Abstract
Background
The mechanisms and immune pathways associated with chronic rhinosinusitis (CRS) are not fully understood. Immunological changes during acute exacerbation of CRS may provide valuable clues to the pathogenesis and perpetuation of the disease.
Objective
To characterize local and systemic immune responses associated with acute worsening of sinonasal symptoms during exacerbation in CRS with nasal polyps (CRSwNP) compared to controls.
Methods
This was a noninterventional prospective study of individuals with CRSwNP and normal controls. Subjects underwent a baseline visit with collection of nasal secretions, nasal washes, and serum specimens. Within 3 days of acute worsening of sinonasal symptoms, subjects underwent a study visit, followed by a post-visit 2 weeks later. The Sinonasal Outcome Test-22 (SNOT-22) scores and immunological parameters in the specimens were analyzed using a novel, unsupervised learning method and by conventional univariate analysis.
Results
Both CRSwNP patients and control subjects showed a significant increase in SNOT-22 scores during acute exacerbation. Increased nasal levels of IL-6, IL-5, and eosinophil major basic protein were observed in CRSwNP patients. A network analysis of serum specimens revealed changes in a set of immunological parameters, which are distinctly associated with CRSwNP but not with controls. In particular, systemic increases in VEGF and GM-CSF levels were notable and were validated by a conventional analysis.
Conclusions
CRSwNP patients demonstrate distinct immunological changes locally and systemically during acute exacerbation. Growth factors VEGF and GM-CSF may be involved in the immunopathogenesis of subjects with CRS and nasal polyps experiencing exacerbation.
Keywords: Chronic rhinosinusitis, GM-CSF, VEGF, exacerbation, nasal polyposis, human rhinovirus
INTRODUCTION
Chronic rhinosinusitis (CRS) is a debilitating, recalcitrant condition secondary to persistent inflammation of the sino-nasal mucosa, resulting in symptoms of nasal congestion, nasal discharge, loss of smell, and facial pressure [1]. Many individuals with CRS continue to suffer lifelong, despite aggressive management with currently available treatments [2]. The pathogenesis of CRS is poorly understood, resulting in no definitive curative therapy at present. Previous cross-sectional studies in CRS have provided valuable information to characterize the differences between patients and controls, and in establishing models to study the pathophysiology of CRS. However, currently key molecule(s) that play pivotal roles at disease inception and/or progression are poorly understood [3]. One way to address this major gap in knowledge may be to study the immunological changes that occur during acute exacerbation of this chronic disease.
Previously, we have performed a pilot study to investigate immunological changes during disease exacerbation in patients with CRS with nasal polyps (CRSwNP). Pre-enrolled patients were asked to contact study personnel within 24 hours of the worsening of their respiratory symptoms (nasal congestion, drainage, sense of smell) for collection of airway specimens. We found that the protocol was feasible and that the levels of IL-5, IL-6, IL-33, and eosinophil major basic protein (MBP) in nasal specimens increased during acute exacerbation of the disease [4]. In the current study, we extended this self-reported disease exacerbation model and explored two major questions: (1) Is the local immunologic response in the upper respiratory tract during acute exacerbation different between patients with CRSwNP and normal controls? (2) Can a unique immunologic response be detected in serum during CRSwNP exacerbation that is distinct from that seen in normal individuals?
In order to do so, we extended analysis of nasal cytokines significant in the pilot experiment [4] and additionally used a novel network graph visualization method to obtain a high-level overview of patterns of cytokines in serum and then performed conventional analyses on notable cytokines.
METHODS
Subjects
Adults participants 18 years and above were recruited from the Allergy-Immunology and Otolaryngology clinical practices at two academic medical centers (Mayo Clinic Minnesota and Mayo Clinic Arizona). Participant recruitment started in the fall of 2012 and extended through spring of 2013; the entire study continued until June 2013. Inclusion criteria along with the subject characteristics are provided in Table 1. CRSwNP was defined by presence of 2 or more of the 4 cardinal symptoms of chronic rhinosinusitis for the preceding 3 months including nasal discharge, nasal obstruction/congestion, facial pain-pressure-fullness, or a decrease in smell, within the past 24 months, along with a sinus CT scan within the past 5 years with a Lund-Mackay score > 5, and nasal polyps documented within the past 3 years on physical exam. All the patients with CRSwNP had been treated with topical corticosteroids; patients who received oral glucocorticoids during the past 3 months were excluded. The study was reviewed and approved by the IRB prior to initiation. All participants provided a written informed consent.
Table 1.
Inclusion criteria and demographics of the participants who experienced acute exacerbations.
| Controls | CRSwNP | |
|---|---|---|
| Inclusion criteria |
|
|
| Number in study group | 10 | 9 |
| Median (IQR) | 26.5 (23.8–54.5) | 53 (44.5–60.5) |
| Gender | ||
| Male | 5 | 5 |
| Female | 5 | 4 |
| Previous Sinus surgery | ||
| No | 10 | 2 |
| Yes | 0 | 7 |
| Asthma Diagnosis | ||
| No | 10 | 4 |
| Yes | 0 | 5 |
| Aspirin Sensitivity | ||
| No | 10 | 8 |
| Yes | 0 | 1 |
| Atopic Status | ||
| Positive | NA | 6 |
| Negative | NA | 2* |
Assessment of atopic status was not performed in one subject.
NA = not available
Study design
The study was a noninterventional prospective design (Fig. 1).
Baseline phase: At the time of enrollment (visit 1, baseline visit), all participants completed a Sinonasal Outcome Test-22 (SNOT-22) questionnaire and demographic factors were recorded. Nasal secretions and serum specimens were collected and stored frozen at −20°C. Two additional SNOT-22 questionnaires were obtained in this baseline phase after visit 1.
Exacerbation phase: Participants were instructed to contact the study team immediately if they experienced an exacerbation. A natural exacerbation was defined as patient-reported worsening of sinonasal symptoms (i.e., runny nose, nasal congestion, and nasal obstruction). The day that a participant first noticed an exacerbation or onset of upper respiratory symptoms was denoted “day 0”. Visit 2 (i.e., exacerbation visit) occurred within 3 days after day 0.
Resolution phase: A final visit (Visit 3) occurred approximately 14 days after day 0.
SNOT-22 scores and nasal secretions were collected on visits 1–3 while additionally serum was collected for visit 1 and 2. Serum specimens were collected at only the Mayo Clinic, Rochester site.
Figure 1.
Study design and measurement scheme. See the Methods section for details.
SNOT-22 questionnaire
The SNOT-22 questionnaire was completed by participants based on a 2-week recall. The SNOT-22 has been validated in a large United Kingdom pre- and postsurgical sample [5–7]. The tool comprises of 22 questions with each item scored on a 6-point scale (0–5 scale). The maximum possible score is 110.
Analyses of nasal secretions and serum specimens
Nasal secretions were collected as previously described at each visit [4]. Briefly, nasal secretions were obtained from the right nasal cavity using a sterile sinus secretion collector (Xomed Surgical Products). All the samples were subjected to a uniform protocol for extracting secretions by mixing with a 3-fold volume of 0.9% sterile NaCl. A cocktail of protease inhibitors (HALT™, Thermo Scientific) was added immediately to the mucus in a 1:100 ratio of volume to weight of nasal secretions collected. Supernatants of nasal secretions were collected and stored at −20°C until analyses. (See Online Repository for Further Information).
Network analysis of cytokine data
Multiplex data from serum samples was analyzed and out of range (OOR) values less than the limit of detection were assigned 1/10 of the lower limit of detection for the assay. To allow for a simultaneous comparison of unrelated biomarkers, expression values for each specific biomarker were normalized across all subjects. Normalization was performed using the following algorithm: Vnorm = (Vact − Vm) / SD. Vnorm is the normalized value, Vact is the raw value of the biomarker expression, Vm is the mean raw value, and SD is the standard deviation across subjects. Since distances have no meaning in negative values, the minimum Vnorm value for a biomarker was scaled to zero and reminder were scaled by same measure preserving the relationship. This algorithm preserves the proportional relationship of a biomarker across patients allowing parallel comparisons in a simultaneous manner for different biomarkers. Data was then converted into a format suitable for generating network layouts, and graphs were generated using Gephi 0.8.2 (software for visualizing and analyzing network graphs; www.gephi.org) [8]. Graphical representation of all subjects and biomarkers was denoted by respective nodes (subject nodes and biomarker nodes) and their relationships were denoted by linked line connections called edges. The transformed Vnorm values formed the basis of these edges. A force-directed algorithm (Force Atlas) [8] which pulls similar nodes together and pushes dissimilar nodes apart was applied to reveal emergent associations. Clusters seen in the network were corroborated, and boundaries of cluster membership defined using unsupervised agglomerative hierarchical clustering. Node size was set between a range of 15 to 60 units prior to quantitative analysis. Quantitative analysis of total biomarker expression was performed using weighted-degree centrality [where the size of a node is the aggregate of all connections made with the node, i.e., number of connections and strength of connections]. Biological inference of molecular clusters and discovery of predicted functional partners and pathways were done in Cytoscape (version 3.1.0 http://www.cytoscape.org/) using the geneMANIA plugin [9, 10].
RT-PCR for nasal virus detection
Viral detection for human rhinovirus/enterovirus (HRV/EV) was performed on nasal washes at visit 2 by RT-PCR in the laboratory of Dr. James Gern, MD (University of Wisconsin-Madison).
Statistical analysis
The study sample size was determined based on expected changes in immunologic parameters from a pilot study [4]. The study sample was calculated based on a hypothesis that IL-33 increases in nasal secretions during exacerbation phase in CRSwNP patients compared to controls: Fifteen total subjects (nine subjects with exacerbation) would allow for 99% power to detect a 3-fold difference in nasal secretion IL-33 levels between groups. We chose IL-33 as the basis for our sample size, based on evidence for this cytokine to initiate a Th2-type immune response [11]. The SNOT-22 scores were compared between visits for CRSwNP and normal control participants as well as between CRSwNP and normal control participants using the Kruskal-Wallis test, with p less than 0.05 considered a significant difference. The significance of the differences between two proportions was calculated, with two-tailed p less than 0.05 considered significant. For network analysis, comparison of controls and cases, or baseline and exacerbation, was done using an unpaired or paired Wilcoxon-Rank test with p less than 0.05 considered significant.
RESULTS
Demographics of the participants who experienced acute exacerbation
Twenty-two controls and twenty-three CRSwNP participants were recruited to the study. Of which, nine CRSwNP participants and 10 control participants had an acute exacerbation or onset of sinonasal symptoms during the study period and returned for the exacerbation phase. The proportions of controls and CRSwNP patients who experienced ‘exacerbation’ were 45% and 39%, respectively. Table 1 describes the clinical history and demographic of the participants who experienced exacerbation. The participants in the CRSwNP group were older, but there were no gender differences between the groups. Most of those in the CRSwNP group had previous surgery and asthma. No significant differences were observed in the rate of nasal HRV/EV detection among participants with exacerbation between CRSwNP patients and controls (43% vs. 55%, p=0.61). Of these, 7 (of 9) CRSwNP patients and 9 (of 10) controls were from Mayo clinic Rochester site.
Global SNOT-22 score discriminates CRSwNP cases from controls at baseline but not in exacerbation
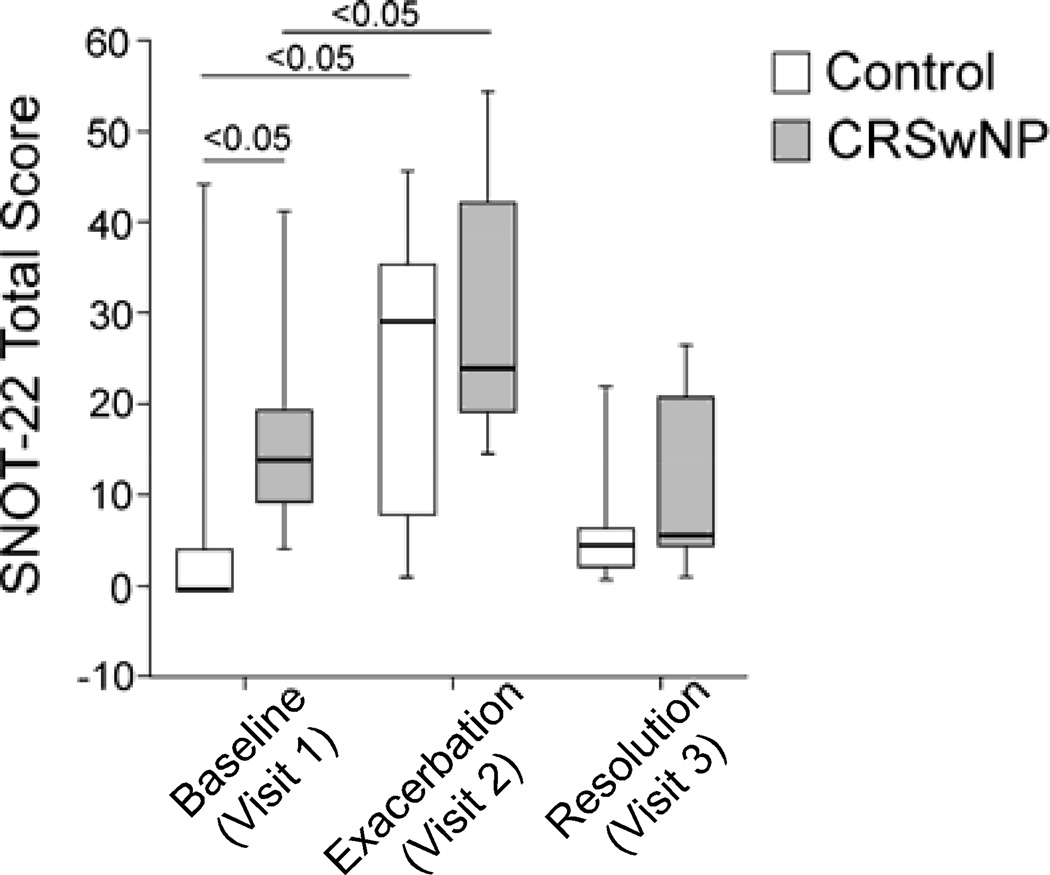
To assess the self-reported measures for tracking CRSwNP symptoms, we compared the SNOT-22 scores for the CRSwNP and control groups at baseline (visit 1), exacerbation (Visit 2), and in resolution phase (visit 3). Testing consistency of self-reporting by patients was done by measuring SNOT-22 two additional times in baseline, and scores were consistent. At visit 1, CRSwNP subjects had significantly higher SNOT-22 scores than did normal controls (p<0.05, Fig. 2). During exacerbation, the SNOT-22 scores increased significantly in both control and CRSwNP groups (i.e., Visit 2 vs. Visit 1, p<0.05). The change in median SNOT-22 scores from baseline to exacerbation exceeded the threshold for significant difference in SNOT-22 tool for both the CRSwNP and control groups [6]. SNOT-22 scores at visit 2 between CRSwNP and normal control subjects showed no significant differences. In both control and CRSwNP groups, the SNOT-22 scores returned to baseline values in resolution phase (Visit 3). Thus, visit 2 scores reflect the clinical exacerbation in CRSwNP cases and controls during the natural course of the illness.
Figure 2.
Changes in SNOT-22 scores before (Visit 1), during (Visit 2), and after (Visit 3) acute exacerbation. Data are presented as median (horizontal bars), quartiles (boxes), and range (vertical lines); n=10 for controls, n=9 for CRSwNP.
Specific local inflammatory parameters are associated with exacerbation in CRSwNP
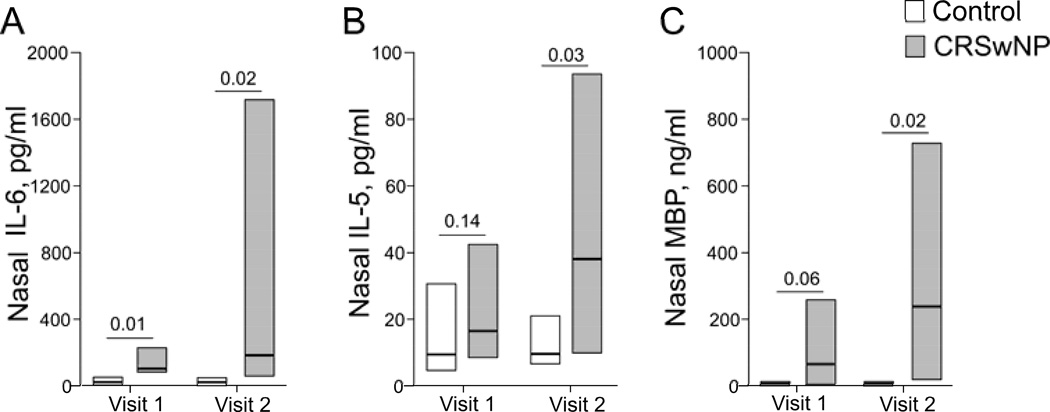
Our prior observations suggested that local inflammatory markers, such as IL-6 and MBP, were elevated in nasal samples of CRSwNP cases during exacerbation compared to baseline. To investigate whether these changes were specific for CRSwNP, we measured selected cytokines in nasal secretions from CRSwNP cases and compared them to normal controls. At Visit 1 (baseline), nasal IL-6 levels were significantly elevated in CRSwNP cases compared to controls (Fig. 3A); the difference was not significant for IL-5 or MBP between cases and controls (Fig. 3B and C). During exacerbation however, IL-6, IL-5, and MBP levels significantly increased in CRSwNP patients compared to controls. Other cytokines, including IL-33, INF-γ, IL-13, IL-17F, IL-17A, IL-17E, TNF-α, and TNF-β, did not show significant differences at visit 2 (exacerbation phase) between CRSwNP patients and controls.
Figure 3.
Nasal levels of IL-6, IL-5 and MBP before (Visit 1) and during (Visit 2) acute exacerbation. Nasal secretions were collected before and during exacerbation and processed as described in the Methods section. Concentrations of inflammatory markers in the supernatants were analyzed by multiplex assay and radioimmunoassay. Data are presented as median (horizontal bars) and quartiles (boxes); n=10 for controls, n=9 for CRSwNP.
Network analyses of serum biomarkers reveal distinct clusters
Analysis of specific inflammatory parameters in nasal secretions confirmed enhanced local inflammatory responses in CRSwNP cases during exacerbation. Since examining global (systemic) response in comparison to local (nasal) immune response offers a unique insight and collection of local specimens has practical limitations (collection, processing, and handling), we next focused our efforts on the biomarker analysis of serum during exacerbation phase. We performed multiplex analysis of a battery of 39 cytokines/biomarkers and analyzed the data using multivariable network visualization to assess for associations and to obtain a high-level overview of the cytokine patterns in sera. Visit 1 and visit 2 serum specimens from patient recruited at the Minnesota site were used for this analysis.
An initial bipartite network depicts a random distribution of set nodes (Fig. 4A, set of white nodes: subjects and a set of grey nodes: Cytokines/biomarker). Each subject node is connected to a biomarker node by an edge (i.e., lines). Once a force-directed algorithm (such as Force Atlas), which brings similar nodes together and pushes dissimilar nodes are apart and weighted degree centrality are applied, the same network reveals visual associations (Fig. 4B). The edge length is inversely proportional to the normalized biomarker expression (i.e., the higher the value, the shorter the length), and the size of each node is proportional to the aggregate value of the connecting edges (i.e., the larger subject nodes have higher, aggregated biomarker values compared to smaller subject nodes). The network topology exhibits a core-periphery layout in regards with larger, more associated nodes in the center, and the smaller, less densely associated nodes at the periphery.
Figure 4.
Unsupervised network analysis of patients and serum biomarkers during exacerbation. An initial bipartite network of 16 subjects (white nodes) and their 39 biomarkers (gray nodes) showing a random layout with subjects in center and biomarkers in periphery (Panel A). Once force-directed and weighted degree centrality algorithm was applied, the network revealed presence of structure and visual association of nodes (Panel B). Closer nodes represent strongly associated nodes. Node size was set between 15 to 60.
CRSwNP is associated with a distinct serum immune signature during exacerbation
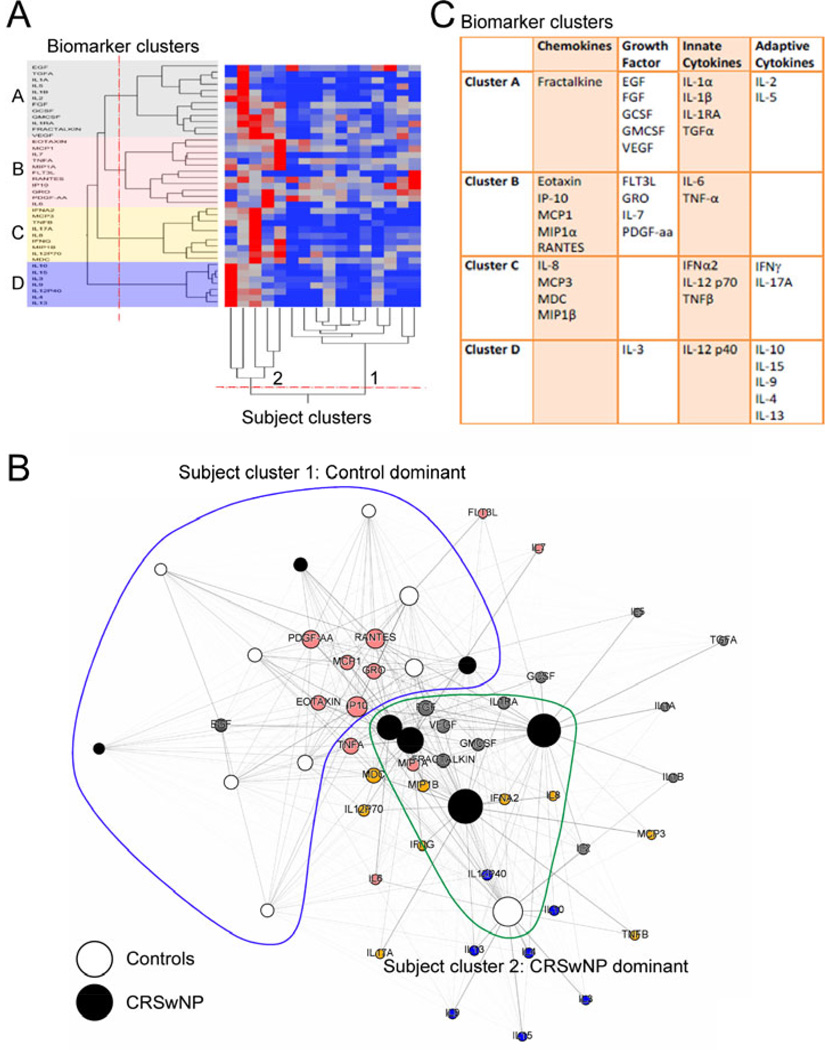
To corroborate the patterns seen in network graph and to define cluster boundaries, we employed an independent, unsupervised learning method of agglomerative hierarchical clustering (Fig. 5A). Rather than a subjectively defining clusters, we objectively calculated significant breaks in the dendrogram using the joining distance function to measure disjointedness (p<0.05), Using this objective approach, unsupervised analysis suggested formation of two subject clusters and four biomarker clusters (Fig. 5A). The two subject clusters revealed a significant difference in membership of clusters (p<0.05), with cluster 1 being control predominant and cluster 2 being CRSwNP predominant (Fig. 5B). Interestingly, the analysis also revealed the presence of three CRSwNP subjects in the control-dominant cluster (cluster 1) and one control subject in the case-dominant cluster (cluster 2), suggesting heterogeneity within the subject groups. Molecules in four biomarker clusters are shown in Figure 5C. Functional pathway analysis and predictive modeling involved with the cluster members is depicted in Supplementary Figure 1A and 1B. Cluster A was enriched for biological function of cell growth and proinflammatory cytokines (p=3.7e-30), cluster B was enriched for cell migration and chemotaxis as biological function (p=5.9e-17). Cluster C was suggestive for cytokine receptor binding, response to virus (p=5e-13), Th17 and Th1 response, while cluster D was suggestive for cytokine regulation (p=2.4e-08) and Th2 response.
Figure 5.
Unsupervised clustering of subject and biomarker nodes in the network during acute exacerbation. (Panel A) Hierarchical agglomerative clustering with heat map and dendrogram based on normalized biomarker values is shown. Breaks in dendrograms (red lines) were assigned by calculating significant breaks in joining distance of clusters (p<0.05). Four clusters of biomarkers (A through D) and two clusters of subjects (1 and 2) were revealed. (Panel B) Subject clusters and membership of biomarkers are overlaid on the network as shown Figure 4B. The nodes for CRS cases (black), controls (white) and biomarker clusters (gray, pink, orange and blue) are color-coded. Subject cluster 1 (blue line) and cluster 2 (green line) define boundaries of these subject clusters. (Panel C) Membership of individual biomarker clusters is presented.
VEGF and GM-CSF significantly change in CRSwNP cases during exacerbation
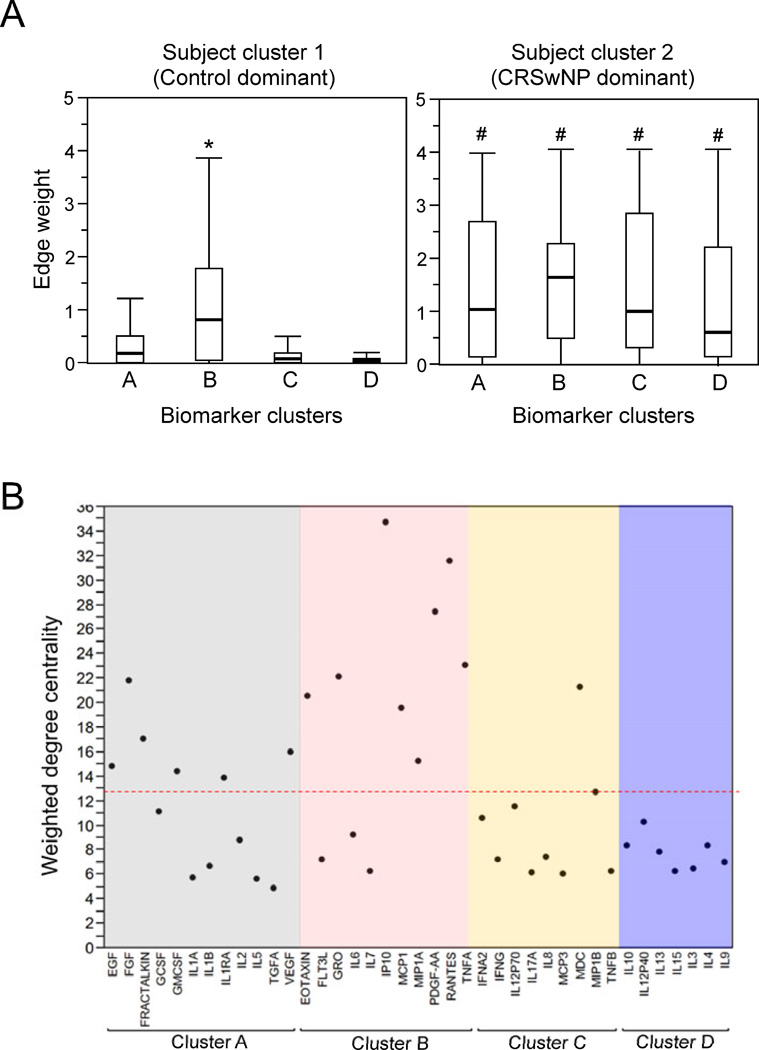
Since the layout suggested presence of four clusters, we further analyzed which of these four clusters is associated closer to the control predominant or CRSwNP predominant cluster. Using aggregate edge weight (biomarker cluster – edge – subject cluster) as a surrogate for inter-cluster distance, we compared the degree of association between subject clusters (1 and 2) to biomarker clusters (A–D). Control-predominant cluster 1 is closer to biomarker cluster B than other biomarker clusters, whereas no such proximity to any specific biomarker clusters is observed in CRSwNP-dominant cluster 2 (Fig. 6A). On the other hand, subject cluster 2 is closer (i.e., higher edge weight values) to all biomarker clusters (A to D) compared to cluster 1, suggesting that CRSwNP-dominant cluster is generally associated more strongly with measured biomarkers as compared to control-dominant cluster. To examine which molecules were most expressed in the network, we compared the weighted-degree centrality (WDC) of each biomarker node (an aggregate measure of the number and the strength of connections made with a node. As seen in Fig. 6B, the biomarkers belonging predominantly to two biomarker clusters A and B were greater than mean WDC. EGF, FGF, fractalkine, GMCSF, IL-1RA, and VEGF from cluster A and Eotaxin, GRO, IP-10, MCP-1, MIP1-α, PGDF, RANTES, and TNF-α from cluster B formed the ‘core’ of the network. In contrast, only one molecule from cluster C (MDC) is represented and no biomarkers from cluster D were involved.
Figure 6.
Association between patient clusters and biomarker clusters during acute exacerbation is presented (Panel A). Horizontal bars, boxes and vertical lines are median, quartiles and range, respectively, of aggregate edge-weights of the subject clusters (1 and 2) to biomarker clusters (A to D). Comparison is made between control-dominant (cluster 1) and CRSwNP-dominant (cluster 2) clusters and four biomarkers. *; p<0.05 for inter cluster comparison per patient cluster while #; p<0.05 for biomarker cluster to biomarker cluster comparison between patient clusters. In Panel B, comparison of weighted degree centrality (node size) of biomarkers. Node size of each biomarker during exacerbation is presented; dotted line represents mean of all the biomarkers.
Thus, we focused our next analysis on these 15 candidate markers and examined which of them show the strongest association with CRSwNP during exacerbation as compared to controls and at baseline. Serum cytokine data from baseline visit was transformed as described in the Materials and Methods and a network graph generated (supplementary figure 2). Using the candidate markers as revealed by analysis of the exacerbation network, we assessed for spatial shifts in associations of the individual cytokines to controls and CRSwNP between exacerbation and baseline (Fig. 7). Amongst the candidates, four markers, FGF, GMCSF, fractalkine, and VEGF from cluster A were significantly closer to CRSwNP at visit 2 (exacerbation) compared to controls (p<0.05), suggesting these markers are more strongly associated with CRSwNP during exacerbation than controls. None of the other ‘core’ markers showed a significant proximity compared to controls.
Figure 7.
Change in association between selected serum biomarkers and subjects before (Visit 1) and during exacerbation (Visit 2). Edge weight of serum biomarkers (FGF, GM-CSF, Fractalkine, and VEGF) in relation to control and CRSwNP subjects was calculated for Visit 1 and Visit 2. Data are presented as median (horizontal bars), quartiles (boxes) and range (vertical lines) of edge weight values. Significant differences are depicted by horizontal lines.
Finally, to confirm the results of the network analysis, we compared the raw untransformed serum protein concentration values (in pg/ml) of these four biomarkers. Serum concentrations of FGF, fractalkine, VEGF, and GM-CSF were significantly higher in CRSwNP patients compared to controls in exacerbation phase (p<0.05, Fig. 8). Furthermore, in CRSwNP subjects, only serum levels of GM-CSF and VEGF were elevated in CRSwNP baseline and exacerbation phases (p<0.05).
Figure 8.
Serum concentrations of FGF, GM-CSF, Fractalkine, and VEGF before (Visit 1) and during (Visit 2) acute exacerbation. Median (horizontal bars), quartiles (boxes) and range (vertical lines) are presented for each visit and each subject group. Significant differences between patient groups or between visits are depicted by horizontal lines.
DISCUSSION
CRS is a recalcitrant disease that is associated with multiple acute exacerbations during the patient’s lifetime. These contribute to considerable morbidity and constitutes a major burden on health care resources. Unlike asthma, where airway challenge and exacerbation models in humans and animals have provided valuable information to understand the pathophysiology of the disease [12], triggers for CRS exacerbation have not been fully understood. Therefore, we used a self-reported, acute, natural exacerbation model to investigate the mechanisms of immune responses in CRS. Similar comparative self-reported models have been used successfully in asthma studies [13, 14].
Using a prospective controlled study design, we detected a local immune response in nasal secretions that distinguished CRSwNP patients from normal controls during acute exacerbation. These data extend our previous findings in CRSwNP patients and verify that IL-5, IL-6, and MBP are elevated locally in nasal secretions of CRSwNP patients during exacerbation. Increased levels of IL-6 in CRSwNP patients at baseline may reflect dysregulated proinflammatory innate immunity, which further increases upon exposure to an inciting stimulus, such as viral infection. Potential roles for the IL-6 and STAT3 pathways in CRSwNP were reported previously by other investigators [15]. Furthermore, increased levels of MBP and IL-5 during exacerbation may point to the involvement of local type 2 immune responses and eosinophilic inflammation. The results are also consistent with observations in previous cross-sectional studies that describe the potential roles of Th2-cytokines and eosinophils in CRSwNP patients in the U.S [16–18]. Altogether, IL-6, IL-5, and eosinophils may play important roles in the pathogenesis of CRSwNP under steady-state conditions and during exacerbation of the disease.
In this study, significant differences in SNOT-22 scores were observed between baseline and exacerbation in both controls and CRSwNP patients. The differences were greater than the minimum significant difference [6], and therefore, the SNOT-22 tool may be suitable for use for research studies that employ self-reported natural exacerbation. On the other hand, the overall SNOT-22 scores did not differ between CRSwNP patients and normal controls during exacerbation (Fig. 2), suggesting that this tool may not be able to discriminate between these groups at the height of exacerbation. The SNOT-22 questionnaire has been used recently for measuring and predicting outcomes in CRS patients, including endoscopic sinus surgery [19] and clinical trials with surfactant [20], corticosteroid irrigation [21], long-term azithromycin [22], and saline irrigation [23]. The current study adds to this growing body of literature, supporting the use of questionnaires such as SNOT-22 as indicators for disease exacerbation, but also suggests potential caution when interpreting the SNOT-22 data in normal control subjects experiencing an acute exacerbation of upper respiratory symptoms.
While nasal and sinus samples are extremely important for understanding the local immune responses in CRS, serum provides supplementary information. Serum markers are useful because they are easily accessed by established and noninvasive standardized collection procedures and serum specimens are near homogeneous. Indeed, serum biomarkers, such as periostin, have been used recently in efforts to predict treatment benefit in patients with asthma [24]. Furthermore, we found that the biomarker spectrum in nasal secretion and serum was not identical, suggesting that both systemic and local specimens need to be examined in an effort to identify best biomarkers for clinical practice and research. We undertook a methodical screening approach using network visualization tool to study serum parameters in CRS exacerbation. Biological parameters can be analyzed by pre-selected variables based on pre acquired, scientific knowledge or by using methods that aid in discovery of patterns in larger sets of data. Conventional analyses employ measurements of central tendency, such as median, mean, or mode, for a limited number of variables, which allows organization and interpretation of data; however, in the process may compress outliers and individual variations, and thus potentially risk missing important clues of information. In contrast, unsupervised simultaneous variable analysis methods allow for the concurrent overview of a high number of variables without compression of information. In particular, certain novel multivariate methods, such as network analysis employ unsupervised learning, allowing the data to reveal patterns without a priori assumptions in comparator classes (e.g., cases and controls) leading to discovery of associations and correlations without preconceived distinctions. The potential advantage of employing such methods is to identify high-level patterns during an initial step, which can then be further focused and substantiated using conventional analyses.
Therefore, in this study, we undertook a step-wise approach to identify serum biomarkers associated with CRSwNP exacerbation, beginning with 39 analytes, identifying high-level patterns of association (clusters) and then focusing on specific markers (candidate markers). A key finding in this study is the distinct biomarker signatures of VEGF and GM-CSF that were detected in serum specimens of CRSwNP patients during their natural disease exacerbation. By focused analysis, raw protein concentration of VEGF and GM-CSF was also significantly increased during exacerbation in CRSwNP compared to controls or to their baseline values. Local production of both VEGF and GM-CSF in polyp tissues has been documented previously [25–27] and their presence in serum may be an important marker for specific exacerbation. Further investigation is necessary to examine whether detection of these factors in blood represents a ‘spillover’ from local production (outside-in effect). As judged by their biological functions, VEGF and GM-CSF may play pivotal roles in tissue remodeling, and innate and adaptive immunity of nasal and sinus mucosa, respectively. Larger studies focusing on these molecules will be necessary to investigate their roles in pathological changes in CRSwNP and other chronic airway diseases.
Several other important findings of the network graph merit discussion. The objective separation of study participant clusters during exacerbation phase revealed “cross-over” subjects whose biomarker signature mimicked their cluster mates but clinically were included in the opposite group. Thus, based on molecular data, the presence of ‘case-like’ controls and ‘control-like’ cases could represent the heterogeneity of human subjects with differing levels of immune dysregulation, which may escalate during acute exacerbation. Indeed, heterogeneity in regard to the involvement of different immunological pathways and differences in responses likely exists in CRSwNP patients [28]. Also, all the biomarker clusters were closer to the CRSwNP-predominant cluster than to the control-predominant cluster indicating an elevated inflammatory state during exacerbation in CRSwNP patients.
There are some limitations in this study. First, the sample size is small, and the power calculation was based on a pilot study that examined the changes in nasal IL-33 levels. Unexpectedly, we found no statistical difference in the IL-33 levels between CRSwNP patients and control subjects during exacerbation. There are a few possible reasons that may explain this finding. Timing of specimen collection could be an important factor, since IL-33 is released within hours in animal models [29] and may have been released acutely prior to the patient reaching the clinic for sample collection. Other explanations may include the potential variability in nasal specimen collection techniques and confounding treatment effects of topical corticosteroids. Alternatively, IL-33 may not be a key factor in the initiation of inflammation in a significant proportion of CRSwNP patients studied. Similarly, other immunologic measurements that were negative in this study share the possible risk of being falsely negative because of the small sample size. Second, we allowed treatment interventions per the usual care for subjects in the study (including continuation of baseline treatments in the CRSwNP group, such as topical corticosteroids), which could have influenced the natural history of disease exacerbation.
There are several strengths of this study. First, the study employed the prospective design in CRSwNP patients that included a normal control group. We demonstrated that an immune signature of increased growth factors is detected systemically and uniquely in CRSwNP patients during exacerbation regardless of nature of exacerbating trigger (allergic versus viral infection etc). This suggests that such a prospective self-reported exacerbation model may provide insights into the underlying pathophysiology of the disease. Second, to the best of our knowledge, this is the first application of an unsupervised learning model to clinical studies in CRS, showing it may be feasible in larger studies. Indeed, the power of the network analysis may lay in the identification of clusters of biomarkers and patients, while elucidating their interactions. Moreover, studies with greater numbers of subjects will also likely reveal novel immunological phenotypes of CRS and specific pathways that operate in each subtype. This supposition brings us closer to the concept of personalized medicine in CRS, leading to appropriate and cost-effective management of a very common but vastly heterogeneous disorder.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Ms. Kay Bachman, Ms. Vardhini Mohan, and Mr. Arcenio Galindo for study coordination, Ms. Diane Squillace for technical assistance, and Ms. LuRaye Eischens for secretarial assistance. The authors also thank Dr. James Gern for analyzing nasal washes for presence of rhinovirus and enterovirus.
Supported by NIH Grant R56 AI49235 and the Mayo Foundation
ABBREVIATIONS
- SNOT-22
Sino-nasal outcome test-22
- CRSwNP
Chronic rhinosinusitis with nasal polyposis
- HRV/EV
Human rhinovirus/Enterovirus
REFERENCES
- 1.Meltzer EO, Hamilos DL, Hadley JA, Lanza DC, Marple BF, Nicklas RA, Bachert C, Baraniuk J, Baroody FM, Benninger MS, Brook I, Chowdhury BA, Druce HM, Durham S, Ferguson B, Gwaltney JM, Kaliner M, Kennedy DW, Lund V, Naclerio R, Pawankar R, Piccirillo JF, Rohane P, Simon R, Slavin RG, Togias A, Wald ER, Zinreich SJ American Academy of Allergy, Asthma and Immunology (AAAAI); American Academy of Otolaryngic Allergy (AAOA); American Academy of Otolaryngology--Head and Neck Surgery (AAO-HNS); American College of Allergy, Asthma and Immunology (ACAAI); American Rhinologic Society (ARS) Rhinosinusitis: establishing definitions for clinical research and patient care. The Journal of allergy and clinical immunology. 2004;114:155–212. doi: 10.1016/j.jaci.2004.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharyya N. Contemporary assessment of the disease burden of sinusitis. American journal of rhinology & allergy. 2009;23:392–395. doi: 10.2500/ajra.2009.23.3355. [DOI] [PubMed] [Google Scholar]
- 3.Payne SC, Borish L, Steinke JW. Genetics and phenotyping in chronic sinusitis. The Journal of allergy and clinical immunology. 2011;128:710–720. doi: 10.1016/j.jaci.2011.05.022. [DOI] [PubMed] [Google Scholar]
- 4.Rank MA, Hagan JB, Samant SA, Kita H. A proposed model to study immunologic changes during chronic rhinosinusitis exacerbations: data from a pilot study. American journal of rhinology & allergy. 2013;27:98–101. doi: 10.2500/ajra.2013.27.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hopkins C, Gillett S, Slack R, Lund VJ, Browne JP. Psychometric validity of the 22-item Sinonasal Outcome Test. Clinical otolaryngology. 2009;34:447–454. doi: 10.1111/j.1749-4486.2009.01995.x. [DOI] [PubMed] [Google Scholar]
- 6.Morley AD, Sharp HR. A review of sinonasal outcome scoring systems - which is best? Clinical otolaryngology. 2006;31:103–109. doi: 10.1111/j.1749-4486.2006.01155.x. [DOI] [PubMed] [Google Scholar]
- 7.Dietz de Loos DA, Segboer CL, Gevorgyan A, Fokkens WJ. Disease-specific quality-of-life questionnaires in rhinitis and rhinosinusitis: review and evaluation. Current allergy and asthma reports. 2013;13:162–170. doi: 10.1007/s11882-012-0334-8. [DOI] [PubMed] [Google Scholar]
- 8.Bastian M, Heymann S, Jacomy M. Gephi: An open source software for exploring and manipulating networks; International AAAI Conference on Weblogs and Social Media; 2009. [Google Scholar]
- 9.Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic acids research. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grotenboer NS, Ketelaar ME, Koppelman GH, Nawijn MC. Decoding asthma: translating genetic variation in IL33 and IL1RL1 into disease pathophysiology. The Journal of allergy and clinical immunology. 2013;131:856–865. doi: 10.1016/j.jaci.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 12.O'Byrne PM, Gauvreau GM, Brannan JD. Provoked models of asthma: what have we learnt? Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2009;39:181–192. doi: 10.1111/j.1365-2222.2008.03172.x. [DOI] [PubMed] [Google Scholar]
- 13.Kistler A, Avila PC, Rouskin S, Wang D, Ward T, Yagi S, Schnurr D, Ganem D, DeRisi JL, Boushey HA. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. The Journal of infectious diseases. 2007;196:817–825. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denlinger LC, Sorkness RL, Lee WM, Evans MD, Wolff MJ, Mathur SK, Crisafi GM, Gaworski KL, Pappas TE, Vrtis RF, Kelly EA, Gern JE, Jarjour NN. Lower airway rhinovirus burden and the seasonal risk of asthma exacerbation. American journal of respiratory and critical care medicine. 2011;184:1007–1014. doi: 10.1164/rccm.201103-0585OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peters AT, Kato A, Zhang N, Conley DB, Suh L, Tancowny B, Carter D, Carr T, Radtke M, Hulse KE, Seshadri S, Chandra R, Grammer LC, Harris KE, Kern R, Schleimer RP. Evidence for altered activity of the IL-6 pathway in chronic rhinosinusitis with nasal polyps. The Journal of allergy and clinical immunology. 2010;125:397–403. doi: 10.1016/j.jaci.2009.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramanathan M, Jr, Lee WK, Spannhake EW, Lane AP. Th2 cytokines associated with chronic rhinosinusitis with polyps down-regulate the antimicrobial immune function of human sinonasal epithelial cells. American journal of rhinology. 2008;22:115–121. doi: 10.2500/ajr.2008.22.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Zele T, Claeys S, Gevaert P, Van Maele G, Holtappels G, Van Cauwenberge P, Bachert C. Differentiation of chronic sinus diseases by measurement of inflammatory mediators. Allergy. 2006;61:1280–1289. doi: 10.1111/j.1398-9995.2006.01225.x. [DOI] [PubMed] [Google Scholar]
- 18.Bradley DT, Kountakis SE. Role of interleukins and transforming growth factor-beta in chronic rhinosinusitis and nasal polyposis. The Laryngoscope. 2005;115:684–686. doi: 10.1097/01.mlg.0000161334.67977.5D. [DOI] [PubMed] [Google Scholar]
- 19.Kennedy JL, Hubbard MA, Huyett P, Patrie JT, Borish L, Payne SC. Sino-nasal outcome test (SNOT-22): a predictor of postsurgical improvement in patients with chronic sinusitis. Annals of allergy, asthma & immunology : official publication of the American College of Allergy, Asthma, & Immunology. 2013;111:246–251. doi: 10.1016/j.anai.2013.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farag AA, Deal AM, McKinney KA, Thorp BD, Senior BA, Ebert CS, Jr, Zanation AM. Single-blind randomized controlled trial of surfactant vs hypertonic saline irrigation following endoscopic endonasal surgery. International forum of allergy & rhinology. 2013;3:276–280. doi: 10.1002/alr.21116. [DOI] [PubMed] [Google Scholar]
- 21.Snidvongs K, Pratt E, Chin D, Sacks R, Earls P, Harvey RJ. Corticosteroid nasal irrigations after endoscopic sinus surgery in the management of chronic rhinosinusitis. International forum of allergy & rhinology. 2012;2:415–421. doi: 10.1002/alr.21047. [DOI] [PubMed] [Google Scholar]
- 22.Videler WJ, Badia L, Harvey RJ, Gane S, Georgalas C, van der Meulen FW, Menger DJ, Lehtonen MT, Toppila-Salmi SK, Vento SI, Hytönen M, Hellings PW, Kalogjera L, Lund VJ, Scadding G, Mullol J, Fokkens WJ. Lack of efficacy of long-term, low-dose azithromycin in chronic rhinosinusitis: a randomized controlled trial. Allergy. 2011;66:1457–1468. doi: 10.1111/j.1398-9995.2011.02693.x. [DOI] [PubMed] [Google Scholar]
- 23.Salib RJ, Talpallikar S, Uppal S, Nair SB. A prospective randomised single-blinded clinical trial comparing the efficacy and tolerability of the nasal douching products Sterimar and Sinus Rinse following functional endoscopic sinus surgery. Clinical otolaryngology : official journal of ENT-UK; official journal of Netherlands Society for Oto-Rhino-Laryngology & Cervico-Facial Surgery. 2013;38:297–305. doi: 10.1111/coa.12132. [DOI] [PubMed] [Google Scholar]
- 24.Arron JR, Choy DF, Scheerens H, Matthews JG. Noninvasive biomarkers that predict treatment benefit from biologic therapies in asthma. Annals of the American Thoracic Society. 2013;10(Suppl):S206–S213. doi: 10.1513/AnnalsATS.201303-047AW. [DOI] [PubMed] [Google Scholar]
- 25.Coste A, Brugel L, Maître B, Boussat S, Papon JF, Wingerstmann L, Peynègre R, Escudier E. Inflammatory cells as well as epithelial cells in nasal polyps express vascular endothelial growth factor. The European respiratory journal. 2000;15:367–372. doi: 10.1034/j.1399-3003.2000.15b24.x. [DOI] [PubMed] [Google Scholar]
- 26.Hamilos DL, Leung DY, Huston DP, Kamil A, Wood R, Hamid Q. GM-CSF, IL-5 and RANTES immunoreactivity and mRNA expression in chronic hyperplastic sinusitis with nasal polyposis (NP) Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 1998;28:1145–1152. doi: 10.1046/j.1365-2222.1998.00380.x. [DOI] [PubMed] [Google Scholar]
- 27.Allen JS, Eisma R, Leonard G, Kreutzer D. Interleukin-3 interleukin-5, and granulocyte-macrophage colony-stimulating factor expression in nasal polyps. American journal of otolaryngology. 1997;18:239–246. doi: 10.1016/s0196-0709(97)90003-x. [DOI] [PubMed] [Google Scholar]
- 28.Van Crombruggen K, Zhang N, Gevaert P, Tomassen P, Bachert C. Pathogenesis of chronic rhinosinusitis: inflammation. The Journal of allergy and clinical immunology. 2011;128:728–732. doi: 10.1016/j.jaci.2011.07.049. [DOI] [PubMed] [Google Scholar]
- 29.Kouzaki H, Iijima K, Kobayashi T, O'Grady SM, Kita H. The danger signal, extracellular ATP, is a sensor for an airborne allergen and triggers IL-33 release and innate Th2-type responses. Journal of immunology. 2011;186:4375–4387. doi: 10.4049/jimmunol.1003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.