Summary
Pathological acid reflux is a common event in patients afflicted with head and neck squamous cell carcinomas (HNSCCs), known to play a role in HNSCC etiology and contribute to complications after surgery or during radiation and chemotherapy. Antacid medications are commonly prescribed in HNSCC patients as part of their cancer treatment, and consist of two classes: histamine 2 receptor antagonist class (H2RA, with cimetidine as its prototypical drug) and proton pump inhibitors class (PPI, with omeprazole as its prototypical drug). Clinical evidences revealed a significant survival benefit of antacid usage in a large cohort of HNSCC patients treated in our Otolaryngology Department, with a median follow-up of over 5 years. Therefore, we postulate that one mechanism by which antacids intake enhance patients’ survival could involve modulation of tumor cells adhesion to endothelium, critical in the initiation of the metastatic dissemination. This study investigates the potential physical interactions between cimetidine and omeprazole with the endothelial E-selectin (E-sel) and its ligand sialyl Lewis X (sLex) using a molecular visualization energy-based program (AutoDock). Docking results were further analyzed with the PyMOL program, which allowed for measurements of the distances between the drugs and the closest interacting atoms or residues on E-sel and sLex molecules. Our model predicts that omeprazole displays a stronger interaction with E-sel than cimetidine, as extrapolated from the calculated overall binding energies. However, the shorter distances existing between interacting atoms in the proposed E-sel/cimetidine complex are suggestive of more stable interactions. Neither antacid/E-sel complex overcame the stronger Autodock-calculated sLex/E-sel interaction, suggesting competitive inhibition was not involved. This study provides the first in silico evidence of omeprazole and cimetidine ability to bind to adhesion molecules involved in tumor dissemination, underlining their therapeutic potential in the HNSCC clinical management.
Keywords: Head and neck squamous carcinoma, in silico modeling, E-selectin, sialyl Lewis x, antacid medication
Introduction
Advances in primary head and neck squamous cell carcinomas (HNSCC) treatment have led to the development of novel therapeutics; however their considerable morbidity and mortality remain a cause for great concern. The HNSCC poor clinical outcome is primarily due to metastasis, the main cause of cancer-related deaths, which remains poorly understood and largely incurable (1, 2). The ability to metastasize requires the active involvement of specific cell adhesion molecules such as selectins and their ligands (3). Tumor cells may obtain a selective advantage in establishing metastatic deposits through altered expression of antigens such as Sialyl Lewis×(sLex), which may affect interactions with selectins such as E-selectin (Endothelial selectin, E-sel), an inducible cell adhesion molecule only expressed by endothelial cells (4). sLeX, which function as a ligand of E-selectin and is principally expressed by leukocytes, is also commonly found on a wide variety of tumor cells and facilitate their binding to lymphatic or vascular endothelium initiating extravasation, a critical step in the process of metastasis via vascular pathways (3–6). In vitro studies have confirmed the ability of the sLex-expressing tumors cells to firmly adhere to endothelial cells via direct binding to the E-selectin, in contrast to non-expressing sLex tumors cells that were unable to (7–12). Expression of sLex has been reported in several cancers (e.g. breast, colorectal, cervical and lung), and its expression was correlated with the malignant phenotype particularly in those from breast and gastro-intestinal (GI) tract (7, 13–18). Circulating levels of E-selectin and its ligand sLex have been found to be predictive for metastasis in colon and gastric carcinoma patients (19–20); they have also been reported to play an important role in lymph node metastasis in invasive breast carcinomas (13). In patients with colorectal cancer sLex expression strongly correlated with advanced stage disease, distant metastasis and poor survival (7); similar prognostic significance has been shown in other cancers, including lung, breast and esophageal cancer (21–23). Studies evaluating sLex in head and neck tumors have provided evidences of sLex significance as negative prognostic marker for cancer-specific survival in HNSCC patients, independent of age, T-stage and alcohol consumption (24). Our previous work have shown that sLex-positive HNSCC tumor cells are able to bind to E-selectin-positive endothelium, and thus through sLex-E-selectin interaction the tumor cells are able to tether and initiate rolling on the endothelium prior to their extravasation (25). Furthermore, our clinical studies have shown that sLex expression associates with poorly differentiated and metastatic tumors, suggesting that its expression may be indicative of HNSCC invasive and metastatic potential (26). The therapeutic implications are potentially very significant because of commonly used and safe agents known to inhibit the expression of E-sel, such as cimetidine (17). In a series of preliminary in vitro experiments we have found that cimetidine treatment resulted in reduced E-selectin expression in endothelial cells and sLex expression in HNSCC cell lines (25, 27–28). Several randomized trials have shown that cimetidine intake correlates with significant survival advantage in gastric and colorectal cancer; in the latter trial the survival advantage was restricted to patients whose tumors expressed these antigens (7, 29). Our clinical evidences have revealed a significant survival benefit of antacid usage in a large cohort of HNSCC patients treated in our Otolaryngology Department at the University of Michigan, with a median follow-up of over 5 years (28). Based on our clinical and laboratory data, we postulate that one mechanism by which antacids intake may contribute to improved HNSCC patients’ survival may involve modulation of tumor cells adhesion to the endothelium, critical in the initiation of the metastatic dissemination. Two classes of antacid medications, histamine 2 receptor antagonists (H2RA, with cimetidine as its prototypical drug) and proton pump inhibitors (PPI, with omeprazole as its prototypical drug), known for their similar ability to decrease and/or inhibit the production of gastric acid, are commonly used for the management of acid reflux and complications from conventional therapies in HNSCC patients.
The objective of this study was to explore if the two prototypical antacid drugs, cimetidine (H2RA class) as compared to omeprazole (PPI class), may physically interact with E-selectin and sLex by using an in silico modeling investigation. This study has allowed visualizing the potential inhibitory ability of the prototypical drugs of the two most commonly used antacid drugs in HNSCC patient population. Our model predicts a stronger interaction with E-sel for omeprazole, as opposed to a more stable interaction with E-sel in the case of cimetidine. Taken together, this study provides the first in silico evidence of the abilities of omeprazole and cimetidine to bind to endothelial adhesion molecules involved in tumor dissemination. This highlights the need for a better understanding of antacids’ biological effects in the context of tumor initiation, progression and dissemination in order to provide comprehensive support for a novel therapeutic approach that can be readily translated into clinical benefit.
Materials and Methods
Antacid drugs
Omeprazole and cimetidine were purchased from Sigma Aldrich (St. Louis, MO) for ongoing in vitro studies conducted in our laboratory. In the protein docking procedures, the chemical structures for the two antacids (illustrated in Fig. 1A-B) were composed based on previously reported chemical structures (30–31) and minimal energy 3D structures were constructed using Trident-3D Analyst software (Wavefunction, Inc.; Irvine, CA, www.trimble.com). The chemical structures of both cimetidine and omeprazole contain imidazole-containing aromatic heterocyclic groups, suggesting that their potential actions as inhibitors of E-selectin or sLex may happen via similar mechanisms. The most striking difference between the chemical structures of the two antacids is the very stable and charged guanidinium group of cimetidine. This side group is hypothesized to be able to affect the binding interaction of cimetidine with either sLex or E-selectin (Fig. 1A). In contrast, omeprazole contains a polar sulfoxide group (Fig. 1B), while cimetidine contains a non-polar thioether. This also may affect the interactions of these antacids with sLex and/or E-selectin. Interestingly, a recent in silico docking study using the crystal structure of an E-selectin-sLex complex reported that potential monosaccharide mimetics, composed of various aromatic and heteroaromatic core structures in combination with (S)-cyclohexyl lactic acid and D-mannose, posed as effective inhibitors of the sLex/E-sel interaction (32). In this study, we employed a similar molecular modeling program to further elucidate the potential effects that antacids may have on tumor cells, specifically regarding disruption of the sLex/E-sel interaction.
Fig. 1.
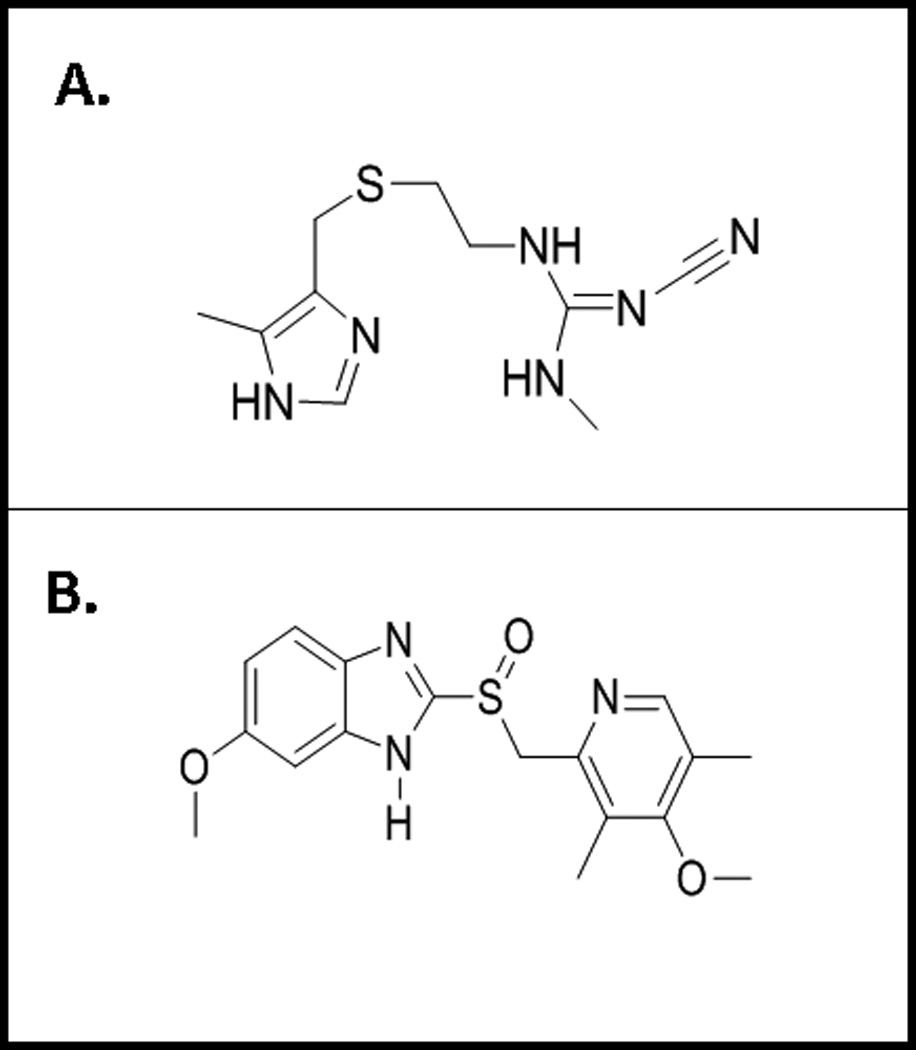
A) Chemical structure of cimetidine, the prototypical drug of histamine 2 receptor antagonists antacid class. This compound contains an imidazole heterocyclic ring, a central sulfur atom and a guanidinium side group. Constructed in ChemDraw with American Chemical Society (ACS) document settings. (http://www.cambridgesoft.com/software/ChemDraw/). B) Chemical structure of omeprazole, the prototypical drug of proton pumps inhibitors antacid class. This compound contains an imidazole heterocyclic ring and a central sulfur atom. Constructed in ChemDraw with American Chemical Society (ACS) document settings. (http://www.cambridgesoft.com/software/ChemDraw/)
Docking procedure and AutoDock Program Description
We used AutoDock Tools MGL-15 software (AutoDock, version 4.2) provided by the Scripps Research Institute (La Jolla, CA, www.autodock.scripps.edu). AutoDock generated a series of ten conformation models, which represents the ten best predicting models of how the antacids could potentially interact with the molecules of interest, E-sel and sLex. AutoDock generated a series of energy values (binding energy, ligand efficiency; inhibition constant; intermolecular energy; Van der Waals, electrostatic and total internal energy) using a Lamarckian program that was then used to analyze the relative strengths of the interactions. Binding energy, ligand efficiency and inhibition constant are the most indicative of the overall strength of a given predicted interaction calculated by AutoDock. Mass-centered grid maps were created using 0.375 angstroms (Å) spacing with the Autogrid program also available from the Scripps Research Institute. In the grid maps the spacing was also set to enclose 99% of the active sites of the macromolecules, and the grids were centered on the sLex/Ca2+ ion. We have identified this as the active site by observing the E-sel/sLex-bound protein as downloaded from the Protein Database website (PDB ID: 1G1T, www.pdb.rcsb.org, 33) using the JMOL molecule viewer software, a free open-source software for chemical structures in 3D (jmol.sourceforge.net). The main amino-acids involved in the interaction, as well as others within a six angstroms sphere surrounding the central calcium atom, were identified and recorded. These residues were later used in the AutoDock experiment as the ‘flexible’ residues of the macromolecule E-sel. A total of three docking experiments were run: 1) the “control”, established when sLex was docked with E-sel, 2) cimetidine was docked with E-sel and 3) omeprazole was docked with E-sel.
Analysis and interpretation of docking results
Docking results were imported into the PyMOL Molecular Visualization program (www.pymol.org) and analyzed. The ten conformations within each run were ordered based on binding energy. For each run, the conformation with the lowest energy was conformation #1 (the strongest binding), while the highest energy designated the conformation #10 (the weakest binding). We have analyzed and interpreted the energy data provided by AutoDock based on previously published studies reporting Autodock-based results (34). According to AutoDock, the binding energy is the sum of the intermolecular forces acting upon the receptor-ligand complex (Equation 1) (35).
| Equation 1 |
Because the binding energy is essentially a calculated Gibb’s Free Energy value, it can be compared to the calculated Gibb’s free energy of the naturally-occurring sLex-Eselectin interaction (Equation 2).
| Equation 2 |
Using the known affinity constant of 0.72 mM (44) for the sLex-Esel interaction, Gibb’s free energy for the complex was found to be −1800 J. The binding energy of the E selectin/sLex ligand is known to be weak, as it is generally accepted for many carbohydrate/receptor interactions (34–36).
Binding substrates (the macromolecule of E-selectin)
Docking the antacids interaction with E-selectin
The structure of E-selectin was downloaded from the Protein Database website (PDB ID: 1ESL, www.pdb.rcsb.org) and modified using the AutoDock software version 4.2. Hydrogen molecules were removed from the structure and Gasteiger charges were added using this energy-based software. Flexible residues isolated from the E-sel active site for the docking procedure were: Asparagine 83 and 105 (Asn83 and Asn105), and Arginine 108 (Arg 108). Potential torsions of E-sel were detected and analyzed in AutoDock. The calcium ion involved in the interaction of sLex was maintained in the docking interactions. The antacids were docked into the active site of the E-sel macromolecule. The binding energy results were compared to the binding energy calculated using the reported dissociation constant.
Results
Docking of cimetidine with E-selectin
The interaction of cimetidine with E-sel was compared to the sLex/E-sel interaction (Fig. 2A-B). The binding energy, ligand efficiency energy and inhibition constant values of the best-predicted conformation model (#1) of cimetidine docked with E-sel had values of −14.54 kcal/mol, −0.86 kcal/mol and 21.84 pM, respectively (Table I). The intermolecular energy, Van der Waals + Hbond+ desolvation energy, electrostatic energy and total internal energy values for the best conformation were −3.15 kcal/mol, −1.07 kcal/mol, 0.71 kcal/mol and −12.76 kcal/mol, respectively (Table I).
Fig. 2.
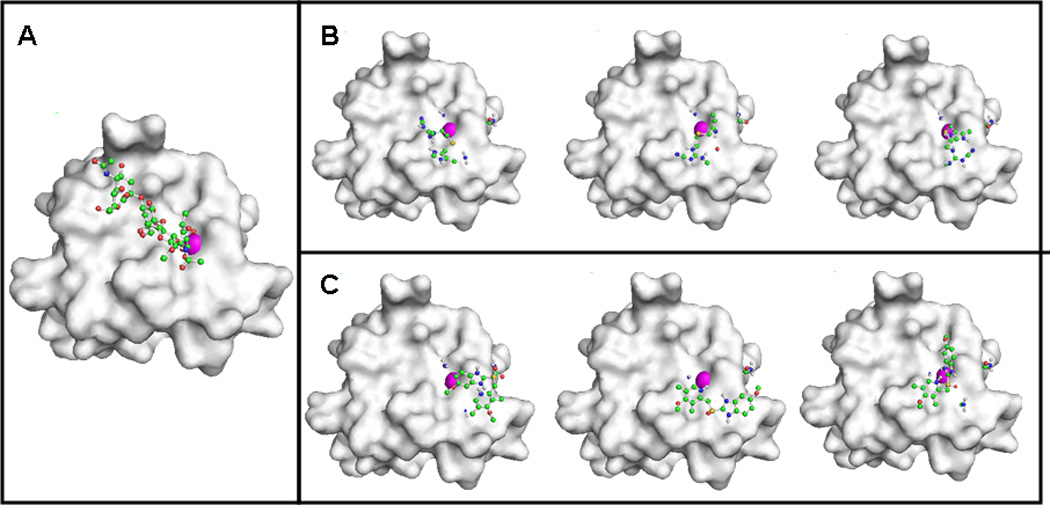
A) Pathological interaction between E-selectin and its ligand sLex. The sLex/E-selectin complex as downloaded from the Protein Database (PDB 1D:1G1T). The E-selectin molecule is shown as a grey tinted model and sLex is represented as a ball-and-stick model. B) Top three models of the interaction between cimetidine and E-selectin. The E-selectin molecule is shown as a grey tinted model and cimetidine is represented as a ball-and-stick model. C) Top three models of the interaction between omeprazole and E-selectin. The E-selectin molecule is shown as a grey tinted model and omeprazole is represented as a ball-and-stick model. Carbon, oxygen, nitrogen and hydrogen atoms, and calcium ion are illustrated (colored green, red, blue and white, respectively; and the calcium ion as a magenta sphere, in the online version).
Table I.
The energy values of ten potential conformational models of cimetidine interaction with E-selectin.
| Conformation | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Binding Energy (kcal/mol) |
−14.54 | −14.19 | −14.18 | −13.65 | −13.58 | −13.41 | −13.26 | −12.37 | −10.89 | −9.99 |
| Ligand Efficiency | −0.86 | −0.83 | −0.83 | −0.8 | −0.8 | −0.79 | −0.78 | −0.73 | −0.64 | −0.59 |
| Calculated Ki (pM) | 21.84 | 39.62 | 40.55 | 99.49 | 110.49 | 146.89 | 191.97 | 850.08 | 10380 | 47670 |
| Intermolecular Energy (kcal/mol) |
−3.15 | −2.58 | −1.89 | −1.67 | −0.77 | −0.31 | −1.69 | −1.42 | −2.41 | −1.35 |
| VDW + Hbond + Desolv Energy |
−1.07 | −1.16 | −0.79 | −1.19 | −0.69 | −0.45 | −1.21 | −0.05 | −1.16 | −0.88 |
| Electrostatic Energy (kcal/mol) |
0.71 | 0.99 | 0.01 | 0.27 | 0.9 | 0.43 | −0.11 | −0.18 | −0.21 | 0.52 |
| Total Internal Energy (kcal/mol) |
−12.76 | −12.98 | −13.66 | −13.34 | −14.19 | −14.48 | −12.94 | −12.32 | −9.86 | −10.0 |
Calculations of binding energy, ligand efficiency, inhibition constant, intermolecular energy, Van Der Waals energy, electrostatic energy and ligand internal energy were generated with the AutoDock program as described in the Materials and Methods. The ten conformations within each run were ordered based on binding energy. The conformation with the lowest energy was conformation #1, and the highest energy was conformation #10 for each run. All energy values have units of kcal/mol. Abbreviations: VDW, Van Der Waals energy; Hbond, Hydrogen Bonding; Desolv, Desolvation.
Docking of omeprazole with E-selectin
Then, we analyzed the interaction of omeprazole with E-sel comparatively with the sLex/E-sel interaction (Figs. 2C&3B). The binding energy, ligand efficiency energy and inhibition constant values of the best-predicted conformation model (#1) of omeprazole docked with E-sel had values of −16.29 kcal/mol, −0.68 kcal/mol and 1.15 pM, respectively (Table II). The intermolecular energy, Van der Waals + Hbond+ desolvation energy, electrostatic energy and total internal energy values for the best conformation were −4.38 kcal/mol, −1.48 kcal/mol, −1.17 kcal/mol and −13.27 kcal/mol, respectively (Table II). The binding energy values were then compared to the cimetidine/E-selectin interaction (Fig. 4 and Table III).
Fig. 3.
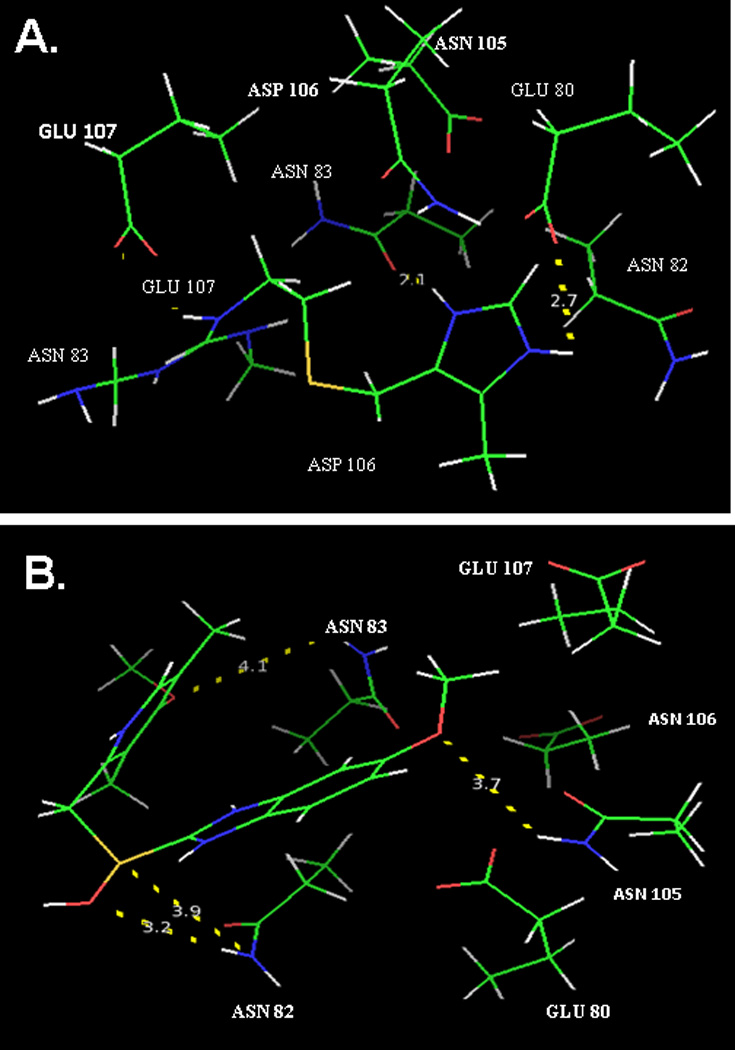
A) Intermolecular characteristics of the specific interactions within the best-predicted conformational model of E-selectin and cimetidine. The most flexible residues within the active site of E-selectin are shown: Asn82, Asn83, Asn105, Glu80, Glu107 and Asp106. The measured distances are 2.1, 2.2 and 2.7 angstroms. B) Intermolecular characteristics of the specific interactions within the best-predicted conformational model (conformation #1) of E-selectin and omeprazole. The most flexible residues within the active site of E-selectin are shown: Asn82, Asn83, Asn105, Glu80, Glu107 and Asp106. The measured distances are 3.2, 3.7, 3.9 and 4.1 angstroms. Longer distances indicate weaker potential interactions between the two molecules. Carbon, oxygen, nitrogen and hydrogen atoms are illustrated as in Fig. 2.
Table II.
The energy values of ten potential conformational models of omeprazole interaction with E-selectin.
| Conformation | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Binding Energy (kcal/mol) |
−16.29 | −16.22 | −16.16 | −15.63 | −15.33 | −15.05 | −14.79 | −13.18 | −12.92 | −11.47 |
| Ligand Efficiency | −0.68 | −0.68 | −0.67 | −0.65 | −0.64 | −0.63 | −0.62 | −0.55 | −0.54 | −0.48 |
| Inhibition Constant (pM) |
1.15 | 1.29 | 1.42 | 3.5 | 5.77 | 9.24 | 14.53 | 219.33 | 337.47 | 3920 |
| Intermolecular Energy |
−4.38 | −3.99 | −4.80 | −3.29 | −3.43 | −3.07 | −1.9 | −3.12 | −2.4 | −2.43 |
| VDW + Hbond + Desolv Energy |
−1.48 | −2.68 | −1.08 | −1.77 | −0.59 | −0.33 | 0.22 | −0.5 | −0.82 | −0.48 |
| Electrostatic Energy |
−1.17 | −0.38 | −1.1 | 0.09 | −0.37 | −0.85 | −1.3 | −0.88 | 0.14 | −0.18 |
| Total Internal Energy |
−13.27 | −13.6 | −13.46 | −13.71 | −13.27 | −13.35 | −14.25 | −11.43 | −11.89 | −10.41 |
Calculations of binding energy, ligand efficiency, inhibition constant, intermolecular energy, Van der Waals energy, electrostatic energy and total internal energy were generated with AutoDock as described in the Materials and Methods section. The ten conformations within each run were ordered based on binding energy. The conformation with the lowest energy was conformation #1, and the highest energy was conformation #10 for each run. Abbreviations: VDW, Van Der Waals energy; Hbond, Hydrogen Bonding; Desolv, Desolvation.
Fig. 4.
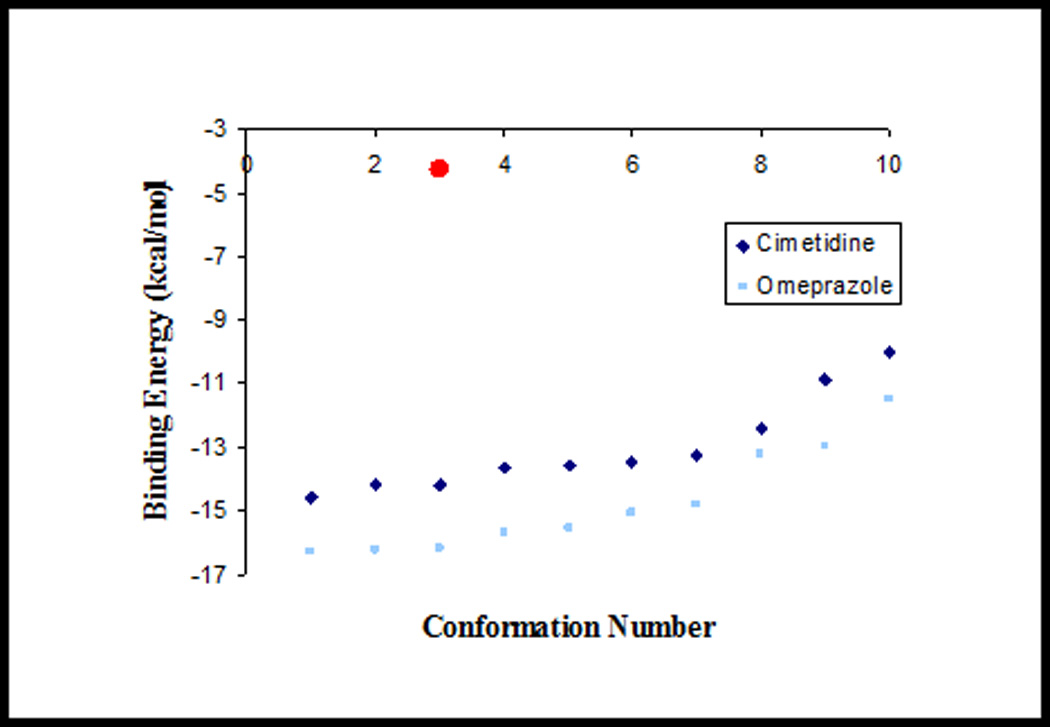
Diagram of the binding energies of the simulated interactions between sLex, omeprazole and cimetidine with E-selectin. Illustrated are the binding energies and relative strengths of interactions as calculated by using AutoDock program when the antacids drugs, cimetidine and omeprazole, were simulated to interact with E-selectin. Dark diamonds and light squares represent cimetidine and omeprazole interactions with E-sel, respectively. The circle represents the binding energy of the pathological sLex/E-sel interaction, as calculated by Gibb’s free energy equation. Omeprazole exhibited greater binding energies than cimetidine, and both antacids had greater binding energies than the sLex/E-sel interaction.
Table III.
Comparison of the binding energy values (kcal/mol) and inhibition constants (mM) of the ten potential conformational models of the two antacid drugs (cimetidine versus omeprazole) interactions with E-selectin.
| Conformation | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Binding Energy (kcal/mol) |
||||||||||
| Cimetidine | −14.54 | −14.19 | −14.18 | −13.65 | −13.58 | −13.41 | −13.26 | −12.37 | −10.89 | −9.99 |
| Omeprazole | −16.29 | −16.22 | −16.16 | −15.63 | −15.55 | −15.05 | −14.79 | −13.18 | −12.92 | −11.47 |
| Inhibition Constants (pM) |
||||||||||
| Cimetidine | 21.84 | 39.62 | 40.55 | 99.49 | 110.49 | 146.89 | 191.97 | 850.08 | 10380 | 47670 |
| Omeprazole | 1.15 | 1.29 | 1.42 | 3.5 | 5.77 | 9.24 | 14.53 | 219.33 | 337.47 | 3920 |
The lowest binding energy and inhibition values indicate a better and stronger overall interaction. Both values show that omeprazole has a stronger interaction with E-selectin than cimetidine.
Observation of physical interactions using PyMOL
The chemical structures of omeprazole and cimetidine (Fig. 1A-B) were used to deduce and predict the specific interactions between the docked amino acids within the active site of E-sel or sLex and these antacids. In the docking results, we consistently identified two hydrogen bonds when both omeprazole and cimetidine were docked with E-selectin; the two bonds were between the asparagine residues at positions 82 and 105 of E-selectin and the antacids. These bonds corresponded to the amide group of the asparagine residue and the availability of hydrogen atoms in either the imidazole rings of the antacids or other available hydroxyl groups.
When the best-predicted conformation (#1) of cimetidine docked with E-sel was analyzed in PyMOL, the closest distance between the antacid molecule and the target protein was reported to be 2.1 Å. Other measured distances were 2.2 Å and 2.7 Å. These values represent hydrogen bonds or other strong interactions. Specifically, the distance of 2.1 Å was from a potential intermolecular interaction between the amine group of the imidazole ring of cimetidine and the carboxyl group of the amide group of the asparagine residue at position 83 on E-sel (Fig. 3A). The second closest distance, 2.2 Å, was between an amine group of the guanidinium group of cimetidine and the hydroxyl group of the carboxylic acid within the glutamate residue at position 80 of E-sel.
When the first conformation produced from the docking study of omeprazole with E-sel was analyzed in PyMOL, four distances were reported. These distances were: i). of 3.2 Å between a hydroxyl group of omeprazole and the amine group in the side chain of asparagine at position 82; ii). of 3.7 Å between one of the ether groups of omeprazole and the amine side chain of asparagine 105; iii). of 3.9 Å between the sulfur of the nonpolar thioether group of omeprazole and the amine group of asparagine 82; and 4). of iv.1 Å between the ether of omeprazole and the amine side chain of asparagine 83 (Fig. 3B).
Discussion
The objective of this current study was to analyze comparatively the interactions between two antacid drugs (cimetidine, H2RAs class versus omeprazole, PPIs class) and E-selectin by using the AutoDock program. This analysis allowed us to explore the potential physical interactions between substrates (these two antacid drugs) and receptors (E-sel), with the goal to investigate potential mechanisms by which antacids may disrupt tumor dissemination and metastasis in HNSCCs, in support of the significant association with improved patient outcome we observed in the clinical settings (27). Two individual docking experiments were run to assess the potential interactions of each antacid drug with E-sel. In each run, the program has generated a series of ten ‘conformations’, or the ten most likely conformational models of how the drugs could potentially interact with their targets. These in silico analyses are predicting that omeprazole, the prototypical proton pump inhibitor, could potentially have a more stable interaction with E-sel receptor than cimetidine, the histamine type 2 receptor antagonist, given that it bound more tightly and formed a more stable complex when docked with E-selectin. When we compared the binding affinity constant of the sLex/E-sel interaction and the binding energies generated by AutoDock, we found that the sLex-E-sel interaction was comparatively weaker. This was expected, since carbohydrates do not usually form very strong complexes with receptors. Using the previously determined (by in vitro experiments, 40) affinity constant of 0.72 mM for the sLex-Esel interaction, the Gibb’s free energy for the complex was found to be −1800 J. We also found that there were 2.6-fold and 3.42-fold increases in binding energy when cimetidine and omeprazole were docked with E-sel, respectively. Given that the sLex/E-sel AutoDock calculated value is lower (the inhibition constant is also lower) than either the omeprazole and cimetidine interactions, it suggests that the natural interaction of sLex/E-sel is stronger than with either drug, and that sLex is more potent. These results suggest that these antacid drugs may have a more downstream/RNA-changing effect on the interaction rather than competitive inhibition; and secondly, that the PPI drug omeprazole, has a greater binding energy to the E-sel than the H2RA drug cimetidine. Comparing the in silico sLex/E-sel data with the in vitro reference values proves difficult because it is very difficult to dock a carbohydrate such as sLex with a large molecule such as E-selectin since the final conformations tend to vary significantly due to water-mediated interactions and the inherent flexibility of the many terminal hydroxyl groups in carbohydrates, as reported by other AutoDock studies (reviewed in 40)
Van der Waals energy, electrostatic and total internal energy values were also calculated with AutoDock to further evaluates the relative strengths of binding. When omeprazole was docked with E-sel, the Van der Waals energy values calculated were consistent, indicating that the relative strength of those intermolecular interactions were dependent on the weak Van der Waals forces. In contrast, when cimetidine was docked with E-sel the Van der Waals energy values were not consistent and these findings were further confirmed; the electrostatic energies of the two antacids docked with E-sel revealed similar results. Finally, the total internal energy values were not consistent in the ten conformations, implying that the total internal energy of the system did not contribute to the overall strength of binding. One plausible explanation is that because the antacid drugs are relatively small ‘ligands’, the AutoDock program may not be able to calculate an accurate value for the total internal energy of the interaction. Therefore, for further confirmation of the resulting physical interactions, the conformations were also observed in the PyMOL program, which in addition also displayed the shortest measurements between the atoms within the docked antacid and the most proximal residues of E-sel. In fact, based on these distances and by observing the actual elements involved in the proximal atomic interactions, the possible intermolecular interactions can be accurately predicted with PyMOL. The results obtained by using PyMOL are very interesting when compared to the energy values generated by AutoDock. It is known that the limits of a hydrogen bond range from approximately 1.87– 2.88 Å (37–39). Surprisingly, there were more potential hydrogen bonds when cimetidine was bound with E-sel than when omeprazole was docked. However, it is interesting to note that the polar sulfoxide group in the structure of omeprazole was involved in the interaction, but, as predicted, the non-polar thio-ester group of cimetidine was not involved. The central location and polarity of this group could account for the increase in binding energy of omeprazole.
Overall, based on these three docking simulations and PyMOL observations, we found that when docked with E-selectin, omeprazole exhibited stronger binding than cimetidine. Thus, our analyses reveal that omeprazole is more likely to form a stronger complex with E-sel than cimetidine, although cimetidine forms stronger intermolecular interactions with E-sel. The binding energy values of these antacids are, however, similar, and are comparatively stronger than the pathological sLex/E-sel interaction, indicating that the antacid drugs could both interact with E-sel, although more likely via different mechanisms. The results of this study are in alignment with our laboratory’s recent in vitro findings that cimetidine affects the sLex/E-sel interaction almost immediately; in contrast, omeprazole requires longer duration treatment with stronger effects (Matossian et al, under review). This study expands previous findings on the antacids’ effects on endothelial adhesion molecules with known roles in cancer dissemination, underlining their therapeutic potential for HNSCC clinical management. Further investigation will require additional analysis by using other algorithms that will allow to generate larger number of conformations, as well as to consider the situation in which a multivalent ligand is interacting with a multivalent receptor, mimicking more closely the clinical situation. Ongoing in vivo investigations in our HNSCC experimental models aims to estimate the necessary amount of antacid in the circulation able to reduce and disrupt the E-sel/sLex interaction as extrapolated from the standard dosage used in HNSCC patients. At this time we do not know the biological mechanisms in all its complexity by which antacid medications may influence patient outcome; we postulate that antacids may alter expression of other factors besides of these key endothelial players. Further evaluation in laboratory and clinical settings of antacids effects on tumor progression may lead to new chemo-preventive strategies for HNSCC patients, which could be extrapolated to other cancer patients.
Acknowledgements
This project was made possible by funding from the NIH NCI NIDCR P50 CA097248 University of Michigan Specialized Program of Research Excellence in Head and Neck Cancer (SPORE, PI: Gregory T. Wolf, MD) and the Research Scholar Grant RSG-13-103-01–CCE from the American Cancer Society (PI: Silvana Papagerakis, MD/PhD) at the University of Michigan Ann Arbor. The authors are grateful for the expert input and guidance of Dr. Laura Furge, Ph.D., Chemistry Professor, Department of Chemistry, Kalamazoo College, Kalamazoo, Michigan. Margarite Matossian has been the 2012 Recipient of the Outstanding Chemistry Student Award from the American Chemical Society and she is currently an MD/PhD student, in the Physician Scientist Program at Tulane University School of Medicine.
Footnotes
Disclosure statement: “None”. The authors do not have any conflict of interest.
References
- 1.American Cancer Society. Global Cancer Facts and Figures. 2011 [Google Scholar]
- 2.Shah J, Johnson NW, Batsakis JG. In: Oral Cancer. 1st ed. Singh B, Shah JP, editors. London: Martin Dunitz, London; 2003. pp. 367–372. [Google Scholar]
- 3.Orr FW, Wang HH, Lafrenie RM, Scherbarth S, Nance DM. Interactions between cancer cells and the endothelium in metastasis. J Pathol. 2000;190:310–329. doi: 10.1002/(SICI)1096-9896(200002)190:3<310::AID-PATH525>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 4.Osborn L. Leukocyte adhesion to endothelium in inflammation. Cell. 1990;62:3–6. doi: 10.1016/0092-8674(90)90230-c. [DOI] [PubMed] [Google Scholar]
- 5.Renkonen J, Paavonen T, Renkonen R. Endothelial and epithelial expression of sialyl Lewis(x) and sialyl Lewis(a) in lesions of breast carcinoma. Int J Cancer. 1997;74:296–300. doi: 10.1002/(sici)1097-0215(19970620)74:3<296::aid-ijc11>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 6.Close LG, Brown PM, Vuitch MF, Reisch J, Schaefer SD. Microvascular invasion and survival in cancer of the oral cavity and oropharynx. Arch Otolaryngol Head Neck Surg. 1989;115:1304–1309. doi: 10.1001/archotol.1989.01860350038011. [DOI] [PubMed] [Google Scholar]
- 7.Matsumoto S, Imaeda Y, Umemoto S, Kobayashi K, Suzuki H, Okamoto T. Cimetidine increases survival of colorectal cancer patients with high levels of sialyl Lewis-X and sialyl Lewis-A epitope expression on tumour cells. Br J Cancer. 2002;86:161–167. doi: 10.1038/sj.bjc.6600048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dejana E, Martin-Padura I, Lauri D, Bernasconi S, Bani MR, Garofalo A, Giavazzi R, Magnani J, Mantovani A, Menard S. Endothelial leukocyte adhesion molecule-1-dependent adhesion of colon carcinoma cells to vascular endothelium is inhibited by an antibody to Lewis fucosylated type I carbohydrate chain. Lab Invest. 1992;66:324–330. [PubMed] [Google Scholar]
- 9.Ichihara T, Sakamoto J, Nakao A, Furukawa K, Watanabe T, Suzuki N, Horisawa M, Nagura H, Lloyd KO, Takagi H. Expression of blood group-related antigens in normal and malignant pancreatic tissue correlated with genotype of the patient defined by saliva glycoprotein. Cancer. 1993;71:71–81. doi: 10.1002/1097-0142(19930101)71:1<71::aid-cncr2820710113>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 10.Irimura T, Nakamori S, Matsushita Y, Taniuchi Y, Todoroki N, Tsuji T, Izumi Y, Kawamura Y, Hoff SD, Cleary KR. Colorectal cancer metastasis determined by carbohydrate-mediated cell adhesion: role of sialyl-LeX antigens. Semin Cancer Biol. 1993;4:319–324. [PubMed] [Google Scholar]
- 11.Nakamori S, Kameyama M, Imaoka S, Furukawa H, Ishikawa O, Sasaki Y, Kabuto T, Iwanaga T, Matsushita Y, Irimura T. Increased expression of sialyl Lewisx antigen correlates with poor survival in patients with colorectal carcinoma: clinicopathological and immunohistochemical study. Cancer Res. 1993;53:3632–3637. [PubMed] [Google Scholar]
- 12.Takada A, Ohmori K, Yoneda T, Tsuyuoka K, Hasegawa A, Kiso M, Kannagi R. Contribution of carbohydrate antigens sialyl Lewis A and sialyl Lewis X to adhesion of human cancer cells to vascular endothelium. Cancer Res. 1993;53:354–361. [PubMed] [Google Scholar]
- 13.Wei J, Cui L, Liu F, Fan Y, Lang R, Gu F, Guo X, Tang P, Fu L. E-selectin and Sialyl Lewis X expression is associated with lymph node metastasis of invasive micropapillary carcinoma of the breast. Int J Surg Pathol. 2010;18:193–200. doi: 10.1177/1066896908320832. [DOI] [PubMed] [Google Scholar]
- 14.Portela SV, Martin CV, Romay LM, Cuevas E, Martin EG, Briera AF. sLea and sLex expression in colorectal cancer: implications for tumourigenesis and disease prognosis. Histol Histopathol. 2011;26:1305–1316. doi: 10.14670/HH-26.1305. [DOI] [PubMed] [Google Scholar]
- 15.Engelstaedter V, Fluegel B, Kunze S, Mayr D, Friese K, Jeschke U, Bergauer F. Expression of the carbohydrate tumour marker Sialyl Lewis A, Sialyl Lewis X, Lewis Y and Thomsen-Friedenreich antigen in normal squamous epithelium of the uterine cervix, cervical dysplasia and cervical cancer. Histol Histopathol. 2012;27:507–514. doi: 10.14670/HH-27.507. [DOI] [PubMed] [Google Scholar]
- 16.St Hill CA, Krieser K, Farooqui M. Neutrophil interactions with sialyl Lewis X on human nonsmall cell lung carcinoma cells regulate invasive behavior. Cancer. 2011;117:4493–4505. doi: 10.1002/cncr.26059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi K, Matsumoto S, Morishima T, Kawabe T, Okamoto T. Cimetidine inhibits cancer cell adhesion to endothelial cells and prevents metastasis by blocking E-selectin expression. Cancer Res. 2000;60:3978–3984. [PubMed] [Google Scholar]
- 18.Yamaguchi A, Ding K, Maehara M, Goi T, Nakagawara G. Expression of nm23-H1 gene and Sialyl Lewis X antigen in breast cancer. Oncology. 1998;55:357–362. doi: 10.1159/000011878. [DOI] [PubMed] [Google Scholar]
- 19.Nakagoe T, Sawai T, Tsuji T, Jibiki MA, Nanashima A, Yamaguchi H, Yasutake T, Ayabe H, Arisawa K, Ishikawa H. Difference in prognostic value between sialyl Lewis(a) and sialyl Lewis(x) antigen levels in the preoperative serum of gastric cancer patients. J Clin Gastroenterol. 2002;34:408–415. doi: 10.1097/00004836-200204000-00005. [DOI] [PubMed] [Google Scholar]
- 20.Wittig BM, Kaulen H, Thees R, Schmitt C, Knolle P, Stock J, Meyer zum Buschenfelde KH, Dippold W. Elevated serum E-selectin in patients with liver metastases of colorectal cancer. Eur J Cancer. 1996;32A:1215–1218. doi: 10.1016/0959-8049(96)00086-x. [DOI] [PubMed] [Google Scholar]
- 21.Shimada Y, Maeda M, Watanabe G, Inamura M. High serum soluble E-selectin levels are associated with post-operative haematogenic recurrence in esophageal squamous cell carcinoma patients. Oncol Rep. 2003;10:991–995. [PubMed] [Google Scholar]
- 22.Hebbar M, Revillion F, Louchez MM, Fournier C, Bonneterre J, Peyrat JP. Prognostic value of circulating soluble e-selectin concentrations in node-negative breast cancer patients. Clin Cancer Res. 1999;5:1427–1433. [PubMed] [Google Scholar]
- 23.Sheen-Chen SM, Eng HL, Huang CC, Chen WJ. Serum levels of soluble E-selectin in women with breast cancer patients. Br J Cancer. 2004;91:1578–1581. doi: 10.1002/bjs.4513. [DOI] [PubMed] [Google Scholar]
- 24.Gunawardena I, Arendse M, Jameson MB, Plank LD, Gregor RT. Prognostic molecular markers in head and neck squamous cell carcinoma in a New Zealand population: matrix metalloproteinase-2 and sialyl Lewis x antigen. ANZ J Surg. 2013 doi: 10.1111/ans.12424. [DOI] [PubMed] [Google Scholar]
- 25.Papagerakis S, Thornhill M. Therapeutic Targets in Oral Cancer. Toxicol Pathol. 2006;34:1009–1009. [Google Scholar]
- 26.Czerwinski MJ, Desiderio V, Shkeir O, Papagerakis P, Lapadatescu MC, Owen JH, Athanassiou-Papaefthymiou M, Zheng L, Papaccio G, Prince ME, Papagerakis S. In vitro evaluation of sialyl Lewis X relationship with head and neck cancer stem cells. Otolaryngol Head Neck Surg. 2013;149:97–104. doi: 10.1177/0194599813482879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papagerakis S, Bellile E, Peterson LA, Pliakas M, Balaskas K, Selman S, Hanauer D, Taylor JMG, Duffy S, Wolf G. Proton pump inhibitors and histamin 2 blockers are associated with improved overall survival in patients with head and neck squamous cell carcinomas. Cancer Prev Res. 2014 doi: 10.1158/1940-6207.CAPR-14-0002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papagerakis S, Bellile E, Hanauer D, Duffy S, Wolf G. Novel therapeutic benefit of proton pump inhibitors and histamine 2 blockers on overall survival in patients with head and neck squamous carcinoma. 8th International Conf Head Neck Cancer. 2012 doi: 10.1158/1940-6207.CAPR-14-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tonnesen H, Knigge U, Bulow S, Damm P, Fisherman K, Hesselfeldt P, Hjortrup A, Pedersen IK, Pedersen VM, Siemssen OJ. Effect of cimetidine on survival after gastric cancer. Lancet. 1988;2:990–992. doi: 10.1016/s0140-6736(88)90743-x. [DOI] [PubMed] [Google Scholar]
- 30.Cooke RM, Hale RS, Lister SG, Shah G, Weir MP. The conformation of the sialyl Lewis X ligand changes upon binding to E-selectin. Biochemistry. 1994;33:10591–10596. doi: 10.1021/bi00201a004. [DOI] [PubMed] [Google Scholar]
- 31.Brimblecombe RW, Duncan WA, Durant GJ, Emmett JC, Ganellin CR, Leslie GB, Parsons ME. Characterization and development of cimetidine as a histamine H2-receptor antagonist. Gastroenterology. 1978;74:339–347. [PubMed] [Google Scholar]
- 32.Weck S, Frank M, Hamann A, Opatz T. Monosaccharidic mimetics of the sialyl Lewis X tetrasaccharide based on 2,7-dihydroxynaphthalene. ARKIVOC. 2012;3:134–148. [Google Scholar]
- 33.Rutgers and UCSD. Protein Data Base: www.rcsb.org/pdb; Updated March 2012. Funded by NSF, NIGMS, DOE, NLM, NCI, NINDS, and NIDDK.
- 34.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J Comp Chem. 1998;19:1639–1662. [Google Scholar]
- 35.Lin HL, Zhang H, Medower C, Hollenberg PF, Johnson WW. Inactivation of cytochrome P450 (P450) 3A4 but not P450 3A5 by OSI-930, a thiophene-containing anticancer drug. Drug Metab Dispos. 2011;39:345–350. doi: 10.1124/dmd.110.034074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poppe L, Brown G, Philo J, Nikrad PV, Shah BV. Conformation of sLex tetrasaccharide, free in solution and bound to E-, P- and L-selectin. J Am Chem Soc. 1997;119:1727–1736. [Google Scholar]
- 37.Taylor R, Kennard O. Hydrogen-bond geometry in organic crystals. Acc Chem Res. 1984;17:320–326. [Google Scholar]
- 38.Jorgensen W. Optimized intermolecular potential functions for liquid alcohols. J Phys Chem. 1986;90:1276–1284. [Google Scholar]
- 39.Torii H, Tatsumi T, Kanazawa T, Tasumi M. Effects of intermolecular hydrogen-bonding interactions on the amide 1 mode of N-methylacetamide: matrix-isolation infrared studies and ab initio molecular orbital calculations. J Phys Chem. 1998;102:309–314. [Google Scholar]
- 40.Wild M, Huang M, Schulze-Horsel U. Affinity, kinetics and thermodynamics of E-selectin binding to E-selectin ligand 1. J Biol Chem. 2001;276:31602–31612. doi: 10.1074/jbc.M104844200. [DOI] [PubMed] [Google Scholar]


