Abstract
The sensitivity of stem cells to environmental perturbations has prompted many studies which aim to characterize the influence of mechanical factors on stem cell morphogenesis and differentiation. Hydrodynamic cultures, often employed for large scale bioprocessing applications, impart complex fluid shear and transport profiles, and influence cell fate as a result of changes in media mixing conditions. However, previous studies of hydrodynamic cultures have been limited in their ability to distinguish confounding factors that may affect differentiation, including modulation of embryoid body size in response to changes in the hydrodynamic environment. In this study, we demonstrate the ability to control and maintain embryoid body (EB) size using a combination of forced aggregation formation and rotary orbital suspension culture, in order to assess the impact of hydrodynamic cultures on ESC differentiation, independent of EB size. Size-controlled EBs maintained at different rotary orbital speeds exhibited similar morphological features and gene expression profiles, consistent with ESC differentiation. The similar differentiation of ESCs across a range of hydrodynamic conditions suggests that controlling EB formation and resultant size may be important for scalable bioprocessing applications, in order to standardize EB morphogenesis. However, perturbations in the hydrodynamic environment also led to subtle changes in differentiation toward certain lineages, including temporal modulation of gene expression, as well changes in the relative efficiencies of differentiated phenotypes, thereby highlighting important tissue engineering principles that should be considered for implementation in bioreactor design, as well as for directed ESC differentiation.
Introduction
New technologies at the interface of stem cell biology and bioprocessing are needed for the production of functional tissues for regeneration or replacement in novel therapeutic applications. The unique capacity of embryonic stem cells (ESCs) to differentiate into all somatic cells1–3 makes them an attractive alternative to cell types for which primary cells are not easily isolated or derived. Strategies for the production of clinically relevant cell yields in vitro (>109 cells)4–6 often employ differentiation protocols based on principals of existing bioprocessing technologies which aim to scale traditional culture formats by producing batch cultures with increased volumes (>100mL).7 However, stem cells are sensitive to environmental perturbations, including manipulation of cell adhesions,8–10 morphogen availability,11,12 and mechanical forces,13–16 all of which may be modulated due to the hydrodynamic environments created in large volume mixed/stirred culture systems.
Fluid shear stress is a critical factor for establishing correct patterning and function during embryonic differentiation in vivo,17–19 and likewise impacts the differentiation of pluripotent and multipotent stem cells toward endothelial and hematopoietic lineages in two-dimensional, adherent monolayer cultures.20–24 Although monolayer cultures permit control and manipulation of the substrate properties, as well as uniform application of fluid shear and soluble factors, ESC differentiation is commonly initiated by the formation of three-dimensional cell aggregates, termed embryoid bodies (EBs); the cell-cell adhesions and signaling within three-dimensional spheroid structure recapitulate many aspects of tissue morphogenesis, as well as several signaling pathways of embryogenesis. Additionally, the differentiation of ESCs as multicellular aggregates in suspension is not constrained by surface area (as in adherent cultures), and therefore is readily scalable to larger volume bioreactors. However, complexity arising from the three-dimensional structure of EBs confounds analysis of environmental perturbations in hydrodynamic cultures.25
Although previous studies have noted that mixed cultures do not inhibit the pluripotent differentiation potential of ESCs,26–29 several groups have reported changes in the relative efficiency of endogenous differentiation to various cell types, including cardiac and hematopoietic phenotypes, when EBs are cultured within hydrodynamic environments compared to those maintained in static suspension culture.30–32 Additionally, various parameters of the hydrodynamic environment (such as mixing speed and bioreactor type) differentially impact ESC commitment.31,33 Scalable ESC cultures, however, generally rely on the spontaneous formation of cell aggregates within stirred bioreactors, which also results in changes in EB formation kinetics and size as a function of bioreactor geometry, configuration and mixing frequency.27–29,34 In rotary orbital cultures, slower rotation frequency (e.g. 25 rpm) resulted in accelerated EB formation and larger EBs, compared to aggregates formed at faster rotary orbital speeds (e.g. 40–55 rpm).31 Recent studies have reported that changes in EB size may influence ESC differentiation, which has been attributed to changes in cell signaling and polarity within the spheroid.35–38 Similarly, the kinetics of cell-cell association during EB formation may alter downstream signaling related to cadherin assembly.39,40 Overall, these studies indicate that hydrodynamic effects on differentiation may be confounded by changes in EB formation kinetics and size in different mixed bioreactor cultures, thus warranting a more systematic study of ESC differentiation in hydrodynamic environments.
Recently, new technologies have been developed which are capable of reproducibly forming large yields of uniform-sized EBs by forcing aggregation of the cells in micro-wells or on micro-patterned substrates.36,37,41–43 Such technologies enable the study of EB differentiation in different hydrodynamic environments, because forced aggregation techniques can standardize the kinetics of EB formation, independent of hydrodynamics. Additionally, pre-forming EBs prior to introduction into mixed environments permits the maintenance of uniform populations throughout the culture period and across different conditions. The objective of this study was to analyze the impact of hydrodynamic environments on differentiation of ESCs, independent of EB size, by developing a method for maintaining uniform EB populations in different mixed suspension cultures. EBs were formed via forced aggregation in polydimethylsiloxane (PDMS) micro-wells, and maintained through the course of differentiation in rotary orbital suspension cultures.30,31,34 The impact of mixing conditions on size-controlled EBs was analyzed by morphometric, morphological, and phenotypic metrics. The results from this study demonstrate that maintaining uniform populations of pre-formed EBs in different hydrodynamic environments overall increases the uniformity of morphogenesis and differentiation within hydrodynamic cultures, with relatively subtle changes in the differentiation toward certain lineages. The differentiation format developed in this study enables a more systematic understanding of the modulation of ESC differentiation within hydrodynamic environments and provides an alternative technique for the standardization of scalable ESC cultures.
Results
Control of EB formation and size by forced aggregation
In order to investigate the ability to control EB formation and size in scalable suspension cultures, EBs were formed via centrifugation of cells into 400 μm diameter PDMS micro-wells (Aggrewellt) prior to transfer into rotary orbital cultures.43 Micro-well aggregation produced large, homogeneous yields of EBs of defined size after 24 h of formation, which were subsequently transferred into bulk suspension cultures (Fig. 1A). Additionally, the EB size was modulated by altering the seeding density of cells in each well (500, 1000, and 2000 cells per well), which produced uniform populations that could be distinguished by morphometric image analysis of EB cross sectional area. Histograms of EB area demonstrated narrow distributions, indicating homogeneity of resulting EBs, as well as distinct peaks, which distinguish the populations formed with different cell seeding densities (Fig. 1B).
Fig. 1. Micro-well formation and rotary maintenance of size-controlled EBs.
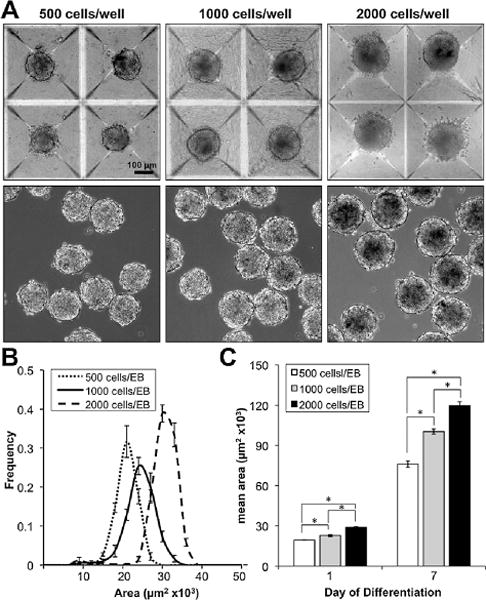
(A) Forced aggregation of ESCs into micro-wells to form size-controlled EBs was accomplished via centrifugation of single cells, followed by 24 h of incubation in wells, and subsequent transfer to rotary orbital suspension (scale bar = 100 μm). (B) After 24 h, the resulting EB populations exhibited narrow size distributions with distinct peaks modulated by ESC seeding density, and (C) relative size differences were maintained over 7 days of differentiation in rotary orbital suspension culture.
EBs formed with different seeding densities were maintained in rotary orbital culture at 45 rpm, in order to analyze the changes in EB morphogenesis during differentiation in mixed suspension. During the first 7 days of differentiation, 500-, 1000-, and 2000-cell EBs increased in size; however, the relative size differences initially exhibited by EBs from different seeding densities were maintained, indicated by statistical differences (P < 0.05) in EB cross sectional area immediately after formation (day 1) and after 7 days of differentiation (Fig. 1C). These results indicate that the size and formation kinetics of EBs can be reproducibly controlled using micro-well technologies, and that distinct populations of pre-formed EBs can be maintained in rotary orbital suspension cultures as a means to regulate and standardize EB size.
To further investigate the feasibility of maintaining preformed EBs in different hydrodynamic conditions, EBs formed with 1000 cells per aggregate in micro-wells were transferred to static or rotary orbital cultures at a range of speeds from 25 to 65 rpm. Visual analysis after 24 h in suspension indicated that agglomeration of individual pre-formed EBs occurred in static cultures and at the lowest (25 rpm) rotary orbital speed (data not shown). In contrast, when maintained at 45 rpm and 65 rpm, EBs maintained uniform spherical populations, and did not exhibit evidence of substantial agglomeration. After 7 days of differentiation, EBs at 45 and 65 rpm remained homogeneous and appeared to be similar in size; however, agglomeration of individual EBs led to spherical, but substantially larger EBs at 25 rpm and large, irregularly-sized EBs, in static cultures (Fig. 2A). The visual observations regarding EB agglomeration were verified by quantification of EB size and yield over 14 days of differentiation (Fig. 2C). In 45 and 65 rpm rotary orbital conditions, EB size significantly increased (P < 0.05) at all time points compared to day 1, (immediately after micro-well formation), but did not exhibit size differences between the hydrodynamic culture conditions at 45 and 65 rpm. Additionally, similar EB yield was maintained in both 45 and 65 rpm rotary conditions; the yield at day 7 of differentiation was not statistically different compared to day 1, and was not different between rotary conditions. Taken together, these results indicate that the hydrodynamic conditions imposed at 45 rpm and 65 rpm prevent agglomeration of pre-formed EBs and enable the culture of uniform EBs in different mixed culture environments. The combination of micro-well formation with hydrodynamic cultures at 45 and 65 rpm was therefore chosen for the further analysis of ESC morphogenesis and differentiation of uniformly sized EBs maintained in different hydrodynamic environments.
Fig. 2. Maintenance of EB populations in different hydrodynamic environments.
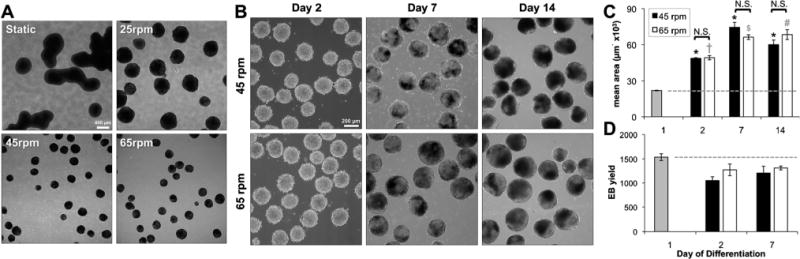
(A) EBs maintained in rotary orbital suspension exhibited changes in size and shape after 7 days of differentiation. The irregular shape and large size of EBs maintained in static and low rotary orbital speed (25 rpm) conditions indicated agglomeration of individual EBs (scale bar = 400 μm). (B) At 45 and 65 rpm, however, EBs maintained uniform, similarly-sized populations through 14 days (scale bar = 200 μm). (C) Although EB size increased during the course of differentiation, the EB area at 45 and 65 rpm was not significantly different. (D) Additionally, the total number of EBs did not change over time or between conditions, indicating minimal agglomeration of individual pre-formed EBs. * = p < 0.05 (45 rpm) compared to all time points; + = p < 0.05 (65 rpm) compared to all time points; $ = p < 0.05 (65 rpm) compared to all time points except day 14; # = p < 0.05 (65 rpm) compared to all time points except day 7; N.S. = no statistical significance.
Impact of hydrodynamic cultures on ESC morphogenesis
Analyses of cell and EB morphology as well as cell viability after 14 days of differentiation suggested similarities between different populations of pre-formed EBs, despite differences in hydrodynamic culture conditions. Morphological features of EBs, examined following H&E staining of histological sections, indicated the appearance of differentiated cell populations, including cells exhibiting tightly packed epithelial-like morphologies, as well as more spread-out mesenchymal-like morphologies in all EB populations; however, no obvious differences were noted between EBs cultured in different hydrodynamic conditions (Supplemental Fig. 1A). Additionally, single cell analysis via flow cytometry indicated similar refractive properties of single cells, with respect to both forward and side scatter metrics, which are indicative of cell size and complexity respectively (Supplemental Fig. 1B). Moreover, there were no differences in the proportion of live and dead cells stained with calcein AM and ethidium homodimer-1, respectively, between EBs from different hydrodynamic conditions and the yield of cells per EB remained similar between hydrodynamic conditions (Supplemental Fig. 1C). Altogether, these results indicate that changes in the hydrodynamic environment did not globally alter the ability of ESCs to undergo morphogenesis within EBs through 14 days of differentiation.
Impact of hydrodynamic cultures on gene expression
Changes in gene expression over the course of differentiation were analyzed using an “embryonic stem cell” PCR array (SABiosciences), which included 84 genes commonly associated with the maintenance of pluripotency and differentiation to the three germ lineages. Heatmap visualization of the gene expression profiles indicated that there were distinct subsets of genes that exhibited increased (Fig. 3A, red) or decreased (Fig. 3A, green) expression in EB cultures compared to the starting population of undifferentiated ESCs. After 7 days of differentiation, 20 genes exhibited a minimum of 3-fold increase compared to ESCs, whereas 15 genes were decreased greater than 3-fold compared to ESCs. Two-way hierarchical clustering of all genes and experimental groups separated the individual time points, reflecting distinct changes in gene expression that arose during the course of differentiation; however, within individual time points, the gene expression profiles of the experimental groups did not cluster separately, which suggested a lack of global distinction between the EB populations subjected to different hydrodynamic conditions. Specific analysis of the subsets of genes that were differentially expressed compared to ESCs (minimum of 1.5 fold change, and p < 0.05 compared to ESCs) indicated that ~93% of the genes (25/27) that were decreased compared to ESCs after 7 days of differentiation were common to both hydrodynamic conditions (Fig. 3B). Of the genes that were significantly decreased compared to ESCs, many were transcription factors (Oct-4, Nanog, Sox2) and signaling molecules (Fgf4, Lefty1, Lefty2, Nodal) known to be associated with the undifferentiated, pluripotent state of ESCs (Supplemental Fig. 2A). Temporal gene expression analyzed using RT-PCR with independent primers verified the decrease in Oct-4 over the time course of differentiation (Supplemental Fig. 2B) and stably transduced cells expressing GFP driven by the Oct-4 promoter exhibited a decrease in the proportion of GFP+ cells (Supplemental Fig. 2C). Additionally, ~44% (12/27) of genes that were increased (minimum of 1.5 fold change, and p < 0.05 compared to ESCs) during EB differentiation were commonly increased in both hydrodynamic conditions (Fig. 3B), including markers of differentiated phenotypes related to epiblast (fibroblast growth factor 5, Fgf5), endoderm (alpha fetoprotein, AFP; forkhead box A2, Foxa2; Sox17), and mesoderm lineages (Gata4, Podxl1; VE-cadherin, Cdh5; Flt1), as well as signaling molecules, such as Noggin (Nog) and factors such as Laminin (Lamb1-1), which are related to the morphogenesis of ESCs in EB differentiation.44,45 Overall, compared to ESCs, the global patterns of gene expression in EBs from both hydrodynamic conditions suggested the progressive loss of pluripotency, with an increase in markers related to differentiation of ESCs.
Fig. 3. Gene expression profiles of size controlled EBs in hydrodynamic environments.
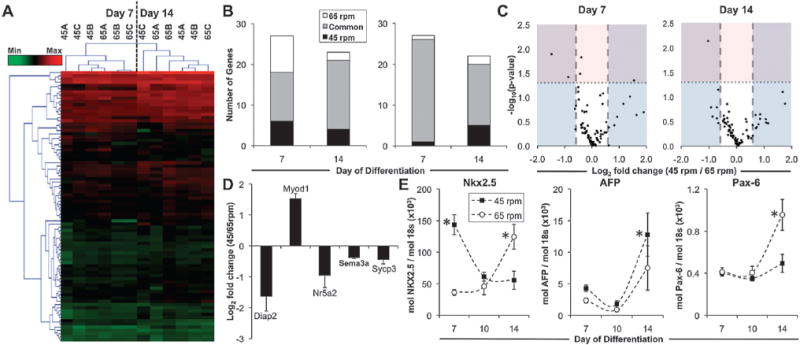
Global gene expression of 84 genes related to ESC pluripotency and differentiation was assessed in EBs from 45 rpm and 65 rpm after 7 and 14 days of differentiation using PCR Arrays. (A) Heatmap visualization of gene expression demonstrated (B) distinct subsets of genes up- and down-regulated compared to ESCs, with (C) several significantly modulated genes being regulated differently between the 45 rpm and 65 rpm conditions. Although several genes exhibited differences in fold change (>1.5 fold, blue; represented as the fold change of 45 rpm/65 rpm), few genes exhibited statistically significant differences (P < 0.05, pink) between the hydrodynamic conditions, (D) including 4 genes increased in 65 rpm, and 1 gene increased in 45 rpm after 7 days of differentiation. (E) Rotary conditions also modulated the temporal expression of genes from the mesoderm (Nkx2.5), endoderm (AFP), and ectoderm (Pax-6) lineages. * P < 0.05.
However, of the genes that were increased compared to ESCs, there were also several genes (45 rpm: 6/27; 65 rpm: 9/27) that were uniquely increased in the specific hydrodynamic conditions, including markers of endoderm (Gata6, Ins2, Iapp, Serpina1a) and mesoderm (Kit, Grb7, Hba-x, Hba-y) lineages, which warranted a direct comparison of the gene expression profiles in different hydrodynamic environments. Comparisons of the expression patterns between the hydrodynamic conditions indicated that the profiles were largely similar at both time points, as evidenced by a direct, positive correlation between data from 45 rpm and 65 rpm (R2 = 0.95, day 7; R2 = 0.95, day 14). Fold changes in gene expression between hydrodynamic conditions (represented as 45 rpm/65 rpm) indicated that a number of genes were modulated greater than 1.5 fold (14 genes at day 7; 19 genes at day 14) (Fig. 3C; blue). However, only 5 genes exhibited statistically significant changes in gene expression between hydrodynamic conditions at day 7 of differentiation, and 1 gene (out of 84) was significantly modulated after 14 days (Fig. 3C; pink). Of the 5 genes significantly changed after 7 days of differentiation, 4 were increased in 65 rpm (2 genes >1.5 fold change), whereas 1 gene was increased in 45 rpm (Fig. 3D). Genes related to pluripotency and germ cell differentiation, Nuclear receptor family 5, group A, member 2 (Nr5a2)46 and synaptonemal complex protein 3 (Sycp3),47 respectively, exhibited overall decreased expression compared to ESCs, and both were significantly increased in 65 rpm compared to 45 rpm. Diaphanous homologue 2 (Diap2), which encodes a downstream effector of the RhoA signaling pathway, and is implicated in cytoskeletal remodeling and stabilization of adherens junctions, was also increased in the 65 rpm rotary orbital condition.48 Additionally, contrasting patterns of gene expression related to muscle (Myod1; increased in 45 rpm) and neural differentiation (Sema3a; increased in 65 rpm), supported the divergent differentiation, albeit modest based on the sub-set of genes examined, of size-controlled EBs cultured in different hydrodynamic conditions (Supplemental Fig. 3).
Individual RT-PCR using independently designed primers also confirmed the temporal modulation of genes from all germ layers. Nkx2.5, a mesoderm marker which is increased during early cardiac differentiation, was significantly increased at day 7 in 45 rpm, and significantly increased in 65 rpm at day 14 (Fig. 3E); the pattern of Nkx2.5 expression indicates a possible temporal shift in the onset and progression of mesoderm differentiation within the hydrodynamic conditions. Further, the gene expression for alpha fetoprotein (AFP), indicative of endoderm differentiation, was significantly increased after 7 days of differentiation in 45 rpm EBs, whereas Pax-6, an ectoderm marker, was increased in 65 rpm rotary conditions after 14 days (Fig. 3E). Overall, gene expression patterns suggested that size-controlled EBs undergo similar changes related to loss of pluripotency and differentiation, with a subset of genes exhibiting subtle changes in temporal kinetics and relative expression levels due to culture in distinct hydrodynamic environments.
Impact of hydrodynamics on visceral endoderm and cardiomyogenic differentiation
EB morphogenesis commonly occurs through the differentiation of the cells at the exterior of the spheroid toward the visceral endoderm lineage.10 Although altered efficiency of ESC differentiation toward visceral endoderm in hydrodynamic environments has been reported,31 the exterior endoderm has also been suggested as a mechanism for changes in differentiation as a result of EB size.38 Therefore, flow cytometry and immunostaining were used to analyze the proportion and localization of visceral endoderm cells within EBs from the hydrodynamic conditions at 45 and 65 rpm. Flow cytometry was performed using cells transduced to express GFP under control of the AFP promoter. Prior to EB formation and differentiation, ESCs expressed low levels of GFP; however, after a week of EB differentiation, increased proportions of cells expressed GFP (>30% GFP+ by day 7 of differentiation), indicating differentiation of ESCs toward the visceral endoderm lineage. Cytometry data from the different hydrodynamic conditions demonstrated both differences in the total number of cells within the gate (45.5 ± 1.6% at 45 rpm compared to 37.6 ± 0.9% at 65 rpm after 7 days of differentiation), as well as differences in the intensity and distribution of expression of GFP expression (geometric mean relative fluorescence values of 956.5 ± 11.6 at 45 rpm compared to 897.0 ± 9.5 at 65 rpm; coefficient of variation of 58.7 ± 1.0 at 45 rpm and compared to 64.7 ± 0.6 at 65 rpm) (Fig. 4A). Quantification of the proportion of GFP+ cells over time also demonstrated a greater number of GFP+ cells after 7, 10 and 14 days of differentiation in the 45 rpm rotary condition compared to 65 rpm (Fig. 4C). Additionally, as the exterior visceral endoderm cells have been reported to direct mesoderm differentiation via paracrine signaling,49 the development of spontaneous contractile foci, indicative of cardiomyocyte differentiation, was quantified (Fig. 4D). Consistent with the increase in visceral endoderm, there was a significant increase in the proportion of EBs maintained at 45 rpm that exhibited contractile foci after 10, 12, and 14 days of differentiation (46.8% at 45 rpm compared to 2.4% at 65 rpm after 10 days of differentiation). Overall, the relative efficiencies of differentiation toward visceral endoderm and cardiomyogenic phenotypes were altered due to perturbations in the hydrodynamic culture environment.
Fig. 4. Modulation of visceral endoderm and cardiac mesoderm differentiation in response to rotary orbital culture.
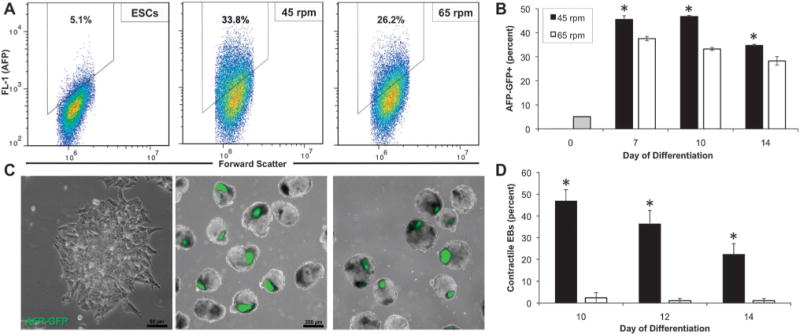
(A) AFP-GFP transduced ESCs exhibited few GFP+ cells when maintained as an undifferentiated monolayer; however after 14 days of differentiation, EBs from rotary orbital cultures increased expression of GFP (AFP), with (B) EBs cultured at 45 rpm consistently exhibiting increased expression of GFP+ cells after 7, 10 and 14 days of differentiation. (C) Within EBs, the GFP+ cells appeared to cluster in distinct areas, which co-localized with dark regions visualized under phase (overlay) (scale bars = 50 μm, 200 μm). (D) The proportion of EBs exhibiting spontaneous contractile activity was increased following culture 45 rpm compared to 65 rpm after 10, 12, and 14 days of differentiation. * p < 0.05.
Discussion
The results of this study demonstrate the ability to systematically decouple various parameters (EB size, formation, and differentiation) which can be modulated by complex hydrodynamic environments, in order to examine the impact on ESC differentiation. This study and others establish the ability to reproducibly control the initial size of EBs by forced aggregation methods in scalable formats.36,37,41–43,50 The combination of micro-well formation of EBs with maintenance in rotary orbital suspension permits the prolonged culture of uniform EBs within a range of hydrodynamic environments, enabling increased standardization of scalable differentiation protocols. Recent studies discuss the modulation of ESC differentiation as a function of EB size,35–38 and are consistent with similar reports demonstrating changes in stem and progenitor cell differentiation as a result of cytoskeletal organization and localization of cells within 3D aggregates.51,52 Bauwens et al. reported that modulation of EB size alters the proportion of endoderm on the exterior surface of the EB, which results in increased mesoderm differentiation within EBs due to paracrine signaling.38 The system developed and discussed in this study permits the formation of EBs with a desired initial size, and maintenance of relative EB size over time, which may be an important factor capable of aiding or hindering differentiation toward distinct cell lineages.
Computational fluid dynamic modeling to determine the shear profiles within rotary orbital cultures previously demonstrated that between cultures at 40 rpm and 55 rpm, there is an increase of approximately 1 dyn/cm2 within the central region of the plate traversed by EBs during the rotation, indicating changes in the fluid shear environment due to modulation of rotary orbital speed.31 The data presented here demonstrate that pre-formed EBs can be maintained at higher rotational frequencies, up to 65 rpm, which were previously reported to not support spheroid formation from single cells.31 Consistent with previous reports, decreased rotary speeds result in larger EBs, due to the agglomeration of individual pre-formed EBs,31,34 indicating that a hydrodynamic threshold (apparently near 45 rpm) may be necessary to maintain homogeneous populations of individual EBs. Similarly, studies have demonstrated decreased size of multicellular aggregates at higher mixing speeds,31,34,53,54 presumably due to modulation of cell collision frequencies, as well as by perturbation of the kinetics of E-cadherin binding.25 Although it remains possible that hydrodynamics may modulate the growth kinetics of ESCs within EBs, the lack of observed differences in EB size and the average number of cells per EBs over 14 days of differentiation suggest that homogeneous populations of EBs can be maintained within different hydrodynamic conditions.
In the context of the present and past experiments, it is difficult to separate the effects of fluid shear and transport on EBs within hydrodynamic environments. In monolayer culture of ESCs, fluid shear has been reported to impact hematopoietic and endothelial differentiation.20–24 However, the response to fluid shear in 3D cultures is more complex and much less defined. The work herein demonstrates that the modulation of hydrodynamic environments in size-controlled, three-dimensional cultures do not appear to dramatically impact overall morphogenesis or the gene expression profile of differentiating ESCs within EBs. It is important to note that, within the context of this study, a relatively small set of genes and phenotypes were analyzed, and it remains possible that there may be differences in other phenotypes that were not directly examined. Additionally, although dramatic changes in global expression patterns were not apparent, the subtle changes in gene expression exhibited due to hydrodynamic conditions may significantly impact cell specification. Interestingly, there was a change in the relative quantities of visceral endoderm cells, with 45 rpm conditions exhibiting an overall increase in visceral endoderm differentiation compared to 65 rpm. As fluid shear is expected to more directly impact the exterior of EBs, it is likely that visceral endoderm differentiation may be responsive to fluid shear mechanotransduction at the EB surface. Additionally, EBs cultured at 65 rpm exhibit significantly increased expression of diaphanous homolog 2 (Diap2), which is involved in microtubule organization and cell-cell contacts, thus indicating changes in cytoskeletal organization, which is a characteristic response to fluid shear in other cell types.55,56
This work also highlights shifts in the temporal kinetics of differentiation in hydrodynamic environments. The mixing conditions in bioreactors may have implications for paracrine signaling and for the delivery of exogenous soluble factors, due to changes in transport and receptor-ligand binding kinetics in hydrodynamic environments. Mixing may potentially impact the transport of nutrients, metabolites, and exogenous factors within the EB microenvironment. It is possible, therefore, that the influence of hydrodynamic cultures may be altered by using chemically defined media formulations, or by the addition of soluble factors to promote the endogenous differentiation of ESCs. Additionally, gradients of oxygen may arise as EBs reach a critical size limit (~300 μm in diameter).57 Hypoxic conditions have been associated with modulation in β-catenin signaling and cardiomyogenesis,58 as well as with expansion of pluripotent cell populations.59 Although there may be changes in fluid transport due to modulation of the mixing speeds, the normalization of multicellular aggregate size may attenuate some of the effects of transport seen in previous experiments where EBs were significantly larger at low mixing speeds.31,34 In the studies presented, EBs were approximately 300 μm in diameter by day 14 of culture, which indicates that few transport limitations likely arose within the system under the experimental conditions used. The decrease in both EB size and cell yield between days 7 and 14 of differentiation may be in part regulated by such transport limitations, indicating that cellular remodeling may dynamically alter transport within EBs. It is, however, difficult to quantitatively assess changes in transport gradients and availability of soluble factors in the EB microenvironment using traditional bioprocessing technologies. As many protocols rely on tightly controlled signaling of factors at different stages in differentiation, shifts in the temporal kinetics of differentiation may alter the efficacy of some directed differentiation approaches. Ultimately, the control of EB size prior to introduction into large scale hydrodynamic cultures may enable increased standardization, which is necessary for the efficacy and reproducibility of directed differentiation protocols in the context of scalable bioprocessing.
Conclusions
We have developed a novel culture platform which enables the study of ESC differentiation as 3D multicellular aggregates in hydrodynamic environments, independent of the effects of mixed cultures on EB formation kinetics and size. The results of this study indicate that, despite subtle changes in temporal differentiation toward certain lineages, EBs maintained under different hydrodynamic conditions exhibit similarities in morphogenesis and the overall differentiation profiles of ESCs. These data suggest that controlling EB formation upstream of hydrodynamic cultures may be amenable to standardization of ESC cultures in scalable suspension formats for bioprocessing applications.
Experimental
Embryonic stem cell culture
Murine ESCs (D3 cell line) were expanded on tissue culture treated polystyrene dishes (Corning Inc., Corning, NY) adsorbed with 0.1% gelatin (Mediatech, Manassas, VA). Undifferentiated ESC media consisted of Dulbecco’s Modified Eagle’s Medium (DMEM; Mediatech) containing 15% fetal bovine serum (Hyclone, Logan, UT), 100 U/mL penicillin (Mediatech), 100 μg μL−1 streptomycin (Mediatech), 0.25 μg Ml−1 amphotericin (Mediatech), 2mM L-glutamine (Mediatech), 1× MEM non-essential amino acid solution (Mediatech), 0.1 mM 2-mercaptoethanol (Fisher Scientific, Fairlawn, NJ), and 103 U/mL leukemia inhibitory factor (LIF; ESGRO, Millipore, Billerica, MA). The media was exchanged every other day, and cells were passaged at approximately 70% confluence.
Embryoid body formation
A single cell suspension of undifferentiated ESCs was obtained by dissociating monolayer cultures using 0.05% trypsin-EDTA (Mediatech). Forced aggregation of ESCs was accomplished by centrifugation (200 rcf) of ESCs into 400 μm diameter polydimethylsiloxane (PDMS) micro-wells (Aggrewellt™, Stem Cell Technologies, Vancouver, Canada) as previously reported.43 The cell inoculation concentration was modulated to seed wells with approximately 500, 1000 and 2000 cells each in differentiation medium, consisting of undifferentiated ESC media without LIF. After incubation for 24 h to allow formation, the resultant EBs were transferred to suspension culture (approximately 2000 EBs in 10 mL of media) in sterile 100 × 15 mm bacteriological grade polystyrene Petri dishes (BD, Franklin Lakes, NJ) and maintained on rotary orbital shakers34 at rotational frequencies of approximately 25, 45, and 65 rpm. For static cultures, EBs were transferred from the micro-wells to dishes coated with sterile 2% agar, which is used to prevent cell attachment to the dish. 90% of the media was exchanged every other day by gravity-induced sedimentation in a 15 mL conical tube. Suspension cultures were maintained through 14 days of differentiation.
Morphometric analysis
Phase images were acquired at days 1, 2, 7, 10 and 14 of differentiation using a Nikon TE 2000 inverted microscope (Nikon Inc., Melville, NY), equipped with a SpotFlex camera (Diagnostic Instruments, Sterling Heights, MI). The cross sectional area of EBs was measured by using a custom-written computational macro in ImageJ software (NIH), which delineates EB boundaries through a combination of thresholding, edge detection, and smoothing, as described previously.31 At least 100 EBs were analyzed for each sample (n = 3 independent samples per experimental condition), and the distribution of EBs was visualized by frequency histograms.
EB yield analysis
The total yield of EBs in each culture at days 1, 2, and 7 was manually counted by dilution of the total population and visual inspection using a Nikon Eclipse TS100 inverted phase microscope, which resulted in approximately 20–50 EBs per well of a 24 well non-tissue culture treated polystyrene plate. 3 separate counts were recorded for each sample (n = 3 independent samples per experimental condition).
Histology and immunohistochemistry
EBs were sampled from rotary cultures at days 7, 10 and 14 of differentiation, rinsed in PBS and fixed in 10% formalin (4% formaldehyde) for 45 min with rotation at room temperature. Fixed EBs were rinsed 3 times in PBS and embedded in Histogel® (Richard-Allen Scientific, Kalamazoo, MI) at 4 1C overnight. Samples were processed via a series of graded ethanol and xylene rinses and embedded in paraffin. Paraffin-embedded samples were sectioned with a thickness of 5 μm, using a rotary microtome (Microtom HM310). For histological analysis, samples were de-paraffinized by rinsing in a series of xylene and graded ethanol concentrations, followed by staining with hematoxylin and eosin (H&E). H&E stained sections were imaged using a Nikon Eclipse 80i microscope (Nikon, Inc.) equipped with a SpotFlex digital camera (Diagnostic Instruments).
PCR array and quantitative real time PCR
EBs were sampled at days 7, 10, and 14 of differentiation for PCR gene expression analysis. For real time (RT)-PCR, total RNA was isolated from EBs using the RNeasy Mini kit (Qiagen Inc, Valencia, CA), followed by synthesis of cDNA from 1 μg of RNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). To prepare cDNA for use in PCR Arrays, the RT2 First Strand Kit (SABiosciences, Frederick, MD) was used with 1 μg of total RNA. RT-PCR was performed with SYBR Green technology and the MyIQiCycler (Bio-Rad), using primers that were designed with Beacon Designer software (Invitrogen) and validated against cell-specific controls. Primers for both pluripotency (Oct-4) and differentiated cell phenotypes (Nkx2.5, AFP, Pax-6) were analyzed by RT-PCR. The relative concentration of each gene transcript was calculated based on a standard curve, and normalized with respect to expression of the housekeeping gene 18s.60
Global gene expression was analyzed using the Mouse Embryonic Stem Cell RT2Profiler™ PCR Array (SABiosciences) which includes 84 genes related to pluripotency and markers of differentiation toward all lineages, as well as 5 housekeeping genes and several PCR controls. RT-PCR was performed on the arrays using a MyIQ iCycler (Bio-Rad) with SYBR green based RT2 qPCR Master Mix (SABiosciences). All sample data were visually inspected post-run, including both the amplification phases and melt curves, to ensure efficacy of the individual primers. To account for changes in PCR efficiency between PCR runs, data from individual runs were normalized with respect to the geometric average of 3 housekeeping genes. The PCR arrays included 5 housekeeping genes (Gusb, Hprt1, Hsp90ab1, Gapdh, Actb), and 3 were chosen based on the calculation of gene stability that yielded a combination with the least variability (Gusb, Hsp90ab1, Actb).61 The normalized Ct values were then used to calculate the relative fold changes in gene expression relative to the group of interest (45 rpm/ESCs, 65 rpm/ESCs, 45 rpm/65 rpm).60 The Genesis software package was used to visualize heatmaps of gene expression, represented as the log2 transform of fold change, and to calculate two-dimensional hierarchical clustering between genes and experimental groups, based on Euclidean distance and average linkage clustering. Statistical differences were calculated using an independent samples T-test for each gene, and genes with P < 0.05 were determined to be statistically significant.
Flow cytometry
EBs were rinsed with PBS and dissociated at 37 °C with rotation, using 0.25% trypsin-EDTA and triturating every 10 min for a total of 30 min in order to obtain a single cell suspension. Cell solutions were centrifuged at 1000 rcf for 5 min, and rinsed 3 times in PBS. Flow cytometry was conducted using an Accuri C6 cytometer (Accuri Cytometers, Ann Arbor, MI), with a minimum of 30 000events within the FSC/SSC gate collected per sample (n = 3 independent samples experimental per condition). After obtaining a single cell suspension, a LIVE/DEAD® kit (Invitrogen) was used to assess cell viability. Briefly, cells were resuspended in 1 mL of PBS and stained for 15 min using 2 μL of 50 μM calcein AM and 4 μL of 2 mM ethidium homodimer-1 (EthD-1). Unstained and single stained populations were used for gating and compensation. Forward scatter (FSC) and side scatter (SSC) were analyzed within the live (calcein AM+, EthD-1−) cell populations.
D3 mESCs were transduced using plasmids encoding EGFP under the AFP (pAFPpHyg/EGFP; provided by Iwao Ikai, M.D., Ph.D., Kyoto University, Kyoto Japan) or Oct-4 (phOCT3-EGFP1; provided by Wei Cui, Ph.D., Imperial College, London, UK) promoters, and stable clones were established following geneticin (G-418) selection. AFP-GFP+ and Oct4-GFP+ cells were analyzed by flow cytometry prior to differentiation, and after 7, 10, and 14 days of differentiation. The dissociated cell population was initially identified by gating on the FSC/SSC plot. Within the FSC/SSC gate, polygonal gating was used on the FSC/FL-1 (480nm excitation; 530 ± 15nm emission) plots, to include 2% of the untransduced (D3 mESCs; negative control) population using FlowJo software (Tree Star, Inc., Ashland, OR).
Analysis of spontaneous contractile activity
After 7 days of differentiation in rotary suspension, single EBs were transferred to individual wells of 0.1% gelatin-coated 48 well tissue culture dishes in a total volume of 500 mL of differentiation media, to promote adherence and spreading of cells (30 EBs per sample, n = 3 independent samples per experimental condition). The development of contractile activity, indicative of cardiomyogenic differentiation, was monitored at days 10, 12, and 14 by visual inspection using a Nikon Eclipse TS100 inverted phase microscope. Blinded counts of contractile activity were conducted by scoring EBs with one or more beating foci were as “contractile”, and representing the contractile EBs relative to the total plated EB population.
Statistical analysis
All experiments were performed with triplicate samples with independent conditions and data is represented as the mean of independent replicates (n = 3) ± standard error. Statistical analysis was performed using an independent samples t-test between the conditions (45 and 65 rpm), assuming either equal or unequal variances, depending on the results from Levene’s equality of variances test. Statistical analysis comparing changes over time was conducted using a one-way ANOVA, with a post-hoc Tukey or Mann-Whitney U test for comparison of individual samples, depending on the equality of variances.
Supplementary Material
Insight, innovation, integration.
Hydrodynamic cultures are often employed in large scale bioprocessing systems, and may impact the differentiation of embryonic stem cells maintained in non-adherent stirred cultures as 3D cell aggregates. The study presented herein demonstrates the utility of forming and maintaining uniform populations of EBs, in order to understand the influence of hydrodynamic environments on three-dimensional ESC differentiation. Although size-controlled embryoid bodies in mixed cultures undergo similar differentiation processes by many phenotypic markers, several subtle yet significant changes are induced by hydrodynamic environments. The results of this study highlight an important role that hydrodynamics may play in regulating cell phenotype, and therefore may ultimately enable increased standardization of large scale bioprocessing applications for ESC expansion and differentiation.
Acknowledgments
The authors thank Dr Wei Cui at Imperial College and Dr Iwao Ikai at Kyoto University for graciously providing Oct-4-EGFP and AFP-EGFP plasmids, respectively. We thank Marissa Cooke at Georgia Institute of Technology for technical assistance, as well as Dr Priya Baraniak and Dr Krista Fridley for assistance with preparation of this manuscript. This work was supported by the National Institutes of Health (NIH R01 EB010061) and by the National Science Foundation (NSF CBET 0939511). M.A.K. is supported by a NSF graduate research fellowship.
References
- 1.Evans MJ, Kaufman MH. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Martin GR. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 4.Lock LT, Tzanakakis ES. Tissue Eng. 2007;13:1399–1412. doi: 10.1089/ten.2007.0047. [DOI] [PubMed] [Google Scholar]
- 5.Jing D, Parikh A, Canty JM, Tzanakakis ES. Tissue Eng, Part B: Rev. 2008;14:393–406. doi: 10.1089/ten.teb.2008.0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tzanakakis ES, Hess DJ, Sielaff TD, Hu WS. Annu Rev Biomed Eng. 2000;2:607–632. doi: 10.1146/annurev.bioeng.2.1.607. [DOI] [PubMed] [Google Scholar]
- 7.Kirouac DC, Zandstra PW. Cell Stem Cell. 2008;3:369–381. doi: 10.1016/j.stem.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Fox V, Gokhale PJ, Walsh JR, Matin M, Jones M, Andrews PW. Stem Cells. 2008;26:715–723. doi: 10.1634/stemcells.2007-0368. [DOI] [PubMed] [Google Scholar]
- 9.Krtolica A, Genbacev O, Escobedo C, Zdravkovic T, Nordstrom A, Vabuena D, Nath A, Simon C, Mostov K, Fisher SJ. Stem Cells. 2007;25:2215–2223. doi: 10.1634/stemcells.2007-0230. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Chen Y, Schéele S, Arman E, Haffner-Krausz R, Ekblom P, Lonai P. J Cell Biol. 2001;153:811–822. doi: 10.1083/jcb.153.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desbordes SC, Placantonakis DG, Ciro A, Socci ND, Lee G, Djaballah H, Studer L. Cell Stem Cell. 2008;2:602–612. doi: 10.1016/j.stem.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nostro MC, Cheng X, Keller GM, Gadue P. Cell Stem Cell. 2008;2:60–71. doi: 10.1016/j.stem.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flaim CJ, Chien S, Bhatia SN. Nat Methods. 2005;2:119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 14.Anderson D, Levenberg S, Langer R. Nat Biotechnol. 2004;22:863–866. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 15.Fu J, Wang YK, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. Nat Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 17.Adamo L, Naveiras O, Wenzel PL, McKinney-Freeman S, Mack PJ, Gracia-Sancho J, Suchy-Dicey A, Yoshimoto M, Lensch MW, Yoder MC, García-Cardeña G, Daley GQ. Nature. 2009;459:1131–1135. doi: 10.1038/nature08073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.North TE, Goessling W, Peeters M, Li P, Ceol C, Lord AM, Weber GJ, Harris J, Cutting CC, Huang P, Dzierzak E, Zon LI. Cell. 2009;137:736–748. doi: 10.1016/j.cell.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hove JR, Köster RW, Forouhar AS, Acevedo-Bolton G, Fraser SE, Gharib M. Nature. 2003;421:172–177. doi: 10.1038/nature01282. [DOI] [PubMed] [Google Scholar]
- 20.Masumura T, Yamamoto K, Shimizu N, Obi S, Ando J. Arterioscler, Thromb, Vasc Biol. 2009 doi: 10.1161/ATVBAHA.109.193185. [DOI] [PubMed] [Google Scholar]
- 21.Zeng L, Xiao Q, Margariti A, Zhang Z, Zampetaki A, Patel S, Capogrossi MC, Hu Y, Xu Q. J Cell Biol. 2006;174:1059–1069. doi: 10.1083/jcb.200605113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Riha GM, Yan S, Li M, Chai H, Yang H, Yao Q, Chen C. Arterioscler, Thromb, Vasc Biol. 2005;25:1817–1823. doi: 10.1161/01.ATV.0000175840.90510.a8. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto K, Sokabe T, Watabe T, Miyazono K, Yamashita JK, Obi S, Ohura N, Matsushita A, Kamiya A, Ando J. Am J Physiol: Heart Circ Physiol. 2004;288:H1915–1924. doi: 10.1152/ajpheart.00956.2004. [DOI] [PubMed] [Google Scholar]
- 24.Ahsan T, Nerem RM. Tissue Eng, Part A. 2010 doi: 10.1089/ten.tea.2010.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinney MA, Sargent CY, McDevitt TC. Tissue Eng, Part B: Rev. 2011 doi: 10.1089/ten.teb.2011.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fok EYL, Zandstra PW. Stem Cells. 2005;23:1333–1342. doi: 10.1634/stemcells.2005-0112. [DOI] [PubMed] [Google Scholar]
- 27.Cameron CM, Hu WS, Kaufman DS. Biotechnol Bioeng. 2006;94:938–948. doi: 10.1002/bit.20919. [DOI] [PubMed] [Google Scholar]
- 28.Yirme G, Amit M, Laevsky I, Osenberg S, Itskovitz-Eldor J. Stem Cells Dev. 2008;17:1227–1241. doi: 10.1089/scd.2007.0272. [DOI] [PubMed] [Google Scholar]
- 29.Gerecht-Nir S, Cohen S, Itskovitz-Eldor J. Biotechnol Bioeng. 2004;86:493–502. doi: 10.1002/bit.20045. [DOI] [PubMed] [Google Scholar]
- 30.Sargent C, Berguig G, McDevitt T. Tissue Eng A. 2009;15:331–342. doi: 10.1089/ten.tea.2008.0145. [DOI] [PubMed] [Google Scholar]
- 31.Sargent CY, Berguig GY, Kinney MA, Hiatt LA, Carpenedo RL, Berson RE, McDevitt TC. Biotechnol Bioeng. 2010;105:611–626. doi: 10.1002/bit.22578. [DOI] [PubMed] [Google Scholar]
- 32.Liu H, Lin J, Roy K. Biomaterials. 2006;27:5978–5989. doi: 10.1016/j.biomaterials.2006.05.053. [DOI] [PubMed] [Google Scholar]
- 33.Fridley KM, Fernandez I, Li MT, Kettlewell RB, Roy K. Tissue Eng, Part A. 2010 doi: 10.1089/ten.tea.2010.0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carpenedo RL, Sargent CY, McDevitt TC. Stem Cells. 2007;25:2224–2234. doi: 10.1634/stemcells.2006-0523. [DOI] [PubMed] [Google Scholar]
- 35.Mohr JC, Zhang J, Azarin SM, Soerens AG, De Pablo JJ, Thomson JA, Lyons GE, Palecek SP, Kamp TJ. Biomaterials. 2010;31:1885–1893. doi: 10.1016/j.biomaterials.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang YS, Chung BG, Ortmann D, Hattori N, Moeller HC, Khademhosseini A. Proc Natl Acad Sci U S A. 2009;106:16978–16983. doi: 10.1073/pnas.0905550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bauwens CL, Peerani R, Niebruegge S, Woodhouse KA, Kumacheva E, Husain M, Zandstra PW. Stem Cells. 2008;26:2300–2310. doi: 10.1634/stemcells.2008-0183. [DOI] [PubMed] [Google Scholar]
- 38.Bauwens CL, Song H, Thavandiran N, Ungrin M, Massé S, Nanthakumar K, Seguin C, Zandstra PW. Tissue Eng, Part A. 2011 doi: 10.1089/ten.TEA.2010.0563. [DOI] [PubMed] [Google Scholar]
- 39.Ueno S, Weidinger G, Osugi T, Kohn AD, Golob JL, Pabon L, Reinecke H, Moon RT, Murry CE. Proc Natl Acad Sci U S A. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.ten Berge D, Koole W, Fuerer C, Fish M, Eroglu E, Nusse R. Cell Stem Cell. 2008;3:508–518. doi: 10.1016/j.stem.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park J, Cho CH, Parashurama N, Li Y, Berthiaume F, Toner M, Tilles AW, Yarmush ML. Lab Chip. 2007;7:1018–1028. doi: 10.1039/b704739h. [DOI] [PubMed] [Google Scholar]
- 42.Mohr JC, De Pablo JJ, Palecek SP. Biomaterials. 2006;27:6032–6042. doi: 10.1016/j.biomaterials.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 43.Ungrin MD, Joshi C, Nica A, Bauwens C, Zandstra PW. PLoS One. 2008;3:e1565. doi: 10.1371/journal.pone.0001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pera MF, Andrade J, Houssami S, Reubinoff B, Trounson A, Stanley EG, Ward-van Oostwaard D, Mummery C. J Cell Sci. 2004;117:1269–1280. doi: 10.1242/jcs.00970. [DOI] [PubMed] [Google Scholar]
- 45.Smyth N, Vatansever HS, Murray P, Meyer M, Frie C, Paulsson M, Edgar D. J Cell Biol. 1999;144:151–160. doi: 10.1083/jcb.144.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heng JCD, Feng B, Han J, Jiang J, Kraus P, Ng JH, Orlov YL, Huss M, Yang L, Lufkin T, Lim B, Ng HH. Cell Stem Cell. 2010;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 47.Toyooka Y, Tsunekawa N, Akasu R, Noce T. Proc Natl Acad Sci U S A. 2003;100:11457–11462. doi: 10.1073/pnas.1932826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sahai E, Marshall CJ. Nat Cell Biol. 2002;4:408–415. doi: 10.1038/ncb796. [DOI] [PubMed] [Google Scholar]
- 49.Mummery C, Ward-van Oostwaard D, Doevendans P, Spijker R, van den Brink S, Hassink R, van der Heyden M, Opthof T, Pera M, de la Riviere A, Passier R, Tertoolen L. Circulation. 2003;107:2733–2740. doi: 10.1161/01.CIR.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- 50.Niebruegge S, Bauwens CL, Peerani R, Thavandiran N, Masse S, Sevaptisidis E, Nanthakumar K, Woodhouse K, Husain M, Kumacheva E, Zandstra PW. Biotechnol Bioeng. 2009;102:493–507. doi: 10.1002/bit.22065. [DOI] [PubMed] [Google Scholar]
- 51.Ruiz SA, Chen CS. Stem Cells. 2008;26:2921–2927. doi: 10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Treiser MD, Yang EH, Gordonov S, Cohen DM, Androulakis IP, Kohn J, Chen CS, Moghe PV. Proc Natl Acad Sci U S A. 2009;107:610–615. doi: 10.1073/pnas.0909597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cormier JT, zur Nieden NI, Rancourt DE, Kallos MS. Tissue Eng. 2006;12:3233–3245. doi: 10.1089/ten.2006.12.3233. [DOI] [PubMed] [Google Scholar]
- 54.Schroeder M, Niebruegge S, Werner A, Willbold E, Burg M, Ruediger M, Field LJ, Lehmann J, Zweigerdt R. Biotechnol Bioeng. 2005;92:920–933. doi: 10.1002/bit.20668. [DOI] [PubMed] [Google Scholar]
- 55.Franke RP, Gräfe M, Schnittler H, Seiffge D, Mittermayer C, Drenckhahn D. Nature. 1984;307:648–649. doi: 10.1038/307648a0. [DOI] [PubMed] [Google Scholar]
- 56.Levesque MJ, Nerem RM. J Biomech Eng. 1985;107:341–347. doi: 10.1115/1.3138567. [DOI] [PubMed] [Google Scholar]
- 57.Sen A, Kallos M, Behie L. Ind Eng Chem Res. 2001;40:5350–5357. [Google Scholar]
- 58.Simon MC, Keith B. Nat Rev Mol Cell Biol. 2008;9:285–296. doi: 10.1038/nrm2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mazumdar J, O’Brien WT, Johnson RS, LaManna JC, Chavez JC, Klein PS, Simon MC. Nat Cell Biol. 2010;12:1007–1013. doi: 10.1038/ncb2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pfaffl MW. Nucleic Acids Res. 2001;29:45e. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Genome Biol. 2002;3:research0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


