Abstract
An accurate measurement of the immune status in patients with immune system disorders is critical in evaluating the stage of diseases and tailoring drug treatments. The functional cellular immunity test is a promising method to establish the diagnosis of immune dysfunctions. The conventional functional cellular immunity test involves measurements of the capacity of peripheral blood mononuclear cells to produce pro-inflammatory cytokines when stimulated ex vivo. However, this “bulk” assay measures the overall reactivity of a population of lymphocytes and monocytes, making it difficult to pinpoint the phenotype or real identity of the reactive immune cells involved. In this research, we develop a large surface micromachined polydimethylsiloxane (PDMS) microfiltration membrane (PMM) with high porosity, which is integrated in a microfluidic microfiltration platform. Using the PMM with functionalized microbeads conjugated with antibodies against specific cell surface proteins, we demonstrated rapid, efficient and high-throughput on-chip isolation, enrichment, and stimulation of subpopulations of immune cells from blood specimens. Furthermore, the PMM-integrated microfiltration platform, coupled with a no-wash homogeneous chemiluminescence assay (“AlphaLISA”), enables us to demonstrate rapid and sensitive on-chip immunophenotyping assays for subpopulations of immune cells isolated directly from minute quantities of blood samples.
1. Introduction
The immune system plays a critical role in protecting a living organism against invasions of viruses, bacteria, and parasitic worms, and distinguishes diseased tissue from healthy one.[1–4] Leukocytes (white blood cells) are important blood constituents that play a major role in innate and adaptive immune responses against pathogenic infections, allergic conditions, and malignancies. Leukocytes are a heterogeneous mixture of multiple cell subsets (granulocytes, lymphocytes, and monocytes) defined by their morphology, surface antigen expression, and production of cytokines – small proteins for intercellular communications between leukocytes of the same type (homotypic) or different types (heterotypic).[5–8] The numbers, proportions and functional responses of leukocyte subsets change drastically in the presence of infections, malignancies, and autoimmune disorders, making analysis of leukocyte subpopulations particularly valuable in the diagnosis and monitoring of diseases.[4,9] For example, human immunodeficiency virus (HIV) infection causes depletion of CD4+ T cells in peripheral blood and other lymphoid tissues.[1,10–12] As a result, the absolute counts of CD4+ T cells and the ratio of CD4+/ CD8+ T cells are commonly used as indicators of the onset of the acquired immunodeficiency syndrome (AIDS) and as benchmarks for the initiation of antiviral therapy to treat AIDS.
Leukocytes orchestrate immune responses by releasing cytokines.[2,9,13–17] Given the heterogeneity of leukocytes and different roles played by different subpopulations of leukocytes in the immune response, measurements of capacity to produce cytokines from different subpopulations of leukocytes become critically important in clinical diagnosis. Production of interferon gamma (IFN-γ) by T cells, for example, correlates with the body's ability to mount a vigorous immune response and is used to identify antigen-specific T cells in diseases such as HIV and tuberculosis.[2,17,18] Sepsis is another serious clinical condition that results from dysregulation of the immune response to infection.[16,19–22] One hallmark of sepsis commonly reported is the decreased capacity of monocytes to release pro-inflammatory cytokines such as tumor necrosis factor-alpha (TNF-α) in response to ex vivo lipopolysaccharide (LPS) challenge.[16]
A significant challenge in profiling cytokine secretion in a heterogeneous cell suspension such as blood is the fact that the same cytokines can be produced by several different subpopulations of leukocytes. For example, IFN-γ is commonly secreted by CD4+ T cells. However, IFN-γ can also be produced by CD8+ T ells, macrophages and neutrophils. In addition, the numbers and proportions of subpopulations of immune cells may vary in individuals and at different stages of diseases. Thus, measurements of the overall capacity of the whole population of leukocytes to produce cytokines may not be informative enough to accurately reveal the immune status of patients, as in these ‘bulk’ assays it is difficult to pinpoint the phenotype or real identity of the reactive cells involved. As a result, the conventional whole blood stimulation assay routinely used in the clinical setting to determine cytokine secretion levels from the whole population of blood leukocytes is insufficient to accurately define and characterize the immune status of patients.
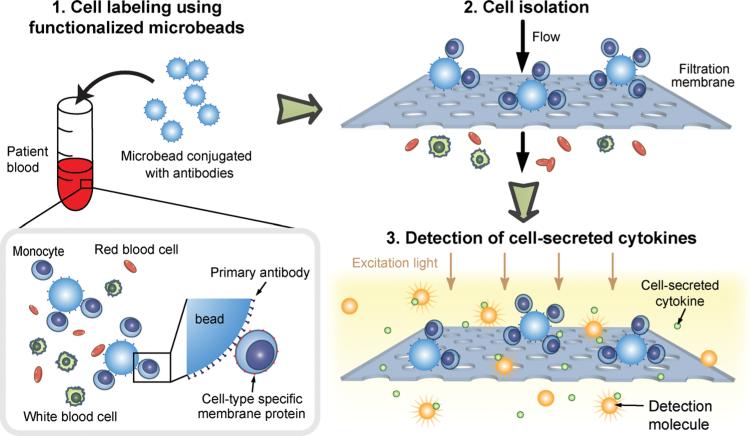
Here we reported a novel poly-dimethylsiloxane (PDMS) microfiltration membrane (PMM) of a large surface area and high porosity for microfluidic isolation and immunopheno-typing of subpopulations of immune cells. The PMM was fabricated using our newly developed surface micromachining technique for soft materials.[23] Recently, we have demosntrated that the PMM could serve as an efficient microfiltration material for bioparticle separation during a sequence of processes entailing cell isolation, enrichment, enumeration, stimulation, incubation, and cytokine secretion detection on a single microfluidic chip for human peripheral blood mononuclear cells (PBMCs).[24] In this study, we further employed a combined use of the PMM and antibody-conjugated polystyrene microbeads for isolation, purification, and functional immunophenotyping of different subpopulations of immune cells directly from blood specimens (Figure 1). Immobilized on the microbead surface were monoclonal antibodies against cell surface antigens specific to the desired subpopulation of immune cells. When mixed with a blood specimen, the microbeads could selectively capture the desired subpopulation of immune cells by recognizing and binding covalently to the specific surface proteins of the cells. The diameter of the through holes in the PMM, which solely defined the effective size cutoff for microfiltration, were designed to be smaller than the size of the microbeads but larger than normal blood leukocytes. Thus when the blood specimen was introduced into the microfiltration device and forced to pass through the PMM, the cell/microbead conjugates were readily trapped and isolated on the PMM, whereas other untargeted leukocytes and erythrocytes (red blood cells) unbound to microbeads could freely pass through the PMM. Owing to its large surface area and high porosity, the PMM permitted a heightened volume throughput (>5 mL min−1) for processing blood specimens without causing clogging of the PMM, a common problem in microfiltration membrane filters.
Figure 1.
Schematic of isolation and immunophenotyping of subpopulations of immune cells from blood specimens. Subpopulations of immune cells were recognized and captured by microbeads conjugated with monoclonal antibodies against specific cell surface antigens. Owing to the increased size of cell-microbead conjugates, the desired subpopulation of immune cells was retained on the PDMS microfiltration membrane (PMM) upon loading of the blood sample, while the other blood cells including undesired immune cells could pass freely through the PMM. On-chip functional immunophenotyping was then performed to examine the capacity of this subpopulation of immune cells to produce pro-inflammatory cytokines following stimulation.
Our PMM-integrated microfiltration device further allowed in situ quantitative immunosensing of cell-secreted cytokines by detecting optical signals resulted from a no-wash homogeneous sandwich chemiluminescence assay. In this homogeneous immunoassay, so called “AlphaLISA”,[25–27] the analyte of interest (i.e., cytokine) binds simultaneously to primary antibody-coated donor and acceptor beads, which brings the beads into a close proximity to each other. In this bead proximal state, singlet oxygen generated by laser excitation (λ = 680 nm) from the donor bead will transfer to the acceptor bead, eliciting a shorter wavelength emission (λ = 615 nm) from the acceptor bead. The AlphaLISA assay provides an efficient way for quantitative detection of cytokine production, without involving complex reagent loading and washing steps as in the conventional fluorescence-based immunoassays (e.g., enzyme-linked immunosorbent assay (ELISA)).[28,29]
In addition, the miniaturized size of the PMM-integrated microfiltration device required only a sample volume as small as 5 μL for each assay. Our device, permitting the homogenous AlphaLISA immunoassay in a microfluidic chamber, could reduce the entire assay time comprising the time for pretreatment, process, and detection down to a few hrs, a nearly 10-fold reduction in total assay time as compared to the conventional whole blood stimulation assay using ELISA or the enzyme-linked immunosorbent spot (ELISpot) assay.[28,29] Given the shortened assay time, enhanced sample efficiency, and the ability to determine the functional status of different subpopulations of immune cells, our microfluidic immunophenotyping technology promises to provide a new approach to diagnosing infectious and inflammatory diseases across a broad patient spectrum.
2. Results and Discussion
2.1. Microfiltration Membrane Fabrication
PDMS is one of the most frequently used structural materials in microfluidics. Yet, a major technical hurdle in micro-fluidics exists owing to the difficulty in achieving high fidelity lithographic patterning on PDMS. Till now, surface patterning techniques of PDMS using conventional photolithography and etching processes have not been achieved. While soft lithography is a popular bulk micromachining technique for rapid fabrication of PDMS structures, thin microstructures such as microfiltration membranes with micron-scale through holes are difficult to fabricate in PDMS by soft lithography.
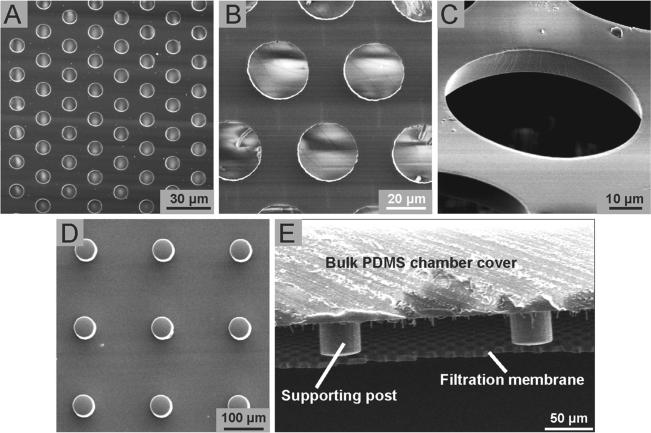
Recently, we have reported a wafer-scale surface micromachining method for PDMS to generate different PDMS thin film microstructures, with their critical dimensions ranging from sub-microns to tens of microns, using a combination of photolithography, reactive-ion etching (RIE), and convenient thin film releasing techniques.[23] Our method to achieve surface patterning of PDMS applies an O2 plasma treatment to PDMS to activate its surface to overcome the challenge of poor photoresist adhesion on PDMS for photolithography. Utilizing this newly developed PDMS surface micromachining technique, in this work we achieved wafer-scale batch fabrication of large PDMS microfiltration membranes (PMMs) containing a regular array of micron-scale through holes with high porosity (see Experimental Section and Figure S1 for the detailed fabrication procedure). Specifically, using our PDMS surface micromachining technique, we successfully fabricated PMMs with an effective membrane area of 3 cm × 3 cm and porosity up to 30%; both factors could contribute to a heightened volume throughput for processing unprocessed blood specimens without causing clogging of the PMM, a common problem in microfiltration membrane filters. For comparison, conventional track-etched polycarbonate filters, which have been used for isolation of blood cells and circulating tumor cells, has porosity of less than 2%, and the other more recently developed parylene-based micropore membrane has porosity of 7% - 15%.[30–32] Since the through holes in the PMM were defined by photolithography, the size and pattern of the through holes in the PMM could be precisely controlled. The PMMs used in this work had a thickness of about 10 μm and contained a regular array of hexagonally arranged through holes with a hole diameter of 6 - 60 μm and a center-to-center distance of 10 – 100 μm (Figure 2A-C). By controlling anisotropic etching of PDMS using RIE, the through holes of the PMM had a near vertical sidewall profile (Figure 2C). The transparent nature of PDMS permitted imaging and enumeration of cells captured on the PMM using standard clinical histopathological stains, in addition to immunofluorescence-conjugated antibodies.
Figure 2.
SEM images showing free-standing PMMs with high porosity. (A-C) Top (A&B) and tilted (C) view SEM images showing PMMs containing a regular array of hexagonally arranged through holes with the hole diameter of either 6 μm (A), 30 μm (B), or 60 μm (C). The PMMs had a thickness of 10 μm. (D) Top view SEM image showing PDMS supporting pillars with a height of 100 μm and a diameter of 60 μm. (E) SEM image showing a cross-section view of a free-standing PMM bound to PDMS supporting posts. In E, the diameter of the PMM through hole was 30 μm.
Our PDMS surface micromachining technique is compatible with soft lithography techniques and other silicon-based micro-fabrication methods. Thus, the PMM can be easily integrated with PDMS supporting structures and other PDMS-based molecular and cellular analysis components while maintaining a large surface area and high porosity (Figure 2D). In this study, using O2 plasma-assisted PDMS-PDMS bonding, the PMM was assembled with two structured PDMS layers serving as the top cell culture chamber and the bottom immunoassay chamber to form a completely sealed PMM-integrated microfiltration device. The cell culture chamber and the immunoassay chamber in the microfiltration devices contained supporting pillar structures to provide the aforementioned mechanical strength for the PMM during cell filtration process (Figure 2E).
2.2. Capture of Subpopulation of Immune Cells Using Microbeads Conjugated with Antibodies
Surface functionalized micro/nano scale beads and particles are widely used for isolations of cells and proteins. Leukocytes are known to produce specific antigens on their surfaces. By using the biotin-streptavidin conjugation chemistry to coat polystyrene microbeads with antibodies directed against specific cell surface antigens, these microbeads could recognize and capture desired subpopulations of immune cells expressing specific surface proteins. Owing to the significant physical size difference between the microbeads and leukocytes, when the blood sample was passing through the PMM, the desired subpopulation of immune cells that were bound to functionalized microbeads could be efficiently isolated by the PMM, while the other blood cells including undesired immune cells would pass freely through the PMM.
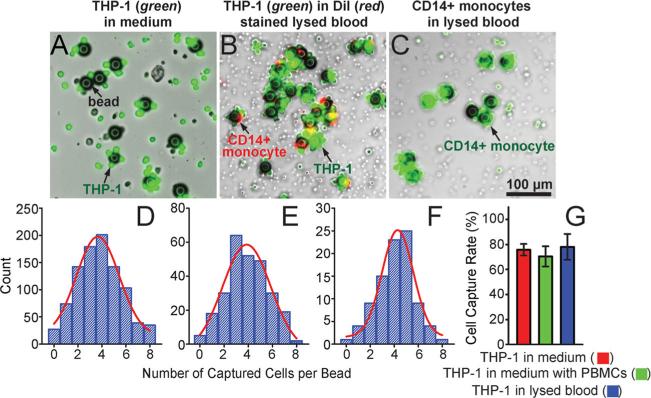
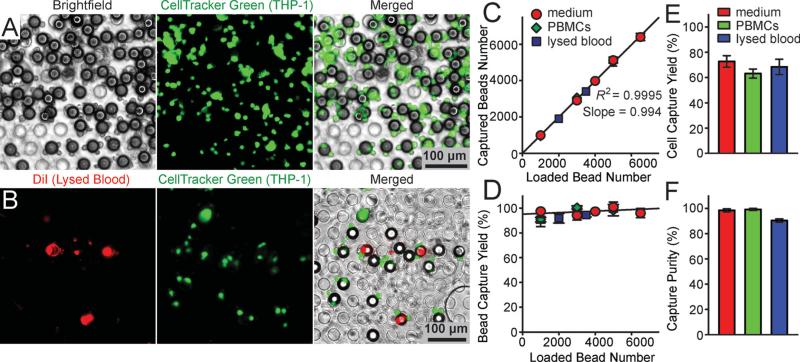
We first characterized the capture efficiency of functionalized microbeads with a diameter of 32 μm and conjugated with monoclonal antibodies directed against CD14, a unique surface protein marker of monocytes. Well-controlled quantities of the microbeads were mixed with known quantities of fluorescence-tagged CD14+ THP-1 cells, a human acute monocytic leukemia cell line as a model for human monocytes. THP-1 cells were suspended in either blank culture media, culture media spiked with known quantities of peripheral blood mononuclear cells (PBMCs) mimicking blood leukocytes, or lysed blood that was pre-stained with DiI to label blood leukocytes (see Experimental Section for details; the concentrations of THP-1 cells and microbeads were 2 × 106 mL−1 and 4 × 105 mL−1 , respectively). We also mixed well-controlled quantities of the microbeads directly with lysed blood that was pre-stained with Calcein AM to label blood leukocytes at a concentration of 10 × 106 mL−1 . The mixture of the microbeads and immune cells were incubated for 2 hrs on a roller mixer before they were examined under florescence microscopy (Figure 3A-C). Our results in Figure 3 D&E showed that for THP-1 cells spiked in growth media or lysed blood, on average, there were about 4 THP-1 cells bound to a single microbead following cell-bead mixing and incubation. Similar to results for THP-1 cells, on average, there were about 4 CD14+ monocytes from lysed blood bound to a single microbead following cell-bead mixing and incubation (Figure 3F). We further quantified the overall capture efficiency of the microbeads for THP-1 cells (defined as the ratio between the total number of THP-1 cells captured by microbeads and the total number of THP-1 cells in the sample). Our results in Figure 3G showed that for THP-1 cells spiked in culture media or lysed blood, the capture efficiency of the microbeads conjugated with anti-CD14 antibodies was all greater than 70%. Together, our data in Figure 3 strongly supported the utility of functionalized microbeads for efficient recognition and capture of desired subpopulations of immune cells from a complex mixture of background cells, such as blood leukocytes.
Figure 3.
Capture of immune cells using microbeads conjugated with anti-CD14 antibodies. (A-C) Representative merged brightfield and fluorescence images showing THP-1 cells (green; A&B) and CD14+ monocytes (green; C) captured by microbeads in either culture media (A) or lysed blood (B&C). The microbeads with a diameter of 32 μm were conjugated with anti-CD14 monoclonal antibodies. THP-1 cells were pre-stained with CellTracker Green. Blood leukocytes in B&C were pre-stained with DiI (B) or CellTracker Green (C). (D-F) Distributions of the number of cells captured per bead for THP-1 cells (D&E) and CD14+ monocytes (F) in either culture media (D) or lysed blood (E&F). For D&E, 2 × 106 THP-1 cells were mixed with 4 × 105 microbeads in 1 mL cell culture media (D) or lysed blood (E). For F, 2 mL lysed blood containing about 2 × 107 leukocytes was mixed with 3 × 105 microbeads. Gaussian functions (red curves) were used for fitting in D-F. (G) Capture rate of functionalized microbeads for THP-1 cells spiked in growth media, in growth media with PBMCs, and in lysed blood as indicated. Error bars represent ± standard error of the mean (s.e.m; n = 4).
2.3. Characterization of Capture Efficiency of the PMM for Microbeads and Cells
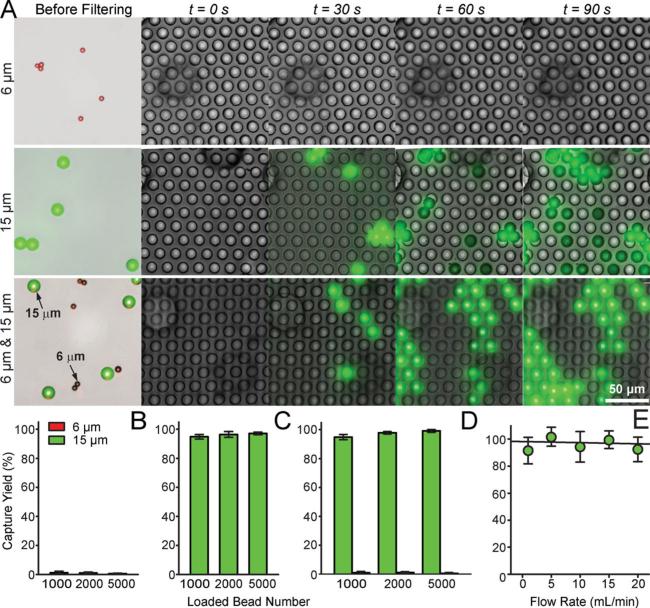
We first characterized the capture efficiency of the PMM using well-controlled quantities of fluorescence-tagged polystyrene microbeads spiked in PBS (Figure 4). The PMM used in these assays contained a regular array of through holes with a hole diameter of 13 μm and a center-to-center distance of 20 μm. The porosity of the PMM was about 25%. Two different sized polystyrene microbeads were assayed with the means of their diameters of 6 μm and 15 μm. The microbeads at a concentration of 2 × 104 mL−1 in the PBS were injected into the PMM-integrated microfiltration device using a syringe infusion pump under a constant flow rate of 100 μL min−1 . The fluorescence-tagged microbeads trapped on the PMM could be visually examined and enumerated under bright-field and fluorescence microscopy (Figure 4A). Since the diameter of the through holes in the PMM was 13 μm, smaller than the 15 μm microbeads but larger than the 6 μm ones, the PMM would capture virtually all the 15 μm microbeads while letting all the 6 μm ones passing through. Our quantitative results in Figure 4B-D demonstrated an extremely high efficiency of the PMM to separate the two different sized microbeads, as evidenced by a capture yield (defined as the ratio of the number of beads captured on the PMM to the total number of beads injected) and purity of 97.2% and 98%, respectively, for the 15 μm microbeads.
Figure 4.
Characterization of capture efficiency of the PMM using florescence-tagged microbeads. (A) Temporal sequence of merged brightfield and fluorescence images showing isolation of 6 μm (top), 15 μm (middle), and a mixture of 6 μm and 15 μm (1:1 ratio; bottom) florescence-tagged microbeads on a PMM with the through hole diameter of 13 μm. The microbead concentration was 2 × 104 mL−1, and they were injected through the PMM with a volume flow rate of 100 μL min−1 . Virtually all 15 μm beads were trapped on the PMM while all 6 μm beads passed through the PMM. (B&C) Captured yield of 6 μm (B) and 15 μm (C) microbeads as a function of the total number of microbeads injected through the PMM. Data was calutlated from the top and middle panels of images shown in A. (D) Captured yield of 6 μm and 15 μm microbeads from a mixture of 6 μm and 15 μm microbeads, as a function of the total number of microbeads injected through the PMM. Data was calculated from the bottom panel of images shown in A. (E) Capture yield of 15 μm microbeads as a function of the volume flow rate. The microbead concentration was 2 × 104 mL−1, and they were injected through the PMM with a varied volume flow rate. Error bars in B-E represent ± s.e.m. (n = 4).
As shown in Figure S2, by integrating different layers of PMMs with supporting pillars in a single microfluidic device, we developed a three-dimensional (3-D) multilayer gradient microfiltration device that could achieve efficient isolation and enrichment of different sized bioparticles simultaneously. The sizes of and the distances between the through holes on each PMM in the 3-D multilayer microfiltration device gradually decreased along the separation path, enabling a gradient filtration scheme. As shown in Figure S2, we could isolate and enrich both 15 μm and 6 μm microbeads simultaneously in an integrated double-layer microfiltration device, in which the diameter of the through holes in the top and bottom PMMs were chosen to be 13 μm and 5 μm, respectively. The 3-D multilayer gradient microfiltration device, when combined with different-sized antibody-conjugated microbeads targeting different surface proteins, could, in principle, provide an effective solution for isolation and enrichment of multiple subpopulations of immune cells simultaneously from blood specimens.
We further examined the effect of flow rate on the capture efficiency of the PMM, by comparing the capture efficiency of the PMM for 32 μm microbeads under different flow rate conditions (from 1 - 20 mL min−1; Figure 4E). The PMM contained an array of hexagonally arranged through holes of 30 μm in diameter and with a center-to-center distance of 50 μm. Our results in Figure 4E suggested that the PMM was able to achieve extremely high capture efficiency for the microbeads even under a flow rate as high as 20 mL min−1, without evidence showing plastic deformation or fracture of the PMM under the microscopy, which might be attributable to the PDMS supporting pillars integrated in the microfiltration device.
We next characterized the performance of the PMM with the through hole diameter of 30 μm for isolation of THP-1 cells bound to 32 μm microbeads functionalized with anti-CD14 antibodies (Figure 5). Here known quantities of THP-1 cells (2 × 106) labeled with CellTracker Green were mixed and incubated with the functionalized microbeads (4 × 105) for 2 hrs in either 1 mL cell culture medium containing 5 × 106 mL−1 PBMCs or 1 mL lysed blood containing about 6 × 106 mL−1 leukocytes and 5 × 105 mL−1 CD14+ monocytes. Different volumes of the culture medium and lysed blood samples were injected into the PMM-integrated microfiltration device at a flow rate of 1 mL min−1. The cells bound to the microbeads trapped on the PMM were examined under fluorescence microscopy to determine the capture efficiencies for the microbeads and THP-1 cells (Figure 5 A&B). As expected, virtually all the 32 μm microbeads (>99%), regardless whether bound with or without cells, were captured on the PMM (Figure 5C&D). The capture yield of THP-1 cells, defined here as the ratio between the number of THP-1 cells bound to microbeads captured on the PMM to the total number of THP-1 cells injected into the microfiltration device, was all greater than 70%, regardless whether THP-1 cells were spiked in culture media, culture media containing PBMCs, or lysed blood. In addition, capture purities of THP-1 cells, defined as the ratio between the number of THP-1 cells bound to microbeads captured on the PMM to the total number of cells captured on the PMM, was on average as high as 97%, regardless whether THP-1 cells were spiked in culture media, culture media containing PBMCs, or lysed blood. Together, our data in Figure 5 convincingly supported our approach of using functionalized microbeads and the PMM for efficient capture and isolation of subpopulations of immune cells with a high purity. From our data in Figure 5, we also concluded that the overall capture yield of our method to isolate subpopulations of immune cells was largely determined by the yield of the cell capture process using the functionalized microbeads, not by the microbead isolation process using the PMM.
Figure 5.
Characterization of capture efficiency of the PMM using samples spiked with THP-1 cells. (A&B) Representative brightfield, fluorescence, and merged microscopic images showing fluorescence-tagged THP-1 cells bound to microbeads isolated on the PMM. THP-1 cells labeled with CellTracker Green were spiked in either cell culture media (A) or lysed blood pre-stained with DiI to label blood leukocytes (B). The microbeads with a diameter of 32 μm were conjugated with anti-CD14 antibodies. The PMM had a through hole diameter of 30 μm. (C) Regression analysis of capture efficiency of the PMM for the functionalized microbeads used for capturing THP-1 cells. The plot represents the number of microbeads (bound with or without cells) captured on the PMM against the total number of microbeads (bound with or without cells) injected through the PMM. (D) Capture yield of microbeads (bound with or without cells) as a function of the total number of microbeads (bound with or without cells) injected through the PMM. (E&F) Capture yield (E) and purity (F) of THP-1 cells on the PMM. For C-F, known quantities of THP-1 cells (2 × 106) were mixed with microbeads (4 × 105) in 1 mL cell culture media (red symbols or bars), 1 mL cell culture media containing 5 × 106 mL−1 PBMCs (green symbols or bars), or 1 mL lysed blood containing about 6 × 106 mL−1 leukocytes (blue symbols or bars), as indicated. The volume flow rate was 1 mL min−1. Solid lines in C&D represent linear fitting. Error bars represent ± s.e.m (n = 4).
2.4. Isolation of Subpopulation of Immune Cells from Human Blood
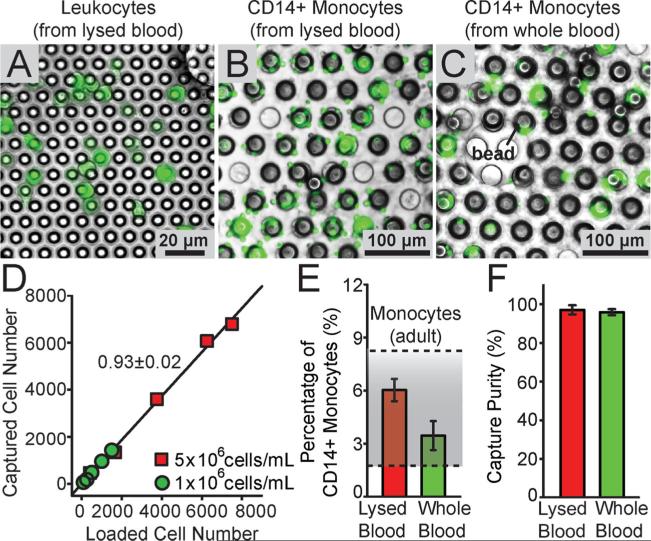
Using the PMM coupled with functionalized microbeads, we demonstrated direct isolation, enrichment, and enumeration of CD14+ monocytes from both lysed as well as whole blood specimens (Figure 6A-C). As a control, we first used a PMM with the through hole diameter of 6 μm to isolate the whole population of leukocytes from lysed blood (Figure 6A). Leukocytes in the lysed blood were stained with Calcein AM for visualization. Lysed blood with the leukocyte concentration of 5 × 106 mL−1 or diluted lysed blood with the leukocyte concentration of 1 × 106 mL−1 was filtered by the PMM with the through hole diameter of 6 μm in the microfiltration device at a flow rate of 1 mL min−1. The number of leukocytes trapped on the PMM increased lineally with the sample injection volume, and our regression analysis of capture efficiency of the PMM for leukocytes in Figure 6D revealed that about 93% of leukocytes in the lysed blood were captured on the PMM under a flow rate of 1 mL min−1.
Figure 6.
Isolation of CD14+ monocytes from blood specimens using the PMM. (A-C) Representative merged brightfield and fluorescence images showing flurescence-tagged leukocytes (A) and CD14+ monocytes (B&C) from lysed blood (A&B) or whole blood (C) isolated on the PMM. The PMM had a through hole diameter of 6 μm (A) or 30 μm (B&C). CD14+ monocytes were first captured by 30 μm microbeads conjugated with anti-CD14 antibodies before the cell-microbead conjugates were isolated on the PMM. Leukocytes in the blood samples were stained with Calcein AM for visualization. (D) Regression analysis of capture efficiency of the PMM with the through hole diameter of 6 μm for blood leukocytes, under a flow rate of 1 mL min−1 . The plot represents the number of leukocytes captured on the PMM against the number of leukocytes injected through the PMM, using undiluted (5 × 106 mL−1) as well as diluted (1 × 106 mL−1) lysed blood samples as indicated. Solid lines represent linear fitting. (E&F) Percentage (E) and purity (F) of CD14+ monocytes captured using functionalized microbeads and the PMM from lysed and whole blood. For E&F, known quantities of microbeads (3 × 105) were mixed with 2 mL lysed and whole blood (containing about 2 × 107 leucocytes). The shaded area in E indicates the normal percentage range of CD14+ monocytes in blood from healthy adults. Error bars represent ± s.e.m. (n = 4).
To isolate CD14+ monocytes from human blood, known quantities of functionalized 32 μm microbeads conjugated with anti-CD14 antibodies were added into blood samples. Prior to this, leukocytes in the blood sample were pre-labeled with Calcein AM. After 2-hr mixing and incubation, the blood specimens were injected through a PMM with the through hole diameter of 30 μm. Our results in Figure 6E showed that about 6.0% and 3.5% leukocytes from the lysed and whole blood samples, respectively, were bound to the 32 μm microbeads and subsequently captured by the PMM with the through hole diameter of 30 μm as CD14+ monocytes. These results are consistent with the fact that the normal percentage range of CD14+ monocytes in blood from healthy adults is from 2% - 8%.[33,34] Compared to the results for lysed blood, the capture yield of CD14+ monocytes from whole blood was slightly lower, likely due to the large number of erythrocytes (red blood cells) and other undesired leukocytes in whole blood blocking effective interactions between CD14+ monocytes and functionalized microbeads.
2.5. Functional Immunophenotyping of Subpopulations of Immune Cells
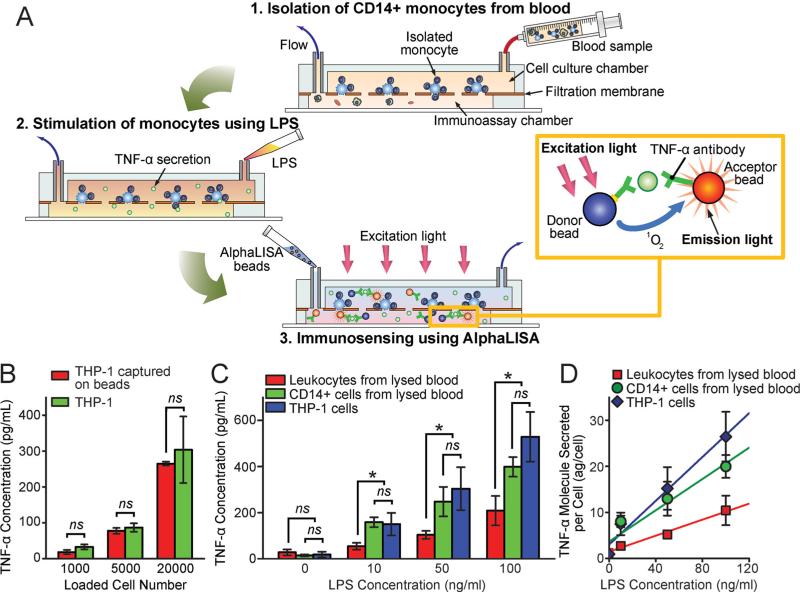
We further demonstrated functional immunophenotyping of subpopulations of immune cells using the PMM-integrated microfiltration device combined with a rapid, no-wash, quantitative immunosensing method, the AlphaLISA,[25–27] using minute quantities of blood. The overall immunophenotyping assay protocol using the micro-filtration device was shown in Figure 7A (see Experimental Section for details). After the desired subpopulation of immune cells (e.g. , CD14+ monocytes) was captured and enriched by functionalized microbeads and the PMM from blood specimens (Step 1 in Figure 7A), the cells trapped on the PMM were stimulated and incubated with LPS loaded from the inlet of the top cell culture chamber of the microfiltration device (Step 2 in Figure 7A). LPS is a canonical endotoxin that can cause septic shock in the human body due to severely pronounced immune response. Pro-inflammatory cytokines secreted from LPS-stimulated immune cells would diffuse rapidly through the PMM to the bottom immunoassay chamber, where the AlphaLISA donor and acceptor beads were loaded and optically detected in situ for quantitative immunosening (Step 3 in Figure 7A). The AlphaLISA signal detected could be converted to the cytokine concentration using standard curves generated using AlphaLISA with samples spiked with known concentrations of cytokines (Figure S3).
Figure 7.
Immunophenotyping of subpopulations of immune cells isolated from human blood. (A) Schematic showing the immunophenotyping assay protocol using the PMM-integrated microfiltration device and the AlphaLISA: (1) isolation and enrichment of CD14+ monocytes on the PMM from blood samples; (2) LPS stimulation of CD14+ monocytes trapped on the PMM to release pro-inflammatory cytokines; (3) Loading and incubation of AlphaLISA beads in the immunoassay chamber; (4) Detection of TNF-α using the AlphaLISA, in which the streptavidin-coated donor (blue) and acceptor beads (orange) were both conjugated with TNF-α antibodies. The beads were brought into a close proximity (<200 nm) through binding simultaneously to TNF-α. Using a 680 nm laser for excitation, the singlet oxygen released by the donor bead would diffuse to the nearby acceptor bead and trigger it to emit the 615 nm fluorescent light. Measurement of this emission light from the acceptor beads using our custom optical setup (Figure S3) would allow a quantitative detection of TNF-α secretion. (B) Comparison of TNF-α secretions from LPS-stimulated free THP-1 cells and THP-1 cells bound to microbeads. (C&D) Comparative study of TNF-α secretion from LPS-stimulated THP-1 cells, leukocytes isolated directly from lysed blood, and CD14+ monocytes isolated from lysed blood. For C&D, about 2 × 104 THP-1 cells, leukocytes, and CD14+ monocytes were trapped on the PMM before quantitative immunosensing. C&D plot the bulk TNF-α concentration and TNF-α molecules secreted per cell, respectively, as a function of LPS concentration. Solid lines in D represent linear fitting. Error bars represent ± s.e.m. (n = 4). P-values were calculated using the paired student's t-test. ns (P > 0.05; not significant), * (P < 0.05).
In this research, we applied the PMM-integrated microfiltration device and the AlphaLISA to detect secretion of TNF-α from LPS-stimulated CD14+ monocytes isolated from blood specimens. TNF-α is a proinflammatory cytokine and a key biomarker associated with host defense and immunosurveillance. Secretion of TNF-α from LPS-stimulated monocytes has been shown to reflect the functional status of the innate immune response. Our data in Figure 7B showed that TNF-α secretion from LPS-stimulated free THP-1 cells and THP-1 cells bound to microbeads were very similar, leading us to conclude that binding to anti-CD14 antibodies on microbeads would not significantly affect TNF-α secretion from CD14+ monocytes.
We next applied the PMM-integrated microfiltration device and the AlphaLISA for quantitative immunosensing of cytokines secreted from LPS-stimulated CD14+ monocytes isolated directly from lysed blood. The results were compared with cytokine secretions from LPS-stimulated leukocytes isolated directly from lysed blood and from LPS-stimulated THP-1 cells. In these assays, equal numbers of THP-1 cells, leukocytes and CD14+ monocytes (about 2 × 104 ) were trapped on the PMM before quantitative immunosensing. Our result in Figure 7C demonstrated that similar to THP-1 cells, TNF-α secreted by CD14+ monocytes isolated directly from lysed blood would increase with LPS concentration. Further, the amount of TNF-α secreted by CD14+ monocytes was very comparable to that from THP-1 cells but significantly higher as compared to TNF-α secretions from leukocytes isolated directly from lysed blood, confirming the functional heterogeneity between different types of immune cells.
3. Conclusions
In summary, here we developed a unique surface-micromachined bioparticle separation membrane made of elastometric organosilicon compounds, named the “PDMS microfiltration membrane (PMM)”. Integrating the PMM in a microfluidic microfiltration device, we successfully demonstrated a simple yet efficient strategy for on-chip isolation and enrichment of subpopulations of immnue cells bound to functionalized microbeads conjugated with antibodies against specific cell surface proteins. Owing to the high porosity and large surface area of the PMM, our PMM-integrated microfiltration device could process lysed and whole blood samples with a high volume throughput (up to 20 mL min−1 ). Our microfiltration device could achieve high capture yield (about 70%, largely depending on the cell-microbead capture process) and excellent capture purity (>97%) for subpopulations of immune cells from blood samples. Further, our microfiltration device could be directly integrated with the AlphaLISA biosensing technique and used for rapid, efficient on-chip immunophenotyping assays for subpopulations of immune cells isolated directly from minute quantities of blood samples. All together, our PMM-integrated microfluidic functional immunophenotyping technology could perform rapid, accurate, and sensitive cellular functional assays on different types or subpopulations of immune cells, which could provide an unprecedented level of information depth on the distribution of immune cell functionalities. Such an innovative tool enabled by the cell separation performance of the PMM will potentially allow comprehensive and systems-level immunomonitoring to define and characterize the “immunotype” of healthy individuals and patients, critical for clinical interventions and managements of patients with immune system disorders.
4. Experimental Section
Surface Micromachining of PMM
The PMM was fabricated using the surface micromachining technique for soft materials we have developed previously (Figure S1). Briefly, a silicon wafer was first activated using the O2 plasma (Plasma Cleaner PDC-001, Harrick Plasma, Ithaca, NY) for 2 min and silanized with (tridecafluoro-1,1,2,2,-tetrahydrooctyl)-1-trichlorosilane vapor (United Chemical Technologies, Bristol, PA) for 1 hr under vacuum to facilitate subsequent release of patterned PDMS layers. PDMS prepolymer (Sylgard-184, Dow Corning, Midland, MI) was prepared by thoroughly mixing the PDMS curing agent with the PDMS base monomer (wt: wt = 1: 10). PDMS prepolymer was then spin-coated on the silanized silicon wafer at a spin speed of 7,000 rpm and completely cured after baking at 110 °C for 4 hrs. The PDMS surface was activated using the O2 plasma for 5 min to allow a uniform photoresist coating for photolithography. After the O2 plasma activation, photoresist (AZ 9260, AZ Electronic Materials, Branchburg, NJ) was spin-coated on PDMS, soft-baked at 90 °C for 10 min, and then patterned using contact photolithography. The silicon wafer was then processed with reactive ion etching (RIE; LAM 9400, Lam Research, Fremont, CA) using SF6 and O2 gas mixtures to transfer patterns from patterned photoresist to the underlying PDMS layer. During RIE, reactive gas ions would etch exposed PDMS regions anisotropically. Photoresist was then striped using organic solvents, leaving patterned PDMS thin fi lms on the silicon wafer.
Microfluidic Device Fabrication and Assembly
The structure of the PMM-integrated microfluidic immunosensing device consisted of three different PDMS layers. The top and bottom PDMS layers were the cell culture and immunoassay chambers, respectively, and the middle layer was the PMM. The cell culture and immunoassay chambers were fabricated using soft lithography. Briefly, silicon molds were first fabricated using photolithography and deep reactive ion-etching (DRIE) (Deep Silicon Etcher, Surface Technology Systems, Allentown, PA). The silicon molds were then silanized with (tridecafluoro-1,1,2,2,-tetrahydrooctyl)-1-trichlorosilane vapor (United Chemical Technologies) for 4 hrs under vacuum to facilitate subsequent release of PDMS sturcutures from the silicon molds. PDMS prepolymer with a 1:10 wt ratio of PDMS curing agent to base monomer was poured onto the silicon molds and cured at 110 °C for 4 hrs. Fully cured PDMS structures were peeled off from the silicon molds, and excessive PDMS was trimmed using a razor blade. The O2 plasma-assisted PDMS-PDMS bonding process was then used to assemble the cell culture and immunoassay chambers with the PMM to form a completely sealed microfluidic immunosensing device. Assembly of the microfluidic immunosensing device was performed under eye inspection using some alignment marks on the cell culture and immunoassay chamber layers.
Scanning Electron Microscopy (SEM) Imaging
SEM images were taken to inspect the geometrical features of the PMM and the microfluidic immunosensing device. Briefly, the sample under inspection was mounted on stubs, sputtered with gold palladium, observed and photographed under a Hitachi SU8000 Ultra-High Resolution SEM machine (Hitachi High Technologies America, Inc., Pleasanton, CA).
Cell Culture and Reagents
THP-1 cells (ATCC, Manassas, VA) were cultured in the cell growth medium (RPMI-1640, ATCC, Manassas, VA) supplemented with 0.05 mM 2-mercaptoethanol (Invitrogen, Carlsbad, CA) and 10% (v/v) heat-inactivated fetal bovine serum (FBS; Atlanta Biological, Atlanta, GA). Cells were maintained at 37 °C with 5% CO2 and 100% humidity and passaged every two days.
Human Blood Specimen Collection and Processing
Human blood specimens were obtained from healthy donors and collected into the vacutainer containing ethylenediaminetetraacetic acid (EDTA) according to the standard protocol. Human blood specimens were processed and assayed within 6 hrs after blood collection. To lyse blood, RBC Lysis Buffer (eBioscience, San Diego, CA) was added to whole blood at a 10:1 v/v ratio. After incubation for 10 min at room temperature, the Lysis Buffer was diluted with 20 −30 mL PBS to stop lysing reaction. Then the solution was centrifuged at 300 g for 10 min before the supernatant was removed. Immune cells in the cell pellet were re-suspended in the cell growth medium before they were used for cell capture assays with functionalized microbeads. For whole blood assays, human blood specimen was centrifuged at 300 g for 10 min to remove plasma and supernatant before the specimen was used for cell capture assays with functionalized microbeads.
Cell Capture Using Microbeads Conjugated with Antibodies
For visualization, THP-1 cells were labeled with CellTracker Green (1 μM; Invitrogen) before spiked into cell culture media, while leukocytes in blood specimens were labeled with Δ9-DiI (2 μM; Invitrogen). Then known quantities (4 × 105) of polystyrene microbeads conjugated with anti-CD14 monoclonal antibodies (pluriSelect GmbH, Leipzig, Gemany) were mixed with known quantities of THP-1 cells (2 × 106) in 1 mL cell culture media, or cell culture media containing PBMCs (5 × 106 ) or lysed blood (6 × 106 leukocytes). After the sample and beads were mixed and incubated at room temperature for 1–2 hrs on a roller mixer (Invitrogen), target cells bound to the microbeads and ready for further isolation. For cell capture using real lysed blood or whole blood, known quantity of microbeads (3 × 105) were mixed with the prepared lysed blood sample containing 20 × 106 leukocytes in 2 mL cell culture medium and incubated at room temperature for 1-2 hrs on a roller mixer (Invitrogen). Then the leukocytes in samples were labeled with Calcein AM (working concentration 10 μM; Invitrogen) for visualization.
Capture of Microbeads and Cells Using PMM
To prevent non-specific interactions between the PMM and microbeads or immune cells, the microfluidic device containing the PMM was first filled with 2% w/w Pluronics F127 (P2443-250G, Sigma) in PBS for 10 min. The microfluidic device was then flushed twice with PBS to remove residual Pluronics F127 molecules. For microbead and cell capture assays, known quantities of microbeads or microbeads bound with immune cells were loaded into the microfiltration device using a syringe infusion pump (World Precision Instruments, Sarasota, FL) at a flow rate between 0.2 – 20 mL min−1. Captures of microbeads and immune cells on the PMM were monitored real-time under an inverted microscope (Nikon Eclipse Ti-S, Nikon, Melville, NY) equipped with an electron multiplying charge-coupled device (EMCCD) camera (Photometrics, Tucson, AZ). Sequential brightfield and fluorescence images were taken using a 10× objective (Ph1 ADL, numerical aperture or N.A. = 0.25, Nikon). A 130 W mercury lamp (Intensilight C-HGFIE, Nikon) was used in the microscope for illumination.
Quantification of Capture Efficiency of the PMM
The concentrations of microbeads as well as cells in each sample were first determined using a hemocytometer. The microbeads and cells isolated on the PMM were then imaged using fluorescence microscopy (Nikon Eclipse Ti-S, Nikon). To quantify capture efficiency of the PMM, the whole surface area of the PMM was scanned using a motorized stage (ProScan III, Prior Scientifi c, Rockland, MA). The images were stitched using microscopic analysis software, the NIS-Element BR from Nikon. Image processing software ImageJ (National Institutes of Health, Bethesda, MD) was then used to determine the total number of beads and cells captured on the PMM.
Immunophenotyping Assay using AlphaLISA
The overall immunophenotyping assay protocol is shown in Figure 6A. First, known quantities of immune cells (free-floating or bound to microbeads) in cell culture media, lysed blood or whole blood were loaded into the microfiltration device by a syringe pump (World Precision Instruments). After the cells were uniformly seeded on the PMM, 10 μL solutions of LPS (L5886, Sigma) of different concentrations (10, 50, and 100 ng mL−1) were loaded into the microfiltration device from the inlet of the cell culture chamber using pipette tips. After LPS loading, two pipette tips were inserted into the inlets of both the cell culture and immunoassay chambers to prevent evaporation and provide a shear stress free microenvironment for cell stimulation. The microfiltration device was then incubated at 37 °C with 5% CO2 for 2 hrs. Then, the pipette tip inserted into the inlet of the immunoassay chamber was replaced by another pipette tip filled with 10 μL AlphaLISA acceptor beads (10 μg mL−1; PerkinElmer, Waltham, MA) mixed with 10 μL biotinylated TNF-α antibody (10 nM). AlphaLISA acceptor beads injected into the microfiltration device were incubated with the immune cells at 37 °C with 5% CO2 for 1 hr, before another pipette tip filled with 10 μL AlphaLISA streptavidin-coated donor beads (400 μg mL−1) was loaded into the inlet of the immunoassay chamber. The whole microfiltration device was incubated at 37 °C with 5% CO2 for another 30 min. During the whole 1.5-hr AlphaLISA bead incubation period, TNF-α secreted by LPS-stimulated immune cells would diffuse from the top cell culture chamber through the PMM into the bottom immunoassay chamber to conjugate with antibody-coated donor and acceptor beads. After AlphaLISA bead incubation, the mirofiltration device was placed into our custom optical setup for AlphaLISA signal detection.
Optical Setup for Detection of AlphaLISA Signal
The microfiltration device was placed on a custom optical setup for detection of the AlphaLISA signal (Figure S3). In this setup, a 500 mW 680 nm laser diode (S-67-500C-100-H, Coherent, Clara, CA) was used as an excitation light to induce singlet oxygen from AlphaLISA donor beads. An electronic shutter (SmartShutter, Lambda SC, Shutter Instrument, Novato, CA) was placed in front of the laser to control the timing of laser pulse for excitation. The laser light was guided into the inverted fluorescent microscope (Nikon Eclipse Ti-S, Nikon) and passed through the excitation filter (680/13nm BrightLine, Semrock, Rochester, New York) and dichroic mirror (ZT660dcspxr, Chroma, Bellows Falls, VT). The 2X objective (CFI Plan Achromat, N.A. = 0.06, Nikon) was used to focus the excitation light onto the device. After the excitation, the 615nm AlphaLISA emission signal, which passed through the dichroic mirror and the 660 nm shortpass filter (ET660SP, Chroma, Bellows Falls, VT), was then detected by an electron multiplying charge-coupled device (EMCCD) camera (Photometrics, Tucson, AZ). The microscopic analysis software (NIS-Element BR, Nikon) was used to synchronize the timing of shutter in front of laser and the opening time of the EMCCD. The software simultaneously counted the AlphaLISA emission signal intensity from the EMCCD during the measurement.
Calibration of TNF-α Standard Curve
To generate the TNF-α standard curve, known amounts of TNF-α were spiked in RPMI media (0 – 5,000 pg mL−1). Then, 10 μg mL−1 AlphaLISA acceptor beads, 10 nM biotinylated TNF-α antibody and 400 μg mL−1 streptavidin-coated donor beads were added to the RPMI media. The AlphaLISA signal was measured using our custom optical setup and was plotted as a function of TNF-α concentration to generate the TNF-α standard curve.
Supplementary Material
Acknowledgements
We acknowledge financial support from the National Science Foundation (ECCS 1231826 to Fu, ECCS 0601237 to Kurabayashi), the UM-SJTU Collaboration on Biomedical Technologies (Fu), the UM Comprehensive Cancer Center Prostate SPORE Pilot Project (Fu), the Michigan Institute for Clinical & Health Research (MICHR) Pilot Program (CTSA UL1RR024986 to Fu, Kurabayashi, Cornell, and Shanley), the Coulter Foundation (Kurabayashi), the National Institute of Health (R01HL097361 to Shanley, K08HD062142 and K12HD047349 to Cornell). N. T. Huang was partially supported by the University of Michian Rackham Predoctoral Fellowship. The Lurie Nanofabrication Facility at the University of Michigan, a member of the National Nanotechnology Infrastructure Network (NNIN) funded by the National Science Foundation, is acknowledged for support in microfabrication.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Weiqiang Chen, Department of Mechanical Engineering University of Michigan Ann Arbor, MI 48109 USA.
Nien-Tsu Huang, Department of Mechanical Engineering University of Michigan Ann Arbor, MI 48109 USA.
Boram Oh, Department of Mechanical Engineering University of Michigan Ann Arbor, MI 48109 USA.
Raymond H. W. Lam, Department of Mechanical and Biomedical Engineering City University of Hong Kong, Hong Kong, China
Rong Fan, Department of Biomedical Engineering Yale University New Haven, CT 06511, USA
Timothy T. Cornell, Department of Pediatrics and Communicable Diseases University of Michigan, Ann Arbor, MI 48109, USA
Thomas P. Shanley, Department of Pediatrics and Communicable Diseases University of Michigan, Ann Arbor, MI 48109, USA
Katsuo Kurabayashi, Department of Mechanical Engineering University of Michigan Ann Arbor, MI 48109 USA; Department of Electrical Engineering and Computer Science University of Michigan Ann Arbor, MI 48109 USA.
Jianping Fu, Department of Mechanical Engineering University of Michigan Ann Arbor, MI 48109 USA; Department of Biomedical Engineering University of Michigan Ann Arbor, MI 48109 USA.
References
- 1.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. Nature. 2002;417:95. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 2.Pantaleo G, Harari A. Nat. Rev. Immunol. 2006;6:417. doi: 10.1038/nri1840. [DOI] [PubMed] [Google Scholar]
- 3.Reece WH, Pinder M, Gothard PK, Milligan P, Bojang K, Doherty T, Plebanski M, Akinwunmi P, Everaere S, Watkins KR, Voss G, Tornieporth N, Alloueche A, Greenwood BM, Kester KE, McAdam KP, Cohen J, Hill AV. Nat. Med. 2004;10:406. doi: 10.1038/nm1009. [DOI] [PubMed] [Google Scholar]
- 4.Revzin A, Maverakis E, Chang HC. Biomicrofl uidics. 2012;6:21301. doi: 10.1063/1.4706845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gordon S, Taylor PR. Nat. Rev. Immunol. 2005;5:953. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 6.O'Shea JJ, Hunter CA, Germain RN. Nat. Immunol. 2008;9:450. doi: 10.1038/ni0508-450. [DOI] [PubMed] [Google Scholar]
- 7.Kaech SM, Wherry EJ. Immunity. 2007;27:393. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Re F, Strominger JL. Immunobiology. 2004;209:191. doi: 10.1016/j.imbio.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Salgame P, Abrams JS, Clayberger C, Goldstein H, Convit J, Modlin RL, Bloom BR. Science. 1991;254:279. doi: 10.1126/science.254.5029.279. [DOI] [PubMed] [Google Scholar]
- 10.Siliciano RF, Lawton T, Knall C, Karr RW, Berman P, Gregory T, Reinherz EL. Cell. 1988;54:561. doi: 10.1016/0092-8674(88)90078-5. [DOI] [PubMed] [Google Scholar]
- 11.Veazey RS, Marx PA, Lackner AA. Trends Immunol. 2001;22:626. doi: 10.1016/s1471-4906(01)02039-7. [DOI] [PubMed] [Google Scholar]
- 12.Harari A, Cellerai C, Pantaleo G. Curr. Opin. HIV AIDS. 2006;1:22. doi: 10.1097/01.COH.0000194103.28063.35. [DOI] [PubMed] [Google Scholar]
- 13.Romagnani S. Clin. Immunol. Immunopathol. 1996;80:225. doi: 10.1006/clin.1996.0118. [DOI] [PubMed] [Google Scholar]
- 14.Clerici M, Hakim FT, Venzon DJ, Blatt S, Hendrix CW, Wynn TA, Shearer GM. J. Clin. Invest. 1993;91:759. doi: 10.1172/JCI116294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. J. Exp. Med. 1993;178:2249. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boomer JS, To K, Chang KC, Takasu O, Osborne DF, Walton AH, Bricker TL, Jarman SD, 2nd, Kreisel D, Krupnick AS, Srivastava A, Swanson PE, Green JM, Hotchkiss RS. J. Am. Med. Assoc. 2011;306:2594. doi: 10.1001/jama.2011.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harari A, Dutoit V, Cellerai C, Bart PA, Du Pasquier RA, Pantaleo G. Immunol. Rev. 2006;211:236. doi: 10.1111/j.0105-2896.2006.00395.x. [DOI] [PubMed] [Google Scholar]
- 18.Bart PA, Pantaleo G. AIDS. 2006;20:617. doi: 10.1097/01.aids.0000199827.79983.66. [DOI] [PubMed] [Google Scholar]
- 19.Remick DG. Am. J. Pathol. 2007;170:1435. doi: 10.2353/ajpath.2007.060872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy TJ, Paterson HM, Mannick JA, Lederer JA, Leukocyte Biol J. 2004;75:400. doi: 10.1189/jlb.0503233. [DOI] [PubMed] [Google Scholar]
- 21.Cohen J. Nature. 2002;420:885. doi: 10.1038/nature01326. [DOI] [PubMed] [Google Scholar]
- 22.Hotchkiss RS, Karl IE, Engl N. J. Med. 2003;348:138. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 23.Chen W, Lam RH, Fu J. Lab Chip. 2012;12:391. doi: 10.1039/c1lc20721k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang NT, Chen W, Oh BR, Cornell TT, Shanley TP, Fu J, Kurabayashi K. Lab Chip. 2012;12:4093. doi: 10.1039/c2lc40619e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bielefeld-Sevigny M. Assay Drug. Dev. Technol. 2009;7:90. doi: 10.1089/adt.2009.9996. [DOI] [PubMed] [Google Scholar]
- 26.Poulsen F, Jensen KB. J. Biomol. Screen. 2007;12:240. doi: 10.1177/1087057106297566. [DOI] [PubMed] [Google Scholar]
- 27.Leister KP, Huang R, Goodwin BL, Chen A, Austin CP, Xia M. Curr. Chem. Genomics. 2011;5:21. doi: 10.2174/1875397301105010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cox JH, Ferrari G, Janetzki S. Methods. 2006;38:274. doi: 10.1016/j.ymeth.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 29.Guerkov RE, Targoni OS, Kreher CR, Boehm BO, Herrera MT, Tary-Lehmann M, Lehmann PV, Schwander SK, Immunol J. Methods. 2003;279:111. doi: 10.1016/s0022-1759(03)00240-0. [DOI] [PubMed] [Google Scholar]
- 30.Hofman VJ, Ilie MI, Bonnetaud C, Selva E, Long E, Molina T, Vignaud JM, Flejou JF, Lantuejoul S, Piaton E, Butori C, Mourad N, Poudenx M, Bahadoran P, Sibon S, Guevara N, Santini J, Venissac N, Mouroux J, Vielh P, Hofman PM. Am. J. Clin. Pathol. 2011;135:146. doi: 10.1309/AJCP9X8OZBEIQVVI. [DOI] [PubMed] [Google Scholar]
- 31.Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schutze K, Capron F, Franco D, Pazzagli M, Vekemans M, Lacour B, Brechot C, Paterlini-Brechot P. Am. J. Pathol. 2000;156:57. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng S, Lin H, Lu B, Williams A, Datar R, Cote R, Tai Y-C. Biomed. Microdevices. 2011;13:203. doi: 10.1007/s10544-010-9485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Górska E, Demkow U, Pińkowski R, Jakubczak B, Matuszewicz D, Gawȩda J, Rzeszotarska W, Wa̧sik M. Centr. Eur. J. Immunol. 2006;31:1. [Google Scholar]
- 34.Toner M, Irimia D. Annu. Rev. Biomed. Eng. 2005;7:77. doi: 10.1146/annurev.bioeng.7.011205.135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.