Abstract
Linking neural microcircuit function to emergent properties of the mammalian brain requires fine-scale manipulation and measurement of neural activity during behavior, where each neuron’s coding and dynamics can be characterized. We developed an optical method for simultaneous cellular-resolution stimulation and large-scale recording of neuronal activity in behaving mice. Dual-wavelength two-photon excitation allowed largely independent functional imaging with a green fluorescent calcium sensor (GCaMP3, λ = 920 ± 6 nm) and single-neuron photostimulation with a red-shifted optogenetic probe (C1V1, λ = 1,064 ± 6 nm) in neurons coexpressing the two proteins. We manipulated task-modulated activity in individual hippocampal CA1 place cells during spatial navigation in a virtual reality environment, mimicking natural place-field activity, or ‘biasing’, to reveal subthreshold dynamics. Notably, manipulating single place-cell activity also affected activity in small groups of other place cells that were active around the same time in the task, suggesting a functional role for local place cell interactions in shaping firing fields.
The development of recent optical sensors, probes and methods for imaging or perturbing activity in the behaving mammalian brain is a promising step toward the functional characterization of brain dynamics on a large scale, at high resolution (thousands of individual neurons during a behavior). For example, population dynamics in behaving rodents may be measured at cellular resolution using two-photon excitation (TPE) fluorescence imaging1–4 or perturbed on a scale of genetically defined populations using optogenetic stimulation5–7. Combining the different advantages of these two approaches would create a new class of experiments to examine behavioral substrates in neural microcircuits by allowing cell-specific perturbation of activity in neurons on the basis of patterns of natural activity during behavior.
Several experimental challenges have hindered the combined use of cellular-resolution photostimulation and imaging in behaving rodents. The first problem is that existing optogenetic probes, calcium sensors and microscope fluorescence detectors are all sensitive to visible-wavelength light. In practice, this introduces substantial crosstalk in all-optical experiments using visible-light illumination, as imaging light may perturb cellular activity and photostimulation may interrupt fluorescence detection8–14. Second, although wide-field optogenetic stimulation (for example, with unfocused blue light) only requires illuminating enough total membrane area to recruit large photocurrents from many cells, spatially resolved photostimulation also requires confining the illuminated area to the cell(s) of interest. Experimentally, cellular-resolution stimulation can be achieved with low-intensity excitation of a volume around the size of a cell soma in an opsin-photocycle time constant15–19. For visible-wavelength illumination, which is the most commonly adopted approach in optogenetic experiments, this is difficult to achieve in intact brain tissue at depths much below a mean-free light-scattering path (50–100 μm20,21), after which visible-wavelength light becomes defocused. Moreover, scattered or ballistic visible-light single-photon excitation, which is linearly proportionate to the incident intensity, may also stimulate photocurrents in dendrites, axons or somas of many other cells away from the plane of focus. We developed an approach that addresses these challenges, combining cellular-resolution photostimulation and simultaneous imaging in a densely labeled population of neurons in awake mice, and demonstrate how this approach can be used to mimic or modify activity of individual neurons during a behavior.
RESULTS
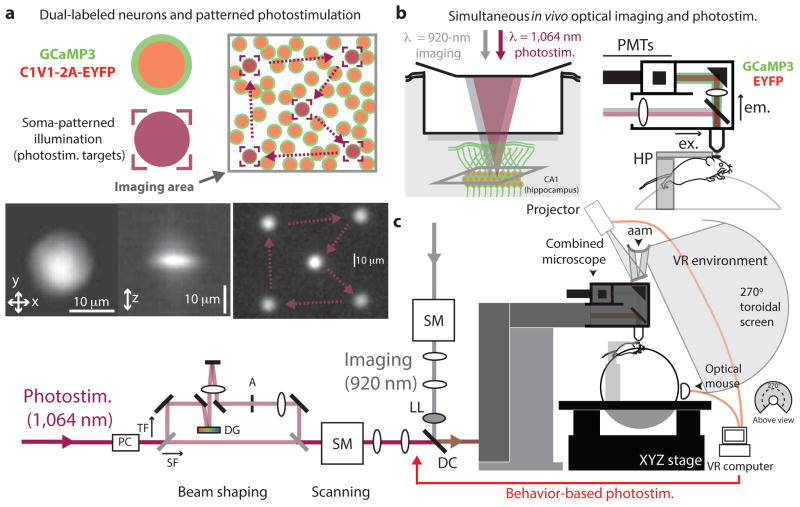
Our approach is based on the combined application of two spectrally separated infrared TPE sources for both fluorescence imaging and photostimulation, and the use of a green calcium sensor (GCaMP3)22 and a red-shifted optogenetic probe (C1V1)18,23 that are coexpressed in a population of neurons (Fig. 1). Neurons were visualized in head-restrained, mobile mice through a sealed optical window3 using TPE fluorescence imaging at sub-micron wavelengths (typically, λ = 920 ± 6 nm) that preferentially excited GCaMP3 fluorescence over C1V1 photocurrents, and wide-field raster-scanning acquisition that did not concentrate imaging light on any one cell. Neurons of interest were selected as targets for photostimulation, which used a second TPE source operating at longer wavelengths (λ = 1,064 ± 6 nm) to preferentially excite C1V1 photocurrents over GCaMP3 fluorescence, and temporal focusing optics16,24,25 to generate an illumination spot patterned after the dimensions of a pyramidal neuron soma (≈10–15 μm diameter, ≈6 μm depth) that concentrated TPE on single target neurons (Fig. 1). Scanning mirrors allowed rapid repositioning of the stimulation spot across different target locations on a physiological timescale (1–2 kHz; Fig. 1a). By combining the system with an apparatus for virtual reality (VR)-based training and behavior, where neuronal activity can be observed and characterized during behavior, single neurons or groups of neurons with behavior-correlated activity could be optically stimulated while we simultaneously measured evoked changes in dynamics of those neurons and others in the population.
Figure 1.
Schematic for simultaneous cellular-resolution photostimulation and functional calcium-imaging in awake, behaving mice. (a) Neurons expressing a green calcium sensor (GCaMP3) and a red-shifted optogenetic probe (C1V1(E122T/E162T)-2A-EYFP; red) visualized in awake mice using TPE imaging were selected as targets for TPE photostimulation. Each target cell was stimulated by transient illumination with a temporally focused ‘spot’ around the size of a soma (10–15 μm). Bottom left, CCD images of TPE fluorescence illustrating in-plane (xy) and projected axial (xz) illumination profiles of the photostimulation spot. Bottom right, image of a patterned photostimulation scan (dwell time of 0.5 ms per location; CCD integration time of 1,000 ms). Arrows indicate the repeat scan trajectory between targets. (b) Two laser-scanning TPE systems (imaging and photostimulation) were combined in a custom microscope to image (λ = 920 nm) and stimulate (λ = 1,064 nm) coexpressing hippocampal CA1 neurons in awake, mobile or behaving mice. ex., excitation path; em., emitted fluorescence detection path; PMTs, photomultiplier tubes; HP, headplate holders. (c) The combined instrument (optical path schematic, left) used a VR system to create a virtual environment in which mice could be trained to perform visually guided behaviors, and large-scale optical recordings were used to characterize neuron population activity. Single neurons of interest were selected and stimulated at designated times in the behavior, synchronized using custom VR software29. PC, Pockels cell; TF, temporal focusing path; SF, spatial focusing path; DG, diffraction grating; A, aperture; SM, scanning mirrors; LL, liquid lens; DC, dichroic mirror; aam, angular amplification mirror.
Coexpression, separable detection, and independent optical excitation of an optogenetic probe and a calcium sensor in awake mice
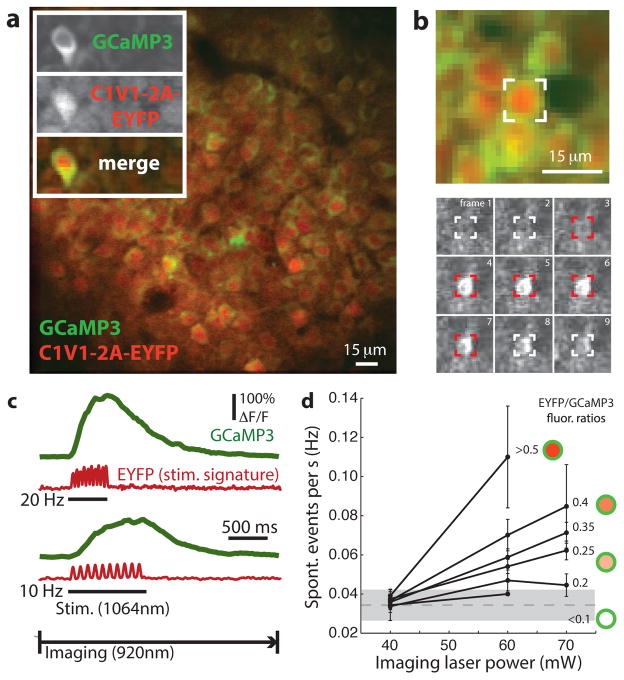
As a first step toward realizing this approach, we screened combinations of genetically encoded optogenetic probes and calcium sensors to identify a probe-sensor pair that could be independently excited, separably detected and functionally coexpressed in a high density of neurons in vivo. We identified one pair (C1V1 and GCaMP3) with well-separated absorption properties under infrared TPE in vitro (Supplementary Fig. 1) that could also be coexpressed in a population of neurons in vivo through virus-mediated expression of the probe (adeno-associated virus, AAV2/5-Camk2a::C1V1(E122T/E162T)-p2A-EYFP-WPRE18) and transgenic expression of the sensor (transgenic strain gp2.11; Janelia Research Campus, Howard Hughes Medical Institute). Beginning 2 weeks after virus injection and optical window implantation3, neurons expressing the probe (visualized using the volume-filling EYFP fluorescence), the sensor (GCaMP3, primarily in the cytoplasm) or both (Fig. 2) were readily distinguished using TPE fluorescence imaging and spectral unmixing26 in awake mice. A high density of neurons expressed both the probe and the sensor (Fig. 2a) and exhibited stable morphology and expression profiles over the duration of these experiments (2–4 weeks post-injection).
Figure 2.
All-optical stimulation and recording of neural activity in awake mice. (a) TPE fluorescence image of CA1 hippocampal neurons expressing GCaMP3 (green) and C1V1(E122T/E162T)-2A-EYFP (red) in an awake mouse. Inset, images of unmixed GCaMP3 and EYFP (top panels) and a pseudocolor merge (bottom panel; image sizes 25 × 65 μm). Somatic GCaMP3 appeared to be annular from nuclear exclusion, whereas EYFP was diffuse. (b) Simultaneous optical stimulation and imaging of activity in a targeted neuron (indicated in top panel). Bottom, GCaMP3 fluorescence images during one stimulationand imaging time series (frame interval of 0.125 s, target is shown in red during a 10 Hz × 0.05-s stimulation pulse-train). 1,064-nm stimulation evoked a GCaMP3 transient in the targeted cell that was detected using 920-nm imaging. (c) Somatic ΔF/F traces (GCaMP3, green) recorded during photostimulation using two-channel fluorescence detection and linear unmixing (five-trial average, 10 × 0.05-s pulses at 20 and 10 Hz during underlined periods). Evoked GCaMP3 transients were separable from optical stimulation artifacts (EYFP fluorescence, red traces), allowing continuous imaging during stimulation. (d) Estimate of activity induced by imaging opsin-expressing neurons. Rates of spontaneous activity versus imaging laser power for several neuron populations with increasing C1V1 expression (estimated by EYFP/GCaMP3 intensity and depicted by soma cartoons). Low-power imaging (≈40 mW) produced minimal changes in spontaneous activity in coexpressing neurons compared withneurons with little or no detectable C1V1 -EYFP expression (error bars indicate mean ± s.d.; mean ± s.d. of lowest-expressing group at 40 mW is shown in shaded area).
To test whether activity in neurons could be both stimulated and detected optically, we selected coexpressing neurons as targets and then stimulated them: in awake mice, 1,064-nm TPE stimulation of targeted CA1 neurons evoked somatic GCaMP3 transients that were detectable using simultaneous 920-nm TPE imaging (Fig. 2b,c). TPE-evoked transients were similar to responses evoked using conventional visible-light single-photon excitation (SPE, λ = 473 nm; Fig. 3a and Supplementary Fig. 2), required coexpression of C1V1 and GCaMP3 (Fig. 2d), and reached peak amplitudes that increased with TPE scanning rate using a diffraction-limited focus or with pulse number in short stimulus trains (Supplementary Fig. 3). These observations are consistent with the time-dependent molecular properties of C1V1 depolarizing currents23,27 and GCaMP3 fluorescence transients reporting accumulating calcium during electrically evoked trains of action potentials (APs) in vivo22. These photostimulation-triggered fluorescence transients therefore likely represent optically evoked trains of APs, demonstrating that this approach allows all-optical stimulation and simultaneous detection of activity in neurons in awake mice.
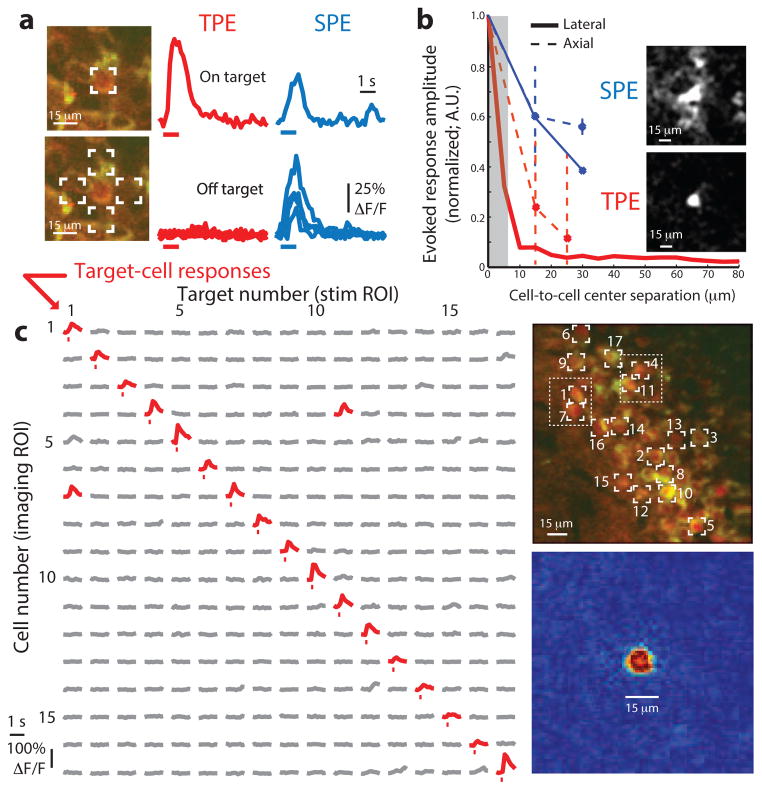
Figure 3.
Cellular-resolution photostimulation in awake mice. (a) Specificity of cell-targeted photostimulation using focused single-photon excitation (SPE) versus TPE. Left, images of an isolated coexpressing CA1 neuron with the 1,064- or 473-nm target positions overlaid. Right, GCaMP3 ΔF/F traces measured during stimulation targeting those locations using TPE or SPE (all traces are averages of >3 trials). TPE and SPE both evoked responses when the cell was targeted, whereas only SPE evoked responses when it was not. (b) Spatial resolution of TPE and SPE photostimulation. Evoked response amplitudes for different displacements between target cells and neighboring cells laterally (as in a; solid lines) or axially (dashed lines; error bars indicate mean ± s.d.). Lateral resolution measurements (solid lines) include trials based on cellular fluorescence changes in nearby cells in densely labeled tissue (five cells, SPE; 101 cells, including 27 and 74 temporal and spatial focusing targets, TPE). Inset, representative responses from one cell (centered in images) included in this measurement. Images represent post-stimulation minus pre-stimulation GCaMP3 fluorescence in each case (three-trial average). TPE and SPE evoked similar responses in targeted neurons, but responses were better confined to the target cell using TPE. (c) Matrix of TPE-stimulated responses from 17 cellular targets (indicated at right), with significant responses shown in red (Online Methods). Each entry shows activity in one cell (given by row number) during stimulation targeting one cell (column number). Most responsive cells could be stimulated independently (two exceptions here are indicated in the image). Bottom right, TPE stimulation–triggered response profile (analogous to a point-spread function in imaging), shown as an image (normalized post- minus pre-stimulation GCaMP3 fluorescence, averaged across stimulation trials targeting 101 different cells).
Signal fluorescence (GCaMP3) was well-isolated from optical artifacts during stimulation (Fig. 2c): 1,064-nm light was completely shielded from photodetectors using infrared light-blocking filters (Online Methods) and excited negligible GCaMP3 fluorescence (Fig. 2c and Supplementary Fig. 1). While 1,064-nm excitation produced low-intensity EYFP fluorescence during stimulation pulses, this EYFP background (associated with cellular opsin expression) was readily separated from GCaMP3 signal fluorescence (Fig. 2c), making it unnecessary to omit stimulation blocks8–13 or estimate and subtract8,14 optical stimulation artifacts from recordings. It was therefore possible to record neural activity continuously during stimulation.
Because neurons expressing light-sensitive optogenetic probes may respond to imaging illumination, in effect perturbing activity by observation, we next sought to determine whether 920-nm imaging under typical conditions (≈200 × 100 μm field of view, ≈15-Hz frame acquisition rate) affected spontaneous activity levels in coexpressing populations of neurons. This form of crosstalk should be most apparent in neurons with high opsin-EYFP expression levels, when imaging at high laser power. To define a regime for low-crosstalk imaging, we measured how the rate of spontaneous activity in neuron populations varied as a function of laser power and opsin expression level (estimated using cellular EYFP intensity; 220 neurons across 4 fields of view were analyzed). As expected, when imaging at high laser power (≈60–70 mW), neurons with high probe expression showed slightly elevated levels of spontaneous activity compared with imaging the same neurons at lower power (≤40 mW) or compared with other neurons expressing the probe at lower levels (Fig. 2d). This helped to define a range for imaging in our experiments (≈30–40 mW) where this effect was small (<1 additional calcium event in total from 50 coexpressing neurons during 3 min of imaging; Fig. 2d). Using this approach, neural activity can be measured in a population of cells expressing both a light-sensitive probe and a sensor without substantially changing underlying activity (see Discussion).
Cellular-resolution stimulation of neurons in awake mice
The main anticipated advantage of infrared TPE over visible-light SPE photostimulation was the ability to stimulate nearby cells separably in densely labeled tissue. To test this, we first compared the response profiles of single cells targeted for photostimulation using either focused visible-light SPE or patterned TPE. SPE produced responses that were broad in space (Fig. 3b), exciting other nearby cells as well as the targeted cell (N = 5 targeted cells, both in-plane and along the optical axis). By contrast, TPE-stimulated responses were limited to the independently responsive cells that were targeted in 99% of cell-cell pairs tested (101 neurons in total, 1,237 of 1,248 pairwise combinations of independently responsive neurons, median separation of 56 μm between targeted somas). In most cases in which photostimulation of one targeted cell also evoked activity in another independently responsive cell (≈1%; 11/1,248), those cells were very close in space and had some spatial overlap between the photostimulation intensity pattern and the non-targeted cell soma (median separation 11 μm; Fig. 3c).
Optogenetic stimulation can also occur through light focused away from a neuron soma (for example, through out-of-focus excitation15 or illumination of neuronal processes18,19,28), potentially causing diffuse responses that limit the separability of single-cell stimulation. We therefore asked, in a separate analysis, what fraction of experiments produced a response in any cell that was not targeted (regardless of that cell’s ability to respond when targeted) using stimulation that produced robust responses in target neurons. In 82% of experiments targeting one soma, no single other neuron in the same field of view (FOV) responded (16 FOVs, 37–123 neurons per FOV, including 101 responsive target neurons of 182 tested). We also estimated out-of-plane activation in a subset of these experiments (5 FOVs, including 36 responsive target neurons), repeating stimulation after refocusing the 1,064-nm path by 15 or 25 μm to target locations directly above or below the original target neurons (≈1–2 cell body separation). At most of these out-of-focus locations, stimulation elicited no detectable responses in any neurons (83 and 86%, 79 of 95 locations at 15 μm out of focus, 95 of 110 at 25 μm).
The spatial resolution of this approach, as defined using the half-width extinction distance of excited fluorescence amplitudes in targeted neurons, was approximately 10 μm (along optical axis; Fig. 3b) and <10 μm (lateral). It was therefore possible, using TPE at low power (typically <100 mW) and subsaturating exposure trains (illumination periods shorter than, and separated by intervals longer than, the C1V1 integration time constant of ≈50 ms), to evoke activity in single neurons without activating other, immediately adjacent neurons.
Optical perturbation of hippocampal place cells
We next trained mice expressing the probe-sensor pair to perform a visually guided spatial behavior in a VR environment, navigating a virtual linear track (Fig. 4a), to determine whether it was possible to measure and then manipulate patterns of task-modulated activity in single neurons. In these experiments, task-modulated activity in CA1 pyramidal neuron populations was optically mapped using GCaMP3 imaging and neurons were classified as either place cells or silent cells (firing fields at specific locations on the track, or no place-specific firing fields; Online Methods). Then, using custom VR software to compute ‘gate’ signals in real time29, we stimulated neurons of interest at designated times in the behavior while simultaneously recording evoked and task-driven dynamics in the local neuron population (Online Methods).
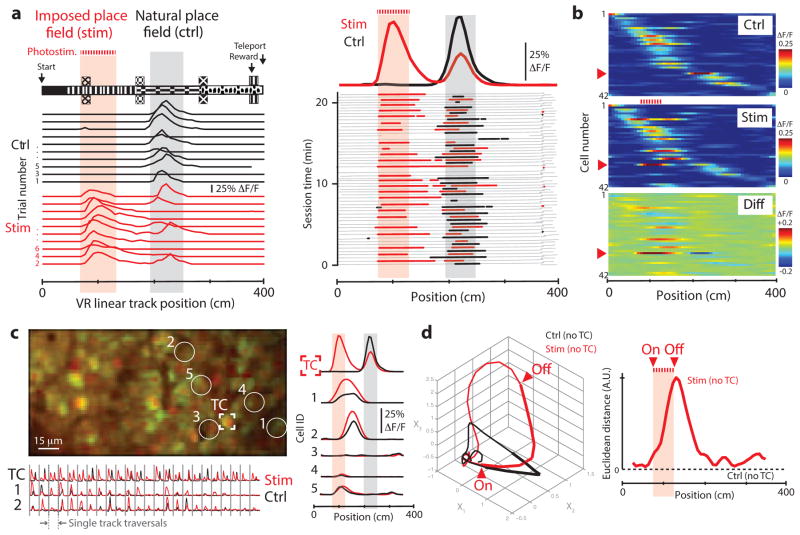
Figure 4.
Optical perturbation of a place cell during virtual navigation. (a) Schematic and experimental examples of place cell perturbation. A trained mouse ran along a 400-cm VR track (upper left). A neuron with a place field in this environment (gray shaded region) was stimulated while the mouse ran through a different part of the track (red shaded region). Single-trial examples of place-cell activity (ΔF/F traces) are shown below for imaging-only and stimulation traversals. Right, activity in the targeted place cell throughout the behavior session (alternating control (ctrl) and stimulation (stim) traversals are shown in black and red, respectively). Position in the environment (gray) and periods of significant transients (colored dots) are shown below, with session averages above (bold lines). Place-specific stimulation mimicked the activity observed in the place field. (b) Intensity maps of spatially modulated activity in neurons from the recorded population (shown in c). Red arrowhead indicates the targeted cell. (c) Secondary effects of stimulation. Left, image of the neurons recorded in this session (target cell, TC). Right, spatial activity profile in the TC, two other cells that showed significantly increased in-field activity during stimulation trials (cells 1 and 2, P = 0.015 and P = 0.028) and three other cells (no difference). Stimulating one place cell increased activity in other neurons with nearby place fields. Concatenated single-trial ΔF/F traces for three cells are shown below. (d) Neuronal circuit trajectories. Left, mean state-space trajectories of population activity during stimulation and control trials (41 cells, TC excluded), visualized using the first three common factors (Online Methods). Right, Euclidean distance between control and stimulation trial trajectories. Stimulating this place cell perturbed activity in other place cells during navigation.
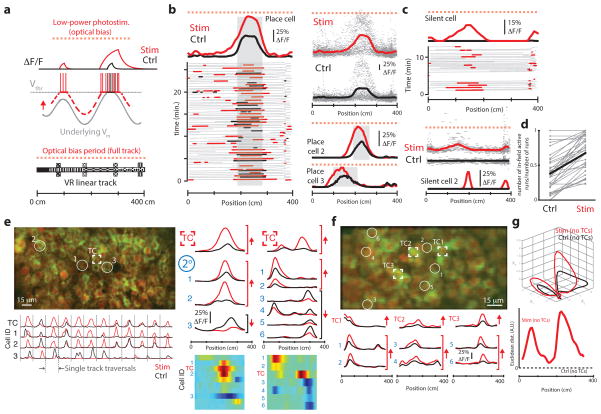
Using this approach, it was possible to impose spatially defined activity that matched the overall width and amplitude of natural place field activity by stimulating single place cells or silent cells as mice traversed a designated part of the VR environment (Fig. 4). Illuminating a place cell with stimulation pulses in one region of the track (75–125 cm on a 400-cm-long track) that preceded the cell’s natural firing field (centered around 225 cm) robustly produced place field-like activity in that region of the track (similar responses were observed in three other experiments in two mice; Fig. 4a). Notably, stimulation trials that produced ‘imposed field’ activity in this example also suppressed activity in the natural place field of that cell (traversed 1.4 ± 0.2 s later). These results indicate that simultaneous imaging and photostimulation can be used to impose patterns of activity in single cells that mimic natural patterns of activity observed during the same task.
Although stimulation of this type typically did not evoke activity in closely neighboring cells, a small number of other, anatomically distributed place cells did in fact show significantly elevated or suppressed activity during stimulation trials (range = 1–4 other neurons in 4 experiments, using bootstrap resampling and a threshold of P = 0.05; Fig. 4c and Online Methods). To quantify the effects of this perturbation on the local microcircuit, we analyzed the activity of the place-cell population, excluding the target cell, as a neural circuit trajectory through a state space describing all traversals. In one example (Fig. 4), a population activity vector consisting of trial-to-trial activity in 41 place cells was identified (using factor analysis to reduce the relevant state space to three dimensions) that described the circuit trajectory during both stimulation and control (no stimulation) traversals. At each point in VR space, the Euclidean distance between the mean trajectory in control trials and stimulation trials was computed to identify periods in stimulation trials in which population activity deviated substantially from control trials. Because this analysis excluded the target cell, the trajectory reported secondary (indirect) effects of stimulation on the network. Stimulation trials that evoked activity in this place cell produced a significant change in the task-modulated activity of other cells, both during the stimulation epoch and later in the trial (just beyond the target cell’s natural place field, P = 0.004; Fig. 4d).
Low-power optical stimulation
Beyond artificially controlling the activity of individual neurons, we reasoned that illuminating single cells with low-power stimulation pulses could function as a strategy to bias and detect subthreshold activity. In this application, a neuron or a group of neurons is illuminated with a very low-intensity pulse train throughout a behavior trial. The goal is to generate a train of small depolarizing pulses in the target cell(s), which act to amplify existing membrane potential depolarizations or excitatory inputs and make them visible as fluorescence transients, allowing all-optical probing of otherwise concealed properties of active neural circuits.
For example (Fig. 5a), an optical bias should drive a neuron above AP threshold during periods of natural membrane potential elevation (for example, during increased excitatory input), producing fluorescence transients in that cell that can be detected optically. Averaged across trials, a cell’s activity profile during optical bias stimulation should differ from the receptive field profile (as seen in imaging-only trials) in that it should reflect the membrane potential profile. We tested this approach by attempting to form an all-optical estimate of membrane potential in place cells, illuminating these cells with targeted, low-power trains during full traversals of the track. In three of four different cells tested, the optical bias of a single place cell (8–10 Hz × 0.01–0.05-s duration, <50 mW) produced an asymmetric increase in fluorescence leading up to the natural firing field compared to imaging-only runs (shift in field center of mass, 9.2 ± 5.8 cm, P = 0.007; Fig. 5b). The emergence of this asymmetric ramp in fluorescence is consistent with CA1 place cell membrane potential recordings under similar behavior conditions, which have reported that an asymmetric depolarization (a ramp) leading up to a typically symmetric firing field is a subthreshold signature of place cells during linear track navigation30. This illustrates that calcium imaging, combined with low-power optical stimulation, can be used to estimate membrane potential dynamics in neurons during an ongoing behavior.
Figure 5.
Low-power biasing to measure underlying dynamics in neurons and networks. (a) Schematic. Low-power stimulation biases a neuron, producing ΔF/F transients preferentially when Vm is near Vthr. (b) Biasing place cells. Left, place cell activity (field shaded gray) during imaging versus low-power biasing traversals (black and red). Averaged across trials (above), biasing increased activity asymmetrically, leading up to the place field. Top right, activity overlaid across all trials (gray dots, lines are averages). Bottom right, two additional examples of place cells biased by low-power stimulation. (c) Biasing silent cells. Data are presented as in b, but show activity arising from biasing two silent cells (no spatial field during imaging trials). Biasing was able to reveal spatial receptive fields (here, centered around 140 and 190 cm). (d) Fraction of receptive field traversals with in-field activity (30 cells) in imaging versus biasing traversals (field locations for silent cells determined using stimulation trials). Thick lines are the group average. (e) Secondary responses to biasing. Left, FOV from one place-cell biasing experiment (target cell and three others are indicated). Right, ΔF/F traces of neurons with significantly different activity in bias versus control trials (arrows indicate sign of the change). Difference maps (below) represent stimulation minus control for all affected cells in the population. A separate example (including all affected cells) is presented in the second column. Biasing one place cell affected activity in others with nearby fields. (f) Data are presented as in e, but low-power stimulation was applied to three cells at once (TC1–3). Average traces (below) are selected from 24 non-targeted cells differing significantly in bias versus control trials. (g) Neuronal circuit trajectories. Data are presented as in Figure4 d, but comparing imaging-only versus bias traversals (54 cells analyzed, 3 target cells excluded). Biasing three place cells perturbed population activity around the firing fields of those cells.
Similarly, other recent reports using intracellular techniques have observed that injecting small somatic depolarizing currents can cause a step-like emergence of spatially tuned fields in CA1 neurons31. Here, optical bias stimulation of silent cells similarly yielded spatially modulated activity fields (N = 2 cells; Fig. 5c). This illustrates that low-power stimulation may also reveal subthreshold dynamics in neurons, possibly by gating or amplifying distal inputs through perisomatic depolarization31. Notably, optically biasing place cells or silent cells (either individually or in groups, 1–12 neurons per experiment) led to an increased probability of in-field activity on a run-by-run basis compared with imaging-only trials (Fig. 5d). This suggests that one source of the trial-to-trial variability in CA1 firing patterns during stereotyped running trajectories, a phenomenon reported previously with extracellular electrode recordings (excess variance)32, is variability in the degree of subthreshold membrane depolarization during specific behavior epochs, rather than the complete absence of spatially modulated inputs.
To our surprise, optically biasing single place cells also led to substantial changes in the activity of other, anatomically distributed cells (Fig. 5e–g). These secondary responses, which included cells with either elevation or suppression of in-field activity, occurred primarily in cells with place fields correlated with the place field of the targeted neuron (for example, correlation coefficient of 0.40–0.89 versus 0.19–0.30 for all other neurons, in imaging-only trials; Fig. 5e). Secondary responses occurred in neurons distributed over large distances (Fig. 5e,f) compared with the spatial resolution of this method (≈10 μm; Fig. 3), whereas affected firing fields of these cells were narrowly distributed (Fig. 5f,g). This suggests that these responses were not a result of collateral stimulation (such as afferent or efferent axons, or dendrites), which might be expected to cause more diffuse effects. Using multiple-cell biasing to drive increased in-field activity in three place cells (Fig. 5g), we found that deviations of the population from the normal sequence of activity (excluding targeted neurons) approximately followed the largest perturbations in targeted neurons. Across six experiments (four biasing and two directly stimulating cells in a location in the environment), secondary responses were observed in an average of five non-targeted place cells for each directly targeted place cell (range = 2–8 cells per targeted cell; 43 cells in total, ≈750 visualized neurons).
DISCUSSION
Our results demonstrate that mammalian neuronal activity can be characterized and then perturbed during a behavior at cellular resolution. Our approach combines genetic expression of an optogenetic probe and a calcium sensor; to the best of our knowledge, this is the first use of combined probe-sensor expression in intact mammalian tissue. GCaMP3 and C1V1-2A-EYFP may be separably detected and near-independently excited in a densely dual-labeled neuron population using spatially patterned and spectrally separated near-infrared sources providing TPE. The success of this approach corroborates and builds on an existing framework15 in which the high two-photon absorption cross-section15 and long integration time constant of many opsin-based ion channels (approximately tens of milliseconds) are conducive to single-neuron stimulation using fast15,18,19 or patterned16,17 illumination of the soma within this interval. Because the opsin photocycle time constant is very long (on the millisecond scale) compared with the excited-state lifetime of a fluorophore (nanoseconds), the combined use of these different types of molecular probes in a single preparation represents a fundamental advance over existing approaches that require either omission or estimation of stimulation artifacts8–13. Our approach instead allows continuous optical recording of activity during arbitrary patterns of photostimulation.
As in previous functional imaging studies of CA13,33–35, we inferred changes in neural activity from changes in somatic calcium-dependent fluorescence. These changes were reported using GCaMP3, a calcium sensor that has been applied widely, but which also has a relatively high detection threshold in vivo8,22. Naturally, this limited our ability to identify, at any instant in a time-series recording, very small changes in neural activity (such as subthreshold membrane depolarization or isolated APs) because such changes are unlikely to produce instantaneous increases in GCaMP3 fluorescence intensity. We anticipate that other, improved sensors reporting calcium36 or membrane potential37 will be compatible with this method and should be explored as additional molecular strategies are developed to achieve functional and stable coexpression with optogenetic probes.
Physiological effects of the imaging beam
The imaging laser could bias the membrane potential of coexpressing neurons; in an intracellular recording, this might appear as small depolarizing pulses from frame-to-frame. Although GCaMP3 does not report subthreshold membrane potential changes, we reasoned that such a depolarizing bias might be estimated and then reduced to acceptable levels by recording changes in the time-averaged rates of spontaneous neuronal activity induced at different imaging laser powers. Increasing the imaging laser power monotonically increased detectable baseline activity of neurons expressing both GCaMP3 and C1V1 as compared with GCaMP3-only neurons (which should not be affected by the imaging beam; Fig. 2d). At low imaging power, these changes became very small (<1 additional spontaneous event in total for every 3 min of imaging 50 coexpressing neurons).
Because GCaMP3 does not typically report single APs8,22, we cannot rule out the possibility that a depolarizing bias from the imaging laser increases the rate of spontaneous isolated APs more rapidly than detectable multiple-AP events; this idea could be tested using improved calcium sensors36. However, given that CA1 pyramidal neurons have low average firing rates and, when active, often fire APs in temporally correlated groups (for example, high-frequency theta bursts)30, it is possible that this analysis, even in the absence of single AP detection, captures most of the changes in neuronal activity caused by a bias from the imaging laser. It has also been shown that CA1 pyramidal neurons respond to subthreshold current injections (that is, a depolarizing bias) with monotonically increasing rates of spontaneous AP bursts38. This effect may explain the small increase in the rate of spontaneous events at high laser power (Fig. 2d). On the basis of the reported relationship between membrane potential depolarization and spontaneous bursting rate38 (≈0.025 additional complex spikes mV−1-s−1), we estimate that high-power imaging (60–70 mW; Fig. 2d) may have depolarized some high-opsin expression neurons by as much as 3 mV, whereas the low-power imaging (<40 mW) used in our subsequent proof-of-principle applications would have produced, on average, <1 mV of depolarization.
Spatial resolution in densely labeled tissue
TPE allows single CA1 pyramidal neuron somas to be stimulated at cellular resolution in awake mice (Fig. 3). We demonstrated this under conditions commonly used for large-scale, cellular-resolution optical recordings in awake, behaving rodents: namely, where brain motion, variable background activity, neuromodulatory state and the orientation of principal cell major dendrites (along the microscope axis) are qualitatively different from preparations in culture15 or physiological slices16–19,39,40. The favorable performance of this approach in a region of very densely packed somas is promising for applications in other brain regions (such as neocortex) where somas are generally separated by larger distances.
We used trains of closely spaced light pulses designed to stimulate trains of closely spaced APs that could be detected using GCaMP3. Consistent with this idea, ΔF/F amplitude scaled with the number of pulses in short stimulus trains (Supplementary Fig. 3a). Given that GCaMP3 does not report all APs reliably, it is possible that some stimulation experiments also produced smaller changes (such as single APs) in nearby neurons. For studies seeking to manipulate activity on the single-AP scale, such potential effects could be characterized using improved calcium indicators with higher sensitivity. However, we anticipate that, because saturating conditions (such as higher power or increased AP number) tend to degrade rather than improve the spatial resolution of stimulation15, experiments to stimulate even lower numbers of APs (for example, using only one or two illumination pulses) will achieve similarly well-resolved stimulation.
Behavior-based stimulation experiments
Perturbation of hippocampal CA1 place cell activity (Figs. 4 and 5) provides experimental proof-of-concept that activity in single neurons associated with an ongoing behavior can be optically measured and manipulated. In these experiments, we used two stimulation procedures: direct stimulation and low-power optical bias stimulation. Direct stimulation can be used to mimic neural activity (for example, spatial receptive fields), imposing new patterns of activity as perturbations to the natural patterns exhibited during a behavior. In these experiments, to mimic place-cell activity during navigation, we repeatedly activated single neurons in association with a place in the VR environment. In principle, these experiments might be extended to test models of potentiation (for example, through Hebbian plasticity) by pairing stimulation with periods in a behavior. Although no changes were observed in the natural firing fields of neurons stimulated repeatedly in our experiments (>25 times in a session, 3 cells), other conditions (such as synchronizing stimulation epochs with measurements of the theta cycle) could be explored.
Although the GCaMP3 signal alone may not provide the exact number of APs induced in these experiments, it is possible to estimate the range of AP firing changes by reference to independent electrical recordings of APs from the same cell population, during the same behavior (CA1 place cells during VR linear track navigation30). For example, around an imposed place field (Fig. 4), optogenetic stimulation drove spatially defined activity similar in overall width and amplitude to the cell’s natural place field activity (width of 60 cm versus 45 cm; trial-averaged peak ΔF/F = 0.37 versus 0.42), and in the range reported using the same calcium sensor for typical CA1 place cells on a VR linear track (width 50 ± 19 cm; mean peak ΔF/F = 0.35 ± 0.29)3. Thus, these responses are typical of full place field responses, which, for mice running on a VR linear track, correspond to average firing rates of 7.3 ± 1.4 Hz30. If a 50-cm field is traversed at a typical running speed of ≈30 cm s−1 in 1.7 s, this should correspond to an average activity of 7.3 × 1.7 = 12 spikes per traversal (range of 10–15 spikes). We therefore estimate the optogenetic-induced spikes shown in Figure 4 to be in this range (10–15 spikes). In the same experiment, stimulation trials also reduced activity in the target cell’s natural place field. For the low number of APs expected in the brief interval of each field traversal (around ten APs in 1–2 s), the peak ΔF/F amplitude of GCaMP3 transients should increase roughly linearly with AP number22 (Supplementary Fig. 3a). As such, for this cell’s natural place field (45-cm width or around 9–13 spikes per traversal; Fig. 4), the observed reduction in peak ΔF/F value (0.45 times the value in control trials) should correspond to an altered AP number of 4–6 spikes per traversal. This approach could also provide the range of induced (or suppressed) APs per traversal in other experiments (Fig. 5), although future studies with improved calcium sensors may be able to visualize these AP changes directly.
We also found that the second type of manipulation (low-power optical bias) could be used to detect subthreshold activity in single neurons, consistent with results obtained with intracellular electrode recordings or manipulations (such as asymmetric membrane potential ramps30 or masked firing fields31). Although optical biasing does not provide the high temporal bandwidth of intracellular electrical recordings, it does confer certain advantages, such as advance knowledge of anatomical location and functional cell type and the ability to repeat measurements in a specific cell across multiple days or to perform recordings from multiple cells simultaneously (Fig. 5f,g). Future experiments might use this approach to further examine the anatomical distribution of spatial information in these cells3 or to map both firing fields and subthreshold inputs31 that may contribute to long-term changes33 in receptive fields.
Secondary-cell responses
CA1 is classically viewed as a region of low auto-associative computation42. It was therefore surprising that both direct stimulation and biasing, when used to perturb single place cell activity during a behavior, also perturbed the spatial firing fields of other place cells. Our interpretation of these effects as functional properties of this brain region, as opposed to optical artifacts (for example, from spatially diffuse stimulation), was based on the following observations. First, although spatially diffuse optogenetic stimulation could depolarize afferent inputs28 or processes18,19 of non-targeted neurons, stimulating one targeted neuron sometimes suppressed activity in other neurons (an effect that was visible around those neurons’ normal receptive fields). This sign flip is inconsistent with diffuse stimulation and it is also unlikely to be a result of photoactivation of inhibitory interneurons, as Camk2a-driven expression of the optogenetic probe should be limited to excitatory neurons. Second, biasing single target neurons drove population activity changes that were largest when biasing also drove the largest changes in the target neurons themselves. Even if these secondary effects had been purely excitatory (for example, through diffuse stimulation), it is likely that they would have been distributed across cells with different place fields, rather than concentrated in the places where the target neurons had fields.
For each stimulated place cell, the firing fields of two to eight other place cells were affected (five on average). In these experiments, which included recordings from around 100 neurons per field of view, this value represents approximately 5% of the local population of principal cells. Given that these experiments sampled only cells in a TPE optical section and reported only changes above the GCaMP3 detection threshold, it is possible that the fraction of local place cells affected by single-cell stimulation estimated here (5%) represents a lower bound on the actual effects. Based on excitatory connectivity alone, the fraction of CA1 principal cells that might be expected to be affected by additional drive to a single place cell is much lower (1 in 100)43 than the fraction observed here (at least 5 in 100). This discrepancy, and the observation of suppressed in-field activity, suggest that place cell–to–place cell interactions in CA1 during a behavior involve more than one synapse. For example, stimulating a single CA1 pyramidal neuron could potentially drive fast or asynchronous changes in firing patterns of one or more interneurons44–46, which in turn might either suppress47 or elevate46 activity in other place cells. It should be possible to test such ideas by augmenting our methods with targeted expression of probes or sensors in different genetically defined populations35. Secondary effects of single-cell stimulation may be even more prevalent in brain regions with higher recurrent connectivity48.
Broadly, our proof-of-principle results with stimulation of place cells adds to the growing evidence that local circuit interactions not only affect properties of a place cell’s firing around its field (such as the precise spike timing49), but may also define the place fields themselves, determining which cells show firing fields and where they are located. Further applications of this approach to probe interactions among single cells while the brain is in its normal operating regime (that is, during a behavior) could contribute to a more complete description of functional connectivity than is currently possible. For example, single-cell stimulation could be combined with methods to identify structural correlates50 between individual cells to reveal multi-synaptic interactions among groups of functionally related neurons.
METHODS
Methods and any associated references are available in the online version of the paper.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
Supplementary Material
Acknowledgments
We thank D. Kim and C. Guo (Genetically Encoded Neuronal Indicator and Effector Project, Janelia Research Campus) for transgenic mice, D. Aronov for VR software, B. Scott for discussions, and C. Domnisoru, A. Miri, F. Collman and S. Wang for comments on the manuscript. This work was supported by the US National Institutes of Health (R01-MH083686; P50-GM071508) and a National Science Foundation Graduate Research Fellowship to J.P.R.
Footnotes
AUTHOR CONTRIBUTIONS
J.P.R. and D.W.T. designed the study. K.D. contributed reagents. J.P.R. and D.W.T. performed the experiments. J.P.R. analyzed data with strategy and methods contributions from D.W.T. J.P.R. and D.W.T. wrote the paper with comments from K.D.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg DS, Houweling AR, Kerr JN. Population imaging of ongoing neuronal activity in the visual cortex of awake rats. Nat Neurosci. 2008;11:749–751. doi: 10.1038/nn.2140. [DOI] [PubMed] [Google Scholar]
- 3.Dombeck DA, Harvey CD, Tian L, Looger LL, Tank DW. Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat Neurosci. 2010;13:1433–1440. doi: 10.1038/nn.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petreanu L, et al. Activity in motor-sensory projections reveals distributed coding in somatosensation. Nature. 2012;489:299–303. doi: 10.1038/nature11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450:420–424. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tye KM, et al. Amygdala circuitry mediating reversible and bidirectional control of anxiety. Nature. 2011;471:358–362. doi: 10.1038/nature09820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter ME, et al. Tuning arousal with optogenetic modulation of locus coeruleus neurons. Nat Neurosci. 2010;13:1526–1533. doi: 10.1038/nn.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akerboom J, et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front Mol Neurosci. 2013;6:2. doi: 10.3389/fnmol.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang YF, Arai Y, Nagai T. Optogenetic activation during detector “dead time” enables compatible real-time fluorescence imaging. Neurosci Res. 2012;73:341–347. doi: 10.1016/j.neures.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Husson SJ, et al. Optogenetic analysis of a nociceptor neuron and network reveals ion channels acting downstream of primary sensors. Curr Biol. 2012;22:743–752. doi: 10.1016/j.cub.2012.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin JY, Lin MZ, Steinbach P, Tsien RY. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys J. 2009;96:1803–1814. doi: 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson NR, Runyan CA, Wang FL, Sur M. Division and subtraction by distinct cortical inhibitory networks in vivo. Nature. 2012;488:343–348. doi: 10.1038/nature11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 14.Guo ZV, Hart AC, Ramanathan S. Optical interrogation of neural circuits in Caenorhabditis elegans. Nat Methods. 2009;6:891–896. doi: 10.1038/nmeth.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rickgauer JP, Tank DW. Two-photon excitation of channelrhodopsin-2 at saturation. Proc Natl Acad Sci USA. 2009;106:15025–15030. doi: 10.1073/pnas.0907084106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrasfalvy BK, Zemelman BV, Tang J, Vaziri A. Two-photon single-cell optogenetic control of neuronal activity by sculpted light. Proc Natl Acad Sci USA. 2010;107:11981–11986. doi: 10.1073/pnas.1006620107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papagiakoumou E, et al. Scanless two-photon excitation of channelrhodopsin-2. Nat Methods. 2010;7:848–854. doi: 10.1038/nmeth.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prakash R, et al. Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nat Methods. 2012;9:1171–1179. doi: 10.1038/nmeth.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Packer AM, et al. Two-photon optogenetics of dendritic spines and neural circuits. Nat Methods. 2012;9:1202–1205. doi: 10.1038/nmeth.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yaroslavsky AN, et al. Optical properties of selected native and coagulated human brain tissues in vitro in the visible and near infrared spectral range. Phys Med Biol. 2002;47:2059–2073. doi: 10.1088/0031-9155/47/12/305. [DOI] [PubMed] [Google Scholar]
- 21.Oheim M, Beaurepaire E, Chaigneau E, Mertz J, Charpak S. Two-photon microscopy in brain tissue: parameters influencing the imaging depth. J Neurosci Methods. 2001;111:29–37. doi: 10.1016/s0165-0270(01)00438-1. [DOI] [PubMed] [Google Scholar]
- 22.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yizhar O, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oron D, Tal E, Silberberg Y. Scanningless depth-resolved microscopy. Opt Express. 2005;13:1468–1476. doi: 10.1364/opex.13.001468. [DOI] [PubMed] [Google Scholar]
- 25.Zhu G, van Howe J, Durst M, Zipfel W, Xu C. Simultaneous spatial and temporal focusing of femtosecond pulses. Opt Express. 2005;13:2153–2159. doi: 10.1364/opex.13.002153. [DOI] [PubMed] [Google Scholar]
- 26.Zimmermann T, Rietdorf J, Pepperkok R. Spectral imaging and its applications in live cell microscopy. FEBS Lett. 2003;546:87–92. doi: 10.1016/s0014-5793(03)00521-0. [DOI] [PubMed] [Google Scholar]
- 27.Mattis J, et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat Methods. 2012;9:159–172. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petreanu L, Huber D, Sobczyk A, Svoboda K. Channelrhodopsin-2–assisted circuit mapping of long-range callosal projections. Nat Neurosci. 2007;10:663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- 29.Aronov D, Tank DW. Engagement of the neural circuits underlying 2D spatial navigation in a rodent virtual reality system. Neuron. 2014;84:442–256. doi: 10.1016/j.neuron.2014.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harvey CD, Collman F, Dombeck DA, Tank DW. Intracellular dynamics of hippocampal place cells during virtual navigation. Nature. 2009;461:941–946. doi: 10.1038/nature08499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee D, Lin BJ, Lee AK. Hippocampal place fields emerge upon single-cell manipulation of excitability during behavior. Science. 2012;337:849–853. doi: 10.1126/science.1221489. [DOI] [PubMed] [Google Scholar]
- 32.Fenton AA, Muller RU. Place cell discharge is extremely variable during individual passes of the rat through the firing field. Proc Natl Acad Sci USA. 1998;95:3182–3187. doi: 10.1073/pnas.95.6.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ziv Y, et al. Long-term dynamics of CA1 hippocampal place codes. Nat Neurosci. 2013;16:264–266. doi: 10.1038/nn.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaifosh P, Lovett-Barron M, Turi GF, Reardon TR, Losonczy A. Septo-hippocampal GABAergic signaling across multiple modalities in awake mice. Nat Neurosci. 2013;16:1182–1184. doi: 10.1038/nn.3482. [DOI] [PubMed] [Google Scholar]
- 35.Lovett-Barron M, et al. Dendritic inhibition in the hippocampus supports fear learning. Science. 2014;343:857–863. doi: 10.1126/science.1247485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.St-Pierre F, et al. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat Neurosci. 2014;17:884–889. doi: 10.1038/nn.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grienberger C, Chen X, Konnerth A. NMDA receptor-dependent multidendrite Ca2+ spikes required for hippocampal burst firing in vivo. Neuron. 2014;81:1274–1281. doi: 10.1016/j.neuron.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 39.Papagiakoumou E, et al. Functional patterned multiphoton excitation deep inside scattering tissue. Nat Photonics. 2013;7:274–278. [Google Scholar]
- 40.Losonczy A, Zemelman BV, Vaziri A, Magee JC. Network mechanisms of theta related neuronal activity in hippocampal CA1 pyramidal neurons. Nat Neurosci. 2010;13:967–972. doi: 10.1038/nn.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dana H, et al. Thy1-GCaMP6 transgenic mice for neuronal population imaging in vivo. PLoS ONE. 2014;9(9):e108697. doi: 10.1371/journal.pone.0108697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amaral DG, Witter MP. The three-dimensional organization of the hippocampal formation: a review of anatomical data. Neuroscience. 1989;31:571–591. doi: 10.1016/0306-4522(89)90424-7. [DOI] [PubMed] [Google Scholar]
- 43.Deuchars J, Thomson AM. CA1 pyramid-pyramid connections in rat hippocampus in vitro: dual intracellular recordings with biocytin filling. Neuroscience. 1996;74:1009–1018. doi: 10.1016/0306-4522(96)00251-5. [DOI] [PubMed] [Google Scholar]
- 44.Marshall L, et al. Hippocampal pyramidal cell-interneuron spike transmission is frequency dependent and responsible for place modulation of interneuron discharge. J Neurosci. 2002;22:RC197. doi: 10.1523/JNEUROSCI.22-02-j0001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galarreta M, Hestrin S. Spike transmission and synchrony detection in networks of GABAergic interneurons. Science. 2001;292:2295–2299. doi: 10.1126/science.1061395. [DOI] [PubMed] [Google Scholar]
- 46.Jonas P, Bischofberger J, Fricker D, Miles R. Interneuron diversity series: fast in, fast out–temporal and spatial signal processing in hippocampal interneurons. Trends Neurosci. 2004;27:30–40. doi: 10.1016/j.tins.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 47.Freund TF, Buzsaki G. Interneurons of the hippocampus. Hippocampus. 1996;6:347–470. doi: 10.1002/(SICI)1098-1063(1996)6:4<347::AID-HIPO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 48.Ko H, et al. Functional specificity of local synaptic connections in neocortical networks. Nature. 2011;473:87–91. doi: 10.1038/nature09880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Royer S, et al. Control of timing, rate and bursts of hippocampal place cells by dendritic and somatic inhibition. Nat Neurosci. 2012;15:769–775. doi: 10.1038/nn.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Briggman KL, Helmstaedter M, Denk W. Wiring specificity in the direction-selectivity circuit of the retina. Nature. 2011;471:183–188. doi: 10.1038/nature09818. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.