Abstract
Neuropathic and inflammatory pain promote a large number of persisting adaptations at the cellular and molecular level, allowing tissue or nerve damage, even if only transient, to elicit changes in cells that contribute to the development of chronic pain and associated symptoms. There is evidence that injury-induced changes in chromatin structure drive stable changes in gene expression and neural function, which may cause several symptoms, including allodynia, hyperalgesia, anxiety, and depression. Recent findings on epigenetic changes in the spinal cord and brain during chronic pain may guide fundamental advances in new treatments. In this review, we provide a brief overview of epigenetic regulation in the nervous system and then discuss the still-limited literature that directly implicates epigenetic modifications in chronic pain syndromes.
Keywords: histone modifications, inflammatory pain, neuropathic pain, HDAC, miRNA, methylation, animal models
Chronic pain: a major clinical and socioeconomic problem
Chronic pain is characterized by persistent nociceptive hypersensitivity [1, 2], experienced by patients as a marked reduction in thresholds required to induce pain, such that innocuous stimuli cause pain (allodynia), and an amplification of responses to noxious stimuli at the site of injury (primary hyperalgesia) and surrounding tissues (secondary hyperalgesia) [3]. Chronic pain is often a result of peripheral tissue damage and persistent inflammation (inflammatory pain), or of pathological adaptations in the peripheral or central nervous system (neuropathic pain). In the USA, the number of adults with chronic pain is estimated to be 100 million, with an economic annual cost of up to $635 billion [4]. Unfortunately, current pain management interventions are insufficient, providing inadequate pain relief and multiple health and societal adverse effects. In a 2006 survey of chronic pain patients undergoing treatment, over half reported having little or no control over their pain [4]. The ineffectiveness of current therapeutic strategies is at least partly due to an incomplete understanding of the mechanisms involved in chronic pain conditions. The development and maintenance of chronic pain involve long-term changes in multiple areas of the central nervous system (CNS), which are characterized by adaptations at the cellular and molecular levels. Here we focus on recent studies that highlight the involvement of epigenetic mechanisms in the CNS in chronic pain conditions.
Rodent models of chronic pain
Several animal models have been used to investigate the molecular and cellular adaptations of nociceptive pathways. Inflammatory pain models use subcutaneous injections of inflammatory agents, such as formalin, capsaicin, or complete Freund’s adjuvant (CFA), usually into the hind or forepaw, whereas neuropathic pain models typically involve surgical injury of a spinal or peripheral nerve [3, 5]. These models result in reliable nociceptive sensitization; mimicking key components of the chronic pain experience in humans (see Box 1). In the spinal cord, for example, neuropathic injuries induce long-lasting abnormal neural activity along primary afferent pathways [6], and in vitro recordings of dorsal horn neurons have shown potentiated excitatory responses and decreased firing thresholds in animals with neuropathic pain [7, 8]. Moreover, evidence from multiple studies suggests that chronic pain development is correlated with changes in gene expression in the spinal cord [2, 9, 10] and cerebral cortex [11, 12], and activity-dependent changes in gene expression have long been understood to be important for long-term alterations in neural activity [13–15].
Box 1. Chronic pain behaviors.
Chronic pain resulting from peripheral tissue damage and inflammation (inflammatory pain), or from pathological adaptations in the peripheral or CNS (neuropathic pain) is characterized by persistent nociceptive hypersensitivity [2, 67]. Inflammatory pain models use subcutaneous injections of inflammatory agents such as formalin or complete Freund’s adjuvant (CFA) into the hindpaw, forepaw, or masseter muscles, resulting in tissue injury. Neuropathic pain models usually involve surgical injury of a peripheral nerve [3]. These models have helped researchers yield much insight into the molecular mechanisms of chronic pain conditions. Chronic inflammatory and neuropathic pain can lead to changes in behavioral responses to tactile and thermal stimuli that are easily detectable in animal models. Experimenters can reliably detect and measure two major forms of sensitized behavioral responses: allodynia, (the sensation of pain to previously innocuous stimuli) and hyperalgesia (the enhancement of painful perception to noxious stimuli) [67,104]. Mechanical allodynia is reliably assessed in animal models and easily detected as stimuli that would otherwise be innocuous will evoke noxious nociceptive behavior [105]. Mechanical thresholds can be assessed by monitoring the responsiveness of the affected paws to the application of von Frey filaments. This method allows investigators to apply an accurate and constant force to specific areas of the skin. In this manner, von Frey filaments that do not elicit a nociceptive response in naïve mice will elicit nociceptive responses in animals with mechanical allodynia [67,104]. Thermal hyperalgesia can be measured through enhanced behavioral responses to thermal stimuli. The Hargreaves test is a common assay, where a high-intensity heat lamp is directed to the mid-plantar hindpaw of the injured limb [21]. Thermal allodynia can also be observed in the cold plate test, where animals with chronic pain will react to cold stimuli that are below the nociceptive threshold [22]. These represent the most common used tests, however several other assays are also used to assess nociceptive behavior or sensory deficits. Typical behavioral responses to noxious stimuli include licking, flicking, or quick withdrawal of the affected paw. These phenomena reflect similar sensitization of pain seen in humans [1], and may correspond to changes of synaptic transmission in the spinal cord DH [8, 40, 54] or pain related cortical areas [51, 54, 97].
Notably, chronic and intense pain can have effects at the level of gene expression in spinal and supraspinal areas located far from the initial lesion and may include brain areas that are not directly associated with the processing of sensory information. A growing number of studies directly implicate alterations in gene expression with the generation of certain chronic pain conditions, such as neuropathic or rheumatoid arthritis pain. However, until recently, the mechanisms by which acute tissue injury and pain induce changes in gene expression for a prolonged period of time have remained poorly understood. Epigenetic mechanisms provide a plausible process through which stable changes in CNS activity may manifest in response to peripheral injuries.
Epigenetic mechanisms drive long-lasting cellular and behavioral changes
Epigenetic mechanisms enhance or suppress gene expression without alterations of the primary DNA sequence. Epigenetic mechanisms have been shown to be involved in synaptic plasticity, learning, and memory [27, 28] as well as in several neuropsychiatric disorders including depression and drug addiction [29–31]. These mechanisms include DNA methylation, several types of histone modifications (e.g., acetylation, methylation, phosphorylation, and ADP-ribosylation), and expression of microRNAs (miRNAs). Epigenetic mechanisms can be dynamic and responsive to changes in experience, thus representing a complex interplay between an organism and its environment [32, 33]. Acetylation of most histone subunits, at any of several Lysine (Lys) residues, typically promotes gene transcription, while histone methylation can either repress or activate gene transcription depending on the amino acid residue undergoing methylation. For example, methylation of Lys9 or Lys27 of histone H3 is usually associated with gene repression, whereas methylation of Lys4, Lys36, or Lys79 of H3 is usually associated with gene activation [23]. Likewise, 5-methylation of cytosine nucleotides in DNA (which occurs in part at CpG dinucleotides) generally mediates gene repression, while 5-hydroxymethylation of cytosine produces the opposite effect. However, chromatin modifications do not occur in isolation, and we know from work in single cell systems that the activation or repression of a gene typically involves numerous histone and DNA modifications, and the recruitment of perhaps hundreds of chromatin regulatory proteins to the regulated gene.
DNA and histone methylation in spinal systems and their involvement in chronic pain
A widely recognized mechanism through which DNA methylation controls transcription, is through suppression of transcription factor binding at promoter regions by a complex comprising a DNA methylation-dependent DNA binding protein, such as methyl-CpG-binding protein 2 (MeCP2) or methyl DNA-binding domain (MBD) proteins. MeCP2 is best known as a transcriptional repressor that has been implicated in the modulation of activity-dependent gene expression [24]. Specifically, MeCP2 inhibits transcription of specific genes by binding to methyl-CpG sites in the DNA [25]. Phosphorylation of MeCP2 results in its removal from methylated CpG promoter regions, and thus can lead to the enhancement of gene expression [26]. However, MeCP2 is also reported to bind to 5-hydroxymethylcytosine and thereby promote gene expression. In a rat inflammatory pain model, it was reported that CFA injections correspond with increases in phosphorylation of MeCP2 in lamina I [27,28] and II [28] of the superficial dorsal horn as early as 30 min post injection. This phosphorylation was associated with induction of genes suppressed by the MeCP2 complex, and a link was demonstrated between MeCP2 levels and the nociceptive threshold. Accordingly, a recent study found that MeCP2 mRNA expression was significantly increased in the spinal dorsal horn of mice following formalin injections [29].
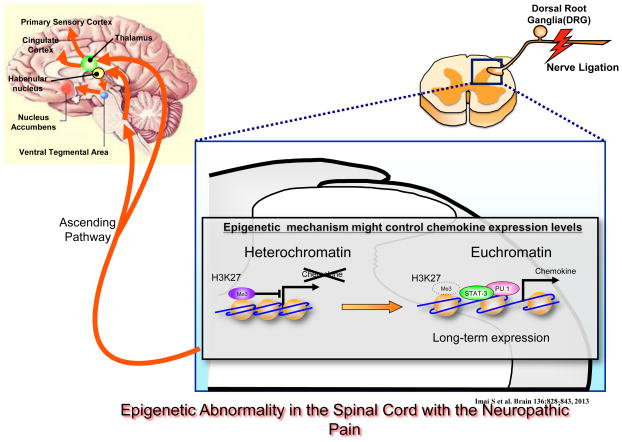
A recent study used a more clinically relevant model, the partial sciatic nerve ligation model of neuropathic pain, to investigate DNA methylation in the spinal cord of mice, and performed a comprehensive analysis focusing on changes in histone modifications in the dorsal horn. Although there was no dramatic change in DNA methylation throughout the whole genome in the spinal cord under this neuropathic pain condition, changes were found in histone methylation at specific genes. Specifically, a robust increase in expression of monocyte chemotactic protein-3 (MCP-3), a pro-inflammatory cytokine, was observed in the spinal cord, which lasted for up to 2 weeks after sciatic nerve ligation, and this increase occurred in astrocytes, but not microglia or neurons (Figure 1) [30]. Coincident with the induction of MCP-3 expression, there were reduced levels of Lys27 H3 trimethylation (H3K27me3) at the Mcp-3 gene promoter. The observed induction of MCP-3 is important for nociceptive sensitization, because intrathecal administration of an anti-MCP-3 antibody significantly reduced neuropathic pain-like behaviors. MCP-3 induction was abolished in interleukin 6 (IL-6) knockout mice, whereas a single intrathecal injection of recombinant IL6 strongly increased MCP-3 expression and decreased H3K27me3 at the MCP-3 gene promoter. These data directly implicate repressive histone methylation in mediating the ability of nerve injury to cause lasting changes in MCP-3 expression and the resulting pain syndrome. It will be important in future studies to examine the time course of this epigenetic modification and to test whether it might outlast the injury itself and help drive long-lasting pain symptoms.
Figure 1. Epigenetic abnormality in the spinal cord during neuropathic pain.
Long-term increases in chemokine expression may cause epigenetic modifications in the spinal cord, and thus may have a crucial role in chronic pain mechanisms. Nerve injury activates primary afferent nociceptors, which transmit information to the dorsal horn of the spinal cord. Activation of secondary neurons in spinal pathways by long-term chemokine expression can induce epigenetic modifications that may produce central sensitization leading to a neuropathic pain-like state. Reproduced from (30).
Cancer is another leading cause of chronic pain [31], and hypermethylation of CpG islands, a common feature of cancer cells [32], has recently been implicated in cancer related chronic pain conditions [31]. Specifically, enhanced secretion of the peptide, endothelin 1, has been observed in multiple malignancies, and it is thought to have a major role in cancer induced pain [33]. The pro-nociceptive effects of this peptide are mediated through endothelin 1A receptors, whereas endothelin 1B receptors have been shown to exert antinoceptive actions [52]. A postmortem study of human carcinoma patients showed increased methylation of a promoter domain of the endothelin 1B receptor gene in painful oral squamous cell carcinoma lesions, but not in non-painful oral dysplasia lesions [31]. In a murine cancer model, restoration of endothelin 1B receptor expression to baseline levels was sufficient to reduce pain behaviors, pointing to pharmacological interventions in DNA methylation as a new approach towards the treatment of cancer-related pain.
Histone acetylation in spinal systems and its involvement in chronic pain
Histone acetylation is thought to be more labile than DNA and histone methylation and to therefore represent a more transient cellular modification that quickly promotes gene expression in response to environmental stimuli [35, 36]. It is driven by histone acetyltransferases (HATs), which catalyze the addition of acetyl groups to histones and promote gene transcription [19,23]. Conversely, histone deacetylases (HDACs) catalyze the removal of acetyl groups from histones, and thereby reduce gene transcription. Eleven HDAC genes have been identified in humans, representing a highly homologous family of enzymes that have been classified into different groups based on their primary and secondary structures. Class I HDACs include 1, 2, 3, and 8; class IIa includes HDACs 4, 5, 7, and 9; class IIb includes HDACs 6 and 10; and class IV comprises HDAC 11[37,38]. Multiple HDAC inhibitors have been observed to upregulate histone acetylation in the brain and spinal dorsal horn [57].
Histone acetylation in the spinal cord has recently been implicated in nociceptive sensitization in animal models of neuropathic pain. Some of the most prominent changes in the DH following spinal nerve ligation include upregulation of HDAC1 expression and reduction of histone H3 acetylation [40]. Such observations led to the hypothesis that modulation of enzyme activity involved in chromatin function may prevent adaptations in the expression of genes that contribute to nociceptive sensitization. Indeed, administration of HDAC inhibitors ameliorated several symptoms of neuropathic pain. Importantly, a single intrathecal dose of baicalin (a non-specific HDAC inhibitor), alleviated spinal nerve ligation-induced hyperalgesia and mechanical allodynia, and reversed injury-related adaptations, such as the reduction of histone H3 acetylation and the increase in HDAC1 expression in the DH [40]. Valproic acid, another non-specific HDAC inhibitor, can also efficiently reduce mechanical allodynia in rat models of neuropathic pain [41]. A more thorough investigation of the mechanisms underlying chromatic modifications under neuropathic pain conditions is expected to provide important information for the development of novel therapeutic interventions. A key consideration is the necessity of targeting specific HDAC isoforms for clinical utility. While the HDAC inhibitors used to date lack specificity, preclinical work suggests that such compounds are effective in treating the symptoms of neuropathic pain: perhaps the ability of an inhibitor to block the activity of several enzymes makes it more beneficial. By contrast, the non-selective inhibition of HDAC activity raises concerns about intolerable drug effects (see Box 3).
Box 3. Outstanding questions.
Current pharmacological interventions for chronic pain have widespread targets (are not pain-specific), are often slow and ineffective at fully reducing pain (neuropathic pain is particularly drug resistant), and cause the development of tolerance (the need for increases in dose in order to achieve the same effect). There is thus a critical need for the development of better pharmacological interventions, and targeting epigenetic mechanisms shows promise in this regard.
Could HDAC inhibitors treat chronic pain?
HDAC inhibition reduces nociceptive sensitization in animal models [24, 57, 59]. HDAC inhibitors are currently marketed for the treatment of cancer, and do not appear to produce tolerance, however reports of various adverse effects have begun to emerge, including nausea, fatigue, blood platelet and bone marrow abnormalities.
Can we target mGlu2 receptors?
Activation of mGlu2 receptors at primary afferent and DH synapses robustly inhibits pain transmission [40], but animal models show that tolerance develops quickly [43]. A promising strategy involves the transcriptional enhancement of mGlu2 expression levels in the DH [43], which has been shown to attenuate nociceptive sensitization in mice [46]. L-acetylcarnitine, a drug already prescribed for diabetic and HIV related neuropathies, has been shown to upregulate mGlu2 receptor expression in the DRG and DH of rats in neuropathic pain [47]. Furthermore, antidepressant effects of LAC are evident in rodents with only 3 days of treatment, whereas the antidepressant chlorimipramine requires 14 days [92]. Moreover, the antidepressant effects of LAC appear to be resistant to tolerance.
What role do epigenetic mechanisms play in chronic pain treatments?
The endogenous opioid system is a major target of current pain therapy. Epigenetic mechanisms may be involved in adaptations determining treatment efficacy. For example, nerve injury induces HDAC dependent silencing of the MOR gene and loss of the pharmacological target for peripheral morphine analgesia [48]. Moreover, HDAC inhibitors have been shown to modulate the expression of the MOR gene in cultured neuroblastoma cells [109].
Other forms of chronic pain, including inflammatory pain, have also been linked to changes in histone acetylation. For instance, CFA-induced inflammatory pain in mice results in upregulation of HDAC2 protein in the DH [39]. Consistent with this upregulation, preclinical evidence strongly suggests that HDAC inhibitors alleviate nociceptive sensitization behaviors caused by inflammatory pain [39,42]. A single intrathecal administration of the HDAC inhibitors suberanilohydroxamic acid (SAHA; also known as vorinostat), trichostatin A (TSA), or LAQ824, which target class I and II HDACs, successfully reduce CFA-induced hyperalgesia [39], and administration of any of these three HDAC inhibitors corresponded with enhanced histone acetylation in the superficial layers of the spinal cord. A detailed understanding of the adaptations in histone modifications induced by inflammatory pain will direct drug development efforts to target specific pain-related alterations.
Beyond histones, multiple proteins (including various transcription factors) can be regulated by acetylation and serve as HDAC targets [43], suggesting alternative mechanisms via which HDAC inhibitors modulate nociceptive sensitization. One such example involves acetylation of the p65 subunit of nuclear factor-KB (NF-KB). L-Acetylcarnitine (LAC), a compound currently prescribed for the treatment of diabetic [44] and HIV related [45] neuropathies, has been shown to enhance p65/Rel-A acetylation in cultured DRG neurons [46]. In a rat CCI model of neuropathic pain, repeated administration of LAC induces analgesia by a mechanism that involved upregulation of metabotropic glutamate (mGlu)-2/3 receptors in the DH [47]. This effect was mediated by the NF-κB pathway and dependent on acetylation of NF-κB family transcription factors. Additional evidence for the importance of mGlu2 receptors was provided by a study using a murine model of inflammatory pain, where chronic (but not acute) administration of MS-275 or SAHA reduced nociceptive responses in the second phase of the formalin test. This reduction corresponded with enhanced expression of mGlu2 receptors in the spinal cord and DRG [42]. Analgesia produced by a 5-day treatment with MS275 or SAHA was largely attenuated by a single injection of the preferential mGlu2/3 receptor antagonist LY341495. The selective effect of MS-275 in the second phase of the formalin test (which is related to the development of central sensitization) suggests that, in addition to histone acetylation [39], p65/NFKB acetylation has a key role in long-term changes associated with the phenomenon of central sensitization. Thus, class I HDAC inhibitors, or acetyl donors like LAC, may be useful tools for the treatment of certain inflammatory pain conditions, because they target both histone and non-histone acetylation events at the spinal cord level. Moreover, these strategies take advantage of endogenous analgesic mechanisms by enhancing mGlu2 function, and studies suggest that such agents produce less analgesic tolerance compared with current pharmacological treatments [43] (see Box 3).
Histone acetylation has also been implicated in adaptations of other endogenous pain- controlling networks. For example, the expression of μ-opioid receptors (MORs) and Nav1.8 sodium channels in the mouse DRG is reduced under neuropathic pain states [48], and is thought to be an underlying cause of negative symptoms of chronic pain. After 7 days of nerve injury, reductions in MOR and Nav1.8 expression have been shown to correspond with prominent histone changes, including enhanced H4 acetylation at the NRSF (neuron-restrictive silencer factor, also known as REST) promoter II region, reduction of H3 and H4 acetylation at MOR-NRSE (neuronrestrictive silencer element, also known as RE1) and at Nav1.8- NRSE-2, and enhanced H3 acetylation at Nav1.8-NRSE-1. In cultured cells, NRSF repressed Oprm1 (gene encoding MOR) promoter activity in an HDAC-dependent manner [65]. Specifically, in the presence of NRSF, NRSE repressed Oprm1 transcription, and this action was blocked by treatment with the HDAC inhibitor TSA [49]. Knock-down of NRSF with antisense oligonucleotides prevented the downregulation of Oprm1 and Scn10a (gene encoding Nav1.8 channels), further supporting the hypothesis that NRSF-mediated mechanisms are responsible for injury-induced transcriptional suppression of Oprm1 and Scn10a genes in the DRG [48]. Once again, these data point to new therapeutic strategies for pain relief, and suggest that interventions at the level of histone complexes would revere the maladaptive downregulation of GPCRs or ion channels in the DRG.
miRNA activity in spinal systems and its involvement in chronic pain
With regards to activity-dependent control of gene expression, much attention has recently been devoted to miRNAs, which are non-coding functional RNAs that play critical roles in gene regulation. Typically around 17–24 nucleotides long, miRNAs can bind to protein-coding mRNAs to post-transcriptionally repress protein expression [66]. The mechanisms by which certain primary transcripts are processed into mature miRNAs, and by which miRNAs suppress protein translation, are now well described. The potential of miRNAs to regulate the expression of specific pain-related genes in the CNS [51,52] raises the exciting possibility that miRNAs represent a novel therapeutic avenue for management of chronic pain conditions.
Interventions in miRNA function in sensory neurons were first achieved via conditional deletion of Dicer—an enzyme with a critical role in miRNAs processing. Conditional deletion of Dicer in the DRG, through the use of the Nav1.8-Cre mouse line, corresponded with reduction in the levels of nociception-related mRNA transcripts, such as Nav1.8 sodium channels, P2X3 receptors (a purine receptor), and Runx-1 (a transcription factor). Remarkably, deletion of Dicer also resulted in a robust reduction of inflammatory pain behaviors in mice, while maintaining intact acute nociception [53]. Therefore disruption of miRNA processing in primary afferent pathways is sufficient to inhibit injury-induced long-term development of chronic pain-related behaviors.
More recent work also linked the development of inflammation-induced nociceptive sensitization with adaptations in the expression of miRNA-124a in the mouse spinal DH [29]. Specifically, hindpaw injections of formalin induced a time dependent reduction in miRNA-124a expression in the spinal cord, which peaked between 8- and 24 hr, and returned to baseline by 48 hr post-injection. This reduction seems to facilitate nociceptive transmission, because systemic application of a miRNA-124a inhibitor markedly increased nociceptive behavior in the formalin model, and reduced the expression of inflammatory molecules involved in chronic pain transmission such as IL-1β [54]. By contrast, systemic application of a miRNA-124a mimic caused a modest reduction in formalin-induced nociceptive behavior, confirming the causal role of miRNA-124a in chronic pain, and perhaps offering a novel treatment target. Changes in miRNA expression have also been observed in the trigeminal root ganglion of rats exposed to chronic inflammatory pain. Specifically, significant decreases of several miRNAs, including miR-134, were observed in response to injections of CFA into the masseter muscle [52]. In agreement with these observations, a recent study showed decreases in miR-134 expression in the DRG of rats following CFA injections into the hind paw [55]. Interestingly, this decrease corresponded with upregulation of MOR, which is implicated in endogenous antinociceptive mechanisms and is the main target of opiate analgesics.
Adaptations in miRNAs expression in response to injury appear to also be involved in the actions of inflammatory molecules. Peripheral injury induces the release of multiple pro-inflammatory agents, including chemokines and cytokines, which are implicated in the development of chronic pain [56]. Intrathecal administration of IL-1β for 3 days resulted in allodynia in intact rats, and was accompanied by increases in miR-21 expression in the DRG [54]. Of particular interest is that IL-1β has also been shown to regulate HAT and HDAC activities in certain tissues [57], suggesting that additional epigenetic pathways are downstream of this cytokine. These data show that changes in miRNA activity correspond with multiple inflammatory pain models, and are driven by inflammatory agents. These changes may represent one way in which injury alters gene expression in nociceptive pathways, and may help identify novel targets for therapeutic interventions.
Neuropathic pain models have also revealed time and tissue selective changes in miRNA expression. For example, L5 spinal nerve ligation or chronic constriction injury increased miR-21 expression at late time points (7 and 14 days) after injury, but not in early stages of neuropathic pain (3 days post injury) in the rat DRG [54]. Similarly, miRNA-195 is upregulated in the DRG after sciatic nerve injury [58], and in the DH as early as 2 days after spinal nerve ligation, and persists for at least 14 days [59]. Administration of miRNA-195 mimic compounds induced the expression of pro-inflammatory cytokines IL-1β, tumor necrosis factor-α (TNF-α), and inducible nitric oxide synthase (iNOS) in cultured microglia [59]. Upregulation of miRNA-195 also enhanced mechanical allodynia and cold hyperalgesia after peripheral nerve injury, whereas the opposite patterns were observed after miRNA-195 inhibition. Linking it further with microglia activity, microglial autophagy was decreased by peripheral nerve injury, and enhanced by miRNA-195 inhibition, suggesting that miR-195 increases neuroinflammation and neuropathic pain by inhibiting autophagy activation. These studies highlight a functional link between miRNA activity in spinal networks and nociceptive sensitization, and provide evidence that targeting miRNA activity in these regions is sufficient to attenuate chronic pain-related behaviors.
In the spinal cord, long-term changes associated with pain sensitization are induced by changes in synaptic transmission and require the activation of voltage-gated calcium channels (VGCCs) [3]. Thus, emphasis has been given to understanding the regulation of VGCCs, with the idea that under certain conditions prevention of VGCC activation will ameliorate chronic pain. Recent evidence has revealed functional links between miRNAs and VGCCs in neuropathic pain [60]. Specifically, investigations of the superficial dorsal horn of rats exposed to spinal nerve ligation revealed an upregulation of L-type Cav1.2 subunits of VGCCs that corresponded to a decrease in miR-103 [60]. Furthermore, repeated intrathecal administration (once daily for 4 days) of miR-103 reduced nerve injury induced allodynia.
Similarly, voltage-gated sodium channels have been implicated in spinal mechanisms of neuropathic pain [61] and miRNA modulation of injury-induced changes in sodium channels have been reported. For example, through microarray analysis, miR-7a was found to be robustly reduced in the DRG of rats 14 days after spinal nerve ligation, and miR-7a was observed to target voltage-gated sodium channel β subunits, whereas downregulation of miR-7a resulted in an increase of the channel [62]. Delivery of adeno-associated viruses expressing miR-7a into DRG of injured rats significantly alleviated pain-associated behaviors. The prominent role of miR-7a in sensitized behaviors was revealed in studies showing that inhibition of miR-7a in intact rats is sufficient to produce mechanical allodynia and thermal hyperalgesia. Similarly, miR-96 mRNA expression in the rat DRG was reduced following sciatic nerve ligation, in correspondence with an increase in the expression of the Nav1.3 sodium channels [63]. Moreover, daily intrathecal injections of miR-96 restored Nav1.3 mRNA and protein levels in DRG of injured rats and significantly reduced tactile allodynia. Several drug development efforts for the treatment of chronic pain are directed towards voltage gated calcium and potassium channels. This new information on a functional link between miRNAs and voltage gated ion channel function may provide additional pathways for therapeutic interventions.
Epigenetic mechanisms in brain networks and their involvement in chronic pain
The bulk of chronic pain research focuses on spinal cord studies, and it is thus unsurprising that few reports exist regarding epigenetic mechanisms in supraspinal regions involved in the expression or modulation of chronic pain symptoms. Chronic pain requires two phases: induction and maintenance, which may be in part driven by distinct mechanisms [64]. Given that chronic pain is a pathological condition that often outlasts the presence of peripheral injury, epigenetic modifications in supraspinal sites related to pain perception, learning, motivation, and mood regulation present a novel area of research into long-term changes involved in persistent pain, and represent potentially new treatment targets. Multiple human neuroimaging studies support long-term changes in activity within areas of the pain matrix in correspondence with chronic pain [79, 80]. Evidence from animal models has also indicated modifications of brain circuits and neuronal activity under chronic pain conditions [16, 81]. Epigenetics mechanisms in these regions are well positioned to drive injury induced alterations in gene expression. Acute pain is characterized by activity in multiple brain areas, including the thalamus, insula, anterior cingulate cortex, prefrontal cortex, and the primary and secondary somatosensory cortices [82–84]. However, activity in additional brain regions has been implicated in affective and cognitive components of pain, and appears to be altered in chronic pain conditions [85, 86]. In fact, neuroimaging data show that chronic pain corresponds with changes in the amygdala [74], the anterior cingulate cortex [83, 88], the prefrontal cortex (PFC) [76], the insular cortex [90], and the nucleus accumbens (NAc) [91]. These regions display robust interconnections with each other, and directly or indirectly modulate the activity of other pain modulating networks, such as the periaqueductal grey (PAG) [86, 92] and its projections to the rostral ventromedial medulla (RVM) [80]. Together, these regions form a network capable of exerting top-down regulation of nociceptive input [81], and have been found to undergo long term changes in neural activity in chronic pain states [82].
The raphe magnus forms part of the RVM, and receives robust excitatory input from the PAG [95, 96]. Excitatory activity within the PAG exerts biphasic modulation of raphe magnus neurons [85], which in turn extend serotonergic afferents to deep and superficial layers of the DH [84]. This top-down modulation of pain may be facilitatory or inhibitory depending on the pattern of activity in the raphe nuclei. Alterations in the activity of this pathway of the pain matrix may robustly affect nociceptive experience [41, 93]. Implicating epigenetic mechanisms in such alterations, hind paw injections of CFA promote global histone H3 acetylation in the rat raphe nucleus 1 day after injection [86]. This study also demonstrated a reduction of GAD65 levels at later time points (3 d after CFA injection), but not earlier (4 hr after injection). This change in GAD65 levels corresponded with reductions in H3 acetylation at the promoter region of the GAD65 gene (GAD2) and was reversed by treatment with the HDAC inhibitor TSA. These observations were replicated using a more clinically relevant model, the spinal nerve ligation model of neuropathic pain, further supporting the key role of GAD65 gene acetylation in both inflammatory and neuropathic states. The time dependent histone acetylation and corresponding GAD65 reduction support previous observations of RVM involvement in the maintenance, but not initiation, of descending facilitation [98], and implicate additional changes in activity in upstream processes. Epigenetic mechanisms decreasing GAD65 expression would create an imbalance biasing towards stimulatory control. Given that RVM-induced antinociceptive effects are at least in part mediated via spinal norepinephrine release [88], a long- term shift towards descending facilitation of nociceptive effects may explain the greater clinical efficacy observed with SNRIs (selective serotonin and norepinephrine reuptake inhibitors) compared with drugs targeting only serotonergic activity [89].
Decreases in global DNA methylation have been observed in the mouse PFC and amygdala in the spared nerve ligation model of neuropathic pain [90]. Moreover, global PFC DNA methylation levels correlated with mechanical and thermal sensitivity induced by this model [90]. Changes in miRNA expression in the mouse PFC have also been reported in models of inflammatory pain, where facial injections of carrageenan lead to bilateral enhancement of miR-155 and miR-223 expression [91]. The PFC is part the pain matrix, and is implicated in the pathophysiology of depression and anxiety disorders as well. As with chronic pain, mounting data point to epigenetic adaptations in depression syndromes [91]. LAC robustly reduces depression like behavior in rats and mice, and analogous to observations in the spinal cord [47], it upregulates mGlu2 mRNA and protein levels in the PFC [93]. LAC mediated induction of mGlu2 expression was sensitive to NF- KB inhibition, and corresponded with enhanced acetylation of H3K27 bound to the mGlu2 gene (Grm2) promoter [93]. Notably, HDAC inhibition replicated LAC effects. Thus acetylation driven mechanisms that modulate mGlu2 expression in the PFC provide an epigenetic link between chronic pain and depression. Indeed, an overwhelming number of individuals with chronic pain develop comorbid depression [94] and sleep disturbances [95]. Accordingly, mouse models of chronic pain also result in depression-like behaviors [96] and disrupted sleep wave patterns [68]. Pain-induced epigenetic modifications of neuronal activity in brain regions involved in mood and sleep regulation may disturb normal neuronal function, and such imbalance may result in subsequent depression and sleep disturbances.
Chronic pain-induced epigenetic modifications have also been observed in the brain reward centre. In the mouse NAc, studies have found evidence that sciatic nerve ligation causes a robust decrease of miR-200b and miR-249, while enhancing expression of miR-34c seven days post injury [97]. These adaptations appeared to be brain region-specific because no changes in expression of these miRNAs were detected in other pain related structures. Interestingly the injury-induced reduction in miR-200b and miR-249 expression was correlated with an increase in the levels of DNA methyltransferase 3a (DNMT3a), which is a target of miR-200b/249. Thus, changes in DNA methylation might be downstream of injury-induced miRNA expression adaptations. In addition to forming part of the pain matrix, the NAc is part of the mesolimbic reward pathway, and plays major roles in substance abuse and mood disorders [98]. Epigenetic adaptations in the rat NAc, for example reductions in DNMT3a expression levels, are observed following chronic cocaine exposure. By contrast, social defeat stress promotes DNMT3a expression in the NAc [99]. Thus chronic pain, chronic social stress, and exposure to drugs of abuse promote significant but distinct changes in DNMT3 function in the NAc [100].
Future directions
As is apparent from this review, there are still a relatively few studies that have examined epigenetic changes in chronic pain models (Table 1). Yet, alterations in chromatin structure represent ideal mechanisms by which the experience of tissue injury and acute pain can be converted gradually and progressively into pathological processes of neuroinflammation, central sensitization, and ultimately chronic pain syndromes. Indeed, while in early stages of development, existing data reviewed here support this hypothesis. Changes in DNA methylation, histone acetylation and methylation, and miRNA expression have each been shown to be robustly altered in various CNS sites under chronic pain states and, manipulation of these processes influence chronic pain-related behaviors. Moving forward, investigators need to explore many other modes of epigenetic regulation in advanced animal models, and to use of genome-wide methods such as chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) to map the range of genic and non-genic regions that are affected in chronic nociceptive conditions. To date, few studies have investigated epigenetic adaptations to chronic pain in brain regions associated with central sensitization, motivation, and depression. Given that adaptations in these brain regions play a major role in symptoms associated with chronic pain syndromes, it will be interesting to identify the key epigenetic determinants of nociceptive behaviors in each of these structures. In addition, most of the described studies evaluating novel compounds in chronic pain models have focused on sensory components of chronic pain (mainly mechanical allodynia and thermal hyperalgesia). It is important to evaluate the actions of these compounds in affective components of chronic pain, and to develop assays to evaluate spontaneous pain, which is a very prominent symptom of this condition in humans.
Table 1.
epigenetic targets corresponding with chronic pain.
| Epigenetic target | Epigenetic effect | Region | Results | Ref. |
|---|---|---|---|---|
| MicroRNAs | ↑ miR-21 | DRG | Mechanical allodynia and thermal hyperalgesia | 21 |
| ↓ miR-134 | DRG | Up-regulation of μ-opioid receptor | 67 | |
| ↓ miR-7a, ↓ miR-96 | DRG | Nerve injury induced increases in voltage gated sodium channels | 74,75 | |
| ↓ miR-124a | Spinal cord | Regulation pro-inflammatory marker genes | 46 | |
| ↓ miR-200b, ↓ miR-249 | NAc | Dysfunction of mesolimbic motivation circuitry | 23 | |
| ↑ miR-155, ↑ miR-223 | PFC | miRNA changes in the PFC, after inflammatory pain induced by facial carrageenan injection | 24 | |
| DNA methylation | ↓ Global DNA Methylation | PFC, AMY | Correlation with the magnitude of nerve injury-induced nociceptive sensitization | 99 |
| ↑ phosphorylation of MeCP2 | Spinal cord | Correlation with magnitude of inflammation induced nociceptive sensitization | 44, 45 | |
| Histone modification | ↓ HistoneH3K27me3 | Spinal cord | Chemokine activation | 36 |
| ↑ HDAC1 | Spinal cord | Hyperalgesia and allodynia in SNL were accompanied by HDAC1 overexpression | 22 | |
| ↑ HDAC1, 2, & 4 | NRM | Inflammatory and nerve injury associated suppression of Gad2 transcription and disrupted GABA synaptic inhibition | 35 | |
| ↑ HDAC2 | Spinal cord | Thermal hyperalgesia | 55 |
DRG: dorsal root ganglion; NAc: nucleus accumbens; PFC: prefrontal cortex; AMY: amygdala; NRM: nucleus raphe magnus.
New information of epigenetic mechanisms governing the induction of chronic pain syndromes is expected to provide a wide range of potential treatment targets but also change the approach towards pain management: Instead of alleviating specific symptoms, targeted epigenetic interventions provide the tools to prevent nociceptive sensitization, mood-related symptoms, and other adaptations that occur in response to injury or inflammation. This approach will greatly facilitate pain management and will improve treatment efficacy in many chronic pain conditions.
Box 2. How epigenetics can mediate chronic pain.
Epigenetics refers to the long lasting changes in gene expression that, although they do not alter the actual DNA sequence, can result in functional alterations of cellular activity [27, 33, 35]. Neurons within the CNS undergo epigenetic modifications in response to an organism’s experience with the external environment [107]. DNA is tightly wound around histones, forming a complex called chromatin. Gene expression is dependent on transcription factors accessing promoter regions on DNA sequences, a process that is regulated by epigenetic modifications to the chromatin structure. Epigenetic mechanisms include DNA methylation, several types of histone modifications (e.g., acetylation, methylation, phosphorylation, ubiquitination, and ADP-ribosylation), and expression of microRNAs (miRNAs) [22]. These adaptations can modify neuronal morphology and activity to produce changes in behavior [107]. Indeed, epigenetic mechanisms have been shown to be involved in synaptic plasticity, learning, and memory [27, 28], as well as in the pathophysiology of several neuropsychiatric disorders, including depression and drug addiction [29–31]. Moreover, recent studies have found multiple epigenetic modifications in spinal and brain regions in correspondence with chronic pain conditions. Mounting evidence indicates that epigenetic modifications highly contribute to sensitized behavioral responses to mechanical and thermal stimuli [25, 37–39]. These modifications may occur in primary afferent, dorsal horn, or spinothalamic tract neurons, and in any of several brain regions of the pain matrix. For example, epigenetic mechanisms could alter receptor expression levels at neuronal synapses in the superficial dorsal horn (DH) [43], and this effect could augment primary afferent activation of spinal systems, facilitating the development of hyperalgesia. Similarly, epigenetic changes in top-down regulatory brain regions (such as the ACC, PFC, PAG, and RVM) [82] could lead to facilitation of stimuli induced activation, perhaps paving the way for the development of allodynia or other sensory deficits. Epigenetic changes may also occur in interneurons [86], causing disinhibition of critical nociceptive pathways that may potentiate allodynia, or even promote hyperalgesia. Glial cells, namely astrocytes in the spinal cord [30] and brain [96], can also undergo injury induced changes in gene expression that are mediated by epigenetic mechanisms and potentially contribute to pain syndromes. Thus, epigenetic mechanisms present dynamic processes for controlling changes in neuronal activity and behavior that may be responsible for the persistent manifestation of a chronic pain state.
Acknowledgments
Supported by the Banting Postdoctoral Fellowships program (GD), NIDA P01DA008227 (EJN, VZ) and NINDS R01NS086444 (VZ), JSPS Grant-in-Aid for Young Scientists (B) 24791627 (DI), JSPS Grant-in-Aid for Scientific Research (B) 26293346 (MN), MEXT-Supported Program for the Strategic Research Foundation at Private Universities, 2014-2018, S1411019 (MN, DI).
References
- 1.Woolf C, Mannion R. Neuropathic pain: aetiology, symptoms, mechanisms, and management. Lancet. 1999;353:1959–1964. doi: 10.1016/S0140-6736(99)01307-0. [DOI] [PubMed] [Google Scholar]
- 2.Ji R, et al. Central sensitization and LTP: do pain and memory share similar mechanisms? Trends Neurosci. 2003;26:696–705. doi: 10.1016/j.tins.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 3.Woolf CJ, Salter MW. Neuronal Plasticity: Increasing the Gain in Pain. Science. 2000;288(5472):1765–1768. doi: 10.1126/science.288.5472.1765. [DOI] [PubMed] [Google Scholar]
- 4.Institute of Medicine; B.o.H.S. Policy, editor. Relieving pain in America A Blueprint for Transforming Prevention, Care, Education, and Research. The National Academies Press; Washington, D.C: 2011. [PubMed] [Google Scholar]
- 5.Mogil JS. Animal models of pain: progress and challenges. Nat Rev Neurosci. 2009;10(4):283–94. doi: 10.1038/nrn2606. [DOI] [PubMed] [Google Scholar]
- 6.Basbaum AI, et al. Cellular and Molecular Mechanisms of Pain. Cell. 2009;139(2):267–284. doi: 10.1016/j.cell.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikeda H, et al. Synaptic amplifier of inflammatory pain in the spinal dorsal horn. Science. 2006;312:1659–1662. doi: 10.1126/science.1127233. [DOI] [PubMed] [Google Scholar]
- 8.Sandkuhler J, Liu; X. Induction of long-term potentiation at spinal synapses by noxious stimulation or nerve injury. Eur J Neurosci. 1998;10:2476–2480. doi: 10.1046/j.1460-9568.1998.00278.x. [DOI] [PubMed] [Google Scholar]
- 9.Ji RR, Rupp; F. Phosphorylation of transcription factor CREB in rat spinal cord after formalin-induced hyperalgesia: Relationship to c-fos induction. Journal of Neuroscience. 1997;17(5):1776–1785. doi: 10.1523/JNEUROSCI.17-05-01776.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narita M, et al. Protease-Activated Receptor-1 and Platelet-Derived Growth Factor in Spinal Cord Neurons Are Implicated in Neuropathic Pain after Nerve Injury. The Journal of Neuroscience. 2005;25(43):10000–10009. doi: 10.1523/JNEUROSCI.2507-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei F, Li P, Zhuo M. Loss of Synaptic Depression in Mammalian Anterior Cingulate Cortex after Amputation. J Neurosci. 1999;19(21):9346–9354. doi: 10.1523/JNEUROSCI.19-21-09346.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alvarado S, et al. Peripheral nerve injury is accompanied by chronic transcriptome-wide changes in the mouse prefrontal cortex. Molecular Pain. 2013;9(1):21. doi: 10.1186/1744-8069-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyman S, Nestler E. The Molecular Foundations of Psychiatry. Washington D.C: American Psychiatric Press, Inc; 1993. [Google Scholar]
- 14.Vignes M, Collingridge GL. The synaptic activation of kainate receptors. Nature. 1997;388(6638):179–182. doi: 10.1038/40639. [DOI] [PubMed] [Google Scholar]
- 15.Kandel E. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 16.Nelson ED, Monteggia LM. Epigenetics in the mature mammalian brain: Effects on behavior and synaptic transmission. Neurobiology of Learning and Memory. 2011;96(1):53–60. doi: 10.1016/j.nlm.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sweatt JD. The Emerging Field of Neuroepigenetics. Neuron. 2013;80(3):624–632. doi: 10.1016/j.neuron.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renthal W, et al. Histone Deacetylase 5 Epigenetically Controls Behavioral Adaptations to Chronic Emotional Stimuli. Neuron. 2007;56(3):517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 19.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12(11):623–37. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vialou V, et al. Epigenetic Mechanisms of Depression and Antidepressant Action. Annual Review of Pharmacology and Toxicology. 2013;53(1):59–87. doi: 10.1146/annurev-pharmtox-010611-134540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraga MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(30):10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 23.Jiang Y, et al. Epigenetics in the Nervous System. The Journal of Neuroscience. 2008;28(46):11753–11759. doi: 10.1523/JNEUROSCI.3797-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bienvenu T, Chelly J. Molecular genetics of Rett syndrome: when DNA methylation goes unrecognized. Nat Rev Genet. 2006;7(6):415–26. doi: 10.1038/nrg1878. [DOI] [PubMed] [Google Scholar]
- 25.Fuks F, et al. The Methyl-CpG-binding Protein MeCP2 Links DNA Methylation to Histone Methylation. Journal of Biological Chemistry. 2003;278(6):4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- 26.Chen WG, et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302(5646):885–9. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 27.Géranton SM, Morenilla-Palao C, Hunt SP. A Role for Transcriptional Repressor Methyl-CpG-Binding Protein 2 and Plasticity-Related Gene Serum- and Glucocorticoid-Inducible Kinase 1 in the Induction of Inflammatory Pain States. The Journal of Neuroscience. 2007;27(23):6163–6173. doi: 10.1523/JNEUROSCI.1306-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geranton S, et al. Descending serotonergic controls regulate inflammation-induced mechanical sensitivity and methyl-CpG-binding protein 2 phosphorylation in the rat superficial dorsal horn. Molecular Pain. 2008;4(1):35. doi: 10.1186/1744-8069-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kynast KL, et al. Modulation of central nervous system–specific microRNA-124a alters the inflammatory response in the formalin test in mice. PAIN. 2013;154(3):368–376. doi: 10.1016/j.pain.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 30.Imai S, et al. Change in MicroRNAs Associated with Neuronal Adaptive Responses in the Nucleus Accumbens under Neuropathic Pain. The Journal of Neuroscience. 2011;31(43):15294–15299. doi: 10.1523/JNEUROSCI.0921-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viet CT, et al. Re-expression of the methylated EDNRB gene in oral squamous cell carcinoma attenuates cancer-induced pain. PAIN. 2011;152(10):2323–2332. doi: 10.1016/j.pain.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freitas Cordeiro-Silva M, et al. Methylation analysis of cancer-related genes in non-neoplastic cells from patients with oral squamous cell carcinoma. Molecular Biology Reports. 2011;38(8):5435–5441. doi: 10.1007/s11033-011-0698-1. [DOI] [PubMed] [Google Scholar]
- 33.Pickering V, et al. Effect of peripheral endothelin-1 concentration on carcinoma-induced pain in mice. European Journal of Pain. 2008;12(3):293–300. doi: 10.1016/j.ejpain.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peters CM, et al. Endothelin and the tumorigenic component of bone cancer pain. Neuroscience. 2004;126(4):1043–1052. doi: 10.1016/j.neuroscience.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 35.Guan Z, et al. Integration of Long-Term-Memory-Related Synaptic Plasticity Involves Bidirectional Regulation of Gene Expression and Chromatin Structure. Cell. 2002;111(4):483–493. doi: 10.1016/s0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- 36.Hart AK, et al. Serotonin-Mediated Synapsin Expression Is Necessary for Long-Term Facilitation of the Aplysia Sensorimotor Synapse. The Journal of Neuroscience. 2011;31(50):18401–18411. doi: 10.1523/JNEUROSCI.2816-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: Functional implications of phylogenetic analysis. Journal of Molecular Biology. 2004;338(1):17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 38.Dekker FJ, van den Bosch T, Martin NI. Small molecule inhibitors of histone acetyltransferases and deacetylases are potential drugs for inflammatory diseases. Drug Discovery Today. 2013;(0) doi: 10.1016/j.drudis.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 39.Bai G, et al. Inhibition of class II histone deacetylases in the spinal cord attenuates inflammatory hyperalgesia. Molecular Pain. 2010;6(1):51. doi: 10.1186/1744-8069-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cherng C-H, et al. Baicalin ameliorates neuropathic pain by suppressing HDAC1 expression in the spinal cord of spinal nerve ligation rats. Journal of the Formosan Medical Association. 2013;(0) doi: 10.1016/j.jfma.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 41.Winkler I, et al. Efficacy of antiepileptic isomers of valproic acid and valpromide in a rat model of neuropathic pain. British Journal of Pharmacology. 2005;146(2):198–208. doi: 10.1038/sj.bjp.0706310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiechio S, et al. Epigenetic modulation of mGlu2 receptors by histone deacetylase inhibitors in the treatment of inflammatory pain. Mol Pharmacol. 2009;75(5):1014–20. doi: 10.1124/mol.108.054346. [DOI] [PubMed] [Google Scholar]
- 43.Chiechio S, et al. Transcriptional regulation of type-2 metabotropic glutamate receptors: an epigenetic path to novel treatments for chronic pain. Trends Pharmacol Sci. 2010;31(4):153–60. doi: 10.1016/j.tips.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 44.Sima AAF, et al. Acetyl-l-Carnitine Improves Pain, Nerve Regeneration, and Vibratory Perception in Patients With Chronic Diabetic Neuropathy: An analysis of two randomized placebo-controlled trials. Diabetes Care. 2005;28(1):89–94. doi: 10.2337/diacare.28.1.89. [DOI] [PubMed] [Google Scholar]
- 44.Chaplan SR, et al. Quantitative assessment of tactile allodynia in the rat paw. Journal of Neuroscience Methods. 1994;53(1):55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 45.Scarpini E, et al. Effect of acetyl-L-carnitine in the treatment of painful peripheral neuropathies in HIV+ patients. J Peripher Nerv Syst. 1997;2(3):250–2. [PubMed] [Google Scholar]
- 46.Chiechio S, et al. Transcriptional regulation of metabotropic glutamate receptor 2/3 expression by the NF-kappaB pathway in primary dorsal root ganglia neurons: a possible mechanism for the analgesic effect of L-acetylcarnitine. Molecular Pain. 2006;2(1):20. doi: 10.1186/1744-8069-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiechio S, et al. l-Acetylcarnitine Induces Analgesia by Selectively Up-Regulating mGlu2 Metabotropic Glutamate Receptors. Molecular Pharmacology. 2002;61(5):989–996. doi: 10.1124/mol.61.5.989. [DOI] [PubMed] [Google Scholar]
- 48.Uchida H, Ma L, Ueda H. Epigenetic Gene Silencing Underlies C-Fiber Dysfunctions in Neuropathic Pain. The Journal of Neuroscience. 2010;30(13):4806–4814. doi: 10.1523/JNEUROSCI.5541-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim CS, et al. Neuron-restrictive Silencer Factor (NRSF) Functions as a Repressor in Neuronal Cells to Regulate the μ Opioid Receptor Gene. Journal of Biological Chemistry. 2004;279(45):46464–46473. doi: 10.1074/jbc.M403633200. [DOI] [PubMed] [Google Scholar]
- 50.Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pohl K-W, Yeol J-F, Ongl W-Y. MicroRNA changes in the mouse prefrontal cortex after inflammatory pain. European Journal of Pain. 2011;15(8):801.e1–801.e12. doi: 10.1016/j.ejpain.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 52.Bai G, et al. Downregulation of selective microRNAs in trigeminal ganglion neurons following inflammatory muscle pain. Molecular Pain. 2007;3(1):15. doi: 10.1186/1744-8069-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao J, et al. Small RNAs Control Sodium Channel Expression, Nociceptor Excitability, and Pain Thresholds. The Journal of Neuroscience. 2010;30(32):10860–10871. doi: 10.1523/JNEUROSCI.1980-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sakai A, Suzuki H. Nerve injury-induced upregulation of miR-21 in the primary sensory neurons contributes to neuropathic pain in rats. Biochemical and Biophysical Research Communications. 2013;435(2):176–181. doi: 10.1016/j.bbrc.2013.04.089. [DOI] [PubMed] [Google Scholar]
- 55.Ni J, et al. Regulation of μ-opioid type 1 receptors by microRNA134 in dorsal root ganglion neurons following peripheral inflammation. European Journal of Pain. 2013;17(3):313–323. doi: 10.1002/j.1532-2149.2012.00197.x. [DOI] [PubMed] [Google Scholar]
- 56.Scholz J, Woolf CJ. The neuropathic pain triad: neurons, immune cells and glia. Nat Neurosci. 2007;10(11):1361–8. doi: 10.1038/nn1992. [DOI] [PubMed] [Google Scholar]
- 57.Ito K, Barnes PJ, Adcock IM. Glucocorticoid Receptor Recruitment of Histone Deacetylase 2 Inhibits Interleukin-1β-Induced Histone H4 Acetylation on Lysines 8 and 12. Molecular and Cellular Biology. 2000;20(18):6891–6903. doi: 10.1128/mcb.20.18.6891-6903.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu B, et al. Profile of microRNAs following rat sciatic nerve injury by deep sequencing: implication for mechanisms of nerve regeneration. PLoS One. 2011;6(9):e24612. doi: 10.1371/journal.pone.0024612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi G, et al. Increased miR-195 aggravates neuropathic pain by inhibiting autophagy following peripheral nerve injury. Glia. 2013;61(4):504–12. doi: 10.1002/glia.22451. [DOI] [PubMed] [Google Scholar]
- 60.Favereaux A, et al. Bidirectional integrative regulation of Cav1.2 calcium channel by microRNA miR-103: role in pain. EMBO J. 2011;30(18):3830–41. doi: 10.1038/emboj.2011.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dib-Hajj SD, et al. Plasticity of sodium channel expression in DRG neurons in the chronic constriction injury model of neuropathic pain. Pain. 1999;83(3):591–600. doi: 10.1016/S0304-3959(99)00169-4. [DOI] [PubMed] [Google Scholar]
- 62.Sakai A, et al. miR-7a alleviates the maintenance of neuropathic pain through regulation of neuronal excitability. Brain. 2013;136(9):2738–2750. doi: 10.1093/brain/awt191. [DOI] [PubMed] [Google Scholar]
- 63.Chen H-P, et al. Intrathecal miR-96 Inhibits Nav1.3 Expression and Alleviates Neuropathic Pain in Rat Following Chronic Construction Injury. Neurochemical Research. 2014;39(1):76–83. doi: 10.1007/s11064-013-1192-z. [DOI] [PubMed] [Google Scholar]
- 64.Li X-Y, et al. Alleviating Neuropathic Pain Hypersensitivity by Inhibiting PKMzeta in the Anterior Cingulate Cortex. Science. 2010;330(6009):1400–1404. doi: 10.1126/science.1191792. [DOI] [PubMed] [Google Scholar]
- 65.Baliki MN, et al. Corticostriatal functional connectivity predicts transition to chronic back pain. Nature neuroscience. 2012;15(8):1117–1119. doi: 10.1038/nn.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garcia-Larrea L, Peyron R. Pain matrices and neuropathic pain matrices: A review. PAIN®. 2013;154(Supplement 1(0)):S29–S43. doi: 10.1016/j.pain.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 67.Zhuo M. Cortical excitation and chronic pain. Trends Neurosci. 2008;31:199–207. doi: 10.1016/j.tins.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 68.Narita M, et al. Sleep disturbances in a neuropathic pain-like condition in the mouse are associated with altered GABAergic transmission in the cingulate cortex. PAIN. 2011;152(6):1358–1372. doi: 10.1016/j.pain.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 69.Apkarian AV, et al. Human brain mechanisms of pain perception and regulation in health and disease. Eur J Pain. 2005;9(4):463–84. doi: 10.1016/j.ejpain.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 70.Hutchison W, et al. Pain-related neurons in the human cingulate cortex. Nat Neurosci. 1999;2(5):403–405. doi: 10.1038/8065. [DOI] [PubMed] [Google Scholar]
- 71.Rainville P, Bushnell M, Duncan G. Representation of acute and persistent pain in the human CNS: potential implications for chemical intolerance. Ann N Y Acad Sci. 2001;933:130–141. doi: 10.1111/j.1749-6632.2001.tb05820.x. [DOI] [PubMed] [Google Scholar]
- 72.Price DD. Psychological and Neural Mechanisms of the Affective Dimension of Pain. Science. 2000;288(5472):1769–1772. doi: 10.1126/science.288.5472.1769. [DOI] [PubMed] [Google Scholar]
- 73.Bushnell MC, Ceko M, Low LA. Cognitive and emotional control of pain and its disruption in chronic pain. Nat Rev Neurosci. 2013;14(7):502–11. doi: 10.1038/nrn3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Neugebauer V, et al. The Amygdala and Persistent Pain. The Neuroscientist. 2004;10(3):221–234. doi: 10.1177/1073858403261077. [DOI] [PubMed] [Google Scholar]
- 75.Talbot J, et al. Multiple representations of pain in human cerebral cortex. Science. 1991;251(4999):1355–1358. doi: 10.1126/science.2003220. [DOI] [PubMed] [Google Scholar]
- 76.Derbyshire SW, Whalley MG, Oakley DA. Fibromyalgia pain and its modulation by hypnotic and non-hypnotic suggestion: an fMRI analysis. Eur J Pain. 2009;13(5):542–50. doi: 10.1016/j.ejpain.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 77.Hofbauer RK, Olausson HW, Bushnell MC. Thermal and tactile sensory deficits and allodynia in a nerve-injured patient: a multimodal psychophysical and functional magnetic resonance imaging study. Clin J Pain. 2006;22(1):104–8. doi: 10.1097/01.ajp.0000149798.93498.7c. [DOI] [PubMed] [Google Scholar]
- 78.Baliki MN, et al. Predicting Value of Pain and Analgesia: Nucleus Accumbens Response to Noxious Stimuli Changes in the Presence of Chronic Pain. Neuron. 2010;66(1):149–160. doi: 10.1016/j.neuron.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Craig AD. PAIN MECHANISMS: Labeled Lines Versus Convergence in Central Processing. Annual Review of Neuroscience. 2003;26(1):1–30. doi: 10.1146/annurev.neuro.26.041002.131022. [DOI] [PubMed] [Google Scholar]
- 80.Tracey I, Mantyh PW. The Cerebral Signature for Pain Perception and Its Modulation. Neuron. 2007;55(3):377–391. doi: 10.1016/j.neuron.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 81.Ossipov MH, Dussor GO, Porreca F. Central modulation of pain. The Journal of Clinical Investigation. 2010;120(11):3779–3787. doi: 10.1172/JCI43766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Porreca F, Ossipov M, Gebhart G. Chronic pain and medullary descending facilitation. Trends Neurosci. 2002;25(6):319–325. doi: 10.1016/s0166-2236(02)02157-4. [DOI] [PubMed] [Google Scholar]
- 83.Behbehani MM, Fields HL. Evidence that an excitatory connection between the periaqueductal gray and nucleus raphe magnus mediates stimulation produced analgesia. Brain Research. 1979;170(1):85–93. doi: 10.1016/0006-8993(79)90942-9. [DOI] [PubMed] [Google Scholar]
- 84.Millan MJ. Descending control of pain. Progress in Neurobiology. 2002;66(6):355–474. doi: 10.1016/s0301-0082(02)00009-6. [DOI] [PubMed] [Google Scholar]
- 85.Zhuo M, Gebhart G. Biphasic modulation of spinal nociceptive transmission from the medullary raphe nuclei in the rat. J Neurophysiol. 1997;78(2):746–758. doi: 10.1152/jn.1997.78.2.746. [DOI] [PubMed] [Google Scholar]
- 86.Zhang Z, et al. Epigenetic suppression of GAD65 expression mediates persistent pain. Nat Med. 2011;17(11):1448–55. doi: 10.1038/nm.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burgess SE, et al. Time-Dependent Descending Facilitation from the Rostral Ventromedial Medulla Maintains, But Does Not Initiate, Neuropathic Pain. The Journal of Neuroscience. 2002;22(12):5129–5136. doi: 10.1523/JNEUROSCI.22-12-05129.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Buhler AV, Proudfit HK, Gebhart GF. Neurotensin-produced antinociception in the rostral ventromedial medulla is partially mediated by spinal cord norepinephrine. Pain. 2008;135(3):280–90. doi: 10.1016/j.pain.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ziegler D. Painful diabetic neuropathy: treatment and future aspects. Diabetes/Metabolism Research and Reviews. 2008;24(S1):S52–S57. doi: 10.1002/dmrr.817. [DOI] [PubMed] [Google Scholar]
- 90.Tajerian M, et al. Peripheral nerve injury is associated with chronic, reversible changes in global DNA methylation in the mouse prefrontal cortex. PLoS One. 2013;8(1):e55259. doi: 10.1371/journal.pone.0055259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nasca C, et al. L-acetylcarnitine causes rapid antidepressant effects through the epigenetic induction of mGlu2 receptors. Proc Natl Acad Sci U S A. 2013;110(12):4804–9. doi: 10.1073/pnas.1216100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bair MJ, et al. Association of Depression and Anxiety Alone and in Combination With Chronic Musculoskeletal Pain in Primary Care Patients. Psychosom Med. 2008;70(8):890–897. doi: 10.1097/PSY.0b013e318185c510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Purushothaman B, et al. Prevalence of insomnia in patients with chronic back pain. J Orthop Surg (Hong Kong) 2013;21(1):68–70. doi: 10.1177/230949901302100118. [DOI] [PubMed] [Google Scholar]
- 94.Yamashita A, et al. Astrocytic activation in the anterior cingulate cortex is critical for sleep disorder under neuropathic pain. Synapse. 2014 doi: 10.1002/syn.21733. p. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- 95.Imai S, et al. Epigenetic transcriptional activation of monocyte chemotactic protein 3 contributes to long-lasting neuropathic pain. Brain. 2013;136(3):828–843. doi: 10.1093/brain/aws330. [DOI] [PubMed] [Google Scholar]
- 96.Russo SJ, Nestler EJ. The brain reward circuitry in mood disorders. Nat Rev Neurosci. 2013;14(9):609–25. doi: 10.1038/nrn3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.LaPlant Q, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13(9):1137–43. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Navratilova E, Porreca F. Reward and motivation in pain and pain relief. Nat Neurosci. 2014;17(10):1304–1312. doi: 10.1038/nn.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chaplan SR, et al. Quantitative assesment og tactile allodynia in the rat paw. J Neurosci Methods. 1994;53:55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 100.Vadakkan KI, et al. A behavioral model of neuropathic pain-induced by ligation of the common peroneal nerve in mice. J Pain. 2005;6:747–756. doi: 10.1016/j.jpain.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 101.Toyoda H, Zhao M, Zhuo M. Enhanced quantal release of excitatory transmitter in anterior cingulate cortex of adult mice with chronic pain. Molecular Pain. 2009;5(4) doi: 10.1186/1744-8069-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barrot M. Tests and models of nociception and pain in rodents. Neuroscience. 2012;211:39–50. doi: 10.1016/j.neuroscience.2011.12.041. [DOI] [PubMed] [Google Scholar]
- 103.Hargreaves K, et al. A new and sensitive method for measuring thermal nociception in cutaneous hyperalgesia. Pain. 1988;32(1):77–88. doi: 10.1016/0304-3959(88)90026-7. [DOI] [PubMed] [Google Scholar]
- 104.Deuis JR, et al. An animal model of oxaliplatin-induced cold allodynia reveals a crucial role for Nav1.6 in peripheral pain pathways. Pain. 2013;154(9):1749–57. doi: 10.1016/j.pain.2013.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kouzarides T. Chromatin Modifications and Their Function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 106.Crepaldi L, Riccio A. Chromatin learns to behave. Epigenetics. 2009;4(1):23–26. doi: 10.4161/epi.4.1.7604. [DOI] [PubMed] [Google Scholar]
- 107.Kress M, et al. microRNAs in nociceptive circuits as predictors of future clinical applications. Front Mol Neurosci. 2013;6:33. doi: 10.3389/fnmol.2013.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lin YC, et al. Effects of trichostatin A on neuronal mu-opioid receptor gene expression. Brain Res. 2008;1246:1–10. doi: 10.1016/j.brainres.2008.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vadakkan KI, Jia YH, Zhuo M. A Behavioral Model of Neuropathic Pain Induced by Ligation of the Common Peroneal Nerve in Mice. The Journal of Pain. 2005;6(11):747–756. doi: 10.1016/j.jpain.2005.07.005. [DOI] [PubMed] [Google Scholar]