Abstract
Signaling by Toll-like receptor 4 (TLR4) is mediated by either of two adaptor proteins: myeloid differentiation marker 88 (MyD88) or Toll–interleukin-1 (IL-1) receptor (TIR) domain–containing adaptor inducing interferon-β (TRIF). Whereas MyD88-mediated signaling leads to proinflammatory responses, TRIF-mediated signaling leads to less toxic immunostimulatory responses that are beneficial in boosting vaccine responses. The hypothesis that monophosphorylated lipid A structures act as TRIF-biased agonists of TLR4 offered a potential mechanism to explain their clinical value as vaccine adjuvants, but studies of TRIF-biased agonists have been contradictory. In experiments with mouse dendritic cells, we found that irrespective of the agonist used, TLR4 functioned as a TRIF-biased signaling system through a mechanism that depended on the autocrine and paracrine effects of type I interferons. The TLR4 agonist synthetic lipid A induced expression of TRIF-dependent genes at lower concentrations than were necessary to induce the expression of genes that depend on MyD88-mediated signaling. Blockade of type I interferon signaling selectively decreased the potency of lipid A (increased the concentration required) in inducing the expression of TRIF-dependent genes, thereby eliminating adaptor bias. These data may explain how high-potency TLR4 agonists can act as clinically useful vaccine adjuvants by selectively activating TRIF-dependent signaling events required for immunostimulation, without or only weakly activating potentially harmful MyD88-dependent inflammatory responses.
INTRODUCTION
Vaccine safety and the transition from whole-pathogen vaccines to protein-subunit vaccine technologies require the development of new vaccine adjuvants to boost immunogenicity. Toll-like receptor 4 (TLR4) has become a promising target for safe immunomodulation, in part because of the success of GlaxoSmithKline’s adjuvant MPL (monophosphoryl lipid A), the first TLR agonist to be approved by the Food and Drug Administration for use in prophylactic vaccination. An ongoing challenge is the development of next-generation adjuvants that minimize inflammatory risk while maintaining or increasing their immunostimulatory effects.
TLR4 initiates the immune response to lipopolysaccharide (LPS), which is found in the cell walls of Gram-negative bacteria. TLR4 is unique among TLRs in its ability to engage both of the major adaptor molecules: myeloid differentiation marker 88 (MyD88) and Toll–interleukin-1 (IL-1) receptor (TIR) domain–containing adaptor inducing interferon-β (TRIF) (1, 2). The interaction between LPS or lipid A with MD-2, the co-receptor of TLR4, at the cell surface stimulates TLR4–MD-2 dimerization, which brings TIR domains in the cytoplasmic tails of both receptors into close proximity with one another (3–6). MyD88 adaptor-like (Mal) and MyD88 are rapidly recruited to the TIR domains, initiating the formation of a helical oligomer of IL-1 receptor–associated kinases (IRAKs) called the myddosome (7). IRAKs are phosphorylated and then released from the myddosome to interact with tumor necrosis factor (TNF) receptor–associated factor 6 (TRAF6), upon which both IRAK1 and TRAF6 are ubiquitylated (8–10). TRAF6 then recruits transforming growth factor–β (TGF-β)–associated kinase 1 (TAK1), the kinase responsible for the rapid downstream activation of mitogen-activated protein kinase (MAPK) and nuclear factor κB (NF-κB) signaling (11, 12). Because of its robust activation of NF-κB and MAPK, the MyD88-dependent pathway is associated with the expression of proinflammatory genes (1, 13, 14).
Several minutes after MyD88-dependent signaling is initiated, TLR4–MD-2 complexes are endocytosed through a CD14-dependent pathway (15). This process stimulates the recruitment of two more signaling adaptors, TRIF-related adaptor molecule (TRAM) and TRIF, to the cytoplasmic TIR domains (16–18). TRIF then recruits TRAF3 and initiates a signaling cascade through TANK-binding kinase 1 (TBK1), which results in the activating phosphorylation of the transcription factor IFN regulatory factor 3 (IRF3) (19, 20). The TRIF signaling pathway also participates in substantial crosstalk with the MyD88 pathway by activating TRAF6, NF-κB, and MAPKs (21, 22). As a result, TRIF deficiency frequently decreases the production of proinflammatory mediators associated with the MyD88 pathway (2, 23–25), a state referred to in this study as MyD88 and TRIF co-dependence.
In general, the TRIF signaling pathway is more associated with the initiation of adaptive immune responses than is the MyD88 signaling pathway. For example, TRIF deficiency in mice substantially impairs T cell priming [the induction of antigen-specific T cell proliferation by antigen-presenting cells (APCs)], whereas MyD88 deficiency has little effect on this process (26, 27). This impairment arises partly because IFN-β production absolutely requires the activation of IRF3 through TRIF (28, 29). IFN-β is essential for the adjuvant effects of several TLR agonists on T cell priming, including TLR4 (26), which may be a result of the ability of IFN-β to stimulate the increased abundance of co-stimulatory molecules, as well as of major histocompatibility complex II (MHCII) on APCs (30, 31). In addition, type I IFN promotes antigen cross-presentation (the presentation of extracellular antigens to CD8+ T cells) (32) and T cell survival (33). TRIF signaling is also linked to adaptive immunity through TRIF-dependent chemokines, such as CXCL10 (also known as IP-10), which promote the recruitment of T cells to dendritic cells (DCs) for priming (34).
Because TLR4 is the only TLR to signal through both MyD88 and TRIF, it is the only TLR with the potential for its signaling to be subject to adaptor modulation (the ability to signal more strongly through one adaptor than through the other). Given the distinct processes primarily controlled by each adaptor, we and others have studied whether defined differences in TLR4 agonist structure can cause MyD88- or TRIF-biased responses at the level of producing distinct gene profiles (27, 35–39). MPL adjuvant is a detoxified derivative of LPS, and it has about 0.1% of the inflammatory toxicity of its parent compound (40). We previously proposed that the success of MPL adjuvant was related to its ability to function as a TRIF-biased agonist of TLR4, which is favorable for vaccination because it limits proinflammatory endpoints more than it limits those pathways required for T cell priming (27). In those studies, we used a research-grade version of MPL adjuvant called MPLA in experiments with mice, and we found that MPLA retained strong induction of the expression of TRIF-dependent genes, but that it was substantially weaker than native LPS in inducing the expression of MyD88-associated genes. The “TRIF bias” hypothesis explained how MPLA could retain efficacy in priming the adaptive immune response (which is often associated with TRIF), but with less inflammatory toxicity (which is often associated with MyD88). In addition to MPLA, which is a mixture of monophosphorylated Salmonella minnesota lipid A species that differ in their numbers of acyl chains, we also studied synthetic MPLA (sMLA), a single hexa-acyl species based on the structure of Escherichia coli lipid A. We concluded that sMLA was also TRIF-biased in mouse cell systems (38, 39).
TLR4 adaptor modulation is a controversial topic. Some have questioned its existence, citing the use of heterogeneous compounds and insufficient doses as confounding factors that make it impossible to draw useful conclusions about structure-activity relationships (41). Here, we compared the potencies of sMLA (not MPLA) and its diphosphorylated counterpart synthetic lipid A (sLipid A) in inducing the expression of a panel of TRIF-dependent or MyD88-associated genes (all of which are MyD88– and TRIF–co-dependent) in mouse bone marrow–derived dendritic cells (BMDCs). None of the analytes that we have tested to date appear to be truly TRIF-independent (MyD88-sufficient) (2, 13, 23). We evaluated IP-10, IFIT-1, and CD86 as representative TRIF-dependent gene products, because TRIF alone is sufficient to induce the expression of the corresponding genes, whereas we evaluated IL-6, cyclooxygenase 2 (Cox-2), and CD80 as MyD88–and TRIF–co-dependent gene products, because optimal expression of the corresponding genes requires both adaptor pathways because of the substantial crosstalk between MyD88 and TRIF. We found that TRIF bias was not unique to sMLA as judged by pharmacological potency. Instead, expression of the TRIF-dependent genes that we tested was induced with substantially less agonist than was required for the expression of MyD88– and TRIF–co-dependent genes, irrespective of the TLR4 agonist used. Together, these data suggest that the TLR4 signaling network is itself biased toward TRIF-dependent events.
RESULTS
sMLA is not a TRIF-biased agonist as assessed by pharmacological measurements
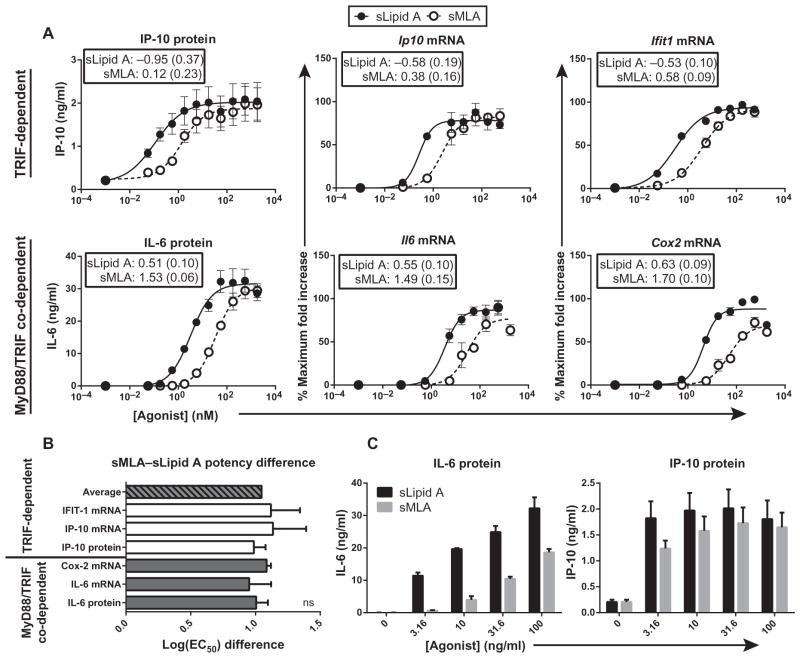
To quantitatively evaluate whether sMLA was a TRIF-biased agonist, we activated mouse BMDCs with an extensive dilution series of sMLA or sLipid A and measured the potencies of these agonists with a panel of TRIF-dependent and MyD88– and TRIF–co-dependent gene products by calculating the log of the EC50 values (the concentration of ligand that gives rise to a half-maximal response), a pharmacological method to determine the potency of a ligand. As expected, sLipid A was more potent than sMLA at inducing expression of the MyD88– and TRIF–co-dependent genes Il6 and Cox2 (Fig. 1A). However, sLipid A was also more potent than sMLA at inducing expression of the TRIF-dependent genes Ip10 and Ifit1 (Fig. 1A). We compared the log(EC50) values of sLipid A and sMLA for each analyte to determine whether adaptor bias was present despite there being obvious differences in agonist potency. We found that sLipid A was about 10-fold more potent than sMLA at inducing the expression of both co-dependent and TRIF-dependent genes, regardless of whether they were measured early (by measuring steady-state mRNA abundance at 4 hours) or late (by measuring amounts of secreted proteins at 18 hours) (Fig. 1, A and B). Therefore, sMLA was not functioning as a true TRIF-biased agonist compared to sLipid A because the lack of potency of sMLA was the same for TRIF-dependent and co-dependent genes.
Fig. 1. sMLA is not a TRIF-biased TLR4 agonist.
(A) BMDCs from C57BL/6 mice were treated with the indicated concentrations of sMLA or sLipid A. After 4 hours, increases in the steady-state abundances of the indicated mRNAs in agonist-treated cells compared to those in vehicle-treated cells were determined by qPCR analysis. After 18 hours, the amounts of the indicated proteins that were secreted into the culture medium were determined by enzyme-linked immunosorbent assay (ELISA). Fold increases in mRNA abundance were converted to percentages of the maximal response measured in each experiment. Data are means ± SEM of three independent experiments. Insets show log(EC50) values (with SE values in parentheses) derived from the goodness of the fit of the nonlinear regression calculation. (B) The log(EC50) values of sLipid A were subtracted from those of sMLA for each analyte measured in (A) within each independent experiment and then were plotted. The log(EC50) difference represents the potency difference between sMLA and sLipid A. Solid gray bars represent MyD88– and TRIF–co-dependent gene products, whereas white bars represent TRIF-dependent gene products. The striped bar represents the average log(EC50) difference between sMLA and sLipid A calculated for all analytes within each independent experiment. Data are mean log(EC50) differences ± SEM from three independent experiments. Differences between the means were not statistically significant when analyzed by one-way analysis of variance (ANOVA) (P > 0.05). (C) The amounts of IL-6 and IP-10 proteins secreted by BMDCs in response to the indicated concentrations of sLipid A and sMLA from the experiments shown in (A) are plotted to show the appearance of TRIF-bias at low concentrations.
To determine whether our experimental system (cells and agonists) had changed since our initial experiments with sMLA, we directly compared agonist concentrations similar to those explored in our previous studies of TRIF bias (38). We found that sMLA-stimulated BMDCs produced lower amounts of the MyD88– and TRIF–co-dependent cytokine IL-6 than did sLipid A–stimulated cells, whereas both agonists stimulated the production of equivalent amounts of the TRIF-dependent chemokine IP-10 (Fig. 1C), consistent with TRIF-biased signaling occurring over these agonist concentrations. Therefore, these results were not fundamentally different from those in our previous studies.
The expression of TRIF-dependent genes is activated with less agonist than is required to activate the expression of MyD88– and TRIF–co-dependent genes
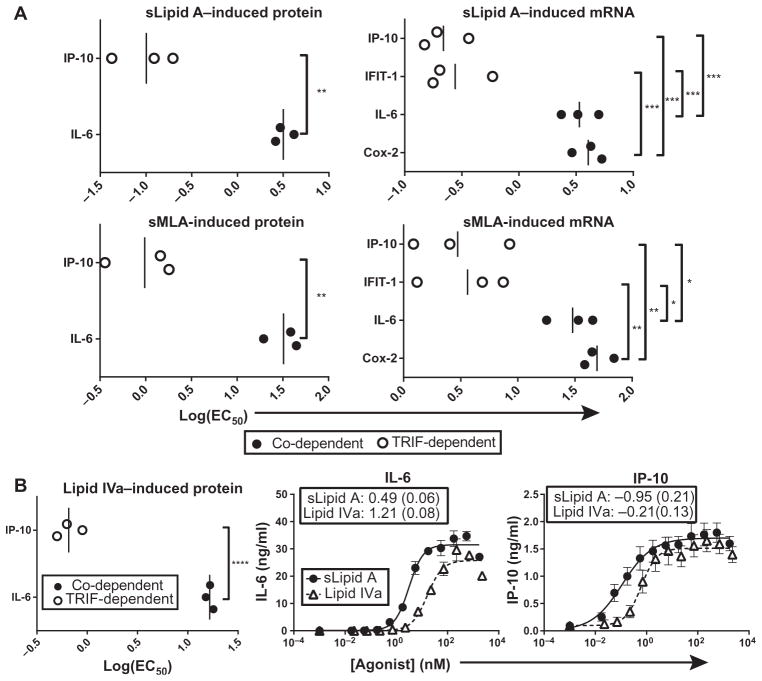
Although sMLA was a weak TLR4 agonist for both co-dependent and TRIF-dependent genes, as measured by log(EC50) values, our earlier data demonstrated that there were distinct doses at which sMLA was equivalent to sLipid A in inducing the expression of TRIF-dependent genes, but that it was weaker than sLipid A in inducing co-dependent gene expression (Fig. 1, A and C). One possible explanation for this trend is that the expression of TRIF-dependent genes is more easily activated (that is, less agonist is required) than is the expression of co-dependent genes. To test this hypothesis, we compared the log(EC50) values of TRIF-dependent and of MyD88– and TRIF–co-dependent signaling outcomes elicited by sLipid A and sMLA. As expected, the log(EC50) values of sMLA and sLipid A for the TRIF-dependent production of IP-10 protein by mouse BMDCs were less than the log(EC50) values for the co-dependent production of IL-6 protein (Fig. 2A). Similarly, the log(EC50) values for the sMLA- and sLipid A–induced, TRIF-dependent increases in the corresponding mRNA abundances were less than the log(EC50) values for the co-dependent increases in mRNA (Fig. 2A). These results suggest that in mouse BMDCs, weak TLR4 agonists, such as sMLA, can appear to be TRIF-biased because the expression of TRIF-dependent genes is induced more efficiently with less agonist than is the expression of MyD88– and TRIF–co-dependent genes.
Fig. 2. The expression of TRIF-dependent genes is activated with lower concentrations of TLR4 agonists than are required for the expression of MyD88– and TRIF–co-dependent genes.
(A) log(EC50) values of the sLipid A– and sMLA-stimulated changes in the abundances of the indicated proteins and mRNAs from the experiments shown in Fig. 1A. Individual log(EC50) values and mean values from three independent experiments (vertical bars) are shown. (B) BMDCs from C57BL/6 mice were activated with half-log dilutions of lipid IVa or sLipid A for 18 hours, after which IP-10 and IL-6 concentrations in culture medium were measured by ELISA. Left: Individual log(EC50) values from three independent experiments were compared. Nonlinear regression analysis of the means ± SEM of those experiments is shown for IL-6 (middle) and IP-10 (right). Insets show log(EC50) values with SE values given in parentheses. Statistical differences were analyzed for the data in (A) and (B) by an unpaired, two-tailed t test or by one-way ANOVA with Tukey’s post-test. *P < 0.05; **P < 0.01; ****P < 0.0001; ns, not significantly different (P > 0.05).
Both sMLA and sLipid A are hexa-acylated lipid A molecules, differing only in the presence or absence of the 1-phosphate of the diglucosamine head group and by two carbon atoms on one of the secondary acyl chains. Because the potential for a lipid A molecule to elicit the production of various cytokines is affected by both the number and the length of its acyl chains (42, 43), it is possible that the efficient activation of TRIF-dependent gene expression at low concentrations of agonist may be unique to these hexa-acylated molecules. To determine whether the relative ease of activating TRIF-dependent gene expression was common to agonists with markedly different structures, we stimulated BMDCs with the tetra-acylated molecule lipid IVa and measured the log(EC50)s for the TRIF-dependent production of IP-10 and the MyD88– and TRIF–co-dependent production of IL-6. Lipid IVa is an agonist of mouse TLR4, but is an antagonist of human TLR4 (44, 45). Similar to sMLA, lipid IVa was less potent than sLipid A in stimulating the production of both IP-10 and IL-6 (Fig. 2B). Similar to hexa-acylated sMLA and sLipid A, lipid IVa stimulated the TRIF-dependent production of IP-10 with a log(EC50) value that was substantially lower than that for the MyD88– and TRIF–co-dependent production of IL-6 (Fig. 2B). This result suggests that the robust activation of TRIF-dependent gene expression at low concentrations of agonist may be intrinsic to TLR4 and less dependent on the structure of the TLR4 agonist.
The abundance of mRNA for the TRIF-dependent costimulatory molecule CD86 is increased with less agonist than is required to increase the abundance of the co-dependent molecule CD80
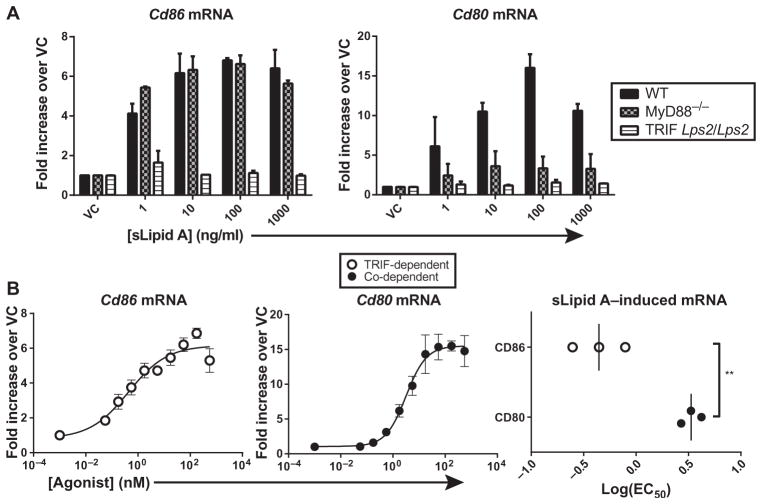
Our characterization of TLR4 agonist potencies for TRIF-dependent and MyD88– and TRIF–co-dependent gene expression was initially focused on genes encoding proinflammatory cytokines and chemokines. In addition to the release of these mediators, DC function is inextricably linked to the increased cell surface abundance of costimulatory molecules that promote adaptive immunity through the activation of T cells. We and others previously described the TLR4 adaptor requirements for the costimulatory molecules CD80 and CD86 in mouse DC subsets in vivo, with increased CD86 abundance appearing to be dependent on TRIF alone, whereas increased CD80 abundance was dependent on both MyD88 and TRIF (26, 46). To confirm the specific adaptors required for CD86 and CD80 in BMDCs, we generated BMDCs from wild-type mice, TRIFLps2/Lps2 mice (which express a nonfunctional TRIF protein), or MyD88 knockout mice, treated them with sLipid A, and measured increases in Cd80 and Cd86 mRNA abundance by quantitative polymerase chain reaction (qPCR) analysis. The ability of sLipid A to induce increases in Cd86 mRNA abundance was reduced in cells in which TRIF signaling was absent, whereas its ability to induce increases in Cd80 mRNA abundance was reduced by the absence of either TRIF or MyD88 signaling (Fig. 3A). Together, these results confirm that in BMDCs, the expression of Cd86 is TRIF-dependent, whereas the expression of Cd80 is MyD88–and TRIF–co-dependent.
Fig. 3. The TRIF-dependent expression of Cd86 is activated with lower TLR4 agonist concentrations than are required for the MyD88– and TRIF–co-dependent expression of Cd80.
(A) BMDCs from wild-type (WT), MyD88−/−, or TRIFLps2/Lps2 mice were treated with vehicle control (VC) or were stimulated with the indicated concentrations of sLipid A for 4 hours. The steady-state abundances of Cd80 and Cd86 mRNAs were then analyzed by qPCR. Data are the mean fold increases in mRNA abundance in sLipid A–stimulated cells compared to those in vehicle-treated cells and are combined from two independent experiments. (B) Log(EC50) values were determined for the sLipid A–stimulated increases in the abundances of Cd80 and Cd86 mRNAs after 4 hours by nonlinear regression analysis. Data are means ± SEM from three independent experiments. Right: log(EC50) values from individual experiments. Statistical differences were determined with an unpaired, two-tailed t test. **P < 0.01.
Because increases in Cd86 mRNA abundance are strictly TRIF-dependent, whereas increases in Cd80 mRNA abundance are MyD88–and TRIF–co-dependent, we hypothesized that sLipid A would be more effective at increasing the expression of Cd86. When BMDCs were treated with increasing concentrations of sLipid A, the log(EC50) value for increased Cd86 mRNA abundance was substantially lower than that for Cd80 mRNA (Fig. 3B), suggesting that the TRIF-dependent expression of Cd86 was more easily induced than was the MyD88– and TRIF–co-dependent expression of Cd80. Therefore, the enhanced potency of TLR4 agonists for TRIF-dependent gene expression may pertain to various gene subsets whose products have different roles in the immune response.
The low log(EC50) values of TLR4 agonists for the expression of TRIF-dependent genes are not explained by TRIF adaptor selectivity
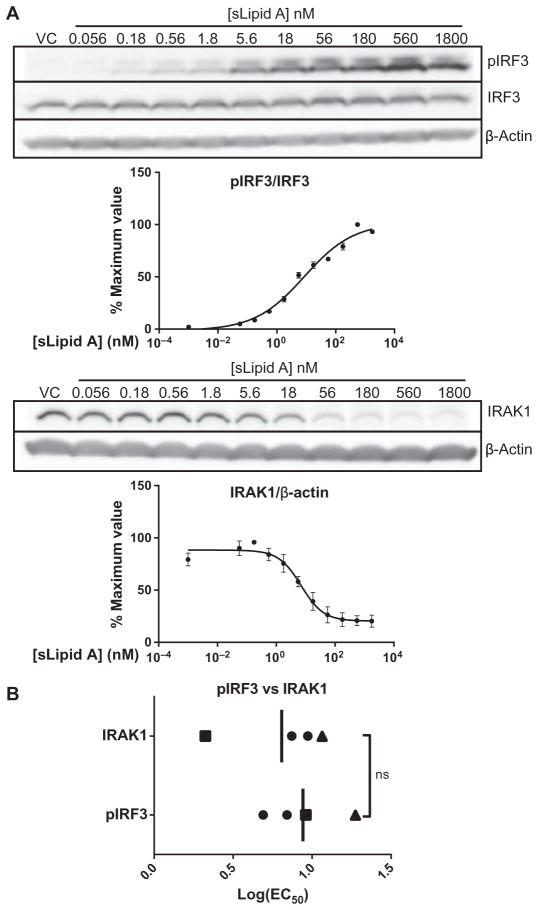
One potential explanation for the low log(EC50) values of the TLR4 agonists sLipid A and sMLA for the expression of TRIF-dependent genes is that comparatively more recruitment of TRIF than of MyD88 to the receptor is stimulated immediately after TLR4 dimerization at limiting concentrations of agonist. To test this hypothesis, we measured the potency of sLipid A in activating IRF3 and IRAK1 (Fig. 4A), two signaling proteins that mediate the TRIF and MyD88 pathways, respectively (7, 18). The log(EC50) values for the activating phosphorylation of IRF3 and the activation-induced disappearance of IRAK1 protein from the viewed region of the Western blot (Fig. 4A), which is likely a result of an increase in its molecular mass because of Lys63-based polyubiquitination upon signaling (8, 47), were not substantially different when measured in four independent experiments (Fig. 4B). In two paired experiments, the log(EC50) values for IRF3 phosphorylation were greater than those for IRAK1 loss (Fig. 4B). These data suggest that initial TRIF-dependent signaling is not stimulated more efficiently than is initial MyD88-dependent signaling at suboptimal concentrations of TLR4 agonist; indeed, MyD88-dependent signaling may be stimulated more easily than is TRIF-dependent signaling. Therefore, the low log(EC50) values of TLR4 agonists for the production of TRIF-dependent gene products cannot be explained by dose-dependent TRIF selectivity at the level of initial adaptor usage.
Fig. 4. Low log(EC50) values for TRIF-dependent genes are not explained by biased adaptor use.
(A) BMDCs from WT mice were treated with vehicle (VC) or were stimulated with a half-log dilution series of sLipid A for 1 hour. Samples were then analyzed by Western blotting with antibodies specific for the indicated proteins. The abundances of IRAK1 and pIRF3 (Ser396) were quantified by densitometric analysis and normalized to those of β-actin and total IRF3, respectively. Representative blots are shown. Normalized values from densitometric analysis were converted to percentage maximal responses for each experiment. Data in graphs are means ± SEM from four independent experiments. (B) Individual log(EC50) values (squares and triangles indicate matched experiments) and means from the experiments shown in (A) were plotted and compared by an unpaired, two-tailed t test. ns, not statistically different (P > 0.05).
The log(EC50) values of a TLR4 agonist for the activation of MAPKs and NF-κB are different
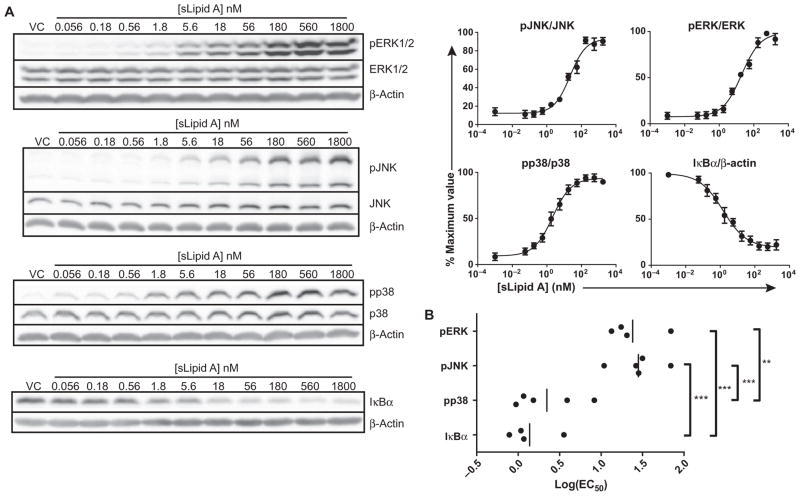
Because a simple TRIF bias of TLR4 did not seem to explain the low log(EC50) values of TLR4 agonists for TRIF-dependent events, we investigated signaling mediators that were activated through both the TRIF and MyD88 pathways. TLR4-induced gene expression is dependent on the transcription factor NF-κB and transcription factors activated by MAPK signaling pathways, and considerable overlap exists between the MyD88 and TRIF signaling pathways in their activation (1, 14, 21). TLR4-induced genes have varying requirements for MAPK and NF-κB signaling for their optimal expression, but the strength of TLR4 signal required to efficiently activate these mediators is unknown. We determined the log(EC50) values for the activation of p38, c-Jun N-terminal kinase (JNK), extracellular signal–regulated kinase 1/2 (ERK1/2), and inhibitor of κB α (IκBα) by treating BMDCs with a range of concentrations of sLipid A and quantifying relative protein abundances by Western blotting analysis (Fig. 5A). Substantially less sLipid A was needed to stimulate the half-maximal degradation of IκBα (an indicator of NF-κB activation) and the phosphorylation of p38 [low log(EC50) values] than was required to stimulate the half-maximal phosphorylation and activation of JNK and ERK1/2 [high log(EC50) values] (Fig. 5B). We were not surprised that NF-κB activation required very little agonist given its prominence as a readily stimulated component of proinflammatory gene regulation; however, the low degree to which the MAPKs JNK and ERK1/2 were activated was striking. Thus, although MAPKs are influenced by both TRIF- and MyD88-dependent pathways, there are considerable differences in the ease with which they are activated by either pathway.
Fig. 5. The log(EC50)s of sLipid A for MAPKs and NF-κB are distinct.
(A) BMDCs from WT mice were treated with vehicle as a control (VC) or were stimulated with a half-log dilution series of sLipid A for 15 min. Samples were then subjected to Western blotting analysis to determine the relative extents of phosphorylation of JNK (Thr183/Tyr185), p38 (Thr180/Tyr182), and ERK1/2 (Thr202/Tyr204), as well as of the degradation of IκBα. Representative Western blots are shown. Densitometric analysis was performed to determine the relative abundances of the indicated phosphorylated proteins normalized to the abundances of their respective total proteins, whereas IκBα abundance was normalized to that of β-actin. Normalized values were converted to percentage maximal responses for each experiment. Data are means ± SEM from the nonlinear regression analysis of at least four independent experiments. (B) Log(EC50) values from the independent experiments shown in (A). Statistical differences were analyzed by one-way ANOVA with Tukey’s post-test. **P < 0.01; ***P < 0.001.
The high log(EC50) values of sLipid A for MyD88– and TRIF–co-dependent events correlate with a dependence on JNK
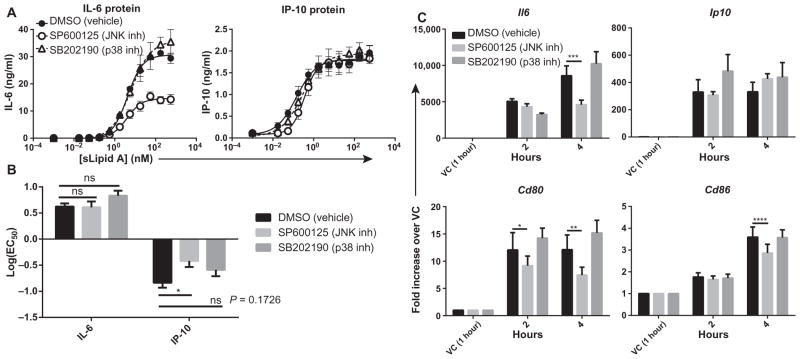
We and others published previously that MAPKs differ in their involvement in transcription factor activation and gene expression downstream of TLR4 (38, 48–52). Although substantial overlap and redundancy in the requirements of MAPKs for gene expression are known, specific TLR4-induced genes may have a greater requirement for certain MAPKs. It is unclear whether specific MAPKs are preferentially required for the expression of TRIF-dependent genes as opposed to MyD88– and TRIF–co-dependent genes. We hypothesized that the high log(EC50) values of agonists for the expression of co-dependent genes might be, in part, a result of their strong dependence on the activity of JNK, which is also associated with high log(EC50) values. Conversely, the low log(EC50) values of agonists for TRIF-dependent genes may be because their expression depends on p38, which has a low log(EC50) value.
To test this hypothesis, we pretreated BMDCs with specific chemical inhibitors of JNK and p38 before measuring the effects of a range of concentrations of sLipid A on TRIF-dependent and co-dependent gene expression. Maximal production of the MyD88-dependent cytokine IL-6 was reduced by the JNK inhibitor, but not the p38 inhibitor (Fig. 6A); however, neither inhibitor substantially changed the log(EC50) of sLipid A in stimulating IL-6 production (Fig. 6B). The amount of the TRIF-dependent IP-10 protein produced was unchanged by the JNK and p38 inhibitors (Fig. 6A). Both inhibitors slightly reduced the amount of IP-10 produced at concentrations of sLipid A less than 1 nM (Fig. 6A). The JNK inhibitor caused a statistically significant increase in the log(EC50) value of sLipid A for IP-10 production, whereas the p38 inhibitor caused an increase in the log(EC50) value of sLipid A for IP-10 that approached statistical significance (Fig. 6B). Neither inhibitor increased the log(EC50) value for IP-10 production to match that for IL-6 production. These data suggest that a varying dependence on specific MAPKs is unlikely to be the cause of the differences in the log(EC50) values between co-dependent and TRIF-dependent events.
Fig. 6. MyD88– and TRIF–co-dependent genes with high log(EC50) values have a greater dependence on JNK for their expression than do TRIF-dependent genes.
(A) BMDCs from WT mice were pretreated with the JNK inhibitor SP600125, the p38 inhibitor SB202190, or dimethyl sulfoxide (DMSO) (vehicle) for 30 min before being stimulated with a half-log dilution series of sLipid A for 18 hours. The concentrations of secreted IL-6 and IP-10 proteins in the culture medium were then measured by ELISA. Data are means ± SEM from the nonlinear regression analysis of at least three independent experiments. (B) Log(EC50) values were calculated from the dose-response curves of the independent experiments shown in (A) and then were compared by two-way ANOVA with Sidak’s post-test. *P < 0.05; ns, not statistically significant. (C) BMDCs from WT mice were pretreated with DMSO or the indicated inhibitors for 30 min before being treated with vehicle (VC) or sLipid A (100 ng/ml) for the indicated times. Increases in the steady-state abundances of the indicated mRNAs in sLipid A–treated cells compared to those in vehicle-treated cells were determined by qPCR analysis. Data are means ± SEM from four independent experiments. Statistical differences between agonist-treated samples and DMSO-treated samples were analyzed with repeated-measures two-way ANOVA with Dunnett’s post-test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Because the efficacy of sLipid A for the production of IL-6 (co-dependent), but not IP-10 (TRIF-dependent), was decreased by JNK inhibition, we explored the role of JNK signaling in the expression of other TRIF-dependent or co-dependent genes. JNK inhibition reduced the sLipid A–stimulated increases in Il6 and Cd80 mRNA abundance (MyD88-dependent) at 4 hours after stimulation (Fig. 6C), but had no effect on increases in Ip10 mRNA abundance (TRIF-dependent). Inhibition of JNK reduced the sLipid A–stimulated increase in Cd86 mRNA abundance (TRIF-dependent) at 4 hours after stimulation, but less effectively than it inhibited the increased abundance in Cd80 mRNA (Fig. 6C). Unexpectedly, inhibition of p38 had no substantial effect on the expression of any gene (Fig. 6C), despite confirmation that SB202190 was effectively taken up by the cells (fig. S1). Therefore, the expression of MyD88– and TRIF–co-dependent genes, such as Il6 and Cd80, correlates with a dependence on the JNK signaling pathway for full induction.
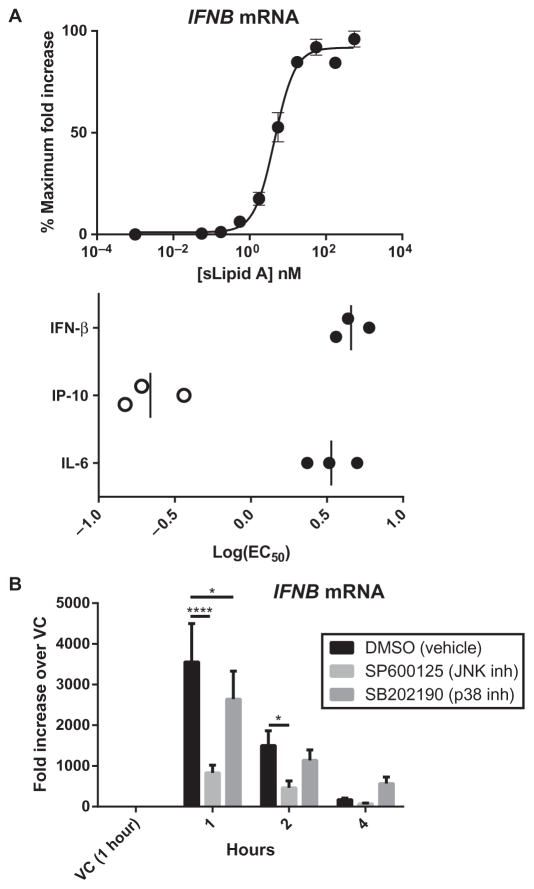
The high log(EC50) value of a TLR4 agonist for IFNB expression correlates with a dependence on JNK
The production of IFN-β in response to TLR4 stimulation absolutely requires TRIF signaling and is downstream of the activation of IRF3 (18, 28, 53). In addition, optimal IFN-β production requires intact MyD88 signaling (46), because the activity of the IFNB promoter requires the activation of MyD88-associated transcription factors, such as AP-1, which is activated downstream of JNK, and NF-κB (54, 55). Because JNK dependence was correlated with MyD88– and TRIF–co-dependent genes with high log(EC50) values, we wondered whether the log(EC50) value for the induction of IFNB expression was also high. We measured IFNB mRNA abundance in BMDCs activated with a range of concentrations of sLipid A and calculated the log(EC50) values at 1 hour, the time at which IFNB mRNA was maximally abundant. We found that the log(EC50) value of sLipid A for IFNB expression was substantially higher than that for the expression of the TRIF-dependent Ip10, and was as high as that for the expression of the co-dependent gene Il6 (Fig. 7A). To confirm that sLipid A–induced IFNB expression was JNK-dependent, we pretreated BMDCs with a JNK inhibitor and measured IFNB mRNA abundance over time. Consistent with previous reports (56, 57), IFNB mRNA abundance was markedly decreased in the absence of JNK signaling (Fig. 7B). In contrast, inhibition of p38 signaling had relatively little effect on IFNB expression; IFNB mRNA abundance was only slightly decreased at 1 hour compared to that in vehicle-treated cells (Fig. 7B). These data suggest that the TLR4-dependent expression of IFNB is characterized by a high log(EC50) value, consistent with it being a co-dependent event.
Fig. 7. The high log(EC50) values for the expression of IFNB correlate with a dependence on JNK signaling.
(A) BMDCs from WT mice were stimulated with a half-log dose series of sLipid A for 1 hour. The steady-state abundance of IFNB mRNA in each of the samples was determined by qPCR analysis, and fold increases in mRNA abundance in agonist-treated cells compared to that in vehicle-treated cells were calculated and then converted to percentage maximal responses for each experiment. Top: Data are means ± SEM of three independent experiments that were subjected to nonlinear regression analysis. Bottom: One-hour IFNB log(EC50)s are compared to those of Il6 and Ip10 (reproduced from Fig. 2A), both measured at 4 hours (bottom). (B) BMDCs from WT mice were pretreated with the indicated inhibitors or DMSO for 30 min before being treated with vehicle (VC) for 1 hour or with sLipid A (100 ng/ml) for the indicated times. The abundance of IFNB mRNA in each sample was determined by qPCR analysis. Data are means ± SEM in the fold increase in IFNB mRNA in agonist-treated cells compared to that in vehicle-treated cells and are from three independent experiments. Statistical differences between inhibitor- and DMSO-treated samples were determined by repeated-measures two-way ANOVA with Dunnett’s post-test. *P < 0.05; ****P < 0.0001.
Autocrine and paracrine signaling by IFN-β determines the low log(EC50) values of TLR4 agonists for TRIF-dependent responses
Although we observed a correlation between JNK dependence and MyD88 and TRIF co-dependence, differential MAPK dependence could not completely explain either the high log(EC50) values of TLR4 agonists for the expression of co-dependent genes or the low log(EC50) values for the expression of TRIF-dependent genes (Fig. 6). Alternatively, the lower log(EC50) values of agonists for TRIF-dependent gene expression could be caused by crosstalk or synergy between TLR4 and other receptors. In addition to inputs directly from TLR4, signaling through the shared receptor for IFN-α and IFN-β (IFNAR) by secreted type I IFN enhances the expression of TRIF-dependent genes through the activation of the IFN-stimulated gene factor 3 (ISGF3) complex, which binds to and activates IFN-stimulated response elements (ISREs) in gene promoters (58, 59).
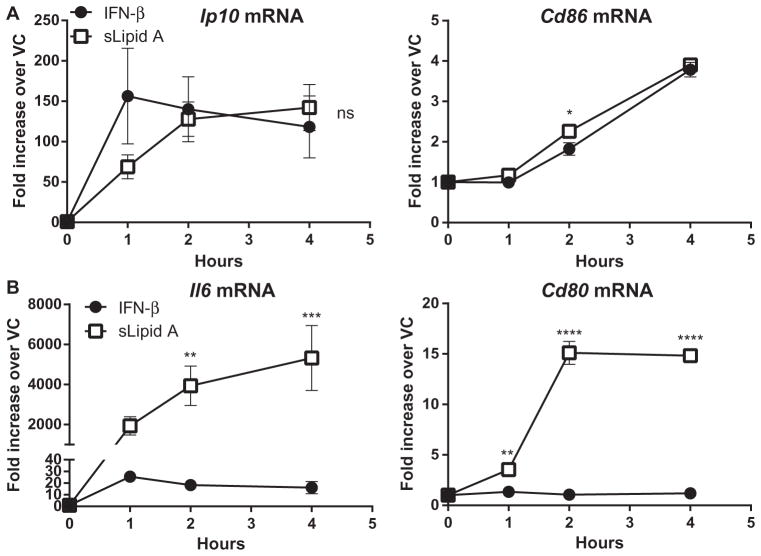
We found that sLipid A induced robust IFNB expression in BMDCs (Fig. 7). Therefore, we hypothesized that the relatively low log(EC50) values of sLipid A for TRIF-dependent gene expression were caused by secondary IFNAR signaling. To determine the extent to which IFN-β alone could induce the expression of TLR4-regulated genes, we incubated BMDCs with recombinant IFN-β or sLipid A and measured increases in the steady-state abundances of the mRNAs of the TRIF-dependent genes Ip10 and Cd86 and compared them to changes in the abundances of the mRNA of the co-dependent genes Il6 and Cd80. As expected, the abundances of Cd86 and Ip10 mRNAs were increased by IFN-β (Fig. 8A). Indeed, the increases in steady-state mRNA abundance by IFN-β were comparable to those caused by sLipid A, suggesting that IFN-β is sufficient to induce the expression of Cd86 and Ip10 (Fig. 8A). In contrast, in comparison to sLipid A, IFN-β caused no measurable increases in Cd80 mRNA abundance, and relatively small increases in Il6 mRNA abundance in stimulated BMDCs (Fig. 8B). Although both Cd80 and Il6 are type I IFN–responsive genes in certain contexts (58, 60–62), IFN-β was insufficient to induce their expression in BMDCs.
Fig. 8. IFN-β is sufficient to induce the maximal expression of genes with low log(EC50) values.
(A and B) BMDCs from WT mice were activated with either IFN-β (1000 U/ml) or sLipid A (100 ng/ml) for the indicated times. Samples were then analyzed by qPCR, and the relative fold increases in the abundances of the mRNAs of (A) TRIF-dependent and (B) MyD88– and TRIF–co-dependent genes in stimulated cells compared to those in DMSO-treated cells were determined. Data are means ± SEM from at least three independent experiments. Statistical differences between sLipid A– and IFN-β–stimulated mRNA abundances at each time point were determined by two-way ANOVA with Sidak’s post-test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not statistically significant (P > 0.05).
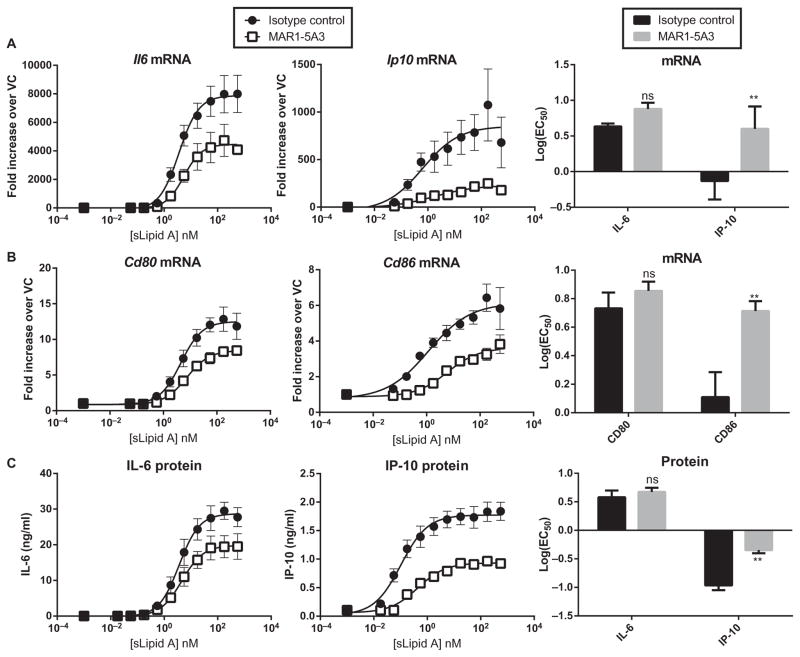
To determine whether autocrine or paracrine signaling by IFN-β was responsible for the low log(EC50) values for the sLipid A–stimulated production of IP-10 and CD86, we blocked IFNAR signaling by pretreating the BMDCs with the IFNAR1 antagonistic antibody MAR1-5A3, or, as a negative control, with isotype control antibody, and then, we measured the log(EC50) values of sLipid A. The maximal extents of the expression of both Ip10 (TRIF-dependent) and Il6 (co-dependent) were decreased in MAR1-5A3–treated cells compared to those in control cells (Fig. 9A), consistent with both of these genes being inducible by type I IFN (Fig. 8, A and B). Despite dampening of global Il6 and Ip10 expression, the log(EC50) value of sLipid A for Ip10 was substantially increased, whereas the log(EC50) value for Il6 expression was unchanged (Fig. 9A). Similarly, sLipid A–induced increases in Cd80 and Cd86 mRNA abundances were dampened by MAR1-5A3, but only the log(EC50) value of sLipid A for the TRIF-dependent Cd86 was substantially increased (Fig. 9B). Further-more, the log(EC50) values of sLipid A for the expression of both TRIF-dependent genes Ip10 and Cd86 were increased to values similar to those for the expression of the co-dependent genes Il6 and Cd80, suggesting that secondary signaling by IFN-β through IFNAR enhances the potency of TLR4 agonists, such as sLipid A, for TRIF-dependent gene expression (Fig. 9, A and B). We also tested whether secondary type I IFN signaling was responsible for the low log(EC50) values of sLipid A for IP-10 protein production. MAR1-5A3 increased the log(EC50) value for the sLipid A– stimulated production of IP-10, but not IL-6, compared to that in stimulated control cells (Fig. 9C). Together, these results suggest that autocrine or paracrine signaling through IFNAR is a determinant of the low log(EC50) values of TLR4 agonists for TRIF-dependent gene expression.
Fig. 9. Autocrine and paracrine signaling by IFN-β contributes to the low log(EC50) values of TRIF-dependent genes.
(A to C) BMDCs from WT mice were pretreated with MAR1-5A3 (an anti-IFNAR1 antibody) or isotype control antibody for 2 hours before being stimulated with a half-log dose series of sLipid A for 4 or 18 hours. (A and B) After 4 hours, samples were analyzed by qPCR to measure the steady-state abundances of the indicated mRNAs in stimulated cells relative to those of vehicle-treated cells. (C) After 18 hours, the concentrations of the indicated proteins in the culture medium were determined by ELISA. Data are means ± SEM from the nonlinear regression analysis of at least three independent experiments. Right: The log(EC50) values derived from each experiment were averaged and are compared in bar graphs. Statistical differences between the log(EC50) values of isotype- and MAR1-5A3–treated cells were determined by repeated-measures two-way ANOVA with Sidak’s post-test. **P < 0.01; ns, not statistically significant (P > 0.05).
DISCUSSION
Several studies are consistent with the idea that modulation of the bifurcated TLR4 signaling pathway can be used to dissociate beneficial immune responses from harmful side effects (26, 27, 39, 63). On one hand, MyD88 signaling is associated with rapid proinflammatory cytokine production and innate immune responses to quickly contain infectious threats (1, 13). Innate immunity is of obvious importance in natural microbial infections, but it is potentially counterproductive in the context of sterile immunization with subunit vaccines. On the other hand, TRIF signaling is more associated with processes that can promote adaptive immune responses essential to effective vaccination. For example, optimal increases in the cell surface abundance of T cell co-stimulators in response to TLR4 agonist require intact TRIF signaling (26, 31). Many chemokines are produced in a TRIF-dependent manner, including IP-10 and CCL5 (RANTES), which play important roles in regulating T cell and DC interactions (34), as well as T cell differentiation (64). Here, we conclude that TLR4 signaling is TRIF-biased in mice because the expression of TRIF-dependent, IFN-β–sufficient genes reached a half-maximal response at lower concentrations of TLR4 agonist than was required for the expression of MyD88–and TRIF–co-dependent genes.
We and others proposed that the modulation of TLR4 adaptor molecules is a function of the structural characteristics of an agonist. Zughaier et al. (36) reported that LPS molecules from E. coli and Vibrio cholerae are MyD88-biased, whereas LPS molecules from Salmonella species are TRIF-biased over a range of concentrations when compared to Neisseria meningitidis lipooligosaccharide as a control. Later, in an effort to understand how MPLA maintains immunostimulatory activity in spite of losing about 99.9% of the inflammatory activity of the LPS from which it is derived (27), we proposed that MPLA acts as a TRIF-biased agonist of mouse TLR4. In our subsequent structural studies with sMLA and sLipid A, we concluded that removal of the 1-phosphate from lipid A was likely responsible for TRIF-biased signaling (38). Bowen et al. (35) also reported TRIF-biased signaling when they demonstrated that the lipid A mimetic CRX-547 was markedly less efficacious than a stereoisomeric compound, CRX-527, in eliciting NF-κB activation and the MyD88– and TRIF–co-dependent production of TNF-α, but that the two compounds were comparable in stimulating production of the TRIF-dependent chemokines IP-10 and RANTES.
Despite this evidence for TRIF-biased signaling by TLR4, other data called its existence into question. Earlier dose-response experiments with mouse peritoneal macrophages demonstrated that MPL adjuvant was much less potent than LPS in stimulating production of both the TRIF-dependent chemokine IP-10 and the MyD88– and TRIF–co-dependent cytokines TNF-α and IL-1β (65). Gaekwad et al. (41) tested preparations of various lipid A structures that were proposed to modulate TLR adaptor signaling. In experiments with mouse macrophage cell lines, the authors found no evidence for the modulation of adaptor usage. Instead, cytokine and transcript EC50 data conformed to an additive model in which changes in agonist structure equally affect the extent of expression of all of the genes tested regardless of their adaptor association. In addition, TLR4 mutagenesis studies were inconsistent with a clear structural basis for TRIF-biased signaling by monophosphorylated agonists (66).
Our results support the conclusion that sMLA is not a TRIF-biased agonist of TLR4 relative to sLipid A, because sMLA was about 10% as potent as sLipid A in stimulating expression of the TRIF-dependent and MyD88– and TRIF–co-dependent genes that we tested (Fig. 1). However, by comparing log(EC50) values, we found that the expression of TRIF-dependent genes was activated with substantially less TLR4 agonist than was required for the expression of MyD88– and TRIF–co-dependent genes (Figs. 2 and 3). This characteristic seemed to be independent of TLR4 agonist structure and potency, because sMLA and lipid IVa, two weaker agonists that differ in phosphorylation status and acyl chain number, also induced the expression of TRIF-dependent genes more effectively than they induced the expression of co-dependent genes. We demonstrated that the log(EC50) value of sLipid A for the increase in abundance of mRNA of the TRIF-dependent T cell costimulatory molecule Cd86 was also low compared to the log(EC50)s for co-dependent genes such as Il6, Cox2 (Fig. 2), and Cd80 (Fig. 3), supporting the idea that enhancement of T cell activation can proceed without the need to produce large amounts of MyD88-associated proinflammatory cytokines, as was seen in our first in vivo adjuvant tests with MPLA (27). Because all TRIF-dependent outcomes were characterized by low log(EC50) values compared to those of MyD88– and TRIF–co-dependent outcomes, we propose that TLR4 itself, and not sMLA, can be considered to be TRIF-biased.
We were able to rule out several possible mechanisms for the TRIF-biased nature of TLR4. It is unlikely that engagement of TRIF occurs at lower concentrations of TLR4 agonist than are required to engage MyD88, because there was no statistically significant difference in the log(EC50) values of sLipid A for the TRIF-dependent activating phosphorylation of IRF3 and the MyD88-dependent loss of IRAK1 (Fig. 4). Indeed, data from two paired experiments suggest that TRIF signaling was initially more difficult to activate, which is not surprising given that MyD88 signaling occurs at the plasma membrane, whereas TRIF signaling requires the comparatively more complicated CD14-dependent endocytosis of TLR4 (15). In addition, differential requirements for JNK are unlikely to explain TRIF-biased signaling, because inhibition of JNK had little effect on the log(EC50) values of sLipid A in its stimulation of the expression of either TRIF-dependent or MyD88– and TRIF–co-dependent genes (Fig. 6).
The expression of many TRIF-dependent genes is also induced by type I IFN, and their activation in response to TLR4 stimulation decreases markedly in the absence of IFN-β within a 3-hour timeframe (58), indicating that autocrine and paracrine signaling by type I IFN may be involved in the TRIF-biased signaling of TLR4. When we blocked type I IFN signaling, we found that the log(EC50) values for the expression of TRIF-dependent genes increased to values similar to those of co-dependent genes, suggesting that the TRIF-biased nature of TLR4 depends on IFNAR. In contrast, blocking IFNAR did not change the log(EC50) values for the MyD88–and TRIF–co-dependent production of IL-6 and increase in Cd80 mRNA abundance; however, IFNAR inhibition led to a decrease in their maximum abundance (Fig. 9), suggesting that IFNAR signaling enhances the expression of their respective genes. TRIF-dependent, IFN-β-sufficient genes appear to be characterized by low log(EC50) values because they are regulated primarily by transcription factors that are activated by autocrine and paracrine IFNAR signaling stimulated by type I IFN released in response to TLR4 activation (58).
Although the gene encoding IFN-β is considered to be a canonical TRIF-dependent gene, it is perhaps more accurately characterized as an MyD88– and TRIF–co-dependent gene because its expression is reduced in the absence of MyD88 (46). Similar to those of other co-dependent transcripts, the log(EC50) values for IFNB mRNA expression were high compared to those of TRIF-dependent genes. Moreover, the expression of IFNB is JNK-dependent, consistent with that of other co-dependent genes (Fig. 7). It is unclear how TRIF-dependent responses characterized by low log(EC50) values could be caused by IFN-β, a gene product characterized by high log(EC50) values. One possibility is that IFN-β is a potent inducer of the expression of TRIF-dependent genes. We have also not ruled out contributions from IFN-α, which may be produced in response to TLR4 stimulation in certain contexts (67). From a teleological perspective, the high log(EC50) value for IFN-β production may be beneficial to the host during infection, because type I IFNs can contribute to endotoxic shock and the production of proinflammatory IL-1β through the activation of caspase-11 (68, 69). These observations suggest that IFN-β may have biphasic functionality, supporting adaptive immune priming at low concentrations and becoming more proinflammatory at higher concentrations.
In conclusion, our results provide further understanding of the TRIF-biased nature of TLR4 signaling. In our culture system, monophosphorylation of lipid A did not cause TRIF-biased TLR4 signaling; rather, TLR4 itself signaled in a TRIF-biased manner through which the expression of TRIF-dependent, IFN-β–sufficient genes whose products are associated with T cell priming was induced with less agonist than was required for the expression of MyD88– and TRIF–co-dependent genes whose products are associated with proinflammatory outcomes. The extent to which TRIF bias can occur will likely depend on the ability of a cell to respond to type I IFN and to the characteristics of the regulation of type I IFN–inducible genes within that cell. These results are relevant to ongoing translational efforts because sMLA is the active component of GLA (glucopyranosyl lipid adjuvant) formulations that are being tested clinically (70). Future studies are needed to determine whether cell types other than mouse BMDCs, including human cells, are capable of intrinsic TRIF-biased TLR4 signaling.
MATERIALS AND METHODS
Mice and reagents
C57BL/6 and TRIFLps2/Lps2 mice were purchased from the Jackson Laboratory. MyD88−/− mice were a gift of S. Akira (through R. Kedl, University of Colorado School of Medicine). Mice were housed in a specific pathogen–free barrier facility at the University of Louisville and cared for according to regulations set forth by its Institutional Animal Care and Use Committee. The compounds sMLA (MW 1763.5, sold as PHAD, cat. no. 699800, Avanti Polar Lipids), sLipid A [MW 1798.4, sold as lipid A (E. coli), cat. no. CLP-24005-s, Peptides International], and synthetic lipid IVa (MW 1405.7, cat. no. CLP-24006-s, Peptides International) were dissolved by vortexing in 100% DMSO (Sigma-Aldrich) at 1 mg/ml and then promptly frozen in single-use aliquots at −80°C. Agonists were serially diluted in culture medium before being added to cell cultures. The DMSO concentration in cell culture was ≤0.32%. A vehicle control corresponding to the highest DMSO concentration in each experiment (0.1 or 0.32%) was always used. Primary antibodies for the Western blotting analysis of the following targets were purchased from Cell Signaling Technology: pIRF3 (Ser396, cat. no. 4947), pJNK (Thr183/Tyr185, cat. no. 4668), pp38 (Thr180/Tyr182, cat. no. 9215), pERK1/2 (Thr202/Tyr204, cat. no. 4370), pcJun (Ser63, cat. no. 9261), pMAPKAPK2 (Thr222, cat. no. 3316), total p38 (cat. no. 9212), total ERK1/2 (cat. no. 4695), total c-Jun (cat. no. 9165), total MAPKAPK2 (cat. no. 3042), and IκBα (cat. no. 4814). Primary antibodies for the Western blotting analysis of the following targets were purchased from Santa Cruz Biotechnology: IRAK1 (cat. no. sc-5288), total IRF3 (cat. no. sc-9082), β-actin (cat. no. sc-1616), and total JNK (cat. no. sc-137018). All horseradish peroxidase (HRP)–conjugated secondary antibodies were purchased from Jackson ImmunoResearch. The JNK inhibitor SP600125 (Sigma-Aldrich) and the p38 MAPK inhibitor SB202190 (Calbiochem) were dissolved in 100% DMSO at concentrations of 25 and 3 mM, respectively, and were frozen at −20°C until needed for use. The anti-mouse IFNAR1 antibody MAR1-5A3 and the functional grade mouse IgG1 isotype control were purchased from Leinco Technologies. Recombinant mouse IFN-β protein was purchased from the PBL Interferon Source.
Generation and culture of BMDCs
BMDCs were prepared with a procedure modified from that of Lutz et al. (71). Briefly, bone marrow plugs were flushed from the femurs and tibiae of mice with sterile Hanks’ balanced salt solution (HBSS) and resuspended in BMDC medium containing R10F [RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum, 2 mM L-glutamine, 1 mM sodium pyruvate, penicillin (50 U/ml), streptomycin (50 μg/ml), 50 μM β-mercaptoethanol, and GM-CSF (granulocyte-macrophage colony-stimulating factor) (5 ng/ml; Miltenyi Biotec or R&D Systems)]. Bone marrow cells (2 × 106, excluding red blood cells) were seeded in 100-mm bacteriological petri dishes in 10 ml of BMDC medium and incubated at 37°C. On days 3 and 8, 10 ml of BMDC medium was added to the cultures. On day 6, 10 ml of BMDC medium was removed and replaced with fresh medium. On day 10, non-adherent BMDCs were collected. BMDCs were typically >85% CD11c+ CD11b+ MHCII+ CD86low CD14+ CD4− CD8− B220− CD19− GR1− as determined by flow cytometric analysis.
Cytokine measurement
BMDCs (1 × 105 per well) suspended in R10F medium were incubated in flat-bottom, 96-well plates for 2 hours at 37°C before TLR4 agonists or DMSO (vehicle control) diluted in R10F was added. In the MAPK inhibition experiments, 10 μM SP600125, 10 μM SB202190, or DMSO (vehicle control) was added 30 min before addition of the TLR4 agonists. In the IFNAR1-blocking experiments, MAR1-5A3 (10 μg/ml) or isotype control antibody was added immediately before plating the BMDCs for a 2-hour preincubation. After 18 hours of stimulation with TLR4 agonists at 37°C, supernatants were collected, and IL-6 (BD Biosciences) and IP-10 (R&D Systems) concentrations were measured by ELISA, according to each manufacturer’s specifications.
Quantitative real-time PCR
BMDCs (5 × 105) suspended in R10F medium were incubated in 5-ml polystyrene round-bottom tubes at 37°C for 2 hours before TLR4 agonists or IFN-β was added. MAPK inhibitors and the IFNAR1 blocking antibody were used in the manner described earlier. After they were stimulated, the cells were washed with ice-cold HBSS. Cell lysis and total RNA isolation were performed with RNeasy Plus Mini Kits (Qiagen) according to the manufacturer’s protocol, and complementary DNA (cDNA) was synthesized with qScript cDNA SuperMix (Quanta Biosciences). qPCR analysis was performed with a Bio-Rad CFX96 real-time system with Power SYBR Green PCR Master Mix (Applied Biosystems) and preoptimized QuantiTect primer pairs (Qiagen). Fold increases in mRNA abundances in treated cells relative to those in vehicle control cells were calculated with the 2−ΔΔCt method, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA abundance was used for normalization.
Western blotting analysis
BMDCs (2.5 × 106 to 3 × 106) in R10F medium were preincubated for 2 hours in 5-ml polystyrene round-bottom tubes at 37°C, which was followed by activation with different concentrations of sLipid A or DMSO. After stimulation for 15 min or 1 hour, BMDCs were washed with ice-cold HBSS containing 50 μM NaF. Cells were lysed with radio-immunoprecipitation buffer [50 mM tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS] containing cOmplete Mini protease inhibitor cocktail tablets (Roche) and phosphatase inhibitor cocktail (Sigma), and the BCA assay (Pierce) was used to determine the protein concentrations of the lysates. Samples normalized for protein content were resolved by 10% SDS–polyacrylamide gel electrophoresis. Proteins were transferred onto nitrocellulose membranes (GE Healthcare). The membranes were blocked with 5% nonfat dry milk or 5% bovine serum albumin (BSA; for the analysis of pIRF3 only) for 1 hour and then were incubated overnight at 4°C with primary antibodies in 5% BSA or 5% non-fat dry milk (for the detection of β-actin, IRAK1, and total IRF3 only). HRP-conjugated secondary antibodies were resuspended in 5% nonfat dry milk and incubated with the Western blots for 1 hour. The blots were developed with ECL Prime (GE Healthcare) or SuperSignal ELISA Femto substrate (Pierce) on a Fujifilm LAS 4000 Mini, and data were quantified with Multi Gauge V3.0 software (Fujifilm).
Statistical analysis and log(EC50) measurement
Log(EC50) values for each agonist-induced response were calculated by generating four-parameter nonlinear fits to dose-response data with GraphPad Prism software using the following equation:
Differences between log(EC50) values were analyzed by unpaired two-tailed t test (for comparisons between two sets of genes) or by ordinary one-way ANOVA with Tukey’s multiple comparison post-test (for comparisons between three or more sets of genes). Statistically significant inhibition of increases in mRNA abundance by MAPK inhibitors was determined with repeated-measures two-way ANOVA with Dunnett’s multiple comparison post-test. Ordinary two-way ANOVA analysis with Sidak’s multiple comparison post-test was used to analyze the effects of MAPK inhibitors on gene log(EC50)s, and to compare IFNβ versus sLipid A–induced time courses. Differences in the log(EC50) values in the IFNAR1-blocking experiments were analyzed with repeated-measures two-way ANOVA with Sidak’s post-test.
Supplementary Material
Acknowledgments
We thank W. Bowen for his expert advice in pharmacological methods.
Funding: This work was supported by the NIH (grant AI071047) and the Commonwealth of Kentucky Research Challenge Trust.
Footnotes
www.sciencesignaling.org/cgi/content/full/7/351/ra108/DC1
Fig. S1. SB202190 and SP600125 inhibit p38 MAPK and JNK signaling, respectively.
Author contributions: J.P.K. designed and performed most experiments and wrote the preliminary manuscript draft; C.R.C. assisted with and performed several MAPK and IkBa Western blotting experiments; S.S. performed all adaptor-deficient BMDC experiments; and C.R.C., P.M.C., and T.C.M. assisted with experimental design, data interpretation, and manuscript revision.
Competing interests: The authors declare that they have no competing interests.
References
- 1.Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 3.Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4–MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 4.Latz E, Visintin A, Lien E, Fitzgerald KA, Espevik T, Golenbock DT. The LPS receptor generates inflammatory signals from the cell surface. J Endotoxin Res. 2003;9:375–380. doi: 10.1179/096805103225003303. [DOI] [PubMed] [Google Scholar]
- 5.Núñez Miguel R, Wong J, Westoll JF, Brooks HJ, O’Neill LA, Gay NJ, Bryant CE, Monie TP. A dimer of the Toll-like receptor 4 cytoplasmic domain provides a specific scaffold for the recruitment of signalling adaptor proteins. PLOS One. 2007;2:e788. doi: 10.1371/journal.pone.0000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim HM, Park BS, Kim JI, Kim SE, Lee J, Oh SC, Enkhbayar P, Matsushima N, Lee H, Yoo OJ, Lee JO. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88–IRAK4–IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conze DB, Wu CJ, Thomas JA, Landstrom A, Ashwell JD. Lys63-linked poly-ubiquitination of IRAK-1 is required for interleukin-1 receptor- and Toll-like receptor-mediated NF-κB activation. Mol Cell Biol. 2008;28:3538–3547. doi: 10.1128/MCB.02098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keating SE, Maloney GM, Moran EM, Bowie AG. IRAK-2 participates in multiple Toll-like receptor signaling pathways to NFκB via activation of TRAF6 ubiquitination. J Biol Chem. 2007;282:33435–33443. doi: 10.1074/jbc.M705266200. [DOI] [PubMed] [Google Scholar]
- 10.Lamothe B, Besse A, Campos AD, Webster WK, Wu H, Darnay BG. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of IκB kinase activation. J Biol Chem. 2007;282:4102–4112. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 12.Gohda J, Matsumura T, Inoue J. Cutting edge: TNFR-associated factor (TRAF) 6 is essential for MyD88-dependent pathway but not Toll/IL-1 receptor domain-containing adaptor-inducing IFN-β (TRIF)-dependent pathway in TLR signaling. J Immunol. 2004;173:2913–2917. doi: 10.4049/jimmunol.173.5.2913. [DOI] [PubMed] [Google Scholar]
- 13.Björkbacka H, Fitzgerald KA, Huet F, Li X, Gregory JA, Lee MA, Ordija CM, Dowley NE, Golenbock DT, Freeman MW. The induction of macrophage gene expression by LPS predominantly utilizes Myd88-independent signaling cascades. Physiol Genomics. 2004;19:319–330. doi: 10.1152/physiolgenomics.00128.2004. [DOI] [PubMed] [Google Scholar]
- 14.Kawai T, Takeuchi O, Fujita T, Inoue J, Mühlradt PF, Sato S, Hoshino K, Akira S. Lipopolysaccharide stimulates the MyD88-independent pathway and results in activation of IFN-regulatory factor 3 and the expression of a subset of lipopolysaccharide-inducible genes. J Immunol. 2001;167:5887–5894. doi: 10.4049/jimmunol.167.10.5887. [DOI] [PubMed] [Google Scholar]
- 15.Zanoni I, Ostuni R, Marek LR, Barresi S, Barbalat R, Barton GM, Granucci F, Kagan JC. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell. 2011;147:868–880. doi: 10.1016/j.cell.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-β. Nat Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanimura N, Saitoh S, Matsumoto F, Akashi-Takamura S, Miyake K. Roles for LPS-dependent interaction and relocation of TLR4 and TRAM in TRIF-signaling. Biochem Biophys Res Commun. 2008;368:94–99. doi: 10.1016/j.bbrc.2008.01.061. [DOI] [PubMed] [Google Scholar]
- 18.Fitzgerald KA, Rowe DC, Barnes BJ, Caffrey DR, Visintin A, Latz E, Monks B, Pitha PM, Golenbock DT. LPS-TLR4 signaling to IRF-3/7 and NF-κB involves the Toll adapters TRAM and TRIF. J Exp Med. 2003;198:1043–1055. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Häcker H, Redecke V, Blagoev B, Kratchmarova I, Hsu LC, Wang GG, Kamps MP, Raz E, Wagner H, Häcker G, Mann M, Karin M. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2006;439:204–207. doi: 10.1038/nature04369. [DOI] [PubMed] [Google Scholar]
- 20.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 21.Cusson-Hermance N, Khurana S, Lee TH, Fitzgerald KA, Kelliher MA. Rip1 mediates the Trif-dependent Toll-like receptor 3- and 4-induced NF-κB activation but does not contribute to interferon regulatory factor 3 activation. J Biol Chem. 2005;280:36560–36566. doi: 10.1074/jbc.M506831200. [DOI] [PubMed] [Google Scholar]
- 22.Sato S, Sugiyama M, Yamamoto M, Watanabe Y, Kawai T, Takeda K, Akira S. Toll/IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-κB and IFN-regulatory factor-3, in the Toll-like receptor signaling. J Immunol. 2003;171:4304–4310. doi: 10.4049/jimmunol.171.8.4304. [DOI] [PubMed] [Google Scholar]
- 23.Hirotani T, Yamamoto M, Kumagai Y, Uematsu S, Kawase I, Takeuchi O, Akira S. Regulation of lipopolysaccharide-inducible genes by MyD88 and Toll/IL-1 domain containing adaptor inducing IFN-β. Biochem Biophys Res Commun. 2005;328:383–392. doi: 10.1016/j.bbrc.2004.12.184. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto M, Sato S, Hemmi H, Uematsu S, Hoshino K, Kaisho T, Takeuchi O, Takeda K, Akira S. TRAM is specifically involved in the Toll-like receptor 4–mediated MyD88-independent signaling pathway. Nat Immunol. 2003;4:1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- 25.Hoebe K, Du X, Georgel P, Janssen E, Tabeta K, Kim SO, Goode J, Lin P, Mann N, Mudd S, Crozat K, Sovath S, Han J, Beutler B. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 26.Gandhapudi SK, Chilton PM, Mitchell TC. TRIF is required for TLR4 mediated adjuvant effects on T cell clonal expansion. PLOS One. 2013;8:e56855. doi: 10.1371/journal.pone.0056855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mata-Haro V, Cekic C, Martin M, Chilton PM, Casella CR, Mitchell TC. The vaccine adjuvant monophosphoryl lipid A as a TRIF-biased agonist of TLR4. Science. 2007;316:1628–1632. doi: 10.1126/science.1138963. [DOI] [PubMed] [Google Scholar]
- 28.Sakaguchi S, Negishi H, Asagiri M, Nakajima C, Mizutani T, Takaoka A, Honda K, Taniguchi T. Essential role of IRF-3 in lipopolysaccharide-induced interferon-β gene expression and endotoxin shock. Biochem Biophys Res Commun. 2003;306:860–866. doi: 10.1016/s0006-291x(03)01049-0. [DOI] [PubMed] [Google Scholar]
- 29.Hiscott J, Pitha P, Genin P, Nguyen H, Heylbroeck C, Mamane Y, Algarte M, Lin R. Triggering the interferon response: The role of IRF-3 transcription factor. J Interferon Cytokine Res. 1999;19:1–13. doi: 10.1089/107999099314360. [DOI] [PubMed] [Google Scholar]
- 30.Hoshino K, Kaisho T, Iwabe T, Takeuchi O, Akira S. Differential involvement of IFN-β in Toll-like receptor-stimulated dendritic cell activation. Int Immunol. 2002;14:1225–1231. doi: 10.1093/intimm/dxf089. [DOI] [PubMed] [Google Scholar]
- 31.Hoebe K, Janssen EM, Kim SO, Alexopoulou L, Flavell RA, Han J, Beutler B. Upregulation of costimulatory molecules induced by lipopolysaccharide and double-stranded RNA occurs by Trif-dependent and Trif-independent pathways. Nat Immunol. 2003;4:1223–1229. doi: 10.1038/ni1010. [DOI] [PubMed] [Google Scholar]
- 32.Le Bon A, Durand V, Kamphuis E, Thompson C, Bulfone-Paus S, Rossmann C, Kalinke U, Tough DF. Direct stimulation of T cells by type I IFN enhances the CD8+ T cell response during cross-priming. J Immunol. 2006;176:4682–4689. doi: 10.4049/jimmunol.176.8.4682. [DOI] [PubMed] [Google Scholar]
- 33.Marrack P, Kappler J, Mitchell T. Type I interferons keep activated T cells alive. J Exp Med. 1999;189:521–530. doi: 10.1084/jem.189.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoneyama H, Narumi S, Zhang Y, Murai M, Baggiolini M, Lanzavecchia A, Ichida T, Asakura H, Matsushima K. Pivotal role of dendritic cell–derived CXCL10 in the retention of T helper cell 1 lymphocytes in secondary lymph nodes. J Exp Med. 2002;195:1257–1266. doi: 10.1084/jem.20011983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bowen WS, Minns LA, Johnson DA, Mitchell TC, Hutton MM, Evans JT. Selective TRIF-dependent signaling by a synthetic Toll-like receptor 4 agonist. Sci Signal. 2012;5:ra13. doi: 10.1126/scisignal.2001963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zughaier SM, Zimmer SM, Datta A, Carlson RW, Stephens DS. Differential induction of the Toll-like receptor 4-MyD88-dependent and -independent signaling pathways by endotoxins. Infect Immun. 2005;73:2940–2950. doi: 10.1128/IAI.73.5.2940-2950.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Ley P, Steeghs L, Hamstra HJ, ten Hove J, Zomer B, van Alphen L. Modification of lipid A biosynthesis in Neisseria meningitidis lpxL mutants: Influence on lipo-polysaccharide structure, toxicity, and adjuvant activity. Infect Immun. 2001;69:5981–5990. doi: 10.1128/IAI.69.10.5981-5990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cekic C, Casella CR, Eaves CA, Matsuzawa A, Ichijo H, Mitchell TC. Selective activation of the p38 MAPK pathway by synthetic monophosphoryl lipid A. J Biol Chem. 2009;284:31982–31991. doi: 10.1074/jbc.M109.046383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Embry CA, Franchi L, Nuñez G, Mitchell TC. Mechanism of impaired NLRP3 inflammasome priming by monophosphoryl lipid A. Sci Signal. 2011;4:ra28. doi: 10.1126/scisignal.2001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans JT, Cluff CW, Johnson DA, Lacy MJ, Persing DH, Baldridge JR. Enhancement of antigen-specific immunity via the TLR4 ligands MPL adjuvant and Ribi.529. Expert Rev Vaccines. 2003;2:219–229. doi: 10.1586/14760584.2.2.219. [DOI] [PubMed] [Google Scholar]
- 41.Gaekwad J, Zhang Y, Zhang W, Reeves J, Wolfert MA, Boons GJ. Differential induction of innate immune responses by synthetic lipid a derivatives. J Biol Chem. 2010;285:29375–29386. doi: 10.1074/jbc.M110.115204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Gaekwad J, Wolfert MA, Boons GJ. Modulation of innate immune responses with synthetic lipid A derivatives. J Am Chem Soc. 2007;129:5200–5216. doi: 10.1021/ja068922a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teghanemt A, Zhang D, Levis EN, Weiss JP, Gioannini TL. Molecular basis of reduced potency of underacylated endotoxins. J Immunol. 2005;175:4669–4676. doi: 10.4049/jimmunol.175.7.4669. [DOI] [PubMed] [Google Scholar]
- 44.Saitoh S, Akashi S, Yamada T, Tanimura N, Kobayashi M, Konno K, Matsumoto F, Fukase K, Kusumoto S, Nagai Y, Kusumoto Y, Kosugi A, Miyake K. Lipid A antagonist, lipid IVa, is distinct from lipid A in interaction with Toll-like receptor 4 (TLR4)-MD-2 and ligand-induced TLR4 oligomerization. Int Immunol. 2004;16:961–969. doi: 10.1093/intimm/dxh097. [DOI] [PubMed] [Google Scholar]
- 45.Akashi S, Nagai Y, Ogata H, Oikawa M, Fukase K, Kusumoto S, Kawasaki K, Nishijima M, Hayashi S, Kimoto M, Miyake K. Human MD-2 confers on mouse Toll-like receptor 4 species-specific lipopolysaccharide recognition. Int Immunol. 2001;13:1595–1599. doi: 10.1093/intimm/13.12.1595. [DOI] [PubMed] [Google Scholar]
- 46.Shen H, Tesar BM, Walker WE, Goldstein DR. Dual signaling of MyD88 and TRIF is critical for maximal TLR4-induced dendritic cell maturation. J Immunol. 2008;181:1849–1858. doi: 10.4049/jimmunol.181.3.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Windheim M, Stafford M, Peggie M, Cohen P. Interleukin-1 (IL-1) induces the Lys63-linked polyubiquitination of IL-1 receptor-associated kinase 1 to facilitate NEMO binding and the activation of Iκβα kinase. Mol Cell Biol. 2008;28:1783–1791. doi: 10.1128/MCB.02380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiang E, Dang O, Anderson K, Matsuzawa A, Ichijo H, David M. Cutting edge: Apoptosis-regulating signal kinase 1 is required for reactive oxygen species-mediated activation of IFN regulatory factor 3 by lipopolysaccharide. J Immunol. 2006;176:5720–5724. doi: 10.4049/jimmunol.176.10.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Navarro L, David M. p38-dependent activation of interferon regulatory factor 3 by lipopolysaccharide. J Biol Chem. 1999;274:35535–35538. doi: 10.1074/jbc.274.50.35535. [DOI] [PubMed] [Google Scholar]
- 50.Wadleigh DJ, Reddy ST, Kopp E, Ghosh S, Herschman HR. Transcriptional activation of the cyclooxygenase-2 gene in endotoxin-treated RAW 264.7 macrophages. J Biol Chem. 2000;275:6259–6266. doi: 10.1074/jbc.275.9.6259. [DOI] [PubMed] [Google Scholar]
- 51.Lasa M, Mahtani KR, Finch A, Brewer G, Saklatvala J, Clark AR. Regulation of cyclooxygenase 2 mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. Mol Cell Biol. 2000;20:4265–4274. doi: 10.1128/mcb.20.12.4265-4274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arndt PG, Suzuki N, Avdi NJ, Malcolm KC, Worthen GS. Lipopolysaccharide-induced c-Jun NH2-terminal kinase activation in human neutrophils: Role of phosphatidylinositol 3-kinase and Syk-mediated pathways. J Biol Chem. 2004;279:10883–10891. doi: 10.1074/jbc.M309901200. [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto M, Sato S, Mori K, Hoshino K, Takeuchi O, Takeda K, Akira S. Cutting edge: A novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-β promoter in the Toll-like receptor signaling. J Immunol. 2002;169:6668–6672. doi: 10.4049/jimmunol.169.12.6668. [DOI] [PubMed] [Google Scholar]
- 54.Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J Biol Chem. 1995;270:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- 55.Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 56.Chu WM, Ostertag D, Li ZW, Chang L, Chen Y, Hu Y, Williams B, Perrault J, Karin M. JNK2 and IKKβ are required for activating the innate response to viral infection. Immunity. 1999;11:721–731. doi: 10.1016/s1074-7613(00)80146-6. [DOI] [PubMed] [Google Scholar]
- 57.Ludwig S, Ehrhardt C, Neumeier ER, Kracht M, Rapp UR, Pleschka S. Influenza virus-induced AP-1-dependent gene expression requires activation of the JNK signaling pathway. J Biol Chem. 2001;276:10990–10998. [PubMed] [Google Scholar]
- 58.Thomas KE, Galligan CL, Newman RD, Fish EN, Vogel SN. Contribution of interferon-β to the murine macrophage response to the Toll-like receptor 4 agonist, lipopolysaccharide. J Biol Chem. 2006;281:31119–31130. doi: 10.1074/jbc.M604958200. [DOI] [PubMed] [Google Scholar]
- 59.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 60.de Weerd NA, Vivian JP, Nguyen TK, Mangan NE, Gould JA, Braniff SJ, Zaker-Tabrizi L, Fung KY, Forster SC, Beddoe T, Reid HH, Rossjohn J, Hertzog PJ. Structural basis of a unique interferon-β signaling axis mediated via the receptor IFNAR1. Nat Immunol. 2013;14:901–907. doi: 10.1038/ni.2667. [DOI] [PubMed] [Google Scholar]
- 61.Gallucci S, Lolkema M, Matzinger P. Natural adjuvants: Endogenous activators of dendritic cells. Nat Med. 1999;5:1249–1255. doi: 10.1038/15200. [DOI] [PubMed] [Google Scholar]
- 62.Santiago-Raber ML, Baccala R, Haraldsson KM, Choubey D, Stewart TA, Kono DH, Theofilopoulos AN. Type-I interferon receptor deficiency reduces lupus-like disease in NZB mice. J Exp Med. 2003;197:777–788. doi: 10.1084/jem.20021996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okemoto K, Kawasaki K, Hanada K, Miura M, Nishijima M. A potent adjuvant monophosphoryl lipid A triggers various immune responses, but not secretion of IL-1β or activation of caspase-1. J Immunol. 2006;176:1203–1208. doi: 10.4049/jimmunol.176.2.1203. [DOI] [PubMed] [Google Scholar]
- 64.Luther SA, Cyster JG. Chemokines as regulators of T cell differentiation. Nat Immunol. 2001;2:102–107. doi: 10.1038/84205. [DOI] [PubMed] [Google Scholar]
- 65.Henricson BE, Manthey CL, Perera PY, Hamilton TA, Vogel SN. Dissociation of lipopolysaccharide (LPS)-inducible gene expression in murine macrophages pre-treated with smooth LPS versus monophosphoryl lipid A. Infect Immun. 1993;61:2325–2333. doi: 10.1128/iai.61.6.2325-2333.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meng J, Drolet JR, Monks BG, Golenbock DT. MD-2 residues tyrosine 42, arginine 69, aspartic acid 122, and leucine 125 provide species specificity for lipid IVA. J Biol Chem. 2010;285:27935–27943. doi: 10.1074/jbc.M110.134668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Richez C, Yasuda K, Watkins AA, Akira S, Lafyatis R, van Seventer JM, Rifkin IR. TLR4 ligands induce IFN-α production by mouse conventional dendritic cells and human monocytes after IFN-β priming. J Immunol. 2009;182:820–828. doi: 10.4049/jimmunol.182.2.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karaghiosoff M, Steinborn R, Kovarik P, Kriegshäuser G, Baccarini M, Donabauer B, Reichart U, Kolbe T, Bogdan C, Leanderson T, Levy D, Decker T, Müller M. Central role for type I interferons and Tyk2 in lipopolysaccharide-induced endotoxin shock. Nat Immunol. 2003;4:471–477. doi: 10.1038/ni910. [DOI] [PubMed] [Google Scholar]
- 69.Rathinam VA, Vanaja SK, Waggoner L, Sokolovska A, Becker C, Stuart LM, Leong JM, Fitzgerald KA. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by Gram-negative bacteria. Cell. 2012;150:606–619. doi: 10.1016/j.cell.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Orr MT, Duthie MS, Windish HP, Lucas EA, Guderian JA, Hudson TE, Shaverdian N, O’Donnell J, Desbien AL, Reed SG, Coler RN. MyD88 and TRIF synergistic interaction is required for TH1-cell polarization with a synthetic TLR4 agonist adjuvant. Eur J Immunol. 2013;43:2398–2408. doi: 10.1002/eji.201243124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lutz MB, Kukutsch N, Ogilvie AL, Rossner S, Koch F, Romani N, Schuler G. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J Immunol Methods. 1999;223:77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.