Abstract
Closely related species of fishes often have different sex chromosome systems. Such rapid turnover of sex chromosomes can occur by several mechanisms, including fusions between an existing sex chromosome and an autosome. These fusions can result in a multiple sex chromosome system, where a species has both an ancestral and a neo-sex chromosome. Although this type of multiple sex chromosome system has been found in many fishes, little is known about the mechanisms that select for the formation of neo-sex chromosomes, or the role of neo-sex chromosomes in phenotypic evolution and speciation. The identification of closely related, sympatric species pairs in which one species has a multiple sex chromosome system and the other has a simple sex chromosome system provides an opportunity to study sex chromosome turnover. Recently, we found that a population of threespine stickleback (Gasterosteus aculeatus) from Japan has an X1X2Y multiple sex chromosome system resulting from a fusion between the ancestral Y chromosome and an autosome, while a sympatric threespine stickleback population has a simple XY sex chromosome system. Furthermore, we demonstrated that the neo-X chromosome (X 2) plays an important role in phenotypic divergence and reproductive isolation between these sympatric stickleback species pairs. Here, we review multiple sex chromosome systems in fishes, as well as recent advances in our understanding of the evolutionary role of sex chromosome turnover in stickleback speciation.
Keywords: Multiple sex chromosomes, Neo-sex chromosome, X1X2Y, Stickleback, Sexual conflict, Speciation
Turnover of sex chromosome in fishes
Sex chromosomes have repeatedly and independently evolved in plant and animal species with genetic sex-determination mechanisms (Bull 1983). Although some taxonomic groups appear to have stable sex chromosomes that have been conserved across hundreds of millions of years of evolution, there are many examples of sex chromosome turnover, where even closely related species have different sex chromosomes (White 1973). Heteromorphic sex chromosomes have been found in about 10% of fish species karyotyped (Devlin and Nagahama 2002), and many independent groups of fishes show evidence for sex chromosome turnover (Devlin and Nagahama 2002; Mank et al. 2006). In fishes, there is evidence that sex chromosome turnover has occurred through multiple mechanisms, including the transposition of an existing male-determination locus to an autosome (Woram et al. 2003), the evolution of a new male-determination locus on an autosome (Kondo et al. 2006; Tanaka et al. 2007), and fusions between an autosome and an existing Y-chromosome (Kitano et al. 2009; Ross et al. 2009). In this review, we focus on the evolution of sex chromosome-autosome fusions, which appear to be quite common in fishes. Then, we discuss the mechanisms that might select for these fusions as well as their potential role in phenotypic evolution and speciation.
Multiple sex chromosome systems in fishes
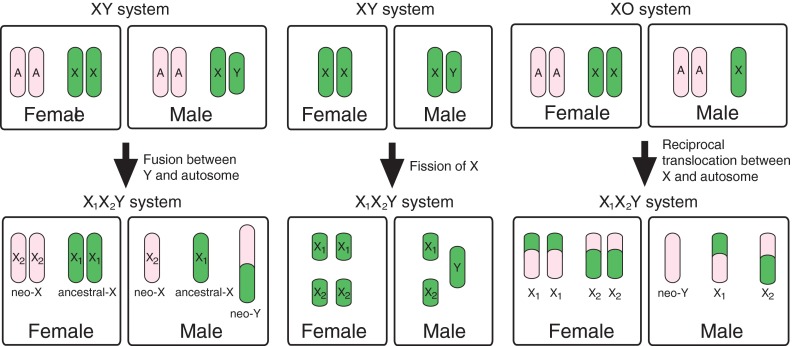
Most species with sex chromosomes either have a simple male heteromorphic (XX female/XY male) or female heteromorphic (ZW female/ZZ male) system. Fusions between an autosome and a sex chromosome (X, Y, Z, or W) create what is commonly referred to as a multiple sex chromosome system. For example, when an autosome fuses to a Y chromosome, this creates an X1X2Y sex chromosome system (White 1973). In this case, males have one neo-Y chromosome, one ancestral X chromosome (X1), and one neo-X chromosome (X 2), while females have two pairs of X chromosomes (X1X1X2X2) (Fig. 1). Similarly, fusions between an autosome and an X, Z, or W chromosome will create an XY1Y2, ZW1W2, or Z1Z2W system, respectively. All of these types of systems have been found in a variety of taxonomic groups. The first multiple sex chromosome system identified in fishes was found by Uyeno and Miller (1971) in a species of Mexican killifish; males have an X1X2Y sex chromosome system, with one large metacentric Y chromosome that is missing in females and two small acrocentric X chromosomes that are also present in females (Uyeno and Miller 1971). Since then, multiple sex chromosome systems have been identified in many fish species across diverse families (Table 1). The presence of multiple sex chromosome systems in diverse groups of fishes suggests that they have evolved independently in multiple lineages.
Fig. 1.
Three proposed mechanisms for the formation of an X1X2Y multiple sex chromosome system, based on White (1983). Autosome, X chromosome, ancestral-X chromosome, neo-X chromosome, and Y chromosome are abbreviated as A, X, X 1, X 2, and Y, respectively
Table 1.
List of fishes that have multiple sex chromosome systems
| Order | Family | Genus | Species | 2n (F/M) | Commentsa | References |
|---|---|---|---|---|---|---|
| X1X1X2X2/X1X2Y | ||||||
| Anguilliformes | Ophichthidae | Muraenichthys | gymnotus | 48/47 | Y-A fusion | (Murofushi and Yosida 1984) |
| Beryciformes | Berycidae | Beryx | splendens | 48/47 | Y-A fusion | (Ojima and Kikuno 1986) |
| Characiformes | Erythrinidae | Erythrinus | erythrinus B b | 54/53 | Y-A fusion; E. erythrinus A population has no obvious sex chromosome (2n = 54) | (Bertollo et al. 2004) |
| Characiformes | Erythrinidae | Erythrinus | erythrinus C b | 52/51 | Y-A fusion; E. erythrinus A population has no obvious sex chromosome (2n = 54) | (Bertollo et al. 2004) |
| Characiformes | Erythrinidae | Erythrinus | erythrinus D b | 52/51 | Y-A fusion; E. erythrinus A population has no obvious sex chromosome (2n = 54) | (Bertollo et al. 2004) |
| Characiformes | Erythrinidae | Hoplias | malabaricus | 40/39 | Y-A fusion; other H. malabaricus karyomorphs are XX/XY (2n = 42) or XX/XY1Y2 (2n = 41/40) | (Bertollo et al. 2000; Rosa et al. 2009) |
| Clupeiformes | Clupeidae | Brevoortia | aurea | 46/45 | Y-A fusion, B. pectinata has no obvious sex chromosome (2n = 46) | (Brum and Galetti 1992) |
| Cypriniformes | Cobitidae | Cobitis | taenia striata | 50/49 | Y-A fusion | (Saitoh 1989) |
| Cyprinodontiformes | Cyprinodontidae | Garmanella | pulchra | 50/49 | Y-A fusion | (Levin and Foster 1972) |
| Cyprinodontiformes | Cyprinodontidae | Megupsilon | aporus | 48/47 | first Y-A fusion identified in fishes | (Uyeno and Miller 1971) |
| Cyprinodontiformes | Goodeidae | Allodontichthys | hubbsi | 42/41 | Y-A fusion; no obvious sex chromosomes in family (2n = 48) | (Uyeno et al. 1983) |
| Cyprinodontiformes | Nothobranchiidae | Nothobranchius | guentheri | 36/35 | Y-A fusion | (Ewulonu et al. 1985) |
| Gasterosteiformes | Gasterosteidae | Gasterosteus | aculeatus (JS) | 42/41 | Y-A fusion; sympatric G. aculeatus population is XX/XY (2n = 42) | (Kitano et al. 2009) |
| Gasterosteiformes | Gasterosteidae | Gasterosteus | wheatlandi | 42/41 | Y-A fusion; closely related, sympatric species G. aculeatus is XX/XY (2n = 42) | (Ross et al. 2009) |
| Gymnotiformes | Gymnotidae | Gymnotus | pantanal | 40/39 | Y-A fusion; two closely related, sympatric species have no obvious sex chromosomes (2n = 40 or 54) | (Margarido et al. 2007) |
| Gymnotiformes | Hypopomidae | Brachyhypopomus | pinnicaudatus | 42/41 | Y-A fusion | (Almeida-Toledo et al. 2000a) |
| Gymnotiformes | Sternopygidae | Eigenmannia | sp.2 | 32/31 | Y-A fusion; closely related, sympatric species E. virescens is XX/XY (2n = 38) | (Almeida-Toledo et al. 2000b; Almeida-Toledo et al. 1984) |
| Perciformes | Blenniidae | Parablennius | tentacularis | 48/47 | Y-A fusion; closely related species have no obvious sex chromosome | (Caputo et al. 2001) |
| Perciformes | Callionymidae | Callionymus | beniteguri | 38/37 | Y-A fusion; closely related species C. punctatus and C. doryssus are XX/XY | (Murofushi et al. 1983) |
| Perciformes | Callionymidae | Callionymus | ornatipinnis | 38/37 | Y-A fusion; closely related species C. punctatus and C. doryssus are XX/XY | (Murofushi et al. 1983) |
| Perciformes | Channichthyidae | Chaenodraco | wilsoni | 48/47 | Y-A fusion; closely related species have no obvious sex chromosome | (Morescalchi et al. 1992) |
| Perciformes | Channichthyidae | Chionodraco | hamatus | 48/47 | Y-A fusion; closely related species have no obvious sex chromosome | (Morescalchi et al. 1992) |
| Perciformes | Channichthyidae | Chionodraco | myersi | 48/47 | Y-A fusion; closely related species have no obvious sex chromosome | (Morescalchi et al. 1992) |
| Perciformes | Channichthyidae | Chionobathyscus | dewitti | 48/47 | Y-A fusion; closely related species have no obvious sex chromosome | (Morescalchi et al. 1992) |
| Perciformes | Channichthyidae | Pagetopsis | macropterus | 48/47 | Y-A fusion; closely related species have no obvious sex chromosome | (Morescalchi et al. 1992) |
| Perciformes | Eleotrididae | Awaous | strigatus | ND | Species in same family Dormitator maculatus is XX/XY | (Oliveira and Almeida-Toledo 2006) |
| Perciformes | Gobiidae | Ctenogobius | shufeldti | 48/47 | Y-A fusion; family has XX/XY, ZW/ZZ and XX/XO sex chromosome systems | (Pezold 1984) |
| Perciformes | Lutjanidae | Lutjanus | quinquelineatus | 48/47 | Y-A fusion; closely related species L. kasmira has no obvious sex chromosome (2n = 48) | (Ueno and Takai 2008) |
| Perciformes | Monodactylidae | Monodactylus | sebae | 48/47 | Y-A fusion; closely related species M. argenteus has no obvious sex chromosome (2n = 48) | (Suzuki et al. 1988) |
| Perciformes | Percidae | Zingel | zingel | 48/47 | Y-A fusion; other species in family are XX/XY | (Halacka et al. 2007) |
| Salmoniformes | Salmonidae | Oncorhynchus | nerka | 58/57 | Y-A fusion; closely related species O. mykiss is XX/XY | (Thorgaard 1978; Ueda and Ojima 1984) |
| Scorpaeniformes | Tetrarogidae | Hypodytes | rubripinnis | 48/47 | Y-A fusion | (Ueno and Kang 1992) |
| Siluriformes | Siluridae | Ompok | bimaculatus | 42/41 | Y-A fusion | (Rishi 1976) |
| Tetraodontiformes | Monacanthidae | Stephanolepis | cirrhifer | 34/33 | Y-A fusion | (Murofushi et al. 1980) |
| Tetraodontiformes | Monacanthidae | Stephanolepis | hispidus | 34/33 | Y-A fusion | (Brum 1996) |
| XX/XY1Y2 | ||||||
| Characiformes | Erythrinidae | Hoplias | malabaricus | 40/41 | Other H. malabaricus karyomorphs are XX/XY or XX/X1X2Y | (Bertollo et al. 2000; Bertollo et al. 1983) |
| Siluriformes | Loricariidae | Ancistrus | sp.1 | 38/39 | Family also has XX/XY, ZW/ZZ, and XX/XO sex chromosome systems | (Alves et al. 2006; Oliveira et al. 2008) |
| Siluriformes | Loricariidae | Harttia | carvalhoi | 52/53 | Closely related species do not have obvious sex chromosomes | (Centofante et al. 2006) |
| Z1Z2W1W2/Z1Z1Z2Z2 | ||||||
| Siluriformes | Loricariidae | Ancistrus | sp.2 | 52/52 | Family also has XX/XY, ZW/ZZ, and XX/XO sex chromosome systems | (Alves et al. 2006; Oliveira et al. 2008) |
| ZW1W2/ZZ | ||||||
| Aulopiformes | Synodontidae | Trachinocephalus | myops | 27/26 | Z-A fusion; other species in family are ZW/ZZ (2n = 48) | (Ueno et al. 2001) |
| Characiformes | Parodontidae | Apareiodon | affinis | 55/54 | Fission of W; sympatric population has no obvious sex chromosome (2n = 54) | (Jesus et al. 1999; Moreira-Filho et al. 1980) |
aWhen mechanisms creating the neo sex chromosomes are suggested in the original paper, they are noted
bThese three karyomorphs are presented as different species in this table, but might constitute a single species
ND = no data available
Interestingly, X1X2Y systems are more common in fishes than the other types of multiple sex chromosome systems (35/41) (Table 2). X1X2Y systems can arise not only through fusions between an autosome and a Y chromosome, but also through centric fission of the X chromosome in species with an XY system or through reciprocal translocations between the X chromosome and an autosome in species with an ancestral XX female/XO male sex chromosome system (Fig. 1) (White 1973). Several lines of evidence support the idea that the multiple sex chromosome systems in fishes arose mainly through fusions between a Y chromosome and an autosome. First, in many species, the neo-Y chromosome is a large metacentric chromosome, while the two X chromosomes are small acrocentric chromosomes, suggesting that the neo-Y chromosome arose through a Robertsonian fusion of two acrocentric chromosomes (Ueno and Takai 2008). Consistent with this hypothesis, the neo-Y is associated with both the X1 and the X 2 chromosomes during meiosis in all species examined (Ueno and Takai 2008).
Table 2.
Number of cases of different multiple sex chromosome systems found in each order
| X1X1X2X2/X1X2Y | XX/XY1Y2 | Z1Z2W1W2/Z1Z2Z1Z2 | Z W1W2/ZZ | |
|---|---|---|---|---|
| Anguilliformes | 1 | |||
| Aulopiformes | 1 | |||
| Beryciformes | 1 | |||
| Characiformes | 4a | 1 | 1 | |
| Clupeiformes | 1 | |||
| Cypriniformes | 1 | |||
| Cyprinidontiformes | 4 | |||
| Gasterosteiformes | 2 | |||
| Gymnotiformes | 3 | |||
| Perciformes | 13 | |||
| Salmoniformes | 1 | |||
| Scorpaeniformes | 1 | |||
| Siluriformes | 1 | 2 | 1 | |
| Tetraodontiformes | 2 |
aThree of these examples are karyomorphs, which might constitute a single species
Second, XX/XO systems are not as common as XY system in fish, with only twelve examples of XX/XO systems documented to date (Devlin and Nagahama 2002; Alves et al. 2006). Furthermore, only one of these species (Gobiodon citrinus) is found in a family (Gobiidae) that also contains a species (Ctenogobius shufeldti) with an X1X2Y system (Pezold 1984). In seven of the known X1X2Y systems, a closely related species in the same genus or other populations of the same species have an XX/XY sex chromosome system, suggesting that this may be the ancestral state (Table 1). In three of these cases, cytogenetic evidence demonstrates that the ancestral Y has indeed fused to an autosome (Kitano et al. 2009; Ross et al. 2009; Cioffi and Bertollo 2010). Although no evidence for heteromorphic sex chromosomes has been found in eleven cases where closely related species have been analyzed cytogenetically (Table 1), it is possible that these species do have an XX/XY sex chromosome system that cannot be distinguished using traditional cytogenetic tools. For example, it was originally reported that the threespine stickleback (Gasterosteus aculeatus) had no heteromorphic sex chromosomes, but later studies using fluorescence in situ hybridization revealed that the X and the Y can be distinguished (Ross and Peichel 2008).
Finally, although centric fission of the X chromosome can result in the formation of an X1X2Y system (Fig. 1), this is unlikely to be the cause in most fish species with this system. When fissions create an X1X2Y system, closely related species with an XY system are expected to have fewer chromosome pairs (2n) than the species with an X1X2Y system. However, there are no such cases (Table 1).
Thus, it is likely that most of the X1X2Y sex chromosome systems in fishes are derived from fusions between an existing Y chromosome (whether cytogenetically visible or not) and an autosome. However, it is still unknown why these Y-autosome fusions are relatively common in fishes, and whether they have any role in phenotypic evolution or speciation. In order to address these questions, it is particularly useful to identify extant species pairs in which one species has the ancestral simple sex chromosome and the other has the derived multiple sex chromosome system. Indeed, for a few of the species with multiple sex chromosome systems listed in Table 1, such species pairs can be found in sympatry or parapatry. For example, in a single watershed, there are sympatric populations of Hoplias aff. malabaricus; some populations have a simple XY system, other populations have an X1X2Y system, and still others have a XY1Y2 system (Bertollo et al. 1983; Bertollo et al. 2000; Rosa et al. 2009). Similarly, two overlapping sub-species of Eigenmannia have non-homologous sex chromosomes; E. virescens has an XY system and E. sp.2 has an X1X2Y system (Almeida-Toledo et al. 1984; Almeida-Toledo et al. 2000b; Henning et al. 2008). In eastern Hokkaido, Japan, an incipient species of threespine stickleback (G. aculeatus) has an X1X2Y system; this Japan Sea form is sympatric with the ancestral Pacific Ocean form, which has a simple XY system (Kitano et al. 2009). In the Japan Sea form, cytogenetic studies and linkage analysis revealed that the X1X2Y system results from a fusion between an autosome (chromosome 9) and the ancestral Y chromosome (chromosome 19) (Kitano et al. 2009). A similar fusion has occurred in the blackspotted stickleback (G. wheatlandi), which is sympatric with the Atlantic Ocean population of threespine stickleback on the eastern coast of North America (FitzGerald and Wootton 1993). Like the Japan Sea form, G. wheatlandi also has an X1X2Y system, but chromosome 12 has fused to the ancestral Y chromosome (Ross et al. 2009). These data suggest that the fusions have occurred independently in the two stickleback lineages.
What drives sex chromosome turnover?
Theoretical work has suggested that the presence of genes with sexually antagonistic effects (i.e. genes with alleles that have differential fitness effects in males and females) on an ancestral autosome might drive the turnover of sex chromosomes (Charlesworth and Charlesworth 1980; van Doorn and Kirkpatrick 2007). Alleles that enhance fitness in one sex will not necessarily increase, and might even reduce, the fitness of the other sex. If such alleles with sexually antagonistic effects are present on autosomes, they will not easily spread within a population, because selection for an increase in allele frequency in one sex will be counteracted by selection against the allele in the other sex. However, if a sexually antagonistic gene is localized on a sex chromosome and tightly linked to the sex-determination locus, both males and females can increase their fitness by having different allele frequencies at the sexually antagonistic locus. Thus, the presence of genes with sexually antagonistic effects on an autosome might select for the evolution of a new sex-determination locus on that autosome, the transposition of an existing sex-determination locus to that autosome, or the fusion of that autosome to an existing sex chromosome.
Although empirically testing this hypothesis presents a major challenge, the identification of closely related species with different sex chromosome systems provides a unique opportunity to test whether sexual antagonism plays a role in sex chromosome turnover. For example, a new female-determination locus has evolved in species of Lake Malawi cichlids, and a gene involved in sexually dimorphic pigmentation is tightly linked to the female sex-determination locus on the invading W chromosome (Roberts et al. 2009). Because the sexually dimorphic pigmentation pattern may be beneficial in females but detrimental in males of these species, sexually antagonistic selection might have played an important role in the evolution of this new ZW sex chromosome system.
Similarly, we have some preliminary evidence that sexually antagonistic traits are found on the neo-X chromosome created by the Y-autosome fusion in the Japan Sea threespine stickleback. Dorsal spine length is sexually dimorphic in the Japan Sea population (Kitano et al. 2007b) and has been suggested as a sexually antagonistic trait in sticklebacks (Reimchen and Nosil 2004). Because variation in dorsal spine length maps to the Japan Sea neo-X chromosome (Kitano et al. 2009), it is possible that the presence of sexually antagonistic variation for spine length on an ancestral autosome might have selected for the fixation of the sex chromosome fusion. However, we have not yet confirmed that spine length is a sexually antagonistic trait or identified any genes with sexually antagonistic alleles on the ancestral autosome (chromosome 9) of the Pacific Ocean population. Further comparative genomic and functional studies of genes on chromosome 9 in the ancestral Pacific Ocean and the derived Japan Sea sticklebacks will provide insights into whether sexually antagonistic selection plays an important role in sex chromosome turnover.
Role of sex chromosome turnover in speciation
It has long been appreciated that sex chromosomes might play a special role in speciation. There is abundant empirical evidence demonstrating that hybrid sterility genes are preferentially localized on sex chromosomes (Coyne and Orr 2004; Presgraves 2008; Qvarnstrom and Bailey 2009). Such an overrepresentation of hybrid sterility genes on the sex chromosomes may be due to multiple factors, such as faster X evolution, accumulation of sexually antagonistic genes, sex ratio meiotic drive, transcriptional suppression of sex chromosomes during early spermatogenesis, and gene traffic between the X and autosomes (Coyne and Orr 2004; Presgraves 2008; Moyle et al. 2010). The relative role of the sex chromosomes in other isolating barriers is still debated (Qvarnstrom and Bailey 2009), but there are empirical data indicating that the sex chromosomes harbor genes that are important for behavioral isolation between closely related species (Prowell 1998; Reinhold 1998; Lindholm and Breden 2002).
Although there is good evidence that the sex chromosomes do play an important role in speciation, the role of sex chromosome turnover in speciation has been less appreciated. However, genes that play a role in reproductive isolation between populations might accumulate on neo-sex chromosomes for many of the same reasons that they are found on ancestral sex chromosomes. In this case, sex chromosome turnover might actually promote speciation. We have tested this idea by performing genetic linkage analysis of the isolating barriers present between the Japan Sea form (X1X2Y) and the Pacific Ocean form (XY) of threespine sticklebacks. These two forms diverged during a period of geographical isolation between the Sea of Japan and the Pacific Ocean about 1.5–2 million years ago. Currently, they are sympatric in eastern Hokkaido, but are reproductively isolated with a low level of hybridization (Higuchi and Goto 1996; Yamada et al. 2001; Kitano et al. 2007a; Yamada et al. 2007). Reproductive isolation between this stickleback species pair results from multiple isolating barriers, including ecogeographical isolation (Kume et al. 2005; Kume et al. 2010), seasonal isolation (Kume et al. 2005), behavioral isolation (Kitano et al. 2007a), reduced fitness of hybrids (Kitano et al. 2009), and hybrid male sterility (Kitano et al. 2007a).
Although the male hybrids resulting from a cross between a Japan Sea female and a Pacific Ocean male are sterile, hybrid males resulting from crosses in the opposite direction and all hybrid females are fertile (Kitano et al. 2007a). Therefore, we were able to make artificial crosses between these two sympatric species and utilize the numerous genomic and genetic tools available for sticklebacks (Kingsley and Peichel 2007) to conduct quantitative trait locus (QTL) mapping of isolating barriers between the species pair (Kitano et al. 2009). Our QTL mapping revealed that hybrid sterility and male courtship display traits mapped to the ancestral X chromosome (chromosome 19) and the neo-X chromosome (chromosome 9) respectively (Kitano et al. 2009). Thus, a neo-X chromosome resulting from a fusion between the ancestral Y chromosome and an autosome contains genes important for the evolution of male courtship displays and can contribute to reproductive isolation between sympatric stickleback species. This is consistent with the idea that the turnover of sex chromosome might promote speciation.
Conclusions
Species with multiple sex chromosome systems are prevalent in fishes. In some cases, they can be found in sympatry or parapatry with closely related species that have the ancestral, simple sex chromosome system. These species pairs provide a unique opportunity to gain insight into the evolutionary forces that drive the evolution of new sex chromosomes and to the consequences of their evolution on phenotypic divergence and speciation. First, by identifying the genomic loci important for reproductive isolation or phenotypic divergence between closely related species, we can investigate whether the new sex chromosomes created by fusions can contribute to speciation, as we found in the Japanese species pair. Because the gaps between model and non-model organisms are decreasing with the advent of high-throughput sequencing technologies (Rokas and Abbot 2009), it should be possible to conduct similar genetic and genomic analyses on other fish species pairs to investigate the generality of our findings. Second, these systems could be used to test the hypothesis that sexual conflict drives the turnover of sex chromosomes. Although challenging, one approach would be to compare the autosome that has fused to the sex chromosome in both the ancestral population and the derived population to search for sexually antagonistic genes that might have driven the fusion. Finally, neo-sex chromosomes created by fusions provide a great system to study the early stages of sex chromosome evolution. Extensive genomic analyses have been conducted on the Drosophila miranda neo-sex chromosome created by a Y-autosome fusion; these studies have provided insights into the evolutionary forces that drive the degeneration of the Y chromosome (Bachtrog and Charlesworth 2002; Bachtrog 2003; Bachtrog 2004; Bachtrog 2006; Bachtrog et al. 2008; Bachtrog et al. 2009). Similar studies will be possible in fishes that have neo-sex chromosomes of a variety of ages, which will deepen our understanding of the process of sex chromosome evolution.
Acknowledgments
This work was supported by the Uehara Memorial Foundation, JST PRESTO Program, a Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science, and Technology (no. 22770075) to JK, and a Burroughs Wellcome Fund Career Award in the Biomedical Sciences and National Institutes of Health grants R01 GM071854 and P50 HG002568 to CLP.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Almeida-Toledo LF, Daniel-Silva MFZ, Lopes CE, Toledo-Filho SA. Sex chromosome evolution in fish. II. Second occurrence of an X1X2Y sex chromosome system in Gymnotiformes. Chrom Res. 2000;8:335–340. doi: 10.1023/A:1009287630301. [DOI] [PubMed] [Google Scholar]
- Almeida-Toledo LF, Foresti F, Daniel MFZ, Toledo SA. Sex chromosome evolution in fish: the formation of the neo-Y chromosome in Eigenmannia (Gymnotiformes) Chromosoma. 2000;109(3):197–200. doi: 10.1007/s004120050428. [DOI] [PubMed] [Google Scholar]
- Almeida-Toledo LF, Foresti H, Toledo-Filho SA. Complex sex chromosome system in Eigenmannia sp. (Pisces, Gymnotiformes) Genetica. 1984;64:165–169. doi: 10.1007/BF00115340. [DOI] [Google Scholar]
- Alves AL, Oliveira C, Nirchio M, Granado A, Foresti F. Karyotypic relationships among the tribes of Hypostominae (Siluriformes: Loricariidae) with description of XO sex chromosome system in a Neotropical fish species. Genetica. 2006;128:1–9. doi: 10.1007/s10709-005-0715-1. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. Accumulation of spock and worf, two novel non-LTR retrotransposons, on the neo-Y chromosome of Drosophila miranda. Mol Biol Evol. 2003;20:173–181. doi: 10.1093/molbev/msg035. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. Evidence that positive selection drives Y-chromosome degeneration in Drosophila miranda. Nat Genet. 2004;36:518–522. doi: 10.1038/ng1347. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. Expression profile of a degenerating neo-Y chromosome in Drosophila. Curr Biol. 2006;16:1694–1699. doi: 10.1016/j.cub.2006.07.053. [DOI] [PubMed] [Google Scholar]
- Bachtrog D, Charlesworth B. Reduced adaptation of a non-recombining neo-Y chromosome. Nature. 2002;416:323–326. doi: 10.1038/416323a. [DOI] [PubMed] [Google Scholar]
- Bachtrog D, Hom E, Wong KM, Maside X, de Jong P. Genomic degradation of a young Y chromosome in Drosophila miranda. Genome Biol. 2008;9:R30. doi: 10.1186/gb-2008-9-2-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Jensen JD, Zhang Z. Accelerated adaptive evolution on a newly formed X chromosome. PLoS Biol. 2009;7:e1000082. doi: 10.1371/journal.pbio.1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertollo LAC, Born GG, Dergam JA, Fenocchio AS, Moreira-Filho O. A biodiversity approach in the neotropical Erythrinidae fish, Hoplias malabaricus. Karyotypic survey, geographic distribution of cytotypes and cytotaxonomic considerations. Chrom Res. 2000;8:603–613. doi: 10.1023/A:1009233907558. [DOI] [PubMed] [Google Scholar]
- Bertollo LAC, et al. Chromosome evolution in the erythrinid fish, Erythrinus erythrinus (Teleostei: Characiformes) Heredity. 2004;93:228–233. doi: 10.1038/sj.hdy.6800511. [DOI] [PubMed] [Google Scholar]
- Bertollo LAC, Takahashi CS, Moreira-Filho O. Multiple sex chromosomes in the genus Hoplias (Pisces: Erythrinidae) Cytologia. 1983;48(1):1–12. doi: 10.1508/cytologia.48.1. [DOI] [Google Scholar]
- Brum MJI. Cytogenetic studies of Brazilian marine fish. Braz J Genet. 1996;19(3):421–427. [Google Scholar]
- Brum MJI, Galetti PM. Multiple sex chromosomes in south Atlantic fish, Brevoortia aurea, Clupeidae. Rev Brazil Genet. 1992;15:547–553. [Google Scholar]
- Bull JJ. Evolution of sex determining mechanisms. Menlo Park: Benjamin Cummings; 1983. [Google Scholar]
- Caputo V, Machella N, Nisi-Cerioni P, Olmo E. Cytogenetics of nine species of Mediterranean blennies and additional evidence for an unusual multiple sex-chromosome system in Parablennius tentacularis (Perciformes, Blenniidae) Chrom Res. 2001;9:3–12. doi: 10.1023/A:1026779314932. [DOI] [PubMed] [Google Scholar]
- Centofante L, Bertollo LA, Moreira-Filho O. Cytogenetic characterization and description of an XX/XY1Y2 sex chromosome system in catfish Harttia carvalhoi (Siluriformes, Loricariidae) Cytogenet Genome Res. 2006;112(3–4):320–324. doi: 10.1159/000089887. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. Sex differences in fitness and selection for centric fusions between sex-chromosomes and autosomes. Genet Res. 1980;35:205–214. doi: 10.1017/S0016672300014051. [DOI] [PubMed] [Google Scholar]
- Cioffi MB, Bertollo LA. Initial steps in XY chromosome differentiation in Hoplias malabaricus and the origin of an X1X2Y sex chromosome system in this fish group. Heredity. 2010;105:554–561. doi: 10.1038/hdy.2010.18. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sunderland: Sinauer; 2004. [Google Scholar]
- Devlin RH, Nagahama Y. Sex determination and sex differentiation in fish: overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208:191–364. doi: 10.1016/S0044-8486(02)00057-1. [DOI] [Google Scholar]
- Ewulonu UK, Haas R, Turner BJ. A multiple sex chromosome system in the annual killifish, Nothobranchius guentheri. Copeia. 1985;1985:503–508. doi: 10.2307/1444868. [DOI] [Google Scholar]
- FitzGerald GJ, Wootton RJ. The behavioural ecology of sticklebacks. In: Pitcher TJ, editor. Behaviour of teleost fishes. London: Chapman and Hall; 1993. pp. 539–572. [Google Scholar]
- Halacka K, Vetesnik L, Lusk S, Mendel J, Papousek I. The X1X1X2X2/X1X2Y multiple sex chromosome system in the Zingel zingel (Pisces: Perciformes) from the Morava river (Czech Republic) Caryologia. 2007;60(3):222–225. [Google Scholar]
- Henning F, Trifonov V, Ferguson-Smith MA, de Almeida-Toledo LF. Non-homologous sex chromosomes in two species of the genus Eigenmannia (Teleostei : Gymnotiformes) Cytogenetic Genome Res. 2008;121(1):55–58. doi: 10.1159/000124382. [DOI] [PubMed] [Google Scholar]
- Higuchi M, Goto A. Genetic evidence supporting the existence of two distinct species in the genus Gasterosteus around Japan. Environ Biol Fish. 1996;47:1–16. doi: 10.1007/BF00002375. [DOI] [Google Scholar]
- Jesus CM, Bertollo LAC, Moreira-Filho O. Comparative cytogenetics in Apareiodon affinis (Pisces, Characiformes) and considerations regarding diversification of the group. Genetica. 1999;105:63–67. doi: 10.1023/A:1003592022927. [DOI] [Google Scholar]
- Kingsley DM, Peichel CL. The molecular genetics of evolutionary change in sticklebacks. In: Ostlund-Nilsson S, Mayer I, Huntingford FA, editors. Biology of the Three-spined Stickleback. Boca Raton: CRC Press; 2007. pp. 41–81. [Google Scholar]
- Kitano J, Mori S, Peichel CL. Phenotypic divergence and reproductive isolation between sympatric forms of Japanese threespine sticklebacks. Biol J Linn Soc. 2007;91:671–685. doi: 10.1111/j.1095-8312.2007.00824.x. [DOI] [Google Scholar]
- Kitano J, Mori S, Peichel CL. Sexual dimorphism in the external morphology of the threespine stickleback (Gasterosteus aculeatus) Copeia. 2007;2007:336–349. doi: 10.1643/0045-8511(2007)7[336:SDITEM]2.0.CO;2. [DOI] [Google Scholar]
- Kitano J, et al. A role for a neo-sex chromosome in stickleback speciation. Nature. 2009;461(7267):1079–1083. doi: 10.1038/nature08441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, et al. Genomic organization of the sex-determining and adjacent regions of the sex chromosomes of medaka. Genome Res. 2006;16:815–826. doi: 10.1101/gr.5016106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume M, Kitamura T, Takahashi H, Goto A. Distinct spawning migration patterns in sympatric Japan Sea and Pacific Ocean forms of threespine stickleback Gasterosteus aculeatus. Ichthyol Res. 2005;52:189–193. doi: 10.1007/s10228-005-0269-3. [DOI] [Google Scholar]
- Kume M, Kitano J, Mori S, Shibuya T. Ecological divergence and habitat isolation between two migratory forms of Japanese threespine stickleback (Gasterosteus aculeatus) J Evol Biol. 2010;23:1436–1446. doi: 10.1111/j.1420-9101.2010.02009.x. [DOI] [PubMed] [Google Scholar]
- Levin B, Foster NR. Cytotaxonomic studies in Cyprinodontidae: multiple sex chromosomes in Garmanella pulchra. Not Nat Acad Nat Sci Philadelphia. 1972;446:1–5. [Google Scholar]
- Lindholm A, Breden F. Sex chromosomes and sexual selection in Poeciliid fishes. Am Nat. 2002;160:S214–S224. doi: 10.1086/342898. [DOI] [PubMed] [Google Scholar]
- Mank JE, Promislow DEL, Avise JC. Evolution of alternative sex-determining mechanisms in teleost fishes. Biol J Linn Soc. 2006;87:83–93. doi: 10.1111/j.1095-8312.2006.00558.x. [DOI] [Google Scholar]
- Margarido VP, Bellafronte E, Moreira-Filho O. Cytogenetic analysis of three sympatric Gymnotus (Gymnotiformes, Gymnotidae) species verifies invasive species in the Upper Parana River basin, Brazil. J Fish Biol. 2007;70:155–164. doi: 10.1111/j.1095-8649.2007.01365.x. [DOI] [Google Scholar]
- Moreira-Filho O, Bertollo LAC, Galetti PM., Jr Evidence for a multiple sex chromosome system with female heterogamety in Apareiodon affinis (Pisces, Parodontidae) Caryologia. 1980;33:83–91. [Google Scholar]
- Morescalchi A, Hureau JC, Olmo E, Ozouf-Costaz C, Pisano E, Stanyon R. A multiple sex-chromosome system in Antarctic ice-fishes. Polar Biol. 1992;11:655–661. doi: 10.1007/BF00237962. [DOI] [Google Scholar]
- Moyle LC, Muir CD, Han MV, Hahn MW. The contribution of gene movement to the “two rules of speciation”. Evolution. 2010;64:1541–1557. doi: 10.1111/j.1558-5646.2010.00990.x. [DOI] [PubMed] [Google Scholar]
- Murofushi M, Nishikawa S, Yosida TH. Cytogenetical studies on fishes. V. Multiple sex chromosome mechanism (XX-Y) found in two dragonet fishes. Proc Jpn Acad B Phys Biol Sci. 1983;59:58–61. doi: 10.2183/pjab.59.58. [DOI] [Google Scholar]
- Murofushi M, Oikawa S, Nishikawa S, Yosida TH. Cytogenetical studies on fishes. III. Multiple sex chromosome mechanism in the filefish, Stephanolepis cirrhifer. Jpn J Genet. 1980;55(2):127–132. doi: 10.1266/jjg.55.127. [DOI] [Google Scholar]
- Murofushi M, Yosida TH. Cytogenetical studies on fishes. VIII. XX-Y sex chromosome mechanism newly found in the snake eel, Muraenichthys gymnotus (Anguilloform, Pisces) Proc Jpn Acad Ser B Phys Biol Sci. 1984;60(2):21–23. doi: 10.2183/pjab.60.21. [DOI] [Google Scholar]
- Ojima Y, Kikuno T. A heteromorphic chromosome of Beryx splendens, Berycidae (Pisces) Proc Jpn Acad B Phys Biol Sci. 1986;62:317–320. doi: 10.2183/pjab.62.317. [DOI] [Google Scholar]
- Oliveira C, Almeida-Toledo LF. Evidence of an XX/XY sex chromosome system in the fish Dormitator maculatus (Teleostei, Eleotrididae) Genet Mol Biol. 2006;29:653–655. doi: 10.1590/S1415-47572006000400013. [DOI] [Google Scholar]
- Oliveira RR, Feldberg E, dos Anjos MB, Zuanon J. Occurrence of multiple sexual chromosomes (XX/XY1Y2 and Z1Z1Z2Z2/Z1Z2W1W2) in catfishes of the genus Ancistrus (Siluriformes, Loricariidae) from the Amazon Basin. Genetica. 2008;134(2):243–249. doi: 10.1007/s10709-007-9231-9. [DOI] [PubMed] [Google Scholar]
- Pezold F. Evidence for multiple sex chromosomes in the freshwater goby, Gobionellus shufeldti (Pisces: Gobiidae) Copeia. 1984;1984:235–238. doi: 10.2307/1445066. [DOI] [Google Scholar]
- Presgraves DC. Sex chromosomes and speciation in Drosophila. Trends Genet. 2008;24:336–343. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prowell DP. Sex linkage and speciation in Lepidoptera. In: Howard DJ, Berlocher SH, editors. Endless forms: Species and speciation. New York: Oxford Univ Press; 1998. pp. 309–319. [Google Scholar]
- Qvarnstrom A, Bailey RI. Speciation through evolution of sex-linked genes. Heredity. 2009;102:4–15. doi: 10.1038/hdy.2008.93. [DOI] [PubMed] [Google Scholar]
- Reimchen TE, Nosil P. Variable predation regimes predict the evolution of sexual dimorphism in a population of threespine stickleback. Evolution. 2004;58:1274–1281. doi: 10.1111/j.0014-3820.2004.tb01706.x. [DOI] [PubMed] [Google Scholar]
- Reinhold K. Sex linkage among genes controlling sexually selected traits. Behav Ecol Sociobiol. 1998;44:1–7. doi: 10.1007/s002650050508. [DOI] [Google Scholar]
- Rishi KK. Mitotic and meiotic chromosomes of a teleost, Callichrous bimaculatus (Bloch) with indications of male heterogamety. Cienc Cult. 1976;28:1171–1173. [Google Scholar]
- Roberts RB, Ser JR, Kocher TD. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science. 2009;326:998–1001. doi: 10.1126/science.1174705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Abbot P. Harnessing genomics for evolutionary insights. Trends Ecol Evol. 2009;24:192–200. doi: 10.1016/j.tree.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Rosa R, Caetano-Filho M, Shibatta OA, Giuliano-Caetano L. Cytotaxonomy in distinct populations in Hoplias aff. malabaricus (Characiformes, Erythrinidae) from lower Paranapanema River basin. J Fish Biol. 2009;75:2682–2694. doi: 10.1111/j.1095-8649.2009.02467.x. [DOI] [PubMed] [Google Scholar]
- Ross JA, Peichel CL. Molecular cytogenetic evidence of rearrangements on the Y chromosome of the threespine stickleback fish. Genetics. 2008;179:2173–2182. doi: 10.1534/genetics.108.088559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, Urton JR, Boland J, Shapiro MD, Peichel CL. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae) PLoS Genet. 2009;5:e1000391. doi: 10.1371/journal.pgen.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh K. Multiple sex-chromosome system in a loach fish. Cytogenet Cell Genet. 1989;52:62–64. doi: 10.1159/000132840. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Taki Y, Takeda M, Akatsu S. Multiple sex chromosomes in a monodactylid fish. Jpn J Ichthyol. 1988;35(1):98–101. [Google Scholar]
- Tanaka K, Takehana Y, Naruse K, Hamaguchi S, Sakaizumi M. Evidence for different origins of sex chromosomes in closely related Oryzias fishes: substitution of the master sex-determining gene. Genetics. 2007;177:2075–2081. doi: 10.1534/genetics.107.075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgaard GH. Sex chromosomes in the sockeye salmon: a Y-autosome fusion. Can J Genet Cytology. 1978;20:349–354. doi: 10.1139/g78-039. [DOI] [PubMed] [Google Scholar]
- Ueda T, Ojima Y. Sex chromosomes in the Kokanee salmon Oncorhynchus nerka. Bull Jpn Soc Sci Fish. 1984;50:1495–1498. doi: 10.2331/suisan.50.1495. [DOI] [Google Scholar]
- Ueno K, Kang J-H. Multiple sex chromosomes in the redfin velvetfish, Hypodytes rubripinnis. Jpn J Ichthyol. 1992;39(2):170–173. [Google Scholar]
- Ueno K, Ota K, Kobayashi T. Heteromorphic sex chromosomes of lizardfish (Synodontidae): focus on the ZZ-ZW1W2 system in Trachinocephalus myops. Genetica. 2001;111:133–142. doi: 10.1023/A:1013749027620. [DOI] [PubMed] [Google Scholar]
- Ueno K, Takai A. Multiple sex chromosome system of X1X1X2X2/X1X2Y type in lutjanid fish, Lutjanus quinquelineatus (Perciformes) Genetica. 2008;132(1):35–41. doi: 10.1007/s10709-007-9146-5. [DOI] [PubMed] [Google Scholar]
- Uyeno T, Miller RR. Multiple sex chromosomes in a Mexican cyprinodontid fish. Nature. 1971;231:452–453. doi: 10.1038/231452a0. [DOI] [PubMed] [Google Scholar]
- Uyeno T, Miller RR, Fitzsimons JM. Karyology of the Cyprinodontoid fishes of the Mexican family Goodeidae. Copeia. 1983;1983:497–510. doi: 10.2307/1444395. [DOI] [Google Scholar]
- van Doorn GS, Kirkpatrick M. Turnover of sex chromosomes induced by sexual conflict. Nature. 2007;449:909–912. doi: 10.1038/nature06178. [DOI] [PubMed] [Google Scholar]
- White MJD. Animal Cytology and Evolution. Cambridge: Cambridge Univ Press; 1973. [Google Scholar]
- Woram RA, et al. Comparative genome analysis of the primary sex-determining locus in salmonid fishes. Genome Res. 2003;13:272–280. doi: 10.1101/gr.578503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Higuchi M, Goto A. Extensive introgression of mitochondrial DNA found between two genetically divergent forms of threespine stickleback, Gasterosteus aculeatus, around Japan. Environ Biol Fish. 2001;61:269–284. doi: 10.1023/A:1010874117127. [DOI] [Google Scholar]
- Yamada M, Higuchi M, Goto A. Long-term occurrence of hybrids between Japan Sea and Pacific Ocean forms of threespine stickleback, Gasterosteus aculeatus, in Hokkaido Island, Japan. Environ Biol Fish. 2007;80:435–443. doi: 10.1007/s10641-006-9142-0. [DOI] [Google Scholar]