Abstract
The mechanisms by which n-3 polyunsaturated fatty acids (PUFAs) decrease colon tumor formation have not been fully elucidated. Examination of genes up- or down-regulated at various stages of tumor development via the monitoring of gene expression relationships will help to determine the biological processes ultimately responsible for the protective effects of n-3 PUFA. Therefore, using a 3 × × × 2 factorial design, we used Codelink DNA microarrays containing ∼9000 genes to help decipher the global changes in colonocyte gene expression profiles in carcinogen-injected Sprague Dawley rats. Animals were assigned to three dietary treatments differing only in the type of fat (corn oil/n-6 PUFA, fish oil/n-3 PUFA, or olive oil/n-9 monounsaturated fatty acid), two treatments (injection with the carcinogen azoxymethane or with saline), and two time points (12 hours and 10 weeks after first injection). Only the consumption of n-3 PUFA exerted a protective effect at the initiation (DNA adduct formation) and promotional (aberrant crypt foci) stages. Importantly, microarray analysis of colonocyte gene expression profiles discerned fundamental differences among animals treated with n-3 PUFA at both the 12 hours and 10-week time points. Thus, in addition to demonstrating that dietary fat composition alters the molecular portrait of gene expression profiles in the colonic epithelium at both the initiation and promotional stages of tumor development, these findings indicate that the chemopreventive effect of fish oil is due to the direct action of n-3 PUFA and not to a reduction in the content of n-6 PUFA.
INTRODUCTION
Second only to lung cancer, colon cancer is a major cause of cancer death in the United States (1). Environmental factors, including diet, are known to influence colon cancer incidence (2). Epidemiologic, clinical and laboratory animal based studies indicate that the type of dietary fat has been shown to modulate colon tumor development. Specifically, diets rich in n-6 polyunsaturated fatty acids (PUFAs) enhance the development of colon tumors (3, 4), whereas n-3 PUFA-containing diets reduce colon cancer incidence (2, 3, 5–7). This is significant because the typical Western diet contains 10 to 20 times more n-6 than n-3 PUFA (8). In addition, diets containing appreciable levels of n-9 monounsaturated fatty acid (MUFA), e.g., olive oil, may also have chemopreventive activity against colon carcinogenesis (9–11). The underlying mechanisms by which dietary fat composition exerts tumor enhancing or inhibiting effects has been under examination (12–21), and some mechanisms at the molecular level are beginning to emerge. For example, rats that receive injections of the carcinogen azoxymethane (AOM) and are fed n-3 PUFA-containing diets have higher levels of apoptosis in colon cells compared with n-6 PUFA-fed rats, possibly due to alteration in mitochondrial membrane composition and hence reactive oxygen species generation (19–21). In addition, n-3 PUFA feeding has been associated with decreased levels of oxidative DNA damage in rats (18). Expression of bcl-2, protein kinase C βII, and transforming growth factor β receptor II have all been shown to be altered by n-3 PUFA feeding, resulting in decreased colonocyte proliferation (17, 20). Additionally, n-3 PUFA feeding of rats and docosahexaenoic acid (DHA; 22:6n-3) treatment of colonocytes in culture has been shown to reduce Rasp21 activation and downstream signaling, cyclooxygenase 2, inducible nitric oxide synthase expression, and suppress the activity of peroxisome proliferator-activated receptors (PPARs; refs. 16, 22–25). These and other mechanisms are likely involved in the role of dietary lipid in colon tumor development.
Because it is difficult to study the subtle global and site-specific epigenetic mechanisms involved in colon tumor development, we used DNA microarrays to help decipher the many changes in gene expression due to fish oil/n-3 PUFA, corn oil/n-6 PUFA, and olive oil/n-9 monounsaturated fat treatment during the tumorigenic process in a highly relevant rat colon cancer model. Because environmental factors may act at either the initiation or promotion/progression phase (3, 26), we examined both time points to delineate the chemopreventive effects of n-3 PUFA at the earliest stage of carcinogenesis, as well as during a later stage when preneoplastic changes are beginning to occur. By comparing the biological properties of three distinct classes of dietary lipids, we show for the first time that the chemopreventive properties of fish oil are due to the specific addition of n-3 PUFA and not indirectly to a reduction in n-6 PUFA content in the diet.
MATERIALS AND METHODS
Animals
Ninety weanling male Sprague Dawley rats (Harlan, Houston, TX) were acclimated for 1 week in a temperature and humidity controlled facility on a 12-hour light/dark cycle. The animal use protocol was approved by the University Animal Care Committee of Texas A&M University and conformed to NIH guidelines. The study was a 3 × 2 × 2 factorial design with three types of dietary fat (n-6 PUFA, n-3 PUFA, or n-9 MUFA), two treatments (injection of AOM or saline), and two time points (12 hours and 10 weeks after first injection). Animals were stratified by body weight after the acclimation period so that mean initial body weights did not differ. Body weight and food intake were monitored during the study.
Diets
After 1-week acclimation on standard pelleted diet, rats were assigned to one of three diet groups which differed only in type of fat. The diets contained (g/100 g diet): dextrose, 51.06; casein, 22.35; D,L-methionine, 0.34; AIN-76 salt mix, 3.91; AIN-76 vitamin mix, 1.12; choline chloride, 0.22; and pectin, 6.00. The total fat content of each diet was 15% by weight as follows: n-6 diet, 15.00 g corn oil/100 g diet; n-3, 11.50 g fish oil/100 g diet; and 3.50 g corn oil/100 g diet; n-9 MUFA, 11.50 g olive oil/100 g diet and 3.5 g corn oil/100 g diet. The fats were chosen based on their different fatty acid content, with the corn oil diet being enriched in n-6 PUFA, the fish oil diet enriched in n-3 PUFA, and the olive oil diet enriched in n-9 MUFA. The fish oil and olive oil diets each contained 3.5% corn oil to ensure that essential fatty acid requirements were met. To prevent formation of oxidized lipids, diets were stored at −20°C and freely provided to the animals fresh each day. Corn oil, ultrarefined marine lipids, and olive oil were obtained from Degussa BioActives (Champaign, IL). To protect against lipid oxidation during storage, 0.025% tertiary butylhydroquinone and mixed tocopherols (MTS-50; ADM, Decatur, IL) were added to the oils. Mixed tocopherols were added to the fish and olive oils so that total tocopherol levels were equivalent to the endogenous level in the corn oil (0.7 mg/g oil).
Carcinogen Treatment
After 3 weeks on the experimental diets, 10 rats per diet group received injections of saline (control), and 20 rats per group received injections of AOM (Sigma, St. Louis, MO) s.c. at 15 mg/kg body weight. Five rats per diet/treatment group were killed 12 hours after the AOM or saline injection. The remaining rats received a second AOM or saline injection 1 week later and were terminated 10 weeks after the first AOM injection.
RNA Isolation
Upon termination, each colon (five per diet/treatment/time group) was resected proximally at the junction between the cecum and colon and distally at the rectum. The colon was opened longitudinally, flushed clean with PBS, and scraped with a glass slide to remove the mucosal layer, which was placed into denaturation solution (Totally RNA kit; Ambion, Austin, TX), homogenized on ice with a Teflon-in-glass homogenizer, and frozen at −80°C until RNA was isolated using the Totally RNA kit followed by DNase treatment (DNA Free; Ambion). After isolation, RNA integrity was assessed using agarose/formaldehyde gels and an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA).
Gene Expression Bioarrays
The Amersham (Piscataway, NJ) CodeLink system using the UniSet Rat I Expression Bioarray, containing 9028 genes probes, was used to generate colonic global transcription profiles. This array contains a broad range of genes derived from publicly available, well-annotated mRNA sequences. The CodeLink array is unique in being capable of detecting minimal differences in gene expression, as low as 1.3-fold with 95% confidence, because of the novel three-dimensional aqueous gel matrix, which the empirically tested 30-mer oligonucleotides are deposited on (27). This substantially reduces background, enhances sensitivity, and allows for the detection and quantification of subtle regulatory relationships among genes in a highly relevant colon cancer model system.
Target Preparation and Array Hybridization
All reagents were provided in the CodeLink expression assay kit (Amersham), except where noted. cRNA synthesis was performed as per manufacturer’s instructions using 10 µg of total RNA. First-strand cDNA was generated using Superscript II reverse transcriptase and a T7 primer. Subsequently, second-strand cDNA was produced using Escherichia coli DNA polymerase I and RNase H. The resultant double-stranded cDNA was purified on a QIAquick column (Qiagen, Valencia, CA) and cRNA was generated via an in vitro transcription reaction using T7 RNA polymerase and biotin-11-UTP (Perkin-Elmer, Boston, MA). cRNA was purified on an RNeasy column (Qiagen), quantified by UV spectrophotometry, and 10 µg were then fragmented by heating at 94°C for 20 minutes in the presence of magnesium. The fragmented cRNA was hybridized overnight at 37°C in hybridization buffer to a UniSet Rat I Bioarray in an Innova 4080 shaking incubator (New Brunswick, Edison, NJ) at 300 rpm.
Posthybridization Processing and Scanning
After hybridization, the arrays were washed in 0.75× TNT buffer [1× TNT: 0.1 mol/L Tris-HCl (pH 7.6), 0.15 mol/L NaCl, and 0.05% Tween 20] at 46°C for 1 hour followed by incubation with streptavidin-Alexa 647 (Molecular Probes, Eugene, OR) at room temperature for 30 minutes in the dark. Arrays were then washed in 1× TNT twice for 5 minutes each followed by a rinse in 0.05% Tween 20 in water. The slides were then dried by centrifugation and kept in the dark until scanning. Images were captured on an Axon GenePix Scanner (Arlington, TX) using CodeLink Expression Scanning Software and were analyzed using CodeLink Expression Analysis Software.
PCR Confirmation of Array Data
To confirm expression levels of a subset of genes, real-time PCR was performed on an ABI 7700 instrument. cDNA was synthesized from 4 µg of total RNA using random hexamers and oligo dT primers with Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). PCR was performed (primer sequences available online)6 using SYBR green master mix (Applied Biosystems, Foster City, CA) and with a predeveloped assay for glycogen synthase 2 (ABI). Expression levels were normalized to 18S expression using a predeveloped assay kit from Applied Biosystems.
Aberrant Crypt Foci Scoring
Immediately after removal, colons (10 per diet group with AOM injection) were flattened between Whatman 1 filter paper and fixed in 70% ethanol for 24 hours. Subsequently, the whole mount colon was stained with 0.5% methylene blue in PBS for 30 seconds, placed on a plastic sheet with a 5-mm grid, and examined under the microscope at ×400. The number of aberrant crypts (putative colon cancer precursors), as singlets and multiples, was determined. Crypts were classified as aberrant using the morphologic characteristics described previously (28).
In vivo Measurement of N7-Methylguanine
Distal colonic sections were fixed in ethanol for an in vivo measurement of N7-methylguanine adducts as previously described (29) using mouse monoclonal anti-N7-methylguanine. The specificity of this monoclonal antibody has been described previously (30). Omission of primary antibody and preadsorption with ligand were used as negative controls (29). At least 20 crypt columns per animal that met architectural criteria were chosen for analysis. The staining intensity was assessed by cell position within the crypt as described previously (29, 31). Images of colonic crypts were captured on a MICROSTAR IV Reichert microscope networked to a Sony DXC-970 MD 3CCD camera and processed using NIH Image, version 1.61. Captured images were adjusted using offset and gain to optimize the image brightness and contrast so that the greatest difference between light- and dark-stained pixels was achieved. Offset and gain were determined by preanalysis of multiple darkly and lightly stained tissues to expand the scale in the necessary region to enhance detection. Once established, the settings remained constant for all samples. Representative photomicrographs have been published (29, 32).
In situ Apoptosis Measurement
Apoptosis was measured in paraformaldehyde-fixed, paraffin-embedded tissues using the terminal deoxynucleotidyl transferase-mediated nick end labeling assay (Intergen, Norcross, GA) as described by Hong et al. (29). Crypt height in number of cells, as well as the number and position of apoptotic cells, were recorded in 100 well-oriented crypts per rat.
Statistics
DNA adduct, apoptosis, and aberrant crypt foci levels were analyzed by ANOVA to determine the effect of fat on outcome of each assay. For the analysis of global transcriptional profiles, we used a linear mixed model (33) for repeated measures to assess gene expression in relation to dietary lipid source (n-3 PUFA, n-6 PUFA, and n-9 MUFA) and treatments (saline, AOM). Genes were ranked by their significance (P values) to contrast differences in gene expressions between dietary fats. Thus, to examine the possible mechanism by which n-3 PUFA decreases colon cancer risk, we used P values from comparing n-3 PUFA with other dietary fats combined. Statistical analyses were performed using the statistical software R7 and SAS Institute, Inc. (Cary, NC).
RESULTS
Food Intake and Weight Gain
Neither carcinogen nor dietary treatment significantly altered food consumption or weight gain (P > 0.05, data not shown).
Dietary Fish Oil Reduces DNA Adduct Formation at the Initiation Stage
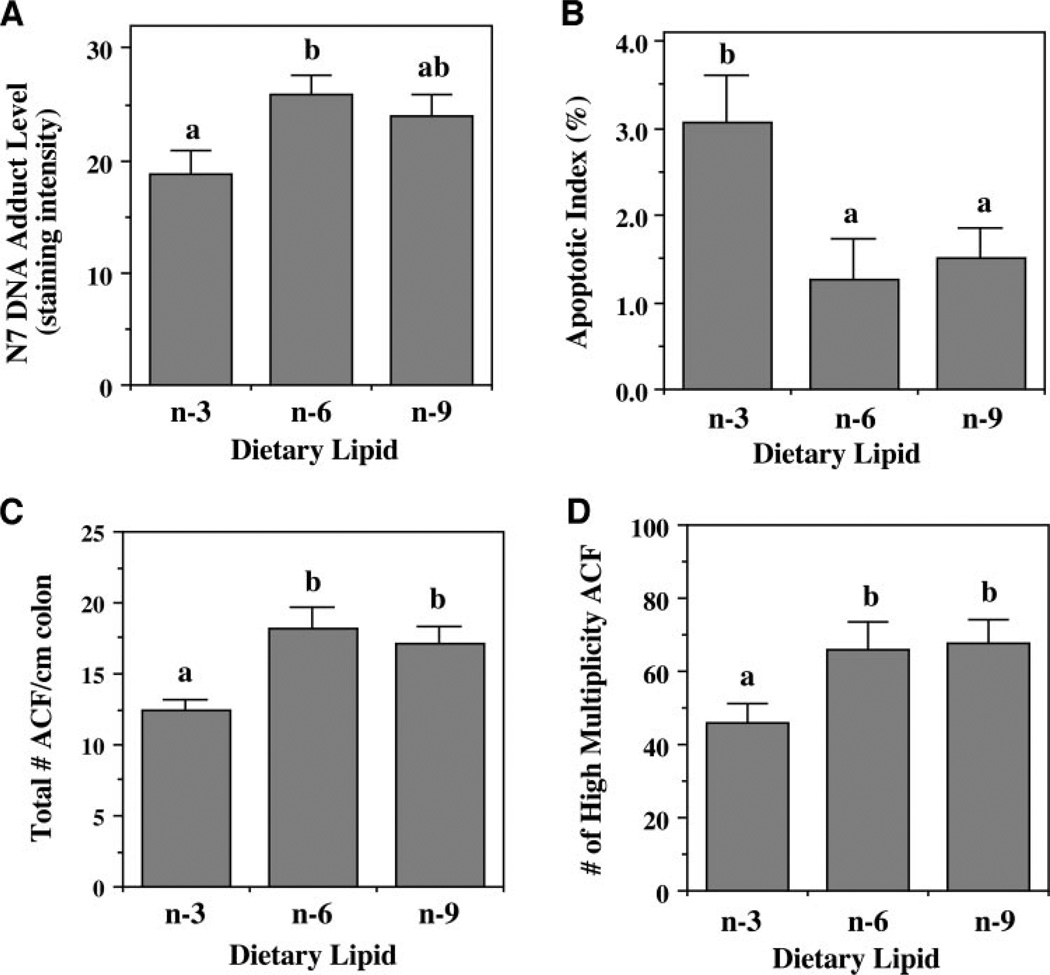
N7-Methylguanine is the major product of DNA alkylation in the colon (34) and is rapidly generated after carcinogen administration (19). Therefore, we examined the effect of dietary lipid source on N7-methylguanine adduct levels in the colon 12 hours after AOM injection. Immunohistochemical staining combined with digital image analysis was used to determine the level and localization of adducts within the crypt epithelial cell nuclei. As shown in Fig. 1A, upon examination of the entire colonic crypt, corn oil-fed animals had 37% more (P = 0.030) DNA adducts compared with fish oil-fed animals. In contrast, a moderate but nonsignificant (P = 0.122) increase (27%) in the n-9 MUFA-fed rats compared with the n-3 PUFA group was observed. These data indicate a suppressive effect of fish oil on adduct levels in the colon.
Fig. 1.
Effect of dietary lipid source on colonocyte phenotype. Rats fed diets containing either n-3, n-6, or n-9 fatty acid-enriched oils received injections of AOM or saline and were terminated 12 hours or 10 weeks later. A. N7-methylguanine DNA adduct levels 12 hours after carcinogen injection were determined by immunohistochemistry followed by analysis of staining intensity. n= 4 rats per treatment, with 20 crypts per animal scored. B. Apoptotic index in the top third (upper tertile) of the colonic crypt was assessed 10 weeks after AOM or saline injection by terminal deoxynucleotidyl transferase-mediated nick end labeling assay on 10 rats per treatment, with 100 crypts per animal scored. AOM- and saline-injected animals were pooled within diet groups because no injection effect was observed (P= 0.79 for injection). Data are expressed as total number of apoptotic cells per 100 crypts. C and D. Aberrant crypt foci (ACF) were scored after methylene blue staining of colons from rats terminated 10 weeks after AOM injection. Total number of ACF per cm of colon (C), as well as number of high multiplicity ACF (more than three aberrant crypts per foci; D) were scored. n= 10 rats per diet group with 20 crypts per animal scored. P for total ACF/cm = 0.010; P for high multiplicity ACF = 0.041
Dietary Fish Oil Enhances Apoptosis at the Promotional Stage
The localization of apoptosis within the crypt is important because there is a distinct hierarchical arrangement of epithelial cells in the colon (31). For example, spontaneous apoptosis generally occurs toward the luminal surface. Therefore, by knowing the position of a cell within the crypt, assumptions can be made regarding the stage of its life cycle. At 10 weeks after the second AOM injection, most of the apoptosis was localized toward the top of the crypt (data not shown). Although there was no overall (combination of all crypt regions) effect of lipid source on apoptosis, fish oil feeding resulted in a significantly higher (P = 0.034) level of apoptosis in the upper one-third (upper tertile) of the crypt compared with corn oil and olive oil treatments (Fig. 1B). This effect was not influenced by carcinogen administration. In contrast to the 10-week time point, there was no significant effect of diet on apoptosis at 12 hours postinjection (data not shown).
Fish Oil Fed Rats Are Less Susceptible to Formation of Carcinogen-Induced Aberrant Crypt Foci
Decreased apoptosis is a significant risk factor for development of colon cancer (5, 35). Hence, as a follow up to this analysis, we assessed to what degree dietary lipid source can modulate aberrant crypt foci formation. Aberrant crypt foci represent well-established preneoplastic colonic lesions in both rodents and humans (28, 36). The number of high multiplicity aberrant crypt foci (with greater than three aberrant crypts in a cluster) is thought to be the best predictor for cancer development (32, 37). Therefore, 10 weeks after the first AOM injection, rats were killed, and their colons were analyzed for the presence of aberrant crypt foci. No aberrant crypt foci were observed in saline-injected animals (data not shown) in agreement with the literature (32, 37). Both the total number of aberrant crypt foci/cm of colon (Fig. 1C), and the number of high multiplicity aberrant crypt foci/colon (Fig. 1D) was significantly lower (P < 0.05) in fish oil compared with corn- and olive oil-fed animals. The total number of high multiplicity aberrant crypt foci per colon, putative colon tumor precursors, was decreased by 30% in the n-3-fed compared with n-6- and n-9-fed rats.
Global Transcriptional Profiling
The mechanisms by which n-3 PUFA decrease colon cancer risk have not been elucidated to date. Examination of genes up- or down-regulated at various stages of the tumorigenic process via the monitoring of gene expression relationships will help to elucidate the biological processes ultimately responsible for the protective effects of n-3 PUFA. Therefore, colonic mucosa RNA from five samples per treatment group was analyzed using the Rat UniSet Expression Bioarray. Results summarized in Table 1–4 indicate that over time, a dietary lipid source had a substantial effect on genes involved in a wide variety of biological processes. Because n-3 PUFA feeding clearly reduced the risk for developing colon cancer (Fig. 1A–D), n-3 PUFAs were contrasted with n-6 PUFA and n-9 MUFA (i.e., n-3 versus other) gene expression profiles at 12 hours and 10 weeks after saline or AOM injection. This allowed us to focus on those genes in which the expression varied nontrivially across samples at two time points.
Table 1.
Relative colonic gene expression in n-3– versus n-6– or n-9 –fed, saline-injected (12 hours) control rats
| Accession no. | P | Relative expression |
Gene name | |
|---|---|---|---|---|
| n-3/n-6 | n-3/n-9 | |||
| Apoptosis | ||||
| U75689 | 0.00529 | 0.708 | 0.613 | DNaseγ |
| Cell cycle/Cell growth | ||||
| D63665 | 0.00173 | 1.610 | 1.362 | Novel G protein-coupled P2 receptor |
| AJ131902 | 0.00247 | 0.669 | 0.592 | GAS-7 |
| NM_021669 | 0.00293 | 0.674 | 0.675 | Ghrelin precursor |
| M31837 | 0.00890 | 0.750 | 0.549 | Insulin-like growth factor binding protein 3 precursor |
| Cytoskeletal/Extracellular matrix | ||||
| NM_012794 | 0.12620 | 0.506 | 0.187 | Glycosylation dependent cell adhesion molecule (Glycam) |
| Inflammation/Immunology | ||||
| NM_017079 | 0.00087 | 1.525 | 1.337 | CD1D antigen (Cd1d) |
| Metabolism/Redox | ||||
| NM_012789 | 0.00030 | 1.662 | 1.434 | Dipeptidyl peptidase 4 (Dpp4) |
| NM_012595 | 0.00107 | 0.721 | 0.560 | Lactate dehydrogenease B (Ldhb) |
| NM_019303 | 0.00136 | 3.471 | 2.701 | Cytochrome P450, subfamily IIF, polypeptide 1 (Cyp2f1) |
| J02997 | 0.00174 | 1.478 | 1.533 | Bile canaliculus domain-specific membrane glycoprotein |
| AB042598 | 0.00224 | 0.720 | 0.587 | Pre-procarboxypeptidase R |
| D87839 | 0.00729 | 2.177 | 1.416 | β-Alanine oxoglutarate aminotransferase |
| X98746 | 0.00766 | 1.619 | 1.310 | Class IV alcohol dehydrogenase |
| AF214568 | 0.03035 | 2.670 | 1.902 | Aminopeptidase A |
| RNA | ||||
| M29293 | 0.00187 | 0.611 | 0.546 | Small nuclear ribonucleoparticle-associated protein (SnRNP) |
| Trafficking and transport | ||||
| U54807 | 0.00897 | 0.724 | 0.680 | GTP-binding protein (rab 3C) |
| Transcription | ||||
| AF003944 | 0.00250 | 1.422 | 1.319 | Ovalbumin upstream promoter beta nuclear receptor rCOUPb |
| X53724 | 0.00256 | 2.274 | 1.882 | MASH-2 (mammalian achaete-scute homologue) |
| NM 021865 | 0.00556 | 0.732 | 0.701 | Jun dimerization protein 1 gene (Jdp1) |
NOTE. Rats were injected with saline and terminated 12 hours later. Genes are grouped by functionality. The relative expression is the expression level in the colon of n-3–fed rats divided by expression in n-6 – or n-9 –fed rats (n= 5 per diet group). The P value is for the effect of n-3 fatty acid feeding versus the other oils. Genes were selected based on a relative expression ≥ 1.3 or ≤0.75 and P< 0.01. A few exceptions to the P value criteria were accepted when relative expression was confirmed by real-time PCR.
Table 4.
Relative colonic gene expression in n-3 versus n-6 or n-9 fed AOM-injected rats at the promotion stage
| Accession no. | P | Relative expression |
Gene name | |
|---|---|---|---|---|
| n-3/n-6 | n-3/n-9 | |||
| Apoptosis | ||||
| X07648 | 0.00836 | 0.585 | 0.731 | Amyloidogenic glycoprotein (rAG) |
| Cell cycle/Cell growth | ||||
| NM 019242 | 0.00270 | 1.369 | 1.374 | IFN-related developmental regulator 1 (Ifrd1) |
| Channels/Transporters | ||||
| NM 017251 | 0.00119 | 0.704 | 0.677 | Gap junction membrane channel protein β 1 (Gjb1) |
| NM 012654 | 0.00465 | 1.356 | 1.336 | Solute carrier family 9, member3 (Slc9a3) |
| D50664 | 0.00498 | 1.665 | 1.411 | Oligopeptide transporter |
| AF029310 | 0.00867 | 1.424 | 1.513 | Vanilloid receptor subtype 1 |
| Cytoskeleton/Extracellular matrix | ||||
| NM 012983 | 0.00035 | 0.509 | 0.497 | Unconventional myosin (Myr4) |
| D14048 | 0.00843 | 1.596 | 1.612 | SP120 |
| Kinases and related | ||||
| D78588 | 0.00319 | 0.692 | 0.734 | Diacylglycerol kinase |
| AF007789 | 0.00678 | 1.405 | 1.487 | Urokinase receptor |
| Metabolism/Redox | ||||
| AF186469 | 0.00240 | 1.461 | 1.400 | TM6P1 |
| Receptors | ||||
| AF090347 | 0.00022 | 0.717 | 0.711 | Putative G-protein coupled receptor GPCR91 |
| Trafficking and transport | ||||
| AF144756 | 0.00566 | 2.507 | 2.077 | Adipocyte lipid-binding protein (ALBP) |
| Transcription | ||||
| L09647 | 0.00239 | 0.649 | 0.682 | Hepatocyte nuclear factor 3a (HNF-3 β) |
| AF003944 | 0.00313 | 0.678 | 0.714 | Ovalbumin upstream promoter β nuclear receptor rCOUPb |
| J04147 | 0.00400 | 0.490 | 0.577 | 1,25-dihydroxyvitamin D-3 receptor |
| AF037199 | 0.00465 | 1.513 | 1.882 | Zinc finger transcription factor REST protein |
NOTE. Rats received injections of AOM and were terminated 10 weeks later. See footnote to Table 1 for additional details.
Dietary Fish Oil Up-Regulates Genes Involved in Apoptosis and Differentiation
For saline-injected animals, only 5 (12 hours, Table 1; 10 weeks, Table 2) of 9028 total genes exhibited a >2-fold change in which fish oil feeding caused a distinct response compared with both the corn and olive oil diets. At 10 weeks, aminopeptidase A (Enpep), a cell surface metallopeptidase capable of mediating cell differentiation (38), was up-regulated in n-3 PUFA-fed rats so that expression was 3.5- and 2.7-fold that of the n-6 and n-9 fatty acid-fed rats, respectively. In contrast, at the earlier time point (12 hours after saline injection), cytochrome P450 (Cyp2f1) expression in fish oil fed rats was 3.5- and 2.7-fold that of corn- and olive oil-fed rats, respectively. Although very little information is available regarding the regulation of colonic cytochrome P450s, this enzyme is clearly involved in the metabolic activation of xenobiotic compounds (39). In addition, mammalian achaete-scute homologous protein-2, a basic-helix-loop-helix transcription factor regulating lineage specification (40), responded similarly. In comparison, glycosylation-dependent cell adhesion molecule 1 was down-regulated in n-3 PUFA-fed animals. Glycosylation-dependent cell adhesion molecule 1 initiates cell binding by presenting carbohydrates to the lectin domain of L-selectin (41).
Table 2.
Relative colonic gene expression in n-3– versus n-6– or n-9–fed, saline-injected (10 weeks) control rats
| Accession no. | P | Relative expression |
Gene name | |
|---|---|---|---|---|
| n-3/n-6 | n-3/n-9 | |||
| Cell cycle/Cell growth | ||||
| AF205717 | 0.00207 | 2.205 | 1.541 | Tetraspan protein LRTM4 |
| Channels and transporters | ||||
| AF029310 | 0.00205 | 1.757 | 1.405 | Vanilloid receptor subtype 1 |
| Cytoskeletal/Extracellular matrix | ||||
| NM 019234 | 0.00222 | 1.493 | 1.357 | Dynein, cytoplasmic, intermediate chain 1 (Dncic1) |
| Metabolism/Redox | ||||
| AF214568 | 0.00486 | 3.539 | 2.692 | Aminopeptidase A (Enpep) |
NOTE. Rats received injections of saline and were terminated 10 weeks later. See footnote to Table 1 for additional details.
Carcinogen Influences the Effect of Diet on Gene Expression Profiles
As shown in Table 3, at 12 hours after AOM injection, fish oil-fed animals had expression levels of adipocyte-lipid binding protein (ALBP) and arginase II (Arg2) 2 to 3-fold times that in corn- and olive oil-fed animals. ALBP is a member of the intracellular lipid binding protein family and associates with fatty acids, retinoids, and other hydrophobic ligands (42). It is noteworthy that ALBP may act as a growth inhibitor in normal epithelial cells, where it is generally highly expressed (43). The mitochondrial Arg2 hydrolyzes arginine to ornithine plus urea and effectively down-regulates nitric oxide synthesis (44). In comparison, glycogen synthase (Gys2) was down-regulated in n-3 PUFA-fed rats to levels 37 and 32% of that in n-6– and n-9–fed animals. Glycogen synthase regulates glycogen synthesis by transferring the glycosyl residue from UDP-glucose to the nonreducing end of α-1,4-glucan. As shown in Table 4, at 10 weeks after AOM injection, similar to the 12-hour time point, ALBP expression in n-3 PUFA-treated rats was more than twice the level in the other rats.
Table 3.
Relative colonic gene expression in n-3 versus n-6 or n-9 fed AOM-injected rats at the initiation stage
| Accession no. | P | Relative expression |
Gene name | |
|---|---|---|---|---|
| n-3/n-6 | n-3/n-9 | |||
| Kinases and related | ||||
| S79760 | 0.00962 | 0.602 | 0.700 | Ink4 |
| Metabolism/Redox | ||||
| NM 019168 | 0.01080 | 2.022 | 2.033 | Arginase type II (Arg2) |
| NM 013089 | 0.04680 | 0.370 | 0.317 | Glycogen synthase 2 (Gys2) |
| Trafficking and transport | ||||
| AF144756 | 0.00065 | 3.010 | 2.027 | Adipocyte lipid-binding protein (ALBP) |
| NM 021590 | 0.00537 | 0.611 | 0.718 | Aryl-hydrocarbon interacting protein-like 1 (Aipl1) |
| L12382 | 0.00578 | 0.624 | 0.731 | ADP-ribosylation factor 3 |
| Transcription | ||||
| U71293 | 0.00293 | 1.337 | 1.522 | Hairless protein |
NOTE. Rats received injections of AOM and were terminated 12 hours later. See footnote to Table 1 for additional details.
Transcriptional Profiles of Genes Altered by AOM Exposure
We also studied the gene expression profiles induced by carcinogen exposure (data available online).6 A large subset of regulated genes may be involved in the epithelial apoptosis and aberrant crypt foci formation induced by AOM at the initiation and postinitiation time points, respectively. Interestingly, little overlap was found between the lists of genes regulated at 12 hours and 10 weeks of AOM treatment. Of note, the majority of genes affected by carcinogen treatment at a significant level (P < 0.002) was down-regulated, indicating a generalized suppression of expression due to AOM treatment.
Real-Time PCR Confirmation of Expression Profiles
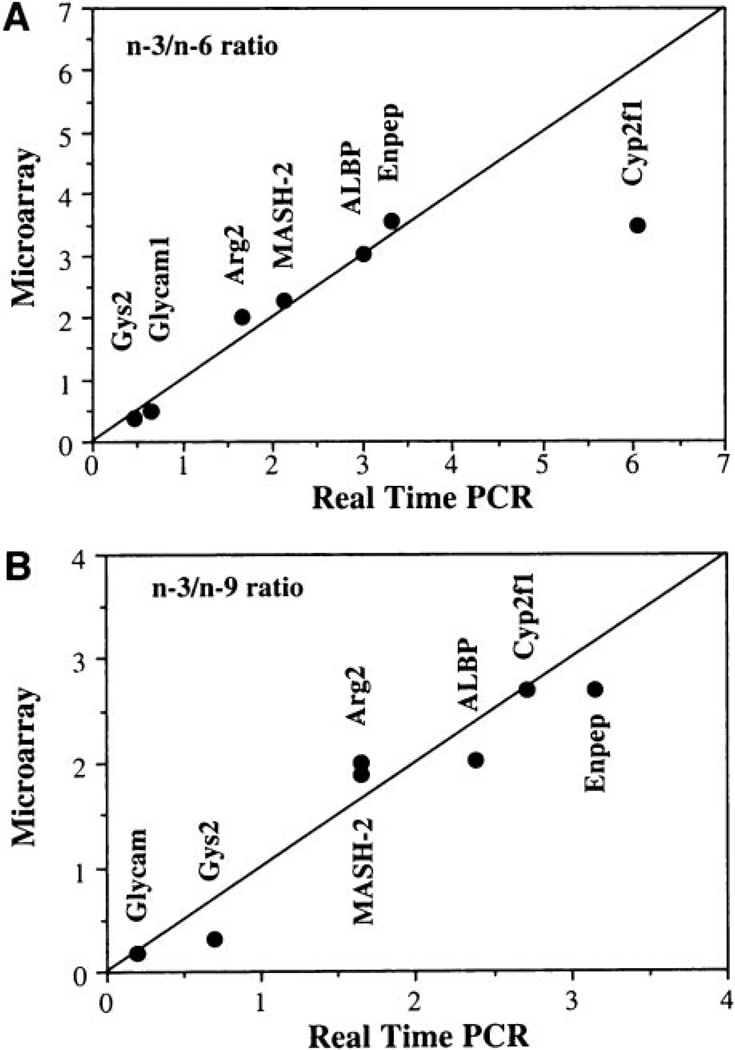
To validate expression patterns, we performed real-time PCR on RNA from four animals per diet/treatment/time point. Genes where expression levels changed most dramatically due to n-3 PUFA feeding were selected for confirmation. In addition, cyclooxygenase 2, which was not present on the array, was quantified by real-time PCR because previous reports implicate its involvement in colon tumorigenesis (45). Data were normalized to the expression of 18S, which was extremely consistent across all samples (cycle threshold values for 18S: 14.24–14.69 across 48 samples). Correlation of array and PCR data are shown in Fig. 2. Data are displayed as the ratio of the expression level in n-3 PUFA-fed rats to that of n-6 or n-9 fatty acid-fed rats.
Fig. 2.
Correlation of microarray and real-time PCR gene expression levels. Genes showing significantly different expression in n-3 versus n-6 or n-3 versus n-9 fatty acid-fed rats were confirmed by real-time PCR. Data are expressed as the level of gene expression in colonic mucosa from n-3 PUFA-fed rats relative to n-6 (top panel) or n-9 (bottom panel) fatty acid-fed rats (n= 5 rats per treatment). The line represents the theoretical correlation between microarray and real-time PCR data
DISCUSSION
The in vivo findings of this study support our postulate that n-3 PUFA have chemoprotective properties and act to decrease carcinogen-induced adduct formation and subsequent aberrant crypt foci formation, thereby mitigating the promotion of tumorigenesis. In addition, apoptosis was elevated only in colonic epithelial cells of n-3 PUFA-fed rats at the promotional stage of tumor development. This is significant because apoptosis is progressively inhibited during the development of colon cancer (46, 47). Our study is unique in that we also addressed the question as to whether the chemopreventive effect of fat is due to the specific action of n-3 PUFA or due indirectly to a reduction of n-6 PUFA content in the diet. This was achieved by comparing the biological properties of three types of commonly consumed dietary fats, i.e., corn oil/n-6 PUFA, fish oil/n-3 PUFA, and olive oil/n-9 MUFA, after carcinogen treatment at both the initiation and postinitiation stages of colon tumor development. We found that only the consumption of n-3 PUFA exerted a protective effect at the initiation (DNA adduct formation) and promotional (aberrant crypt foci levels) stages. Contrary to previous studies (10, 11, 48, 49), dietary n-9 MUFA did not have chemopreventive activity against colon carcinogenesis. This may be related to the fact that a highly fermentable fiber source, pectin, was added to the diets as opposed to poorly fermentable cellulose. This is an important distinction in view of the interactive effect of fat and fiber on colon tumorigenesis (29, 50, 51). Specifically, we have shown that highly fermentable fiber, which generates butyrate in the colon, only exerts a chemoprotective effect when n-3 PUFA, in contrast to n-6 PUFA, is simultaneously provided as the lipid source (51, 52). This may be attributed to the fact that n-3 PUFA can prime colonocytes for butyrate-induced apoptosis by enhancing the unsaturation of mitochondrial phospholipids, e.g., cardiolipin, resulting in an increase in reactive oxygen species and the initiation of the apoptotic cascade (19, 21). Hence, a major finding of this study is that n-9 MUFA consumed in the presence of a highly fermentable fiber source can promote the development of an abnormal epithelial phenotype. Studies are in progress to determine how fiber source influences genomic responses to dietary fat consumption.
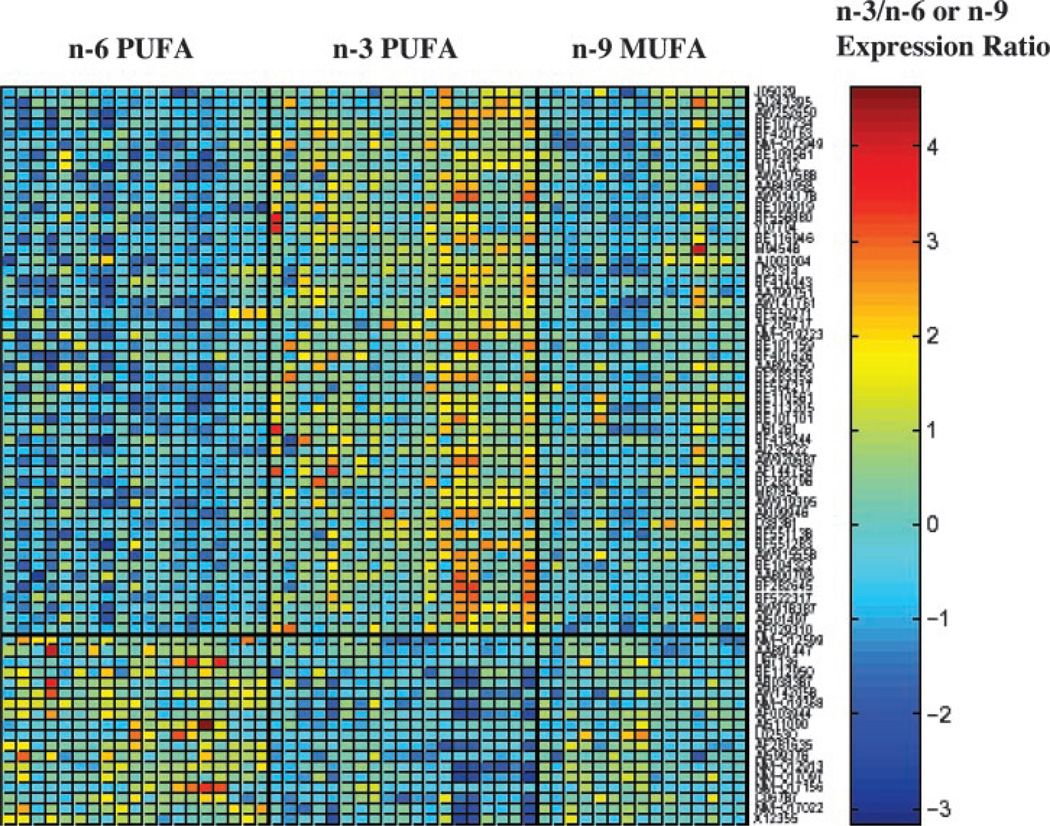
The present study builds upon previous in vitro microarray experiments designed to elucidate the molecular mechanisms by which n-3 PUFA inhibit colon carcinogenesis (25, 53). In this large scale dietary in vivo application of array and quantitative imaging methods, we used the rat AOM colon cancer model for the purpose of determining gene expression profiles over time to characterize transcription profiles associated with experimentally verifiable phenotypic characteristics (i.e., DNA damage, apoptosis, and aberrant crypt foci) relevant to colon tumor initiation, promotion, and progression. For these purposes, rats were exposed to an environmental carcinogen (AOM or saline control) and dietary lipid modifiers (n-3, n-6, and n-9 fatty acids) of tumorigenesis and examined over time (12 hours and 10 weeks after carcinogen exposure). This experimental protocol enabled us to identify and quantify subtle regulatory relationships among genes in a highly relevant colon cancer model system. Our study demonstrates for the first time that n-3 PUFA uniquely alters colonic gene expression profiles in vivo. This is readily observed when the data are expressed as a matrix plot of gene expression values (Fig. 3). The organization of the microarray database in this manner revealed that, in contrast to n-3 PUFA, n-6 PUFA, and n-9 MUFA generate similar patterns of expression. The results suggest a pleiotropic regulatory role for n-3 PUFA. Although it is highly likely that the relatively large number of gene alterations cannot be considered out of context from the overall response, it may be possible to uncover classes of genes that are coregulated. For example, within the trafficking and transport functional class, we have determined that ALBP, which can selectively and noncovalently bind fatty acids, is up-regulated by n-3 PUFA treatment (Tables 3 and 4). Although the physiologic role of this class of binding protein was originally thought to be regulation of intermediary metabolism (54, 55), recent data show that this lipophilic molecule may influence cell signaling by transporting fatty acid ligand to nuclear receptors such as PPARy or to competitively deplete ligands from PPARγ (56, 57). This is significant because PPARγ functions to induce terminal differentiation in the colon (58, 59), thereby suppressing colon carcinogenesis (60). Collectively, these data suggest that ALBP acts as a cytosolic gateway capable of modulating nuclear receptor activation. Although not measured in this study, we have previously shown that the administration of n-3 PUFA increases markers of differentiation in the colon (5, 51). With respect to the precise origin of the signaling molecules, which are available for PPARγ activation in the nucleus, we have previously demonstrated that highly purified dietary eicosapentaenoic acid (20:5n-3) and DHA (22:6n-3) ethyl esters recapitulate the chemoprotective results achieved by fish oil feeding (19, 21). These results unequivocally indicate that the primary bioactive effector molecules are eicosapentaenoic acid and DHA rather than some other component found in fish oil. Whether eicosapentaenoic acid and DHA, or their metabolites, preferentially serve as ALBP binding partners to initiate the cascade of transcriptional events that regulate the expression of target genes, which reduce the development of colonic tumors, requires additional investigation.
Fig. 3.
Expression values of genes with significant differences between n-3 and n-6 PUFA treatment at 10 weeks. Shown is the expression matrix plot of gene expression values (rows). A linear-mixed model was used to assess gene expression in relation to dietary treatment (n-3, n-6 PUFA or n-9 MUFA). Genes were ranked by their significance (P values) by contrasting differences in gene expression between n-3 and n-6 PUFA-fed animals. Displayed in the plot are the observed expression values of the top-ranked 70 genes, of which 52 (top panel) have significantly higher expression (red-orange color) with n-3 versus n-6 PUFA treatment. The bottom panel displays the expression values of 18 genes highly expressed in the n-6 compared with n-3 PUFA treatment. Only animals with no missing data for the 60 displayed genes were included in the figure. A list of the gene descriptions is available online.6
Although a growing body of evidence suggests that DHA is capable of suppressing inducible nitric oxide synthase, cyclooxygenase 2, and enhancing cyclin-dependent kinase inhibitor expression (25, 53), the biological relevance of these observations using in vitro colon cancer cell systems remains unknown. For example, in contrast to the reports using the CaCo-2 model system, in the present study, inducible nitric oxide synthase, cyclooxygenase 2, p21waf1/cip1, and p27 expression levels were unaffected by diet and carcinogen treatment (data not shown). The primary reason for this apparent discrepancy may be that our samples for array analysis were not microdissected and did contain RNA from stromal components. However, because of the recent literature documenting the important contribution of the surrounding stroma to signaling processes, which influence the malignant transformation process (61), we believe that it would lead to a substantial loss of information if only colonic epithelial cells were analyzed.
Recently, we have demonstrated that retinoid X receptor (RXR), an obligatory component of a large number of nuclear receptors, is preferentially activated by DHA in mouse and human colonocytes (62). Because RXR ligands are capable of antagonizing the β-catenin/LEF/TCF signaling pathway (63), it is possible, therefore, that n-3 PUFA may mediate proapoptotic, antitumorigenic effects in the colon through the RXR subunit of nuclear receptor heterodimers. This is noteworthy because we found that RXR mRNA expression is influenced by diet and carcinogen treatment (62). Therefore, it is important to additionally elucidate the role of the n-3 PUFA-RXR-signaling axis in relation to resistance to colon carcinogenesis. This study has also revealed a novel molecular target, achaete-scute homologue 2 (MASH-2), associated with n-3 PUFA feeding. This transcription factor has been shown to be up-regulated in human colon cancer cells treated with 1-α-25-dihydroxy vitamin D3 to induce differentiation (64). Although not examined in this study, we have previously demonstrated that n-3 PUFA supplementation enhances expression of differentiation markers in the rat colon (51).
We have demonstrated that n-3 PUFA beneficially modulate the balance between colonic DNA adduct formation, epithelial cell apoptosis, and aberrant crypt foci multiplicity. In a major step toward understanding why n-3 PUFA suppresses colon cancer, we contrasted the colonic epithelial cell transcription profiles derived from rats fed n-3 PUFA, n-6 PUFA, and n-9 MUFA. In conclusion, our findings indicate that the chemopreventive effect of fish oil is due to the direct action of n-3 PUFA and not to a reduction in the content of n-6 PUFA.
ACKNOWLEDGMENTS
We thank Drs. Yang-Yi Fan, Mee Young Hong, and Stella Taddeo for technical assistance. We also thank Dr. Geoffrey P. Margison for the generous donation of the N7-methylguanine antibody and to Degussa Bioactives for the donation of dietary oils.
Grant support: NIH Grants CA59034, CA57030, and CA61750, National Institute of Environmental Health Sciences Grant P30-ES09106, and Life Sciences Enhancement Funds, Texas A&M University.
Footnotes
Internet address: http://dnguyen.ucdavis.edu/.html/datatest3/main.html
Internet address: http://www.r-project.org/
REFERENCES
- 1.Jemal A, Thomas A, Murray T, Thun M. Cancer statistics, 2002. CA - Cancer J Clin. 2002;52(1):23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- 2.Potter JD, Slattery ML, Bostick RM, Gapstur SM. Colon cancer: a review of the epidemiology. Epidemiol Rev. 1993;15(2):499–545. doi: 10.1093/oxfordjournals.epirev.a036132. [DOI] [PubMed] [Google Scholar]
- 3.Reddy BS, Burill C, Rigotty J. Effect of diets high in omega-3 and omega-6 fatty acids on initiation and postinitiation stages of colon carcinogenesis. Cancer Res. 1991;51(2):487–491. [PubMed] [Google Scholar]
- 4.Bull AW, Soullier BK, Wilson PS, Hayden MT, Nigro ND. Promotion of azoxymethane-induced intestinal cancer by high-fat diet in rats. Cancer Res. 1979;39(12):4956–4959. [PubMed] [Google Scholar]
- 5.Chang WL, Chapkin RS, Lupton JR. Fish oil blocks azoxymethane-induced rat colon tumorigenesis by increasing cell differentiation and apoptosis rather than decreasing cell proliferation. J Nutr. 1998;128(3):491–497. doi: 10.1093/jn/128.3.491. [DOI] [PubMed] [Google Scholar]
- 6.Anti M, Armelao F, Marra G, et al. Effects of different doses of fish oil on rectal cell proliferation in patients with sporadic colonic adenomas. Gastroenterology. 1994;107(6):1709–1718. doi: 10.1016/0016-5085(94)90811-7. [DOI] [PubMed] [Google Scholar]
- 7.Cheng J, Ogawa K, Kuriki K, et al. Increased intake of n-3 polyunsaturated fatty acids elevates the level of apoptosis in the normal sigmoid colon of patients polypectomized for adenomas/tumors. Cancer Lett. 2003;193(1):17–24. doi: 10.1016/s0304383502007176. [DOI] [PubMed] [Google Scholar]
- 8.Spector AA. Essentiality of fatty acids. Lipids. 1999;34(Suppl):S1–S3. doi: 10.1007/BF02562220. [DOI] [PubMed] [Google Scholar]
- 9.Tokudome S, Nagaya T, Okuyama H, et al. Japanese versus Mediterranean diets and cancer. Asian Pac J Cancer Prev. 2000;1(1):61–66. [PubMed] [Google Scholar]
- 10.Takeshita M, Ueda H, Shirabe K, Higuchi Y, Yoshida S. Lack of promotion of colon carcinogenesis by high-oleic safflower oil. Cancer (Phila.) 1997;79(8):1487–1493. doi: 10.1002/(sici)1097-0142(19970415)79:8<1487::aid-cncr7>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 11.Bartoli R, Fernandez-Banares F, Navarro E, et al. Effect of olive oil on early and late events of colon carcinogenesis in rats: modulation of arachidonic acid metabolism and local prostaglandin E(2) synthesis. Gut. 2000;46(2):191–199. doi: 10.1136/gut.46.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pickering JS, Lupton JR, Chapkin RS. Dietary fat, fiber, and carcinogen alter fecal diacylglycerol composition and mass. Cancer Res. 1995;55(11):2293–2298. [PubMed] [Google Scholar]
- 13.Lee DY, Lupton JR, Aukema HM, Chapkin RS. Dietary fat and fiber alter rat colonic mucosal lipid mediators and cell proliferation. J Nutr. 1993;123(11):1808–1817. doi: 10.1093/jn/123.11.1808. [DOI] [PubMed] [Google Scholar]
- 14.Chapkin RS, Gao J, Lee DY, Lupton JR. Dietary fibers and fats alter rat colon protein kinase C activity: correlation to cell proliferation. J Nutr. 1993;123(4):649–655. doi: 10.1093/jn/123.4.649. [DOI] [PubMed] [Google Scholar]
- 15.Davidson LA, Lupton JR, Jiang YH, Chang WC, Aukema HM, Chapkin RS. Dietary fat and fiber alter rat colonic protein kinase C isozyme expression. J Nutr. 1995;125(1):49–56. doi: 10.1093/jn/125.1.49. [DOI] [PubMed] [Google Scholar]
- 16.Davidson LA, Lupton JR, Jiang YH, Chapkin RS. Carcinogen and dietary lipid regulate ras expression and localization in rat colon without affecting farnesylation kinetics. Carcinogenesis (Lond.) 1999;20(5):785–791. doi: 10.1093/carcin/20.5.785. [DOI] [PubMed] [Google Scholar]
- 17.Hong MY, Chapkin RS, Davidson LA, et al. Fish oil enhances targeted apoptosis during colon tumor initiation in part by down-regulating Bcl-2. Nutr Cancer. 2003;46(1):44–51. doi: 10.1207/S15327914NC4601_06. [DOI] [PubMed] [Google Scholar]
- 18.Bancroft LK, Lupton JR, Davidson LA, et al. Dietary fish oil reduces oxidative DNA damage in rat colonocytes. Free Radic Biol Med. 2003;35(2):149–159. doi: 10.1016/s0891-5849(03)00240-5. [DOI] [PubMed] [Google Scholar]
- 19.Hong MY, Chapkin RS, Barhoumi R, et al. Fish oil increases mitochondrial phospholipid unsaturation, up-regulating reactive oxygen species and apoptosis in rat colonocytes. Carcinogenesis (Lond.) 2002;23(11):1919–1925. doi: 10.1093/carcin/23.11.1919. [DOI] [PubMed] [Google Scholar]
- 20.Murray NR, Weems C, Chen L, et al. Protein kinase C betaII and TGFbetaRII in omega-3 fatty acid-mediated inhibition of colon carcinogenesis. J Cell Biol. 2002;157(6):915–920. doi: 10.1083/jcb.200201127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapkin RS, Hong MY, Fan YY, et al. Dietary n-3 PUFA alter colonocyte mitochondrial membrane composition and function. Lipids. 2002;37(2):193–199. doi: 10.1007/s11745-002-0880-8. [DOI] [PubMed] [Google Scholar]
- 22.Collett ED, Davidson LA, Fan YY, Lupton JR, Chapkin RS. n-6 and n-3 polyunsaturated fatty acids differentially modulate oncogenic Ras activation in colonocytes. Am J Physiol Cell Physiol. 2001;280(5):C1066–C1075. doi: 10.1152/ajpcell.2001.280.5.C1066. [DOI] [PubMed] [Google Scholar]
- 23.Singh J, Hamid R, Reddy BS. Dietary fat and colon cancer: modulation of cyclooxygenase-2 by types and amount of dietary fat during the postinitiation stage of colon carcinogenesis. Cancer Res. 1997;57(16):3465–3470. [PubMed] [Google Scholar]
- 24.Lee JY, Hwang DH. Docosahexaenoic acid suppresses the activity of peroxisome proliferator-activated receptors in a colon tumor cell line. Biochem Biophys Res Commun. 2002;298(5):667–674. doi: 10.1016/s0006-291x(02)02530-5. [DOI] [PubMed] [Google Scholar]
- 25.Narayanan BA, Narayanan NK, Simi B, Reddy BS. Modulation of inducible nitric oxide synthase and related proinflammatory genes by the omega-3 fatty acid docosahexaenoic acid in human colon cancer cells. Cancer Res. 2003;63(5):972–979. [PubMed] [Google Scholar]
- 26.Reddy BS. Chemoprevention of colon cancer by dietary fatty acids. Cancer Metastasis Rev. 1994;13(3–4):285–302. doi: 10.1007/BF00666099. [DOI] [PubMed] [Google Scholar]
- 27.Ramakrishnan R, Dorris D, Lublinsky A, et al. An assessment of Motorola CodeLink microarray performance for gene expression profiling applications. Nucleic Acids Res. 2002;30(7):e30. doi: 10.1093/nar/30.7.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLellan EA, Bird RP. Aberrant crypts: potential preneoplastic lesions in the murine colon. Cancer Res. 1988;48(21):6187–6192. [PubMed] [Google Scholar]
- 29.Hong MY, Lupton JR, Morris JS, et al. Dietary fish oil reduces O 6-methylguanine DNA adduct levels in rat colon in part by increasing apoptosis during tumor initiation. Cancer Epidemiol Biomark Prev. 2000;9(8):819–826. [PubMed] [Google Scholar]
- 30.Elder RH, Jansen JG, Weeks RJ, et al. Alkylpurine-DNA-N-glycosylase knockout mice show increased susceptibility to induction of mutations by methyl methanesulfonate. Mol Cell Biol. 1998;18(10):5828–5837. doi: 10.1128/mcb.18.10.5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong MY, Chapkin RS, Wild CP, et al. Relationship between DNA adduct levels, repair enzyme, and apoptosis as a function of DNA methylation by azoxymethane. Cell Growth Differ. 1999;10(11):749–758. [PubMed] [Google Scholar]
- 32.McLellan EA, Medline A, Bird RP. Dose response and proliferative characteristics of aberrant crypt foci: putative preneoplastic lesions in rat colon. Carcinogenesis (Lond.) 1991;12(11):2093–2098. doi: 10.1093/carcin/12.11.2093. [DOI] [PubMed] [Google Scholar]
- 33.Littell RC, Milliken GA, Stroup WW, Wolfinger RD. SAS system for mixed models. Cary, NC: SAS Publishing; 1996. pp. 87–134. [Google Scholar]
- 34.Likhachev AJ, Margison GP, Montesano R. Alkylated purines in the DNA of various rat tissues after administration of 1,2-dimethylhydrazine. Chem Biol Interact. 1977;18(2):235–240. doi: 10.1016/0009-2797(77)90009-6. [DOI] [PubMed] [Google Scholar]
- 35.Tomlinson IP, Bodmer WF. Failure of programmed cell death and differentiation as causes of tumors: some simple mathematical models. Proc Natl Acad Sci USA. 1995;92(24):11130–11134. doi: 10.1073/pnas.92.24.11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takayama T, Katsuki S, Takahashi Y, et al. Aberrant crypt foci of the colon as precursors of adenoma and cancer. N Engl J Med. 1998;339(18):1277–1284. doi: 10.1056/NEJM199810293391803. [DOI] [PubMed] [Google Scholar]
- 37.Pretlow TP, O’Riordan MA, Somich GA, Amini SB, Pretlow TG. Aberrant crypts correlate with tumor incidence in F344 rats treated with azoxymethane and phytate. Carcinogenesis (Lond.) 1992;13(9):1509–1512. doi: 10.1093/carcin/13.9.1509. [DOI] [PubMed] [Google Scholar]
- 38.Fujimura H, Ino K, Nagasaka T, et al. Aminopeptidase A expression in cervical neoplasia and its relationship to neoplastic transformation and progression. Oncology. 2000;58(4):342–352. doi: 10.1159/000012122. [DOI] [PubMed] [Google Scholar]
- 39.Ding X, Kaminsky LS. Human extrahepatic cytochromes P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu Rev Pharmacol Toxicol. 2003;43:149–173. doi: 10.1146/annurev.pharmtox.43.100901.140251. [DOI] [PubMed] [Google Scholar]
- 40.Guillemot F, Nagy A, Auerbach A, Rossant J, Joyner AL. Essential role of Mash-2 in extraembryonic development. Nature (Lond.) 1994;371(6495):333–336. doi: 10.1038/371333a0. [DOI] [PubMed] [Google Scholar]
- 41.Ley K. Sulfated sugars for rolling lymphocytes. J Exp Med. 2003;198(9):1285–1288. doi: 10.1084/jem.20031664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen WJ, Sridhar K, Bernlohr DA, Kraemer FB. Interaction of rat hormone-sensitive lipase with adipocyte lipid-binding protein. Proc Natl Acad Sci USA. 1999;96(10):5528–5532. doi: 10.1073/pnas.96.10.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das R, Hammamieh R, Neill R, Melhem M, Jett M. Expression pattern of fatty acid-binding proteins in human normal and cancer prostate cells and tissues. Clin Cancer Res. 2001;7(6):1706–1715. [PubMed] [Google Scholar]
- 44.Jenkinson CP, Grody WW, Cederbaum SD. Comparative properties of arginases. Comp Biochem Physiol B Biochem Mol Biol. 1996;114(1):107–132. doi: 10.1016/0305-0491(95)02138-8. [DOI] [PubMed] [Google Scholar]
- 45.Gupta RA, Dubois RN. Colorectal cancer prevention and treatment by inhibition of cyclooxygenase-2. Nat Rev Cancer. 2001;1(1):11–21. doi: 10.1038/35094017. [DOI] [PubMed] [Google Scholar]
- 46.Hall PA, Coates PJ, Ansari B, Hopwood D. Regulation of cell number in the mammalian gastrointestinal tract: the importance of apoptosis. J Cell Sci. 1994;107(Pt 12):3569–3577. doi: 10.1242/jcs.107.12.3569. [DOI] [PubMed] [Google Scholar]
- 47.Bedi A, Pasricha PJ, Akhtar AJ, et al. Inhibition of apoptosis during development of colorectal cancer. Cancer Res. 1995;55(9):1811–1816. [PubMed] [Google Scholar]
- 48.Bird RP, Lafave LM. Varying effect of dietary lipids and azoxymethane on early stages of colon carcinogenesis: enumeration of aberrant crypt foci and proliferative indices. Cancer Detect Prev. 1995;19(4):308–315. [PubMed] [Google Scholar]
- 49.Rao CV, Reddy BS. Modulating effect of amount and types of dietary fat on ornithine decarboxylase, tyrosine protein kinase and prostaglandins production during colon carcinogenesis in male F344 rats. Carcinogenesis (Lond.) 1993;14(7):1327–1333. doi: 10.1093/carcin/14.7.1327. [DOI] [PubMed] [Google Scholar]
- 50.Lee DY, Chapkin RS, Lupton JR. Dietary fat and fiber modulate colonic cell proliferation in an interactive site-specific manner. Nutr Cancer. 1993;20(2):107–118. doi: 10.1080/01635589309514277. [DOI] [PubMed] [Google Scholar]
- 51.Chang WC, Chapkin RS, Lupton JR. Predictive value of proliferation, differentiation and apoptosis as intermediate markers for colon tumorigenesis. Carcinogenesis (Lond.) 1997;18(4):721–730. doi: 10.1093/carcin/18.4.721. [DOI] [PubMed] [Google Scholar]
- 52.Zoran DL, Turner ND, Taddeo SS, Chapkin RS, Lupton JR. Wheat bran diet reduces tumor incidence in a rat model of colon cancer independent of effects on distal luminal butyrate concentrations. J Nutr. 1997;127(11):2217–2225. doi: 10.1093/jn/127.11.2217. [DOI] [PubMed] [Google Scholar]
- 53.Narayanan BA, Narayanan NK, Reddy BS. Docosahexaenoic acid regulated genes and transcription factors inducing apoptosis in human colon cancer cells. Int J Oncol. 2001;19(6):1255–1262. doi: 10.3892/ijo.19.6.1255. [DOI] [PubMed] [Google Scholar]
- 54.Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science (WashDC) 2001;294(5548):1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 55.Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc Natl Acad Sci USA. 1997;94(9):4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan NS, Shaw NS, Vinckenbosch N, et al. Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol Cell Biol. 2002;22(14):5114–5127. doi: 10.1128/MCB.22.14.5114-5127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Helledie T, Jorgensen C, Antonius M, et al. Role of adipocyte lipid-binding protein (ALBP) and acyl-coA binding protein (ACBP) in PPAR-mediated transactivation. Mol Cell Biochem. 2002;239(1-2):157–164. [PubMed] [Google Scholar]
- 58.Gupta RA, Sarraf P, Brockman JA, et al. Peroxisome proliferator-activated receptor gamma and transforming growth factor-beta pathways inhibit intestinal epithelial cell growth by regulating levels of TSC-22. J Biol Chem. 2003;278(9):7431–7438. doi: 10.1074/jbc.M208076200. [DOI] [PubMed] [Google Scholar]
- 59.Lefebvre M, Paulweber B, Fajas L, et al. Peroxisome proliferator-activated receptor gamma is induced during differentiation of colon epithelium cells. J Endocrinol. 1999;162(3):331–340. doi: 10.1677/joe.0.1620331. [DOI] [PubMed] [Google Scholar]
- 60.Girnun GD, Smith WM, Drori S, et al. APC-dependent suppression of colon carcinogenesis by PPARgamma. Proc Natl Acad Sci USA. 2002;99(21):13771–13776. doi: 10.1073/pnas.162480299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature (Lond.) 2001;411(6835):375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 62.Fan YY, Spencer TE, Wang N, Moyer MP, Chapkin RS. Chemopreventive n-3 fatty acids activate RXRalpha in colonocytes. Carcinogenesis (Lond.) 2003;24(9):1541–1548. doi: 10.1093/carcin/bgg110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Easwaran V, Pishvaian M, Salimuddin Byers S. Cross-regulation of beta-catenin-LEF/TCF and retinoid signaling pathways. Curr Biol. 1999;9(23):1415–1418. doi: 10.1016/s0960-9822(00)80088-3. [DOI] [PubMed] [Google Scholar]
- 64.Palmer HG, Sanchez-Carbayo M, Ordonez-Moran P, Larriba MJ, Cordon-Cardo C, Munoz A. Genetic signatures of differentiation induced by 1alpha,25-dihydroxyvitamin D3 in human colon cancer cells. Cancer Res. 2003;63(22):7799–7806. [PubMed] [Google Scholar]