; NLAD, nonlesional AD; NLRs, NOD-like receptors.
Fig. 1.
Number of the common and distinct differentially expressed transcripts among the three primary analyses of canine AD microarray data. The differentially expressed transcripts listed in supplemental Tables 2S (ALAD versus Normal), 3S (NLAD versus Normal), and 4S (ALAD versus NLAD) are shown in a Venn diagram format.
As might be expected and consistent with our human AD microarray data (Plager et al., 2007), the comparison of NLAD to Normal skin from dogs identified the least number of differentially expressed transcripts (Fig. 1), suggesting the relative similarity between NLAD and Normal skin. Furthermore, abnormal transcription of IgE-associated (increased FCER1A and FCER1G), Th2-associated (increased IL13RA2), Th1-associated (decreased IL12RB2), and eosinophil-associated (increased CCR3) genes are consistent with the expected Th2 eosinophilic profile for ALAD skin (Tables S2 and/or S4). Thus, the microarray data presented here generally conform to previous targeted molecular and cellular analyses.
Because of the potential value of identifying abnormal transcription early in the inflammatory process, both acute lesional and nearby visibly nonlesional AD skin from dogs were analyzed. While obvious disease-related differences between NLAD and Normal skin can be difficult to discern, several abnormally transcribed genes in NLAD skin may be important. Interleukin 6 (IL6) was markedly under transcribed in NLAD (Table S3; −4.1 Fc, p = 0.035) and over transcribed in ALAD skin (Table S2; 3.3 Fc, p = 0.087; and Table S4; 13.7 Fc, p = 0.017). Given the influence of IL6 on T helper cell subtype development, perhaps decreased IL6 in NLAD adversely effects appropriate T helper cell development in AD? Its increase in ALAD, in contrast, may simply reflect its well-known association with an inflammatory response. Increased transcription of NFAM1 in NLAD (Table S3; 2.9 Fc, p = 0.042) and in ALAD (Table S2; 2.6 Fc, p = 0.031) is another intriguing abnormality. NFAM1 is a membrane receptor capable of activating the calcineurin/NFAT pathway and downstream cytokine gene promoters, like that of the Th2-type cytokine IL13. The recent success of calcineurin inhibitors for controlling human and canine AD further suggests its potential importance. Increased KRT2 and decreased KRT37 transcription in NLAD (Table S3; 2.9 Fc, p = 0.083 and −2.9 Fc, p = 0.059) appear to be further increased and decreased, respectively, in ALAD (Table S2; 4.1 Fc, p = 0.017 and −6.55 Fc, p = 0.012) compared to Normal skin from dogs. Perhaps these and other keratin transcription changes (Table 1) reflect skin barrier abnormalities in atopic skin of dogs? Like KRT37, decreased transcription of other hair-associated keratins, KRT31 and KRT81, is common to both canine and human AD (Table S5). Similarly, decreased transcription of MSRA, which is proposed to repair oxidative damage to proteins, in NLAD and ALAD compared to Normal skin from dogs (Tables S3 and S2; −2.6 Fc, p = 0.046 and −2.0 Fc, p = 0.066) might decrease the ability of atopic skin of dogs to limit tissue damage caused by reactive oxygen species typically generated during an inflammatory or stress response (Nan et al., 2010). Finally, SYK is increased in both NLAD and ALAD compared to Normal skin from dogs (Tables S3 and S2; 2.6 Fc, p = 0.042 and 4.1 Fc, p = 0.004) and increased in human AD and aCRSwNP (Table S5), and SYK inhibitors are being tested for their ability to inhibit atopic disease (Masuda and Schmitz, 2008). Overall, the potential role of these genes, as well as the others listed in Table S3 (NLAD versus Normal), in the early inflammatory changes of canine and possibly human allergic disease are intriguing.
Considering the abnormal transcription observed in ALAD skin from dogs, several groups of functionally related genes were conspicuous (Table 1). Prominent among these were multiple genes reportedly induced by the Th2 cytokines IL4 and/or IL13. This included increased transcription of genes associated with so-called alternatively activated monocyte-derived cells (aaMDCs) such as ARG1, members of the monocyte chemotactic protein (MCP) gene cluster (CCL2, CCL7, and CCL8), CCL17, CHI3L1, CLEC4G, and TFEC (Table 1 and Tables S2 and/or S4), and most of these are also expressed by human aaMDCs (Martinez et al., 2009). While the precise structure and particular genes comprising the MCP gene cluster varies somewhat between dogs (chromosome 9) and humans (chromosome 17q11.2), the MCP gene cluster appears important in the early inflammatory response of human eosinophilic allergic disease (Plager et al., 2010). Most recently, the importance of mouse CCL8 in the recruitment of IL5+ Th2 cells and in cutaneous eosinophilic inflammation also has been demonstrated (Islam et al., 2011). Similarly, a role for CHI3L1 in the initiation and perpetuation of Th2 inflammation has been reported (Lee et al., 2009). Thus, these data importantly point to the likely role of aaMDCs in canine AD and the particular aaMDC-associated genes (e.g. those of the MCP gene cluster) that apparently perpetuate Th2 eosinophilic inflammation in canine AD as well as related human allergic diseases.
A second functional group of likely importance in canine and human AD is the IL1 protein family (Table 1) (Jensen, 2010). Transcription of pro-inflammatory IL1B and IL1F8 was increased while that of anti-inflammatory IL1F10 was decreased in canine AD (Table 2 and Tables S2 and/or S4). This pattern of increased pro-inflammatory and decreased anti-inflammatory IL1 family member transcription is similar to that seen in human AD, although abnormal transcription of different IL1 family member orthologs (e.g. canine IL1F8 and IL1F10, instead of human IL1F9 and IL1F7) and increased IL1B transcription was observed in canine AD (Table 2). Notably, confirmation of a corresponding increase in pro-inflammatory human IL1F9 at the protein level has been reported in human AD (Broccardo et al., 2009). Furthermore, an imbalance in pro- and anti-inflammatory IL1 family member transcription has also been shown in human psoriasis (Blumberg et al., 2007; Guttmann-Yassky et al., 2009); however, there appears to be a pronounced offset in the relative transcription levels of the IL1 family members (IL1Fs) in AD compared to psoriasis (Table 2) (Guttmann-Yassky et al., 2009). That is, the transcription levels of both pro- and anti-inflammatory IL1Fs tend to be markedly lower in AD than in psoriasis. It is interesting to note that the relatively decreased levels of pro-inflammatory IL1Fs in AD are reminiscent of the relatively decreased levels of antimicrobial peptides, like cathelicidin and beta-defensins (Wollenberg et al., 2011), also seen in AD compared to psoriasis. Furthermore, it is intriguing to speculate that the imbalance and relative abundance of the various IL1Fs might contribute to autoinflammatory aspects and skewed T helper cell profiles of inflammatory skin diseases.
Table 2.
Abnormally transcribed IL1 family genes in atopic dermatitis (AD) and psoriasis (Ps).
| Data set | Gene symbol | Probe set ID | AD/Normal |
| Human ADa | IL1F9 ▲b | 220322_at | 3.06 |
| IL1F7 ▼ | 224555_x_at | 0.34 | |
| IL1F7 ▼ | 221470_s_at | 0.10 | |
| IL1RN (IL1ra, IL1F3) ▼ | 216244_at | 0.32 | |
| Canine AD | IL1F10 ▼ | CfaAffx.11766.1.S1_at | 0.49 |
| IL1F8 ▲ | CfaAffx.11760.1.S1_at | 2.77 | |
| IL1B ▲ | Cfa.3554.1.S1_at | 2.78 | |
| Data set | Gene symbol | AD/Normal | Ps/Normal |
| Human AD and psoriasisc | IL1F9 ▲ | 2.06 | 122 |
| IL1F7 ▼ | 0.007 | 0.088 | |
| IL1F5 ▼ | 0.73 | 7.71 | |
| IL1RN ▼ | 0.68 | 30.8 | |
| IL1B (IL1F2)d ▲ | n.s.e | ~4 | |
Putative pro-inflammatory (▲) and anti-inflammatory (▼).
Measured by qRT-PCR.
Not significantly different.
Eosinophils appear to contribute to allergic disease pathology, and our recent analyses of human aCRSwNP showed increased transcription of the G-protein coupled receptors EMR1 and EMR3 (Plager et al., 2010), which are preferentially expressed by eosinophils (Hamann et al., 2007). The orthologous EMR1 and EMR3 genes in dogs also showed increased transcription in canine AD (Table 1 and Tables S2 and S5). Given their unknown function(s) and eosinophil-associated expression, like that of CCR3, the potential contribution(s) of EMR1 and EMR3 genes to canine and human allergic disease pathology are another area of interest.
Ten interferon-induced genes showed increased transcription in canine AD, and a majority of their orthologous human genes also showed elevated transcription in human AD (Table 1 and Table S5). Among these were MX1 and OAS1, which are most closely linked to an anti-viral response. It is intriguing to speculate that this increased interferon-induced gene transcription profile might result from a subclinical or lingering cutaneous viral infection capable of sustaining chronic AD inflammation, particularly given the known susceptibility of humans with AD to certain cutaneous viral infections. However, no viruses are known to preferentially infect the skin of dogs with AD, so perhaps this profile is associated more generally with the AD inflammatory response. Regardless, understanding the cause and consequences of this particular novel gene transcription profile would likely prove informative with respect to AD disease pathology.
Notably, as an attempt to provide additional context to the abnormally transcribed canine AD genes identified here, these genes were analyzed for relevant molecular networks using an Ingenuity Pathways Analysis approach. Because annotation of the dog genome is less complete than that for humans, a high percentage of the over- and under-expressed transcripts were not yet assigned to a particular gene. This incomplete annotation likely contributed to the limited benefit of analyzing for relevant molecular networks using an Ingenuity Pathways Analysis approach at this time.
Other potential limitations of this microarray study also exist. To avoid an excessive enrollment period, precise matching of factors such as gender, age, breed, and biopsy site location was not achieved (Table S1). Similarly, while concurrent medication use was avoided as described in Section 2, 5 of the 13 dogs with AD were taking some form of oral medication at the time of skin biopsy (Table S1). Lesion type or duration are also potential complicating factors. Nonetheless, most of these factors were comparable overall or on average between AD and Normal dogs (see Table S1; e.g. 7/6 and 5/8 female/male, average age of 4.4 and 5.5 years, and 8 of 13 breed matches), or the analysis of a reasonable sample size (n = 13 for both AD and Normal dogs) and of both acute lesional and nearby visibly nonlesional AD skin were undertaken to constrain the potential impact of such factors.
Canine AD is a complex polygenic disease and heterogeneity in environmental triggers and breed-specific genetic makeup (Wood et al., 2010) can be additional complicating factors for identifying disease-specific gene transcription. Indeed, any of the above complicating factors may contribute to incomplete concordance between different studies of canine AD. For example, in a differential transcription study of canine AD using RT-PCR analysis of 20 candidate genes (Wood et al., 2009), 13 genes showed >2 Fc (i.e. >1 true fold change) in transcription (2 genes), differential transcription at p < 0.05 (6 genes), or both (5 genes). And of these 13 genes, only 4 were detected here at ≥2 Fc and p ≤ 0.1 (S100A8, SAA1, SCCA2/SERPINB4, and TIMP1). Nonetheless, while different environmental and polygenic triggers for developing canine and human AD almost certainly exist, some common downstream molecular networks likely lead to the characteristic Th2 eosinophilic inflammation of AD and other related allergic diseases, several of which are highlighted above.
In summary, we provide well-described, publicly accessible microarray data of normal and acute lesional and nearby visibly nonlesional AD skin from dogs, with preliminary analyses and highlighting of several novel and likely relevant abnormal transcription patterns. Numerous abnormally transcribed genes or gene sets were observed only in canine AD while many others are common to related human diseases, human AD and aCRSwNP. These common abnormally transcribed genes should be amenable to testing in dogs with AD for their impact on both canine and human eosinophilic allergic disease pathology.
Supplementary Material
Acknowledgements
We thank the Mayo CTSA Microarray Facility for microarray data generation and The Minnesota Partnership for Biotechnology and Medical Genomics for funding.
Abbreviations
- aaMDCs
alternatively activated monocyte-derived cells
- aCRSwNP
asthmatic chronic rhinosinusitis with nasal polyps
- AD
atopic dermatitis
- ALAD
acute lesional atopic dermatitis
- CADESI-2
Canine Atopic Dermatitis Extent and Severity Index-2
- Fc
fold change
- MCP
monocyte chemotactic protein
- NCBI GEO
National Center for Biotechnology Information Gene Expression Omnibus
- NLAD
nonlesional atopic dermatitis
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.vetimm.2012.06.003.
Footnotes
Conflict of interest
The authors have no conflicts of interest to disclose.
References
- Blumberg H, Dinh H, Trueblood ES, Pretorius J, Kugler D, Weng N, Kanaly ST, Towne JE, Willis CR, Kuechle MK, Sims JE, Peschon JJ. Opposing activities of two novel members of the IL-1 ligand family regulate skin inflammation. J. Exp. Med. 2007;204:2603–2614. doi: 10.1084/jem.20070157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccardo CJ, Mahaffey SB, Strand M, Reisdorph NA, Leung DY. Peeling off the layers: skin taping and a novel proteomics approach to study atopic dermatitis. J. Allergy Clin. Immunol. 2009;124:1113–1115. doi: 10.1016/j.jaci.2009.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favrot C, Steffan J, Wolfgang S, Picco F. A prospective study on the clinical features of chronic atopic dermatitis and its diagnosis. Vet. Dermatol. 2010;21:23–31. doi: 10.1111/j.1365-3164.2009.00758.x. [DOI] [PubMed] [Google Scholar]
- Guttmann-Yassky E, Suárez-Fariñas M, Chiricozzi A, Nograles KE, Shemer A, Fuentes-Duculan J, Cardinale I, Lin P, Bergman R, Bowcock AM, Krueger JG. Broad defects in epidermal cornification in atopic dermatitis identified through genomic analysis. J. Allergy Clin. Immunol. 2009;124:1235–1244. doi: 10.1016/j.jaci.2009.09.031. [DOI] [PubMed] [Google Scholar]
- Hamann J, Koning N, Pouwels W, Ulfman LH, van Eijk M, Stacey M, Lin HH, Gordon S, Kwakkenbos MJ. EMR1, the human homolog of F4/80, is an eosinophil-specific receptor. Eur. J. Immunol. 2007;37:2797–2802. doi: 10.1002/eji.200737553. [DOI] [PubMed] [Google Scholar]
- Islam SA, Chang DS, Colvin RA, Byrne MH, McCully ML, Moser B, Lira SA, Charo IF, Luster AD. Mouse CCL8, a CCR8 agonist, promotes atopic dermatitis by recruiting IL5+ Th2 cells. Nat. Immunol. 2011;12:167–177. doi: 10.1038/ni.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LE. Targeting the IL-1 family members in skin inflammation. Curr. Opin. Investig. Drugs. 2010;11:1211–1220. [PMC free article] [PubMed] [Google Scholar]
- Lee CG, Hartl D, Lee GR, Koller B, Matsuura H, Da Silva CA, Sohn MH, Cohn L, Homer RJ, Kozhich AA, Humbles A, Kearley J, Coyle A, Chupp G, Reed J, Flavell RA, Elias JA. Role of breast regression protein 39 (BRP-39)/chitinase 3-like-1 in Th2 and IL-13-induced tissue responses and apoptosis. J. Exp. Med. 2009;206:1149–1166. doi: 10.1084/jem.20081271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S, Okayama T, Omori K, Masuda K, Sakaguchi M, Ohno K, Tsujimoto H. Expression of CC chemokine receptor 4 (CCR4) mRNA in canine atopic skin lesion. Vet. Immunol. Immunopathol. 2002;90:145–154. doi: 10.1016/s0165-2427(02)00232-5. [DOI] [PubMed] [Google Scholar]
- Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu. Rev. Immunol. 2009;27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- Masuda ES, Schmitz J. Syk inhibitors as treatment for allergic rhinitis. Pulm. Pharmacol. Ther. 2008;21:461–467. doi: 10.1016/j.pupt.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Merryman-Simpson AE, Wood SH, Fretwell N, Jones PG, McLaren WM, McEwan NA, Clements DN, Carter SD, Ollier WE, Nuttall T. Gene (mRNA) expression in canine atopic dermatitis: microarray analysis. Vet. Dermatol. 2008;19:59–66. doi: 10.1111/j.1365-3164.2008.00653.x. [DOI] [PubMed] [Google Scholar]
- Nan C, Li Y, Jean-Charles PY, Chen G, Kreymerman A, Prentice H, Weissbach H, Huang X. Deficiency of methionine sulfoxide reductase A causes cellular dysfunction and mitochondrial damage in cardiac myocytes under physical and oxidative stresses. Biochem. Biophys. Res. Commun. 2010;402:608–613. doi: 10.1016/j.bbrc.2010.10.064. [DOI] [PubMed] [Google Scholar]
- Nuttall TJ, Knight PA, McAleese SM, Lamb JR, Hill PB. T-helper 1, T-helper 2 and immunosuppressive cytokines in canine atopic dermatitis. Vet. Immunol. Immunopathol. 2002;87:379–384. doi: 10.1016/s0165-2427(02)00076-4. [DOI] [PubMed] [Google Scholar]
- Olsson M, Broberg A, Jernås M, Carlsson L, Rudemo M, Suurkula M, Svensson PA, Benson M. Increased expression of aquaporin 3 in atopic eczema. Allergy. 2006;61:1132–1137. doi: 10.1111/j.1398-9995.2006.01151.x. [DOI] [PubMed] [Google Scholar]
- Plager DA, Leontovich AA, Henke SA, Davis MD, McEvoy MT, Sciallis GF, 2nd, Pittelkow MR. Early cutaneous gene transcription changes in adult atopic dermatitis and potential clinical implications. Exp. Dermatol. 2007;16:28–36. doi: 10.1111/j.1600-0625.2006.00504.x. [DOI] [PubMed] [Google Scholar]
- Plager DA, Kahl JC, Asmann YW, Nilson AE, Pallanch JF, Friedman O, Kita H. Gene transcription changes in asthmatic chronic rhinosinusitis with nasal polyps and comparison to those in atopic dermatitis. PLoS One. 2010;5:e11450. doi: 10.1371/journal.pone.0011450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter LV, Torres SM, Wertz PW. Characterization and quantification of ceramides in the nonlesional skin of canine patients with atopic dermatitis compared with controls. Vet. Dermatol. 2009;20:260–266. doi: 10.1111/j.1365-3164.2009.00759.x. [DOI] [PubMed] [Google Scholar]
- Shimada K, Yoon YS, Yoshihara T, Iwasaki T, Nishifuji K. Increased transepidermal water loss and decreased ceramide content in lesional and non-lesional skin of dogs with atopic dermatitis. Vet. Dermatol. 2009;20:541–546. doi: 10.1111/j.1365-3164.2009.00847.x. [DOI] [PubMed] [Google Scholar]
- van Damme CM, Willemse T, van Dijk A, Haagsman HP, Veldhuizen EJ. Altered cutaneous expression of beta-defensins in dogs with atopic dermatitis. Mol. Immunol. 2009;46:2449–2455. doi: 10.1016/j.molimm.2009.05.028. [DOI] [PubMed] [Google Scholar]
- Wollenberg A, Räwer HC, Schauber J. Innate immunity in atopic dermatitis. Clin. Rev. Allergy Immunol. 2011;41:272–281. doi: 10.1007/s12016-010-8227-x. [DOI] [PubMed] [Google Scholar]
- Wood SH, Clements DN, Ollier WE, Nuttall T, McEwan NA, Carter SD. Gene expression in canine atopic dermatitis and correlation with clinical severity scores. J. Dermatol. Sci. 2009;55:27–33. doi: 10.1016/j.jdermsci.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Wood SH, Ollier WE, Nuttall T, McEwan NA, Carter SD. Despite identifying some shared gene associations with human atopic dermatitis the use of multiple dog breeds from various locations limits detection of gene associations in canine atopic dermatitis. Vet. Immunol. Immunopathol. 2010;138:193–197. doi: 10.1016/j.vetimm.2010.07.020. [DOI] [PubMed] [Google Scholar]
- Yoon JS, Nishifuji K, Sasaki A, Ide K, Ishikawa J, Yoshihara T, Iwasaki T. Alteration of stratum corneum ceramide profiles in spontaneous canine model of atopic dermatitis. Exp. Dermatol. 2011;20:732–736. doi: 10.1111/j.1600-0625.2011.01306.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.