Abstract
Familial inheritance of drug abuse is composed of both genetic and environmental factors. Additionally, epigenetic transgenerational inheritance may provide a means by which parental drug use can influence several generations of offspring. Recent evidence suggests that parental drug exposure produces behavioral, biochemical, and neuroanatomical changes in future generations. The focus of this review is to discuss these multigenerational and transgenerational phenotypes in the offspring of animals exposed to drugs of abuse. Specifically, changes found following the administration of alcohol, opioids, cocaine, marijuana, and nicotine will be discussed. In addition, epigenetic modifications to the genome following administration of these drugs will be detailed as well as their potential for transmission to the next generation.
Keywords: Multigenerational, Transgenerational, Epigenetic, Drug Abuse
1 Introduction
Drug abuse impacts critical brain regions resulting in reward-seeking and craving, drug dependence, withdrawal, and alterations in both anxiety and learning and memory (Koob and Volkow, 2009). Clinical findings document familial patterns of drug use and dependence (Nielsen et al., 2012). Genome-wide association studies (GWAS) have identified a number of genes, chromosomal regions, and allelic variants likely to contribute to drug addiction, however not all inherited information is present in DNA sequence. Recently chemical alterations in the genome not related to DNA sequence variations have been identified following drug exposure and these epigenetic modifications may contribute to drug abuse and dependence across familial generations (Hughes, 2014). Specifically, several phenotypic consequences of drug exposure are found across multiple generations of offspring, despite no previous exposure to drug and no allelic or chromosomal variation (Skinner, 2008). Thus, epigenetic modifications could provide a mechanism underlying the longevity of psychiatric conditions such as drug abuse both within, as well as across generations (Nielsen et al., 2012).
The focus of this review is to discuss phenotypic variation (behavior, neurochemical, and structural) in offspring of drug-exposed parents and grandparents in animal models. Inheritance of behavioral changes will be limited to outcomes that measure drug response and reward sensitivity in animals. In addition, epigenetic modifications of gene regulation in response to or mediating the effects of drugs of abuse will be discussed. Finally, the transmission of epigenetic changes through the germ line as a mechanism that may underlie transgenerational inheritance will be reviewed.
2 Epigenetics
In an attempt to rectify the vast differences in use of the term epigenetics, Adrian Bird proposed a definition that encompasses both the chemical mechanisms as well as the necessity for inheritance: epigenetics is “the structural adaptation of chromosomal regions so as to register, signal or perpetuate altered activity states” (Bird, 2007). Therefore, these chromosomal modifications can be sudden or accumulate over time in response to exposure to a stimulus and is, nevertheless, inherited in the absence of the signal or event that initiated the change (Bird, 2007). In addition, chemical alterations to the genome, also referred to as the epigenome when including the DNA packaging, can ultimately change the functional expression of genes (Bird, 2002). Epigenetic inheritance occurs by persistence of the modifications through several generations of cell division or animals (Bird, 2007).
Cellular epigenetic modifications in response to the environment include chromatin remodeling and DNA methylation (Skinner, 2011; Daxinger and Whitelaw, 2012). Further explanation of these modifications can be found in Box 1. There is some evidence that expression of a class of small noncoding RNAs known as micro RNAs (miRNAs) can transmit phenotypes to the next generation (Rassoulzadegan et al., 2006). While miRNAs have been implicated in the inheritance of stress (Gapp et al., 2014; Rodgers et al., 2013), there is no evidence to suggest miRNA mediated effects of drug exposure on offspring. It is quite complex to establish a mechanism of inheritance that could account for altered behaviors, biochemical responses, and cellular morphology in the offspring of F0 exposed parents. Therefore, within each drug class epigenetic changes of interest within the animal exposed will be discussed as well as any evidence of generational inheritance of chemical modifications to the genome.
Box 1. Epigenetic Modifications.
Chromatin Remodeling and Histone Modifications
Histone complexes contain an amino (N) terminal tail that can undergo modifications; these modifications “condense” or “relax” the state of DNA wrapped around the histone. Modifications include acetylation, methylation, phosphorylation, ubiquitylation, and SUMOylation, however for the purposes of this review only acetylation and methylation will be discussed.
Acetylation
Acetylation of histone tail relaxes chromatin and allows for gene transcription. For example acetylation of histone 3 (AcH3) and acetylation of lysine 14 of histone 3 (aceH3K14) are both correlated to increased transcription of target genes.
Methylation
Histone methylation can cause gene activation or repression depending on the residue undergoing modification. Histone 3 lysine 4 trimethylation (H3K4me3) causes the activation of gene transcription while histone 3 lysine 9 dimethylation (H3K9me2) association with a gene is correlated to repression of that gene.
For a thorough review on epigenetic inheritance mechanisms and specifically theories on histone modification transmission through DNA replication see (Martin and Zhang, 2007).
DNA methylation
DNA methylation regulates transcription by acting directly on the genome. In the past research has suggested that the presence of methyl groups suppresses gene expression, however this is not always the case. DNA methyltransferases (DNMTs) catalyze the addition of a methyl group onto a cytosine nucleotide that is usually positioned next to a guanine nucleotide (CpG). Methyl groups are donated by S-adenosyl methionine (SAM); therefore, the addition of a methyl onto the cytosine converts the cytosine to 5-methylcytosine. Gene regulation via DNA methylation is achieved through CpGs in the regulatory regions (promoters, enhancers, insulator) of genes.
3 Epigenetics across generations
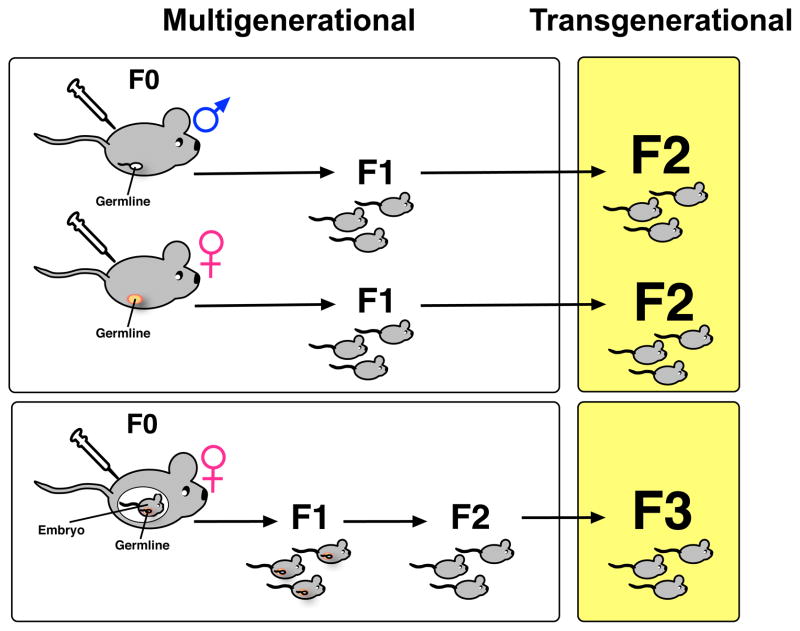
Epigenetic transgenerational inheritance is defined as “germline-mediated inheritance of epigenetic information between generations in the absence of direct environmental influences that leads to phenotypic variation” (Skinner, 2011). In contrast, “multigenerational” phenotypes are those derived from direct exposure to the drug. Thus, if drug exposure occurs in F0 males or females prior to pregnancy, the germ cells, which go on to produce the F1 generation are also “exposed”. Therefore, phenotypes found in F0 and F1 animals are multigenerational and only those present in F2 animals are considered transgenerational as these F2 animals are the first generation whose cells have not been exposed to drug (Figure 1, top).
Figure 1. Multigenerational and Transgenerational Phenotypes Following Drug Exposure.
Paternal or maternal exposure paradigms are used as a model for inheritance in rodents. F0 males or females exposed to drug prior to mating produce F1 offspring with multigenerational phenotypes and F2 offspring with transgenerational phenotypes (top). F0 females exposed to drug during pregnancy produce F1 and F2 offspring with multigenerational phenotypes and F3 offspring with transgenerational phenotypes (bottom).
In contrast, if F0 mothers are exposed to drug during pregnancy, the somatic and germ cells of the F1 offspring receive direct exposure to the drug in utero (Figure 1, bottom). Since the germ cells of the F1 offspring are exposed to drug and the F2 offspring originate from these exposed germ cells, then the F0 parents, F1 and F2 offspring are all exposed to the drug (Skinner, 2008). In this case, F0, F1, and F2 phenotypes are multigenerational and only phenotypes observed in the F3 generation and beyond are transgenerational (Figure 1).
4 Gamete development and reprogramming
Germline reprogramming events may directly facilitate epigenetic inheritance across generations. Genome wide DNA methylation reprogramming occurs at two time points in early embryonic development. Both reprogramming events are well understood in mice. First, genome-wide DNA demethylation occurs post-fertilization in the zygote to erase gamete epigenome methylation in order to promote cellular totipotency in the developing embryo. However, at this time genomic imprints, methylation on imprinted genes such as H19 and Igf2, remain intact. Later, a second major reprogramming event occurs in the germ line where paternal and maternal somatic programming is erased from most genes, including imprinted genes. Parent-specific imprints are subsequently imposed in the germ line and DNA methylation across the genome occurs through the action of DNA methyltransferases (for a complete review see Daxinger and Whitelaw, 2012). During these periods of reprogramming, exposure to a challenge that increases or decreases the activity of the epigenomic machinery may lead to alterations in DNA methylation. Embryonic reprogramming suggests that epigenetic modifications to the DNA made in response to drug exposure may be lost, however, there is evidence that some methylation marks escape complete erasure. The best known example is that of imprinted genes, whose methylation marks are retained in the developing embryo (Bartolomei, 2009). In addition there is growing evidence that non-imprinted genes and repetitive genomic elements escape complete loss of methylation patterning following reprogramming events (Lane et al., 2003; Orozco et al., 2014). The retention of genomic methylation patterns in sperm of exposed parents and brains of offspring may occur following generational stress (Franklin et al., 2010) or drug-exposure (Govorko et al., 2012).
Histone acetylation and methylation, although not as well described as DNA methylation, may also be subject to epigenetic mechanisms of modulation by drug exposure. Although the vast majority of histones are replaced by protamines during spermatogenesis, not all histones are lost and sperm DNA retention within histones has been discovered in both humans and mice (Bench et al., 1996; Gatewood et al., 1987; Hammoud et al., 2009). Furthermore, DNA methylation retention in sperm may be due to the association of DNA to histones. There is an enrichment of histone bound sperm DNA at loci of imprinted genes and developmentally important genes that retain methylation marks in the embryo (Hammoud et al., 2009; Wykes and Krawetz, 2003).
Histone variants as well as the presence of histone methyltransferases have been identified in mature oocytes and differential acetylation and methylation has been implicated in oocyte development (Wang et al., 2014a). In addition, histone marks in the oocyte can be transmitted across generations (Gaydos et al., 2014). Therefore the genome content in sperm and oocytes along with chemical modifications are potential carriers of epigenetic information.
5 Inheritance of drug exposure
Substance abuse is influenced by a genetic component (Kendler et al., 2003; Tsuang et al., 1998), with heritability estimates ranging from 45% to 79% (Agrawal and Lynskey, 2006; Kendler et al., 2003; Tsuang et al., 2001). Individuals with a family history of drug abuse have an 8-fold increase in the likelihood of drug use, suggesting familial transmission of substance abuse disorders (Merikangas et al., 1998). The heritability of substance abuse varies depending on drug class, although polymorphisms altering gene expression and function are associated with drug-taking susceptibility and resilience across all substances of abuse (Goldman et al., 2005).
While genetics plays a role in variation of drug initiation and dependence, progress in identifying genetic factors that are responsible for addiction risk across most drug classes has been modest. The contribution of other factors that are involved in addiction susceptibility must account for the prevalence of drug use disorders. Indeed, in addition to the genetic information carried in the DNA sequence; offspring also receive epigenetic information from parents in the form of chemical modifications of DNA and associated histones. These modifications (see Box 1) can significantly alter gene expression. In the following sections, we will discuss the specific epigenetic modifications (if known) that are associated with multi- and transgenerational inheritance of phenotype for each drug class.
5.1 Alcohol
The effects of alcohol are mediated through several neurotransmitter systems. Alcohol directly reduces glutamate signaling at the N-methyl-D-aspartate (NMDA) receptor and enhances gamma amino butyric acid receptor (GABA) signaling at the GABAA receptor. In addition alcohol mediates increased dopamine and opioid release. Genetic variation in GABRA2, the gene encoding for the alpha 2 subunit of the GABAA receptor has been associated with alcohol dependence in humans and alcohol response in animals (Demers et al., 2014). In addition, polymorphisms in alcohol dehydrogenase and aldehyde dehydrogenase are associated with alcohol consumption; both genes encode for enzymes responsible for metabolism of alcohol. Interestingly, variation in alcohol dehydrogenase and dopamine receptor or dopamine transporter genes has been found to epistatically influence alcohol dependence (reviewed in Demers et al., 2014). Finally, there is evidence to suggest that genetic variation in serotonin, opioid, and other neuromodulatory systems can affect alcohol use (reviewed in Kreek et al., 2004).
Clinical reports have provided evidence of inherited drug use behavior in individuals whose parents engage in alcohol consumption. Episodic drinking in both parents and heavy paternal drinking is correlated with early onset of drinking and amount of alcohol consumed in offspring (Vermeulen-Smit et al., 2012). The heritability of alcohol dependence is well-studied with estimates ranging from 40% to 70% (Agrawal and Lynskey, 2008; Enoch and Goldman, 1999; Nieratschker et al., 2013). Thus, genetics explains some but not all alterations in neurochemical response to alcohol.
The behavioral effects of alcohol are well studied as well as the consequences of parental alcohol use on fetal development. Specifically, F0 mothers exposed to alcohol during pregnancy produce offspring with neurodevelopmental deficits (Lam et al., 2000) and alcohol-exposed F0 fathers produce several generations of offspring with developmental and physiological deficits (Friedler, 1996). Inheritance models in animals have identified changes in reward behavior, neurochemical signaling, and brain morphology in F1 offspring of F0 fathers exposed to alcohol (summarized below and in Table 1). However, to date, no work has described the neurobehavioral effects on offspring of F0 alcohol-exposure in mothers prior to conception.
Table 1.
Multigenerational and Transgenerational phenotypes of F0 drug exposure
| Drug | F0 exposure | Generation affected | Sex of offspring affected | Phenotype | Species | Source | |
|---|---|---|---|---|---|---|---|
| Behavior | Alcohol | Paternal | F1 | Both | Increased sensitivity to amphetamine | Rat | Abel, 1993 |
| Paternal | F1 | Male | Reduced alcohol intake and increased behavioral response to alcohol | Mouse | Finegersh and Homanics, 2014 | ||
| Paternal | F1 | Both | Decreased fear and increased aggression | Mouse | Meek et al., 2007 | ||
| Paternal | F1 | Male | Decreased attention and increased impulsivity | Mouse | Kim et al., 2014 | ||
| Paternal | F1 | Both | Enhanced passive avoidance learning and deficits in T-maze | Mouse | Abel and Lee, 1988 | ||
| Paternal | F1 | Female | Deficits in passive avoidance learning | Rat | Abel and Tan, 1988 | ||
| Paternal | F1 | Both | Deficits in radial arm maze learning | Rat | Wozniak et al., 1991 | ||
| Paternal | F1 | Male | Hyperactivity attenuated by physostigmine treatment | Rat | Abel, 1994 | ||
| Morphine | Maternal | F1 | Both | Increased morphine sensitization (locomotor) | Rat | Byrnes, 2005 | |
| Maternal | F1, F2 | Male | Decreased locomotor response to D1/D2 agonist | Rat | Byrnes et al., 2013 | ||
| Cocaine | Paternal | F1 | Male | Resilience to cocaine self-administration | Rat | Vassoler et al., 2013b | |
| Maternal | F1 | Male | Increased sensitivity to cocaine | Rat | Sasaki et al., 2014 | ||
| Paternal | F1 | Female | Learning deficits | Mouse | He et al., 2006 | ||
| Paternal | F1 | Both | Increase in depression-like phenotype, no change in anxiety or learning and memory | Mouse | Killinger et al., 2012 | ||
| CB1/CB2 agonist (WIN 55, 212-12) | Maternal | F1 | Male | Increased morphine CPP | Rat | Byrnes et al., 2012 | |
| Maternal | F1 | Female | Increased morphine sensitization (locomotor) | Rat | Vassoler et al., 2013a | ||
| Δ9-THC | Both parents exposed and mated together | F1 | Both | Increased heroin self-administration and enhanced heroin withdrawal | Rat | Szutorisz et al., 2014 | |
| Nicotine | Maternal | F1, F2, F3 | Both | Hyperactive, decreased attention | Mouse | Zhu et al., 2014 | |
|
| |||||||
| Neurochemical | Alcohol | Paternal | F1 | Both | Decreased DAT in cortex and striatum | Mouse | Kim et al., 2014 |
| Paternal | F1 | Male | Decreased beta-endorphin in hypothalamus | Rat | Cicero et al., 1990 | ||
| Maternal | F1, F2, F3 | Both | Decreased POMC expression in arcuate nucleus | Rat | Govorko et al., 2012 | ||
| Morphine | Paternal | F1 | Female | Increased beta-endorphin in hypothalamus | Rat | Cicero et al., 1991 | |
| Maternal | F1 | Male | Increased kappa-opioid receptor in response to D1/D2 agonist in nucleus accumbens | Rat | Byrnes et al., 2013 | ||
| Maternal | F1, F2 | Male | Increased D2 receptor in response to D1/D2 agonist in nucleus accumbens | Rat | Byrnes et al., 2013 | ||
| Maternal | F1, F2 | Male | Decreased quinpirole induced cortiscosterone | Rat | Byrnes et al., 2013 | ||
| Cocaine | Maternal | F1 | Male | Increased D1 receptor in medial prefrontal cortex | Rat | Sasaki et al., 2014 | |
| Δ9-THC | Maternal | F1 | Female | Increased Oprm1 in nucleus accumbens | Rat | Vassoler et al., 2013a | |
| Both parents exposed and mated together | F1 | Both | Decreased expression of cannabinoid, dopamine, and glutamate receptors, reduced NMDA binding and enhanced LTD in the dorsal striatum | Rat | Szutorisz et al., 2014 | ||
|
| |||||||
| Structural Physiological | Alcohol | Paternal | F1 | Both | Cortical thickening | Rat | Jamerson et al., 2004 |
| Paternal | F1 | Both | No change in pyramidal cells of hippocampus | Rat | Zajac et al., 1989 | ||
| Morphine | Paternal | F1 | Both | Decreased hippocampal LTP | Rat | Sarkaki et al., 2008 | |
| Maternal | F1 | Both | Decreased hippocampal LTD | Rat | Sarkaki et al., 2008 | ||
| Paternal | F2 | Both | Decreased synaptophysin | Rat | Vyssotski et al., 2011 | ||
5.1a Behavioral, neurochemical, and structural changes in offspring of alcohol-exposed parents
Paternal alcohol exposure alters reward directed behaviors in offspring. Specifically, when F0 fathers are exposed to alcohol, male F1 offspring show increased sensitivity to amphetamine (Abel, 1993) and alcohol (Finegersh and Homanics, 2014) As amphetamine and alcohol modulate dopamine signaling in the mesolimbic pathway, these results suggest that alcohol mediates functional changes in neuronal dopamine response across generations. However, alcohol exposed F0 fathers appear to pass different phenotypes to their sons and daughters. F0 alcohol-exposed fathers produce F1 female offspring with increased sensitivity to amphetamine (Abel, 1993) but not alcohol (Finegersh and Homanics, 2014).
In addition to drug response, cognitive behaviors implicated in drug abuse and dependence have been assayed in F1 offspring of F0 fathers exposed to alcohol (Abel and Tan, 1988; Kim et al., 2014; Meek et al., 2007; Wozniak et al., 1991). Decreased fear and increased aggression is found in F1 mice from F0 fathers exposed to acute alcohol prior to mating (Meek et al., 2007). In addition, F0 alcohol-exposed fathers produce F1 male offspring with deficits in attention and increased impulsivity (Kim et al., 2014). Learning and memory in F1 animals are differentially affected by F0 alcohol exposure based on the cognitive task being assayed and the animal species tested. F0 male mice exposed to alcohol produce F1 offspring with enhanced passive avoidance learning (Abel and Lee, 1988) while, in rats, female F1 offspring of F0 alcohol-exposed fathers perform worse in a passive avoidance learning task (Abel and Tan, 1988). However, learning deficits are found in both mice and rat F1 offspring of F0 alcohol-exposed father in working memory (choice point task in the T-maze) (Abel and Lee, 1988) and spatial learning (radial arm maze) (Wozniak et al., 1991).
Alcohol also mediates changes in offspring within several neurotransmitter systems that include dopamine, acetylcholine, and opioids. Alcohol-exposed fathers produced F1 offspring with decreased dopamine transporter (DAT) expression in the cortex and striatum (Kim et al., 2014) and hyperactivity in F1 male offspring of F0 alcohol-exposed fathers is attenuated by increased levels of acetylcholine through administration of the cholinesterase inhibitor physostigmine (Abel, 1994). In addition, alcohol exposure in F0 male rats during puberty decreases beta-endorphin, a primary endogenous ligand for opioid receptors, in the hypothalamus of F1 male offspring (Cicero et al., 1990) and F0 females exposed to alcohol during pregnancy produce F1 males with decreased POMC (the gene that codes for beta-endorphin and melanocortin) expression in the arcuate nucleus. In addition, this effect is transmitted to F2 and F3 offspring through the paternal line (Govorko et al., 2012). Endocrine signaling from the hypothalamus is essential for stress response and plays a critical role in the acquisition of drug addiction (Koob and Volkow, 2009). Thus, these neurochemical changes may establish a multi- and transgenerational vulnerability to drug use in offspring of alcohol-exposed fathers.
In addition to behavioral and neurochemical phenotypes, F0 fathers exposed to alcohol produce F1 offspring with altered brain morphology. Cortical thickening is found in F1 offspring of F0 fathers exposed to alcohol before or during mating (Jamerson et al., 2004). Specifically, alterations in thickness are found in cortical sections I and V. Layer I receives projections from pyramidal cells of other cortical layers and layer V contains pyramidal cells. Consequently, the structural effects on these layers suggest alterations in pyramidal cell density and construction of the cortex. However, in another study no differences were found in pyramidal cell density of areas CA1 and CA3 in the hippocampus of offspring derived from F0 fathers exposed to alcohol (Zajac et al., 1989). Therefore, additional research must be done to better understand the change in neuronal morphology in F1 offspring of F0 fathers exposed to alcohol.
5.1b Alcohol exposure and changes to the epigenome
Alcohol exposure produces a variety of epigenetic modifications and related gene expression changes in brain reward regions (Robison and Nestler, 2011). Epigenetic modifications have been identified within the F0 generation of animals exposed to alcohol. In addition, evidence of multi- and transgenerational epigenetic changes in animals will be discussed.
Chronic alcohol use upregulates long-term repeat (LTR) transposons through a decrease in methylation (Ponomarev et al., 2012). Long-term repeat (LTR) transposons are transposable elements (TEs) that are typically silenced by the presence of DNA methylation and have been increasingly implicated in epigenomic regulation (Pray, 2008; Slotkin and Martienssen, 2007). Interestingly methylation marks on TEs are stable in gametes and strongly implicated in inheritance (Whitelaw et al., 1999). In addition, chronic alcohol use upregulates gene expression networks composed of gene sequences rich in CG repeats and downregulates gene expression networks composed of low CG repeats (Ponomarev et al., 2012). CG-rich genes involved in synaptic transmission, specifically glutamatergic transmission, are heavily upregulated in the cortex of alcoholics (Ponomarev et al., 2012). Alcohol decreases the activity of DNA methyltransferase 1 (DNMT1), which donates methyl groups to nucleotide residues, thereby providing a possible mechanism for this regulation (Ponomarev et al., 2012). However, alcohol may also mediate reduced methylation through its regulation of the methyl donor cycle. Rats exposed to chronic ethanol have reduced production of the methyl donor S-adenosylmethionine (SAM) and hypomethylation of the entire genome in the liver (Lu and Mato, 2005). Although the same effect has not been found in neurons or gametes, alcohol may mediate methylation changes through similar signaling.
Imprinted genes appear especially vulnerable to epigenetic modification mediated by alcohol exposure. Demethylation is detected at the locus of imprinted gene H19 in gametes of male alcoholics (Ouko et al., 2009). In addition, changes in H19 methylation have been identified in rodent models of multigenerational alcohol exposure. Specifically, F0 mothers exposed to alcohol during pregnancy produce F1 male offspring with decreased methylation at CpGs in H19 in sperm and F2 male offspring with deceased methylation at H19 in brains (Stouder et al., 2011). Hypomethylation at H19 produces alterations in strict imprinting patterns (Knezovich and Ramsay, 2012; Stouder et al., 2011), that normally escape complete methylation erasure during steps of developmental reprogramming. While this suggests transmission of aberrant methylation from F1 sperm to F2 somatic cells in offspring, a second study found hypomethylation at H19 in F1 offspring but no alteration in sperm of alcohol-exposed F0 fathers (Knezovich and Ramsay, 2012).
Alcohol increases and decreases DNA methylation in non-imprinted genes and both multigenerational and transgenerational epigenetic inheritance of such changes have been found. F0 alcohol exposure decreases methylation at the brain derived neurotrophic factor (BDNF) exon IV promoter in the sperm of exposed males. This reduction of methylation is also measured at the exon IV promoter of BDNF in the ventral tegmental area (VTA) of F1 offspring (Finegersh and Homanics, 2014). Consequently, associated with the decrease in BDNF methylation, increased exon IV transcripts are found in the VTA (Finegersh and Homanics, 2014). In contrast, increased methylation is found in the promoter region of DAT and decreased mRNA and protein expression of the dopamine transporter in the cortex and striatum of F1 offspring of F0 fathers exposed to alcohol (Kim et al., 2014). Alcohol-mediated decrease in DNMT1, as was discussed with alcohol-mediated regulation of LTRs and CpG dense regions, is thought to also mediate hypermethylation of DAT in the cortex and striatum of F1 offspring (Kim et al., 2014). Finally, F0 females exposed to alcohol during pregnancy produce F1 male offspring and F2 and F3 offspring with increased POMC methylation in the arcuate nucleus. Interestingly, F1 sperm also has an increase in POMC methylation (Govorko et al., 2012). Therefore alcohol produces differential methylation changes depending on the gene target.
Alcohol can also mediate chromatin remodeling in the animal exposed though histone methylation and acetylation. Upregulation of CG-rich genes following alcohol exposure is correlated with increased levels of trimethylation of histone 3 at lysine 4 (H3K4me3) in the promoters of the same genes within a generation (Ponomarev et al., 2012). In addition, some of the genes that are upregulated promote trimethylation at histone 3 which therefore increases H3K4me3 (Ponomarev et al., 2012). This suggests an interaction between alcohol-mediated DNA methylation and chromatin remodeling. In addition to methylation changes on the histone, genome wide histone acetylation is increased following alcohol administration (Ghezzi et al., 2013). In a rat model of alcohol addiction, decreased histone deacetylase (HDAC) activity and an increase in the acetylation of histones H3 and H4 is found in the amygdala (Pandey et al., 2008). Thus alcohol may change the expression of genes through histone methylation and acetylation within the animal being exposed, however unlike DNA methylation, no studies have been conducted to determine if these changes are also found in offspring.
5.1c Gametes and the effects of alcohol
Alcohol exposure is detrimental to mammalian reproduction in both males and females. The oocyte is sensitive to the effects of alcohol; parthenogenic activation occurs in oocytes bathed in alcohol through intracellular oscillations of free calcium ions (Cuthbertson, 1983). Thus, in female animals alcohol use affects puberty, disrupts menstrual cycling, and alters reproductive function (Emanuele et al., 2002). In addition, chronic alcohol consumption has detrimental effects on male reproductive hormones and sperm quality (Muthusami and Chinnaswamy, 2005). While a direct effect of alcohol on gametic gene expression has not been found, direct or indirect exposure of the gametes to alcohol does decrease fertility. Therefore, it is confirmed that gametes are receptive to the effects of alcohol exposure.
5.2 Opioids
Opioids such as morphine and heroin are powerful analgesics that act on membrane bound Gi-protein coupled receptors; mu-, kappa- and delta. Opioid receptors are distributed throughout the central nervous system and mediate both the rewarding and analgesic properties of this drug class. Genetic vulnerability for opiate abuse is between 43% and 60% (Ho et al., 2010). The most thoroughly studied genetic association being the A118G single nucleotide polymorphism in the mu opioid receptor gene, OPRM1. A complex relationship has been characterized between the A118G polymorphism and the prevalence of opioid abuse in patients (Dick and Agrawal, 2008). However, genetics does not completely explain the inheritance of altered phenotypes in several generations of offspring following initial drug exposure. Recent work has shown that altered morphine response is transmittable to offspring of morphine exposed parents (Byrnes, 2005; Byrnes et al., 2013). Physiological and structural changes in the brains of offspring of morphine-exposed parents have also been characterized (Byrnes et al., 2013; Cicero et al., 1991; Sarkaki et al., 2008; Vyssotski, 2011) (Table 1).
5.2a Behavioral, neurochemical, and structural changes in offspring of opioid-exposed parents
Opioid exposure in F0 parents changes offspring response to drug administration (Byrnes, 2005; Byrnes et al., 2013). Female F0 rats exposed to morphine during adolescence give birth to offspring with altered development of morphine and dopamine-mediated locomotor sensitization. Both male and female F1 offspring from female F0 rats exposed to morphine prior to pregnancy have significantly increased locomotor activity in response to morphine (Byrnes, 2005). In addition, male F1 offspring show attenuated locomotor sensitization to the dopamine receptor 1 and 2 (D1/D2) agonist quinpirole (Byrnes et al., 2013). Interestingly, this phenotype is transmitted through the maternal line as F1 female rats derived from F0 morphine exposure also produce F2 male offspring with attenuated locomotor sensitization to the D1/D2 agonist quinpirole (Byrnes et al., 2013)
Neuroadaptations in dopamine signaling may explain why response to drug challenges are different in F1 male and female offspring and male F2 offspring of morphine-exposed F0 mothers. F0 morphine-exposed mothers produce male F1 offspring with increased D2 receptor mRNA expression in the nucleus accumbens (NAc) in response to chronic D1/D2 agonist exposure. The same changes are found in F2 male offspring of F1 females from F0 morphine-exposed mothers (Byrnes et al., 2013). Therefore, dopamine response is altered due to parent drug history and the trajectory of offspring drug response is shaped accordingly due to these changes in dopamine reward circuitry which are implicated in drug addiction (Wise, 1998).
The endogenous opioid system is also altered in F1 offspring of F0 morphine-exposed males and females. Specifically, morphine-exposed fathers produce F1 female offspring with increased beta-endorphin in the hypothalamus and increased circulating serum cortiscosterone levels (Cicero et al., 1991). Similar changes have been found in additional generations. Morphine-exposed F0 mothers produce male F1 and F2 offspring (through the maternal line) with decreased quinpirole induced cortiscosterone (Byrnes et al., 2013). Dopamine and opioid interactions are also altered in offspring of morphine-exposed F0 mothers. Female F0 rats exposed to morphine during adolescence produce male F1 offspring with increased kappa opioid receptor mRNA expression in the NAc following chronic D1/D2 agonist exposure (Byrnes et al., 2013). Thus, multi- and trans-generational effects of morphine exposure can be seen as changes in opioid neuroadaptations in response to D1/D2 agonist exposure in offspring.
Physiological changes in offspring of morphine-exposed parents have also been identified. Alterations in hippocampal synaptic plasticity have been found in the F1 offspring of F0 morphine-exposed males and females mated together (Sarkaki et al., 2008). Synaptic plasticity is essential to the process of learning and memory as well as learned behaviors that include drug abuse (Hyman et al., 2006). Long-term potentiation (LTP) and long-term depression (LTD) reflect increases and decreases in synaptic strength, respectively. Decreased LTP is found in the hippocampus of F1 offspring of F0 morphine treated fathers. In contrast, F0 morphine-exposed mothers produce F1 offspring with decreased LTD in the hippocampus but intact hippocampal LTP (Sarkaki et al., 2008). These findings suggest that morphine mediates physiological changes that result in impaired synaptic plasticity in the hippocampus of F1 offspring.
Finally, transgenerational effects on cellular morphology have been observed following morphine exposure, suggesting parental morphine use can change neuronal structure. Decreased synaptophysin is found in the brains of F2 offspring derived from morphine-exposed F0 male rats (Vyssotski, 2011). As synaptophysin expression is associated with the density of nerve terminals, decreased synaptic connections exist in the F2 male brains of F0 morphine-exposed fathers.
Together these studies demonstrate that morphine exposure can impact behavior, biochemical signaling, synaptic plasticity, and neuronal structure across multiple generations. While behavioral and biochemical effects are observed in F1 generations, extension of modifications in cellular morphology out to the F2 offspring indicates a transgenerational inheritance of morphine-exposure.
5.2b Opioid exposure and changes to the epigenome
To date no definitive modification to the epigenome has been identified that could explain transgenerational inheritance of morphine exposure, however there have been reports of epigenetic modifications following morphine exposure within a single generation. For example, histone modifications have been measured in the NAc and other brain regions following exposure to morphine in animals (Maze and Nestler, 2011; Sun et al., 2012). Morphine reward and sensitization in mice following chronic morphine administration has been correlated with global levels of histone 3 lysine 9 dimethylation (H3K9me2) in the NAc (Sun et al., 2012). Because H3k9me2 is typically associated with repression of transcription, decreased H3k9me2 results in increased gene expression (Rice and Allis, 2001), and this gene expression may mediate the rewarding effects of morphine.
Histone methylation of long-interspersed nuclear (LINE-1) retrotransposons is decreased in the NAc of mice following direct injection of morphine to the area (Sun et al., 2012). LINE-1 elements are repetitive DNA sequences that make up a large portion of genomic DNA (~20%) (Prak and Kazazian, 2000). The mobilization, integration, and regulation of these elements have been characterized in several brain regions (Singer et al., 2010) and alterations in LINE-1 expression through epigenetic mechanisms have been implicated in drug addiction within the user (Maze et al., 2011; Prak and Kazazian, 2000). Interestingly, transgene mobilization and expression through epigenomic release from silencing were some of the first epigenetic changes to be implicated in transgenerational inheritance (Daxinger and Whitelaw, 2012). While LINE-1 expression is associated with morphine use in somatic cells, it is not known if LINE-1 expression is altered in the germ cells of F0 morphine-exposed parents and if this is maintained in the brains and gametes of F1 offspring or future generations.
Histone acetylation is also altered in response to morphine exposure. In rats, exposure to morphine increases acetylation of histone H3 lysine 14 (aceH3K14) in the NAc (Sheng et al., 2011) and basolateral amygdala (Wang et al., 2014b), a region critically involved in stimulus-reward associations (Everitt et al., 1999). Histone acetylation is associated with transcriptional activation (see Box 1). While these modifications have been found within a generation, there is no evidence of altered histone acetylation in offspring following parental morphine exposure.
In addition to methylation and acetylation modifications of histones, morphine exposure induces methylation changes on DNA directly (Nielsen et al., 2008; Chorbov et al., 2011; Doehring et al., 2013; Trivedi et al., 2014). Morphine modulates cellular oxidative stress and methyl group donation through the antioxidant glutathione and the methyl donor SAM, respectively (Trivedi et al., 2014). Consequently, as the redox state of the cell impacts methylation events, DNA methylation changes can occur following morphine exposure. Just as LINE-1 seems to be vulnerable to histone methylation changes following morphine exposure, the transposable element exhibits decreases in methylation after morphine administration. Hypomethylation at LINE-1 retrotransposons has been identified in leukocytes of chronic heroin users (Doehring et al., 2013) and in neuronal cell lines (Trivedi et al., 2014); consequently, LINE-1 mRNA expression increases (Trivedi et al., 2014). In contrast, increased CpG methylation has been reported in lymphocytes, blood, and sperm of chronic heroin users in the promoter region of the OPRM1 gene (Chorbov et al., 2011; Nielsen et al., 2008). Thus, morphine administration can lead to both increased and decreased DNA methylation in a gene-dependent manner in the individual user. To date no studies have determined if these chemical modifications are inherited by offspring.
5.2c Gametes and opioid receptors
Localization of the mu opioid receptor as well as endogenous expression of beta-endorphin in the male reproductive tract suggests that paternal gametes are receptive to endogenous and exogenous opioids (Albrizio et al., 2006). Mu-, kappa- and delta opioid receptors are present in oocytes, most likely for mediating oocyte maturation (Agirregoitia et al., 2012). Thus the presence of opioid receptors on gametes not only maintains proper function and development, but may mediate transgenerational inheritance.
5.3 Cocaine
Cocaine increases dopamine by binding to the dopamine transporter and blocking the reuptake of dopamine into the presynaptic cell. The resulting increase in dopamine in the NAc is associated with the rewarding effects of cocaine (Wise, 1998). Cocaine use heritability is 44% based on Vietnam Era Twin Registry studies (Tsuang et al., 1996). Inter-individual differences such as baseline dopamine signaling (Volkow et al., 1999) and allelic variation in genes including dynorphin and dopamine beta-hydroxylase (DBH) (LaForge et al., 2000) have been correlated with risk of cocaine use and abuse. However, as genetics accounts for less than 50% of heritability for cocaine use, other factors likely contribute to cocaine use and abuse across generations. Current research has identified multigenerational effects on behavior, neurochemical signaling (Table 1), and the epigenome following cocaine administration in animal studies.
5.3a Behavioral and neurochemical changes in offspring of cocaine-exposed parents
In utero exposure to cocaine is well-studied; in animals cocaine exposed offspring show behavioral, biochemical, and structural alterations in the brain (Vassoler et al., 2014). While, there are currently no reports on the transgenerational inheritance of cocaine exposure there is evidence of multigenerational effects.
Studies employing F0 parental cocaine exposure have found changes in behavior in F1 male and female offspring. For example, F1 male offspring of F0 cocaine-exposed fathers find cocaine less rewarding; male rats acquire self-administration more slowly and administer less drug, while F1 female rats are not affected (Vassoler et al., 2013b). In addition, male F1 offspring of F0 mothers exposed to cocaine prior to pregnancy are more sensitive to the psychomotor effects of cocaine administration (Sasaki et al., 2014). However, contrasting results have been reported for behaviors that may influence drug abuse in offspring derived from paternal cocaine exposure; some have found learning deficits in female F1 offspring of F0 fathers exposed to cocaine (He et al., 2006) while others report no learning effects in either male or female F1 offspring of F0 cocaine-exposed fathers (Killinger et al., 2012). While these results demonstrate some multigenerational effects of cocaine exposure, examination of these phenotypes in the F2 generation will determine transgenerational inheritance.
In addition to changes in behavior, alterations in neurotransmitter systems have been found in F1 offspring of F0 cocaine-exposed mothers. Specifically, F0 cocaine-exposed mothers produce F1 male offspring with increased D1 receptor expression in the medial prefrontal cortex (mPFC) (Sasaki et al., 2014). An increase in the number of dopamine receptors in the mPFC could cause enhanced cocaine response due to the ability of these receptors to activate the mesocortical dopamine pathway and activity at the D1 receptor mediates the psychomotor response to cocaine in rodents (Steketee and Kalivas, 2011; Xu et al., 1994).
5.3b Cocaine exposure and changes to the epigenome
Multigenerational changes in histone 3 acetylation (AcH3), commonly associated with chromatin remodeling and increased transcription of target genes has been identified in response to cocaine exposure. F0 fathers that self-administer cocaine have increased AcH3 in the testes and greater AcH3 association with exon IV of BDNF in sperm (Vassoler et al., 2013b). F1 offspring of these fathers also show increased AcH3 association with exon IV of BDNF and increased transcription of BDNF in the mPFC (Vassoler et al., 2013b). BDNF and its modulation through chromatin remodeling has been implicated in both chronic and generational cocaine exposure (Kumar et al., 2005; Sasaki et al., 2014; Vassoler et al., 2013b). Functional antagonism of increased BDNF in the F1 male offspring of F0 cocaine-exposed fathers increases cocaine self-administration to that of control animals (Vassoler et al., 2013b). Thus epigenetic changes can occur across generations and suggest that chromatin remodeling in response to drug use may be an important mechanism of inheritance and cocaine response. Additional evidence supports that drug reinforcement may converge on a common epigenetic pathway. Like morphine (Sun et al., 2012), direct cocaine exposure in mice reduces H3K9me2 in the NAc and these changes mediate cocaine preference and cellular plasticity (Maze et al., 2010). However, multigenerational or transgenerational inheritance of this modification has not been examined.
Epigenomic machinery, specifically methyl-CpG binding protein (MeCP2) and DNA methyltransferase 3a (DNMT3a), are altered within a generation following exposure to cocaine. Increased expression of MeCP2 is found after cocaine self-administration (Host et al., 2011); MeCP2 acts as a repressor or activator of transcription when it binds to the promoter region of a target gene (Chahrour et al., 2008). MeCP2 is involved in learning and memory and therefore may be implicated in drug abuse behaviors (Feng and Nestler, 2010). Cocaine stimulation of MeCP2 also represses the transcription of two small non-coding RNAs, miR-212 and miR-312 (Im et al., 2010). Decreased transcription of these miRNAs releases BDNF from miRNA transcriptional repression resulting in increased BDNF (Im et al., 2010) which has been shown to mediate cocaine use (Sadri-Vakili et al., 2010).
DNA methylation can be altered following cocaine exposure through regulation of DNA methyltransferase 3a (DNMT3a) (LaPlant et al., 2010). While, DNMT3a regulates cocaine response it is also important in modulating de novo methylation of differentially methylated regions of paternal imprinted genes and some repetitive elements in male germ cell development. For example, DNMT3a is responsible for methylation of both the imprinted gene H19 and the repetitive element LINE-1 (Kato et al., 2007). Similar to morphine, cocaine alters levels of the antioxidant glutathione that can lead to impaired methylation of the genome (Lee et al., 2001). Thus, cocaine mediates changes in the activity of molecules responsible for the methylation status of DNA. However, it remains to be determined if these changes are found in F1 and subsequent generations of offspring.
Although little has been published on transgenerational changes in DNA methylation following cocaine exposure, DNA methylation is altered in F1 offspring with in utero exposure to cocaine. F0 mothers exposed to cocaine during pregnancy produce F1 offspring with altered global patterns of hippocampal DNA methylation. These changes seem to be functionally significant as they are accompanied by corresponding alterations in gene transcription (Novikova et al., 2008).
5.3c The effects of cocaine on gametes
Testes contain high affinity binding sites for cocaine (Sweet and Murdock, 1987), exceeded in number only by the brain (Mulé and Misra, 1977). In addition, the presence of DAT protein in sperm has been characterized and dopamine signaling is implicated in sperm viability, mobility, and capacitation (Ramírez et al., 2009). Interestingly, cocaine remains bound to human sperm at traceable levels for up to 24 hours, and there is history of concern for cocaine transmission via “piggybacking” on sperm to the oocyte (Cone et al., 1996; Yazigi et al., 1991). Although cocaine presence in early embryonic development is known to be detrimental, the more likely process of transmission of phenotypes associated with cocaine exposure from F0 father to F1 offspring is through the epigenetic mechanisms previously discussed. Thus, changes in DNA methylation, and chromatin state in sperm could be an important mechanism for transgenerational inheritance of behavioral consequences of cocaine exposure. To date, it is unclear if oocytes have binding sites for cocaine and therefore, any maternally-derived effects of cocaine exposure may be due to indirect mechanisms within the oocyte.
5.4 Marijuana
The active component in marijuana, Δ9-tetrahydrocannabinol (Δ9-THC), is responsible for the drug’s biological effects. Δ9-THC binds to and activates Gi-coupled cannabinoid type 1 and 2 (CB1 and CB2) receptors throughout the central nervous system. A meta-analysis of cannabis initiation and problematic cannabis use among twins suggest that the heritability of cannabis initiation is 40–48% and chronic problematic use is 51–59% depending on gender (Verweij et al., 2010). However, higher genetic associations for cannabis use have been found in female twin studies (Kendler and Prescott, 1998).
Polymorphic variation in the gene that codes for cannabinoid receptor 1 (CNR1) has been implicated in marijuana craving during times of abstinence and withdrawal (Demers et al., 2014). In addition, polymorphic variation in CYP2C9, a member of the cytochrome family of enzymes responsible for the metabolism of Δ9-THC, results in greater active metabolite and heightened sensitivity to the sedative effects of marijuana (Sachse-Seeboth et al., 2008). More recently clinical studies suggest an interaction between genetics and epigenetics that alters response to marijuana use. In adolescents homozygous for the catechol-o-methyltransferase (COMT) Val108/158 Met polymorphism, methylation at the promoter of this gene is associated with lower risk of marijuana use (van der Knaap et al., 2014). Greater methylation at the promoter of COMT reduces expression of the enzyme. In addition the homozygous Met/Met genotype is associated with decreased enzyme activity. Together these changes result in decreased degradation of COMT substrates such as dopamine. Therefore, individuals with hypermethylation and the polymorphism have increased dopamine levels. Conversely, low brain dopamine results in an attenuated reward system that manifests into anhedonia. It is hypothesized that substance use is an attempt to alleviate this unfavorable anhedonic state (Markou and Koob, 1991; Volkow et al., 2010). Hence, Met/Met individuals with hypermethylation may be less vulnerable to substance abuse due to increases in dopamine. While genetic interactions explain some of the risk for marijuana use, other factors must also affect administration in individuals. Currently, multigenerational effects of cannabinoid exposure have been identified (Table 1).
5.4a Behavioral and neurochemical changes in offspring of cannabinoid-exposed parents
Endocannabinoid and opioid systems closely interact as cannabinoid exposure can cause cross-sensitization to the effects of opioids in dopaminergic neurons (Pistis et al., 2004). To date, studies have only examined the consequence of cannabinoid exposure on morphine response in offspring. Response to morphine administration is altered in F1 offspring of mothers exposed to the CB1/CB2 receptor agonist WIN 55, 212-2. Specifically, F0 mothers exposed to WIN 55, 212-2 during adolescence produce male offspring with increased morphine conditioned place preference (Byrnes et al., 2012) and female offspring with increased morphine locomotor sensitization (Vassoler et al., 2013a). Therefore, both drug response and reinforcement is altered in the F1 offspring of WIN 55, 212-2-exposed mothers. In addition, male and female rats exposed to Δ9-THC and then mated together produce F1 offspring that show increased motivation to self-administer heroin as well as enhanced withdrawal behaviors during periods of opioid withdrawal (Szutorisz et al., 2014). Therefore, while cross-sensitization between cannabinoids and opioids has been identified in the individual user (Cadoni et al., 2001), the studies discussed above are the first examples of cross-sensitization of cannabinoids and opioids across a generation for reinforcement, drug response, and self-administration. These results suggest multigenerational effects of cannabinoid use; altered response to morphine and heroin occurred exposure to CB1/CB2 agonist or Δ9-THC. However, no data is available for these phenotypes in the F2 generation. In addition, no studies have examined the effects in F1 or F2 offspring of F0 fathers exposed to cannabinoids.
Multigenerational alterations in gene expression and neurotransmission have been found following exposure to cannabinoids. F0 females exposed to CB1 agonist produce F1 female offspring with increased Oprm1 expression in the NAc (Vassoler et al., 2013a). Male and female rats exposed to Δ9-THC and then mated together produce offspring with decreased expression of cannabinoid, dopamine, and glutamate receptors, reduced NMDA binding and enhanced LTD in the dorsal striatum (Szutorisz et al., 2014). Further analysis of these systems is necessary to determine if any of these phenotypes are inherited in additional generations.
5.4b Cannabinoid exposure and changes to the epigenome
There is currently no evidence of multi- or transgenerational epigenetic changes following marijuana exposure. However, activation of the immediate early gene FosB occurs following direct Δ9-THC administration (Fürst et al., 2013; Porcella et al., 1998). Cocaine activation of FosB has been correlated to increased histone 4 acetylation in the promoter region of the gene (Kumar et al., 2005) and Δ9-THC may mediate similar chromatin remodeling and gene activation. Indeed, Δ9-THC causes genome wide changes in histone methylation (Yang et al., 2014) although other histone modifications, including acetylation, have not been thoroughly explored. Changes in DNA methylation have not been examined following exposure to marijuana.
5.4c Gametes and cannabinoid receptors
CB1 receptors and the endogenous endocannabinoid anandamide are expressed in the ovaries and uterine endometrium (Bari, 2011). Endocannabinoid signaling has been implicated in oocyte maturation, ovulation, and fertility signals (Battista et al., 2012). In addition, cannabinoid receptor localization has been characterized in testes (Schuel et al., 2002) and sperm (Aquila et al., 2010). Furthermore, anandamide analog administration alters the PI3K/Akt cell survival pathway in sperm (Aquila et al., 2010). Further research is required to identify the direct or indirect epigenetic changes caused by marijuana administration in gametes.
5.5 Nicotine
Nicotine is the active ingredient in tobacco and is responsible for the positive experience associated with tobacco use. Nicotine binds to nicotinic acetylcholine receptors (nAChRs) which are pentameric ligand-gated ion channels (Benowitz, 1999). The relative contribution of genetic influence is 44% for smoking initiation and 75% for nicotine dependence (Vink et al., 2005). Work from human GWAS as well as animal research identified several neurotransmitter systems that may play a role in the likeliness of nicotine use and dependence. Genetic variation in dopamine receptor 2 and 4 (DRD2 and DRD4) have been associated with smoking behavior. In addition, genetic variation in the dopamine transporter gene, SLC6A3, has been associated with smoking behavior and is also known to interact in an allele-specific epistatic manner with DRD2 allele expression. Allelic variation in genes that regulate norepinephrine (DBH and MAOA) and serotonin (5HTTLPR and TPH) have also been implicated in nicotine use (reviewed in Lerman and Berrettini, 2003). In addition, polymorphic variation in the gene that codes for choline acetyltransferase (ChAT) (Ray et al., 2010) as well as variation in the primary enzyme that metabolizes nicotine CYP2A6 and gene clusters coding for nicotinic acetylcholine receptor (nAChR) subunits (Demers et al., 2014) have been associated with sensitivity to nicotine, tobacco use, and success for smoking cessation. However, association studies have suggested that genetic predisposition accounts for less than half of the likelihood of smoking initiation and three-fourths of nicotine dependence (Vink et al., 2005). Therefore, environmental influence, including epigenetic modifications, may account for remaining risk. Recent evidence suggests the transgenerational inheritance of behavior and multigenerational neurochemical changes following parental nicotine exposure (Table 1).
5.5a Behavioral and neurochemical changes in offspring of nicotine-exposed parents
While no studies have examined drug response and reward behavior in F1 or F2 offspring from F0 nicotine-exposed parents, there is evidence that nicotine exposure can cause multigenerational changes in cognition and dopamine in offspring. Zhu and colleagues found that F1 male and female offspring of F0 mothers exposed to nicotine during pregnancy are hyperactive with decreased attention. In addition, this phenotype is transmitted to F2 offspring and F3 offspring through the maternal line (Zhu et al., 2014). Hyperactivity in the F2 offspring is attenuated by methylphenidate-induced dopamine increase (Zhu et al., 2014), thereby implicating a hypodopaminergic state as the mechanism by which nicotine alters offspring response. Thus, F0 maternal exposure to nicotine during pregnancy produces transgenerational changes in behavior and multigenerational changes in dopamine signaling (Zhu et al., 2014). Studies have examined additional neurochemical changes following exposure to nicotine, however, these are only found in offspring of F0 mothers exposed to nicotine during pregnancy (Zhu et al., 2012, Yochum et al., 2014). To date, no neurochemical changes have been identified in animals not directly exposed to nicotine.
5.5b Nicotine exposure and changes to the epigenome
The epigenome is vulnerable to modification by nicotine exposure. Global DNA methylation patterns in leukocytes are similar between non-smoking offspring and non-smoking fathers, while there are differences in methylation if offspring are smokers and fathers are non-smokers (Hillemacher et al., 2008). In addition, nicotine-induced variation in DNA methylation has been identified in several genes implicated in drug abuse. For example, nicotine exposure is associated with DNA methylation changes in the gene coding for monoamine oxidase A (MAOA), a key enzyme in the metabolism of dopamine and other monoamines. Decreased methylation of the MAOA promoter is found in the blood and lymphoblasts of current smokers (Philibert et al., 2010). This modification may be mechanistically important, as it has been proposed by Zhu and colleagues that nicotine induced hypomethylation of MAOA decreases dopamine synthesis in animals exposed to nicotine in utero (Philibert et al., 2010; Zhu et al., 2012). Chronic nicotine administration produces an increase in expression of DNMT1, resulting in methylation of glutamate decarboxylase (GAD67), and reduced GAD67 mRNA (Satta et al., 2008). Consequently, changes in GAD67 mRNA can influence cortical GABAergic signaling. In addition, evidence that methylation and expression of GAD67, a gene implicated in the phenotype of patients with schizophrenia, is modulated by chronic nicotine administration suggests that the co-morbidity that exists between the disorder and smoking addiction may have an epigenetic basis. Interestingly, DNA methylation has been associated with multigenerational exposure to nicotine. F0 mothers exposed to nicotine during pregnancy produce F1 offspring with increased methylation at BDNF in blood (Toledo Rodriguez et al., 2010) and decreased BDNF mRNA and protein in the frontal cortex (Yochum et al., 2014). As discussed previously, decreases in levels/activity of BDNF has been implicated in the self-administration of drugs including cocaine (Sadri-Vakili et al., 2010), as well as methylphenidate (Cadet et al., 2014) and alcohol (Jeanblanc et al., 2012).
In addition to changes in DNA methylation, nicotine can remodel chromatin through histone modifications. Following nicotine administration in mice there is an increase in the acetylation of histone H3 in the striatum (Levine et al., 2011) and a decrease in H3K9me2 at promoter regions of target genes (Chase and Sharma, 2013). Taken together, these studies suggest that nicotine reduces epigenetic histone marks that promote a restrictive genomic state, thereby opening normally repressed genes to enhanced transcription.
5.5c Gametes and nicotinic receptors
Nicotine mediated increase in the production of cholesterol, triglycerides, phospholipids and free fatty acids in the testes can be blocked by administration of the nAChR antagonist mecamylamine suggesting binding sites for both drugs in the testes (Kavitharaj and Vijayammal, 1998). Indeed, the mRNA of subunits that compose the pentameric nAChR (i.e. α7, α9, α3, α5, and β4) are expressed in sperm (Kumar and Meizel, 2005). Furthermore, functional nAChRs have been found in sperm (Kumar and Meizel, 2005). Acetylcholine mediated spikes in calcium through α7 homomeric receptors occurs in the sperm head during the acrosomal reaction (Bray et al., 2005). Finally, nicotine, likely acting on nAChRs has detrimental effects on sperm viability. Maternal smoking has been linked to male offspring infertility through gonadal toxicity (Ratcliffe et al., 1992). While it is well-established that nicotine exposure produces reproductive challenges in females and the early developing embryo (Omotoso et al., 2013), nicotine binding has not been identified in oocytes therefore inheritance through the maternal line may be through indirect effects of nicotine exposure.
6 Conclusion and Perspectives
Although the idea that parental experience can influence behavior in offspring is not new, an understanding of drug-mediated changes to the epigenome may provide an avenue to identify mechanisms of transgenerational inheritance in drug addiction. While stress (Franklin et al., 2010) and diet (McGowan et al., 2008) have long been examined as environmental challenges that impact the behavior and physiology of future offspring, drug abuse remains to be thoroughly examined for the same features. Several instances of multigenerational inheritance has been reported across drug types, however there is a deficit of studies that examine transgenerational inheritance of phenotypes following F0 exposure to drugs of abuse (Table 1). To date, transgenerational phenotypes have only been reported following parental exposure to morphine (Brynes et al., 2013; Vyssotski, 2011), alcohol (Govorko et al., 2012), and nicotine (Zhu et al., 2014).
This review has examined changes in behavior, neurochemical signaling, and cellular structure found in the F1 and F2 offspring of drug-exposed parents. While some studies have implicated epigenetic inheritance of these phenotypes, fewer have identified epigenetic transgenerational inheritance. In addition, alterations to the epigenome in response to drug-exposure have been discussed but no evidence of a specific mechanism for transgenerational epigenetic inheritance of drug exposure has been determined. While there are some common players that are epigenetically modified following drug exposure, such as BDNF and DNMT1, a complex epigenomic network may be uniquely mediated by different classes of drugs of abuse. Identifying novel changes made to the epigenome in response to drug helps to select candidate genes to examine in future generations. The inheritance of these changes can then be identified and characterization of the functional relevance of epigenetic changes in offspring can be determined.
In translating research, drug exposure and epigenetic regulation of the genome becomes quite complex. Animal studies allow for careful control over the environment and drug exposure variables. Thus, such studies can provide the best evidence of the existence of multigenerational and transgenerational inheritance of drug abuse and the possible epigenetic mechanisms that may underlie these phenotypes. Thus, future studies aimed at identification of functional epigenetic modifications following drug in the somatic and germ cells of F0, F1, and F2 animals after F0 drug exposure are critical.
The scope of current work in transgenerational drug inheritance should focus on extending observations of behavioral phenotypes to F2 and F3 generations and molecular analysis to include brains of offspring and gametes of parents. With advancements in techniques to evaluate epigenetic changes to the genome the field of drug abuse and inheritance is promising. Future work may start to identify the vulnerability of individuals to epigenomic inheritance of drug abuse and provide evidence for mechanisms of genomic regulation through drug exposure.
Acknowledgments
This work was supported by the National Institutes of Health National Institute on Drug Abuse [Grants T32-DA28874 and R01-DA033646].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Nicole L. Yohn, Department of Systems Pharmacology and Translational Therapeutics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA
Marisa S. Bartolomei, Department of Cell and Developmental Biology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA
Julie A. Blendy, Department of Systems Pharmacology and Translational Therapeutics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA
References
- Abel EL. Paternal alcohol exposure and hyperactivity in rat offspring: Effects of amphetamine. Neurotoxicol Teratol. 1993;15:445–449. doi: 10.1016/0892-0362(93)90063-t. [DOI] [PubMed] [Google Scholar]
- Abel EL. Effects of Physostigmine on Male Offspring Sired by Alcohol-Treated Fathers. Alcoholism: Clinical and Experimental Research. 1994;18:648–652. doi: 10.1111/j.1530-0277.1994.tb00925.x. [DOI] [PubMed] [Google Scholar]
- Abel EL, Lee JA. Paternal Alcohol Exposure Affects Offspring Behavior but not Body or Organ Weights in Mice. Alcoholism: Clinical and Experimental Research. 1988;12:349–355. doi: 10.1111/j.1530-0277.1988.tb00205.x. [DOI] [PubMed] [Google Scholar]
- Abel EL, Tan SE. Effects of paternal alcohol consumption on pregnancy outcome in rats. Neurotoxicol Teratol. 1988;10:187–192. doi: 10.1016/0892-0362(88)90016-5. [DOI] [PubMed] [Google Scholar]
- Agirregoitia E, Peralta L, Mendoza R, Expósito A, Ereño ED, Matorras R, Agirregoitia N. Expression and localization of opioid receptors during the maturation of human oocytes. Reprod Biomed Online. 2012;24:550–557. doi: 10.1016/j.rbmo.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Lynskey MT. The genetic epidemiology of cannabis use, abuse and dependence. Addiction. 2006;101:801–812. doi: 10.1111/j.1360-0443.2006.01399.x. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Lynskey MT. Are there genetic influences on addiction: evidence from family, adoption and twin studies. Addiction. 2008;103:1069–1081. doi: 10.1111/j.1360-0443.2008.02213.x. [DOI] [PubMed] [Google Scholar]
- Albrizio M, Guaricci AC, Calamita G, Zarrilli A, Minoia P. Expression and immunolocalization of the mu-opioid receptor in human sperm cells. Fertility and Sterility. 2006;86:1776–1779. doi: 10.1016/j.fertnstert.2006.04.037. [DOI] [PubMed] [Google Scholar]
- Aquila S, Guido C, Santoro A, Perrotta I, Laezza C, Bifulco M, Sebastiano A. Human sperm anatomy: ultrastructural localization of the cannabinoid1 receptor and a potential role of anandamide in sperm survival and acrosome reaction. Anat Rec (Hoboken) 2010;293:298–309. doi: 10.1002/ar.21042. [DOI] [PubMed] [Google Scholar]
- Bari M. The manifold actions of endocannabinoids on female and male reproductive events. Front Biosci. 2011;16:498. doi: 10.2741/3701. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS. Genomic imprinting: employing and avoiding epigenetic processes. Genes & Development. 2009;23:2124–2133. doi: 10.1101/gad.1841409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battista N, Meccariello R, Cobellis G, Fasano S, Di Tommaso M, Pirazzi V, Konje JC, Pierantoni R, Maccarrone M. The role of endocannabinoids in gonadal function and fertility along the evolutionary axis. Molecular and Cellular Endocrinology. 2012;355:1–14. doi: 10.1016/j.mce.2012.01.014. [DOI] [PubMed] [Google Scholar]
- Bench GS, Friz AM, Corzett MH, Morse DH, Balhorn R. DNA and total protamine masses in individual sperm from fertile mammalian subjects. Cytometry. 1996;23:263–271. doi: 10.1002/(SICI)1097-0320(19960401)23:4<263::AID-CYTO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Benowitz NL. Nicotine Addiction. Primary Care: Clinics in Office Practice. 1999;26:611–631. doi: 10.1016/s0095-4543(05)70120-2. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & Development. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Bray C, Son J-H, Kumar P, Meizel S. Mice deficient in CHRNA7, a subunit of the nicotinic acetylcholine receptor, produce sperm with impaired motility. Biol Reprod. 2005;73:807–814. doi: 10.1095/biolreprod.105.042184. [DOI] [PubMed] [Google Scholar]
- Byrnes EM. Transgenerational consequences of adolescent morphine exposure in female rats: effects on anxiety-like behaviors and morphine sensitization in adult offspring. Psychopharmacology. 2005;182:537–544. doi: 10.1007/s00213-005-0122-4. [DOI] [PubMed] [Google Scholar]
- Byrnes JJ, Johnson NL, Schenk ME, Byrnes EM. Cannabinoid exposure in adolescent female rats induces transgenerational effects on morphine conditioned place preference in male offspring. Journal of Psychopharmacology. 2012;26:1348–1354. doi: 10.1177/0269881112443745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes JJ, Johnson NL, Carini LM, Byrnes EM. Multigenerational effects of adolescent morphine exposure on dopamine D2 receptor function. Psychopharmacology. 2013;227:263–272. doi: 10.1007/s00213-012-2960-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet JL, Brannock C, Jayanthi S, Krasnova IN. Transcriptional and Epigenetic Substrates of Methamphetamine Addiction and Withdrawal: Evidence from a Long-Access Self-Administration Model in the Rat. Mol Neurobiol. 2014:1–22. doi: 10.1007/s12035-014-8776-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadoni C, Pisanu A, Solinas M, Acquas E, Chiara G. Behavioural sensitization after repeated exposure to Δ9-tetrahydrocannabinol and cross-sensitization with morphine. Psychopharmacology. 2001;158:259–266. doi: 10.1007/s002130100875. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong STC, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase KA, Sharma RP. Nicotine induces chromatin remodelling through decreases in the methyltransferases GLP, G9a, Setdb1 and levels of H3K9me2. Int J Neuropsychopharmacol. 2013;16:1129–1138. doi: 10.1017/S1461145712001101. [DOI] [PubMed] [Google Scholar]
- Chorbov VM, Todorov AA, Lynskey MT, Cicero TJ. Elevated levels of DNA methylation at the OPRM1 promoter in blood and sperm from male opioid addicts. J Opioid Manag. 2011;7:258–264. doi: 10.5055/jom.2011.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicero TJ, Adams ML, Giordano A, Miller BT, O’connor L, Nock B. Influence of morphine exposure during adolescence on the sexual maturation of male rats and the development of their offspring. J Pharmacol Exp Ther. 1991;256:1086–1093. [PubMed] [Google Scholar]
- Cicero TJ, Adams ML, O’connor L, Nock B, Meyer ER, Wozniak D. Influence of chronic alcohol administration on representative indices of puberty and sexual maturation in male rats and the development of their progeny. J Pharmacol Exp Ther. 1990;255:707–715. [PubMed] [Google Scholar]
- Cone EJ, Kato K, Hillsgrove M. Cocaine Excretion in the Semen of Drug Users. J Anal Toxicol. 1996;20:139–140. doi: 10.1093/jat/20.2.139. [DOI] [PubMed] [Google Scholar]
- Cuthbertson KS. Parthenogenetic activation of mouse oocytes in vitro with ethanol and benzyl alcohol. J Exp Zool. 1983;226:311–314. doi: 10.1002/jez.1402260217. [DOI] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nature Reviews Genetics. 2012;13:153–162. doi: 10.1038/nrg3188. [DOI] [PubMed] [Google Scholar]
- Demers CH, Bogdan R, Agrawal A. The Genetics, Neurogenetics and Pharmacogenetics of Addiction. Curr Behav Neurosci Rep. 2014;1:33–44. doi: 10.1007/s40473-013-0004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Agrawal A. The Genetics of Alcohol and Other Drug Dependence. Alcohol Res Health. 2008;31:111–118. [PMC free article] [PubMed] [Google Scholar]
- Doehring A, Oertel BG, Sittl R, Lötsch J. Chronic opioid use is associated with increased DNA methylation correlating with increased clinical pain. Pain. 2013;154:15–23. doi: 10.1016/j.pain.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Emanuele MA, Wezeman F, Emanuele NV. Alcohol’s effects on female reproductive function. Alcohol Research and Health. 2002 [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Goldman D. Genetics of alcoholism and substance abuse. Psychiatric Clinics of North America. 1999 doi: 10.1016/s0193-953x(05)70077-0. [DOI] [PubMed] [Google Scholar]
- Everitt BJ, Parkinson JA, Olmstead MC, Arroyo M, Robledo P, Robbins TW. Associative processes in addiction and reward. The role of amygdala-ventral striatal subsystems. Annals of the New York Academy of Sciences. 1999;877:412–438. doi: 10.1111/j.1749-6632.1999.tb09280.x. [DOI] [PubMed] [Google Scholar]
- Feng J, Nestler EJ. MeCP2 and drug addiction. Nat Neurosci. 2010;13:1039–1041. doi: 10.1038/nn0910-1039. [DOI] [PubMed] [Google Scholar]
- Finegersh A, Homanics GE. Paternal alcohol exposure reduces alcohol drinking and increases behavioral sensitivity to alcohol selectively in male offspring. PLoS ONE. 2014;9:e99078. doi: 10.1371/journal.pone.0099078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin TB, Russig H, Weiss IC, Gräff J, Linder N, Michalon A, Vizi S, Mansuy IM. Epigenetic Transmission of the Impact of Early Stress Across Generations. Biological Psychiatry. 2010;68:408–415. doi: 10.1016/j.biopsych.2010.05.036. [DOI] [PubMed] [Google Scholar]
- Friedler G. Paternal exposures: impact on reproductive and developmental outcome. An overview Pharmacol Biochem Behav. 1996;55:691–700. doi: 10.1016/s0091-3057(96)00286-9. [DOI] [PubMed] [Google Scholar]
- Fürst Z, Riba P, Al-Khrasani M. New approach to the neurobiological mechanisms of addiction. Neuropsychopharmacol Hung. 2013;15:189–205. [PubMed] [Google Scholar]
- Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E, Mansuy IM. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci. 2014;17:667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatewood JM, Cook GR, Balhorn R, Bradbury EM, Schmid CW. Sequence-specific packaging of DNA in human sperm chromatin. Science. 1987;236:962–964. doi: 10.1126/science.3576213. [DOI] [PubMed] [Google Scholar]
- Gaydos LJ, Wang W, Strome S. Gene repression. H3K27me and PRC2 transmit a memory of repression across generations and during development. Science. 2014;345:1515–1518. doi: 10.1126/science.1255023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi A, Krishnan HR, Lew L, Prado FJ, III, Ong DS, Atkinson NS. Alcohol-Induced Histone Acetylation Reveals a Gene Network Involved in Alcohol Tolerance. PLoS Genet. 2013;9:e1003986. doi: 10.1371/journal.pgen.1003986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nature Reviews Genetics. 2005;6:521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- Govorko D, Bekdash RA, Zhang C, Sarkar DK. Male germline transmits fetal alcohol adverse effect on hypothalamic proopiomelanocortin gene across generations. Biological Psychiatry. 2012;72:378–388. doi: 10.1016/j.biopsych.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud S, Liu L, Carrell DT. Protamine ratio and the level of histone retention in sperm selected from a density gradient preparation. Andrologia. 2009;41:88–94. doi: 10.1111/j.1439-0272.2008.00890.x. [DOI] [PubMed] [Google Scholar]
- He F, Lidow IA, Lidow MS. Consequences of paternal cocaine exposure in mice. Neurotoxicol Teratol. 2006;28:198–209. doi: 10.1016/j.ntt.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Hillemacher T, Frieling H, Moskau S, Muschler MAN, Semmler A, Kornhuber J, Klockgether T, Bleich S, Linnebank M. Global DNA methylation is influenced by smoking behaviour. European Neuropsychopharmacology. 2008;18:295–298. doi: 10.1016/j.euroneuro.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Ho MK, Goldman D, Heinz A, Kaprio J, Kreek MJ, Li MD, Munafò MR, Tyndale RF. Breaking barriers in the genomics and pharmacogenetics of drug addiction. Clin Pharmacol Ther. 2010;88:779–791. doi: 10.1038/clpt.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander JA, Im HI, Amelio AL, Kocerha J, Bali P, Lu Q, Willoughby D, Wahlestedt C, Conkright MD, Kenny PJ. Striatal microRNA controls cocaine intake through CREB signalling. Nature. 2010;466:197–202. doi: 10.1038/nature09202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Host L, Dietrich JB, Carouge D, Aunis D, Zwiller J. Cocaine self-administration alters the expression of chromatin-remodelling proteins; modulation by histone deacetylase inhibition. J Psychopharmacol (Oxford) 2011;25:222–229. doi: 10.1177/0269881109348173. [DOI] [PubMed] [Google Scholar]
- Hughes V. Epigenetics: The sins of the father. Nature. 2014;507:22–24. doi: 10.1038/507022a. [DOI] [PubMed] [Google Scholar]
- Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci. 2006;29:565–598. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- Im H-I, Hollander JA, Bali P, Kenny PJ. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat Neurosci. 2010;13:1120–1127. doi: 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamerson PA, Wulser MJ, Kimler BF. Neurobehavioral effects in rat pups whose sires were exposed to alcohol. Brain Res Dev Brain Res. 2004;149:103–111. doi: 10.1016/j.devbrainres.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Jeanblanc J, Logrip ML, Janak PH, Ron D. BDNF-mediated regulation of ethanol consumption requires the activation of the MAP kinase pathway and protein synthesis. European Journal of Neuroscience. 2012;37:607–612. doi: 10.1111/ejn.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet. 2007;16:2272–2280. doi: 10.1093/hmg/ddm179. [DOI] [PubMed] [Google Scholar]
- Kavitharaj NK, Vijayammal PL. Nicotine Administration Induced Changes in the Gonadal Functions in Male Rats. Pharmacology. 1998;58:2–7. doi: 10.1159/000028262. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Prescott CA. Cannabis Use, Abuse, and Dependence in a Population-Based Sample of Female Twins. Am J Psychiatry. 1998;155:1016–1022. doi: 10.1176/ajp.155.8.1016. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Jacobson KC, Prescott CA, Neale MC. Specificity of Genetic and Environmental Risk Factors for Use and Abuse/Dependence of Cannabis, Cocaine, Hallucinogens, Sedatives, Stimulants, and Opiates in Male Twins. Am J Psychiatry. 2003;160:687–695. doi: 10.1176/appi.ajp.160.4.687. [DOI] [PubMed] [Google Scholar]
- Killinger CE, Robinson S, Stanwood GD. Subtle biobehavioral effects produced by paternal cocaine exposure. Synapse. 2012;66:902–908. doi: 10.1002/syn.21582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P, Choi CS, Park JH, Joo SH, Kim SY, Ko HM, Kim KC, Jeon SJ, Park SH, Han S-H, et al. Chronic exposure to ethanol of male mice before mating produces attention deficit hyperactivity disorder-like phenotype along with epigenetic dysregulation of dopamine transporter expression in mouse offspring. J Neurosci Res. 2014;92:658–670. doi: 10.1002/jnr.23275. [DOI] [PubMed] [Google Scholar]
- Knezovich JG, Ramsay M. The effect of preconception paternal alcohol exposure on epigenetic remodeling of the h19 and rasgrf1 imprinting control regions in mouse offspring. Epigenomics and Epigenetics. 2012 doi: 10.3389/fgene.2012.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Volkow ND. Neurocircuitry of Addiction. Neuropsychopharmacology. 2009;35:217–238. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreek MJ, Nielsen DA, LaForge KS. Genes associated with addiction. Neuromol Med. 2004;5:85–108. doi: 10.1385/NMM:5:1:085. [DOI] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DEH, Truong HT, Russo SJ, LaPlant Q, Sasaki TS, Whistler KN, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Kumar P, Meizel S. Nicotinic acetylcholine receptor subunits and associated proteins in human sperm. J Biol Chem. 2005;280:25928–25935. doi: 10.1074/jbc.M502435200. [DOI] [PubMed] [Google Scholar]
- LaForge KS, Yuferov V, Kreek MJ. Opioid receptor and peptide gene polymorphisms: potential implications for addictions. European Journal of Pharmacology. 2000;410:249–268. doi: 10.1016/s0014-2999(00)00819-0. [DOI] [PubMed] [Google Scholar]
- Lam MKP, Homewood J, Taylor AJ, Mazurski EJ. Second generation effects of maternal alcohol consumption during pregnancy in rats. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2000;24:619–631. doi: 10.1016/s0278-5846(00)00097-x. [DOI] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iñiguez SD, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YW, Hennig B, Fiala M, Kim KS, Toborek M. Cocaine activates redox-regulated transcription factors and induces TNF-α expression in human brain endothelial cells. Brain Research. 2001;920:125–133. doi: 10.1016/s0006-8993(01)03047-5. [DOI] [PubMed] [Google Scholar]
- Lerman C, Berrettini W. Elucidating the role of genetic factors in smoking behavior and nicotine dependence. Am J Med Genet B Neuropsychiatr Genet. 2003;118B:48–54. doi: 10.1002/ajmg.b.10003. [DOI] [PubMed] [Google Scholar]
- Levine A, Huang Y, Drisaldi B, Griffin EA, Pollak DD, Xu S, Yin D, Schaffran C, Kandel DB, Kandel ER. Molecular mechanism for a gateway drug: epigenetic changes initiated by nicotine prime gene expression by cocaine. Sci Transl Med. 2011;3:107ra109–107ra109. doi: 10.1126/scitranslmed.3003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SC, Mato JM. Role of methionine adenosyltransferase and S-adenosylmethionine in alcohol-associated liver cancer. Alcohol. 2005;35:227–234. doi: 10.1016/j.alcohol.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Markou A, Koob GF. Postcocaine anhedonia. An animal model of cocaine withdrawal. Neuropsychopharmacology. 1991;4:17–26. [PubMed] [Google Scholar]
- Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Current Opinion in Cell Biology. 2007;19:266–272. doi: 10.1016/j.ceb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Maze I, Nestler EJ. The epigenetic landscape of addiction. Annals of the New York Academy of Sciences. 2011;1216:99–113. doi: 10.1111/j.1749-6632.2010.05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Covington HE, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Feng J, Wilkinson MB, Sun H, Shen L, Nestler EJ. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc Natl Acad Sci USa. 2011;108:3035–3040. doi: 10.1073/pnas.1015483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Meaney MJ, Szyf M. Diet and the epigenetic (re)programming of phenotypic differences in behavior. Brain Research. 2008;1237:12–24. doi: 10.1016/j.brainres.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek LR, Myren K, Sturm J, Burau D. Acute paternal alcohol use affects offspring development and adult behavior. Physiology & Behavior. 2007;91:154–160. doi: 10.1016/j.physbeh.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Merikangas KR, Stolar M, Stevens DE, Goulet J, Preisig MA, Fenton B, Zhang H, O’Malley SS, Rounsaville BJ. Familial Transmission of Substance Use Disorders. Arch Gen Psychiatry. 1998;55:973–979. doi: 10.1001/archpsyc.55.11.973. [DOI] [PubMed] [Google Scholar]
- Mulé SJ, Misra AL. Cocaine: Distribution and Metabolism in Animals 1977 [Google Scholar]
- Muthusami KR, Chinnaswamy P. Effect of chronic alcoholism on male fertility hormones and semen quality. Fertility and Sterility. 2005;84:919–924. doi: 10.1016/j.fertnstert.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Nielsen DA, Utrankar A, Reyes JA, Simons DD, Kosten TR. Epigenetics of drug abuse: predisposition or response. Pharmacogenomics. 2012;13:1149–1160. doi: 10.2217/pgs.12.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen DA, Yuferov V, Hamon S, Jackson C, Ho A, Ott J, Kreek MJ. Increased OPRM1 DNA Methylation in Lymphocytes of Methadone-Maintained Former Heroin Addicts. Neuropsychopharmacology. 2008;34:867–873. doi: 10.1038/npp.2008.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieratschker V, Batra A, Fallgatter AJ. Genetics and epigenetics of alcohol dependence. Gene. 2013 doi: 10.1186/2049-9256-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova SI, He F, Bai J, Cutrufello NJ, Lidow MS. Maternal cocaine administration in mice alters DNA methylation and gene expression in hippocampal neurons of neonatal and prepubertal offspring. PLoS ONE. 2008 doi: 10.1371/journal.pone.0001919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omotoso GO, Ibitolu JO, Femi-Akinlosotu OM, Akinola OB, Enaibe BU. Morphological and neurohistological changes in adolescent rats administered with nicotine during intrauterine life. Niger J Physiol Sci. 2013;28:147–151. [PubMed] [Google Scholar]
- Orozco LD, Rubbi L, Martin LJ, Fang F, Hormozdiari F, Che N, Smith AD, Lusis AJ, Pellegrini M. Intergenerational genomic DNA methylation patterns in mouse hybrid strains. Genome Biology. 2014;15:R68. doi: 10.1186/gb-2014-15-5-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouko LA, Shantikumar K, Knezovich J, Haycock P, Schnugh DJ, Ramsay M. Effect of Alcohol Consumption on CpG Methylation in the Differentially Methylated Regions of H19 and IG - DMR in Male Gametes—Implications for Fetal Alcohol Spectrum Disorders. Alcoholism: Clinical and Experimental Research. 2009;33:1615–1627. doi: 10.1111/j.1530-0277.2009.00993.x. [DOI] [PubMed] [Google Scholar]
- Pandey SC, Ugale R, Zhang H, Tang L, Prakash A. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci. 2008;28:3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Beach SRH, Gunter TD, Brody GH, Madan A, Gerrard M. The effect of smoking on MAOA promoter methylation in DNA prepared from lymphoblasts and whole blood. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2010;153B:619–628. doi: 10.1002/ajmg.b.31031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistis M, Perra S, Pillolla G, Melis M, Muntoni AL, Gessa GL. Adolescent exposure to cannabinoids induces long-lasting changes in the response to drugs of abuse of rat midbrain dopamine neurons. Bps. 2004;56:86–94. doi: 10.1016/j.biopsych.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32:1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcella A, Gessa GL, Pani L. Delta9-tetrahydrocannabinol increases sequence-specific AP-1 DNA-binding activity and Fos-related antigens in the rat brain. Eur J Neurosci. 1998;10:1743–1751. doi: 10.1046/j.1460-9568.1998.00175.x. [DOI] [PubMed] [Google Scholar]
- Prak ET, Kazazian HH. Mobile elements and the human genome. Nature Reviews Genetics. 2000;1:134–144. doi: 10.1038/35038572. [DOI] [PubMed] [Google Scholar]
- Pray L. Transposons, or jumping genes: Not junk DNA. Nature Education. 2008;1:32. [Google Scholar]
- Ramírez AR, Castro MA, Angulo C, Ramió L, Rivera MM, Torres M, Rigau T, Rodríguez-Gil JE, Concha II. The presence and function of dopamine type 2 receptors in boar sperm: a possible role for dopamine in viability, capacitation, and modulation of sperm motility. Biol Reprod. 2009;80:753–761. doi: 10.1095/biolreprod.108.070961. [DOI] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- Ratcliffe JM, Gladen BC, Wilcox AJ, Herbst AL. Does early exposure to maternal smoking affect future fertility in adult males? Reproductive Toxicology. 1992;6:297–307. doi: 10.1016/0890-6238(92)90192-v. [DOI] [PubMed] [Google Scholar]
- Ray R, Mitra N, Baldwin D, Guo M, Patterson F, Heitjan DF, Jepson C, Wileyto EP, Wei J, Payne T, et al. Convergent evidence that choline acetyltransferase gene variation is associated with prospective smoking cessation and nicotine dependence. Neuropsychopharmacology. 2010;35:1374–1382. doi: 10.1038/npp.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, Allis CD. Gene regulation: Code of silence. Nature. 2001;414:258–261. doi: 10.1038/35104721. [DOI] [PubMed] [Google Scholar]
- Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers AB, Morgan CP, Bronson SL, Revello S, Bale TL. Paternal stress exposure alters sperm microRNA content and reprograms offspring HPA stress axis regulation. J Neurosci. 2013;33:9003–9012. doi: 10.1523/JNEUROSCI.0914-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse-Seeboth C, Pfeil J, Sehrt D, Meineke I, Tzvetkov M, Bruns E, Poser W, Vormfelde SV, Brockmöller J. Interindividual Variation in the Pharmacokinetics of Δ9-Tetrahydrocannabinol as Related to Genetic Polymorphisms in CYP2C9. Clin Pharmacol Ther. 2008;85:273–276. doi: 10.1038/clpt.2008.213. [DOI] [PubMed] [Google Scholar]
- Sadri-Vakili G, Kumaresan V, Schmidt HD, Famous KR, Chawla P, Vassoler FM, Overland RP, Xia E, Bass CE, Terwilliger EF, et al. Cocaine-induced chromatin remodeling increases brain-derived neurotrophic factor transcription in the rat medial prefrontal cortex, which alters the reinforcing efficacy of cocaine. J Neurosci. 2010;30:11735–11744. doi: 10.1523/JNEUROSCI.2328-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkaki A, Assaei R, Motamedi F, Badavi M. Effect of parental morphine addiction on hippocampal long-term potentiation in rats offspring. Behavioural Brain Research. 2008;186:72–77. doi: 10.1016/j.bbr.2007.07.041. [DOI] [PubMed] [Google Scholar]
- Sasaki A, Constantinof A, Pan P. Cocaine exposure prior to pregnancy alters the psychomotor response to cocaine and transcriptional regulation of the dopamine D1 receptor in adult male offspring. Behavioural Brain Research. 2014;265:163–170. doi: 10.1016/j.bbr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- Satta R, Maloku E, Zhubi A, Pibiri F, Hajos M, Costa E, Guidotti A. Nicotine decreases DNA methyltransferase 1 expression and glutamic acid decarboxylase 67 promoter methylation in GABAergic interneurons. Proc Natl Acad Sci USa. 2008;105:16356–16361. doi: 10.1073/pnas.0808699105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuel H, Burkman LJ, Lippes J, Crickard K, Forester E, Piomelli D, Giuffrida A. N-Acylethanolamines in human reproductive fluids. Chem Phys Lipids. 2002;121:211–227. doi: 10.1016/s0009-3084(02)00158-5. [DOI] [PubMed] [Google Scholar]
- Sheng J, Lv ZG, Wang L, Zhou Y, Hui B. Histone H3 phosphoacetylation is critical for heroin-induced place preference. NeuroReport. 2011;22:575. doi: 10.1097/WNR.0b013e328348e6aa. [DOI] [PubMed] [Google Scholar]
- Singer T, McConnell MJ, Marchetto MCN, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends in Neurosciences. 2010;33:345–354. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK. What is an epigenetic transgenerational phenotype? Reproductive Toxicology. 2008;25:2–6. doi: 10.1016/j.reprotox.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK. Environmental epigenetic transgenerational inheritance and somatic epigenetic mitotic stability. Epigenetics. 2011;6:838–842. doi: 10.4161/epi.6.7.16537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nature Reviews Genetics. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Steketee JD, Kalivas PW. Drug Wanting: Behavioral Sensitization and Relapse to Drug-Seeking Behavior. Pharmacological Reviews. 2011;63:348–365. doi: 10.1124/pr.109.001933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouder C, Somm E, Paoloni-Giacobino A. Prenatal exposure to ethanol: A specific effect on the H19 gene in sperm. Reproductive Toxicology. 2011;31:507–512. doi: 10.1016/j.reprotox.2011.02.009. [DOI] [PubMed] [Google Scholar]
- Sun H, Maze I, Dietz DM, Scobie KN, Kennedy PJ, Damez-Werno D, Neve RL, Zachariou V, Shen L, Nestler EJ. Morphine epigenomically regulates behavior through alterations in histone H3 lysine 9 dimethylation in the nucleus accumbens. J Neurosci. 2012;32:17454–17464. doi: 10.1523/JNEUROSCI.1357-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet F, Murdock GL. Affinity labeling of hormone-specific proteins. Endocr Rev. 1987;8:154–184. doi: 10.1210/edrv-8-2-154. [DOI] [PubMed] [Google Scholar]
- Szutorisz H, DiNieri JA, Sweet E, Egervari G, Michaelides M, Carter JM, Ren Y, Miller ML, Blitzer RD, Hurd YL. Parental THC exposure leads to compulsive heroin-seeking and altered striatal synaptic plasticity in the subsequent generation. Neuropsychopharmacology. 2014;39:1315–1323. doi: 10.1038/npp.2013.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo Rodriguez M, Lotfipour S, Leonard G, Perron M, Richer L, Veillette S, Pausova Z, Paus T. Maternal smoking during pregnancy is associated with epigenetic modifications of the brain-derived neurotrophic factor-6 exon in adolescent offspring. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2010;153B:1350–1354. doi: 10.1002/ajmg.b.31109. [DOI] [PubMed] [Google Scholar]
- Trivedi M, Shah J, Hodgson N, Byun H-M, Deth R. Morphine induces redox-based changes in global DNA methylation and retrotransposon transcription by inhibition of excitatory amino acid transporter type 3-mediated cysteine uptake. Mol Pharmacol. 2014;85:747–757. doi: 10.1124/mol.114.091728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Eisen SA, Goldberg J, True W, Lin N, Meyer JM, Toomey R, Faraone SV, Eaves L. Genetic influences on DSM-III-R drug abuse and dependence: a study of 3,372 twin pairs. Am J Med Genet. 1996;67:473–477. doi: 10.1002/(SICI)1096-8628(19960920)67:5<473::AID-AJMG6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Bar JL, Harley RM, Lyons MJ. The Harvard Twin Study of Substance Abuse: What We Have Learned. Harv Rev Psychiatry. 2001;9:267–279. [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, True W, Lin N, Toomey R, Eaves L. Co-occurrence of Abuse of Different Drugs in Men: The Role of Drug-Specific and Shared Vulnerabilities. Arch Gen Psychiatry. 1998;55:967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- van der Knaap LJ, Schaefer JM, Franken IHA, Verhulst FC, van Oort FVA, Riese H. Catechol-O-methyltransferase gene methylation and substance use in adolescents: the TRAILS study. Genes Brain Behav. 2014:n/a–n/a. doi: 10.1111/gbb.12147. [DOI] [PubMed] [Google Scholar]
- Vassoler FM, Byrnes EM, Pierce RC. The impact of exposure to addictive drugs on future generations: Physiological and behavioral effects. Neuropharmacology. 2014;76:269–275. doi: 10.1016/j.neuropharm.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassoler FM, Johnson NL, Byrnes EM. Female adolescent exposure to cannabinoids causes transgenerational effects on morphine sensitization in female offspring in the absence of in utero exposure. J Psychopharmacol (Oxford) 2013a;27:1015–1022. doi: 10.1177/0269881113503504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassoler FM, White SL, Schmidt HD, Sadri-Vakili G, Pierce RC. Epigenetic inheritance of a cocaine-resistance phenotype. Nat Neurosci. 2013b;16:42–47. doi: 10.1038/nn.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen-Smit E, Koning IM, Verdurmen JEE, Van der Vorst H, Engels RCME, Vollebergh WAM. The influence of paternal and maternal drinking patterns within two-partner families on the initiation and development of adolescent drinking. Addictive Behaviors. 2012;37:1248–1256. doi: 10.1016/j.addbeh.2012.06.005. [DOI] [PubMed] [Google Scholar]
- Verweij KJH, Zietsch BP, Lynskey MT, Medland SE, Neale MC, Martin NG, Boomsma DI, Vink JM. Genetic and environmental influences on cannabis use initiation and problematic use: a meta-analysis of twin studies. Addiction. 2010;105:417–430. doi: 10.1111/j.1360-0443.2009.02831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink JM, Willemsen G, Boomsma DI. Heritability of Smoking Initiation and Nicotine Dependence. Behav Genet. 2005;35:397–406. doi: 10.1007/s10519-004-1327-8. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Fowler JS, Logan J, Gatley SJ, Gifford A, Hitzemann R, Ding YS, Pappas N. Prediction of Reinforcing Responses to Psychostimulants in Humans by Brain Dopamine D2 Receptor Levels. Am J Psychiatry. 1999;156:1440–1443. doi: 10.1176/ajp.156.9.1440. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Fowler JS, Tomasi D, Telang F, Baler R. Addiction: decreased reward sensitivity and increased expectation sensitivity conspire to overwhelm the brain’s control circuit. Bioessays. 2010;32:748–755. doi: 10.1002/bies.201000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyssotski DL. Transgenerational epigenetic compensation. Evolocus 2011 [Google Scholar]
- Wang X, Gao W, Ma X, Wang X, Song C, Huang X, Liu H. Dot1L mediated histone H3 lysine79 methylation is essential to meiosis progression in mouse oocytes. Neuro Endocrinol Lett. 2014a;35:523–530. [PubMed] [Google Scholar]
- Wang Y, Lai J, Cui H, Zhu Y, Bin Zhao Wang W, Wei S. Inhibition of Histone Deacetylase in the Basolateral Amygdala Facilitates Morphine Context-Associated Memory Formation in Rats. J Mol Neurosci. 2014b:1–10. doi: 10.1007/s12031-014-0317-4. [DOI] [PubMed] [Google Scholar]
- Whitelaw E, Morgan HD, Sutherland HGE, Martin DIK. Epigenetic inheritance at the agouti locus in the mouse. Nature Genetics. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Wise RA. Wise: Drug-activation of brain reward pathways - Google Scholar (Drug and alcohol dependence) 1998 doi: 10.1016/s0376-8716(98)00063-5. [DOI] [PubMed] [Google Scholar]
- Wozniak DF, Cicero TJ, Kettinger L, III, Meyer ER. Paternal alcohol consumption in the rat impairs spatial learning performance in male offspring. Psychopharmacology. 1991;105:289–302. doi: 10.1007/BF02244324. [DOI] [PubMed] [Google Scholar]
- Wykes SM, Krawetz SA. The structural organization of sperm chromatin. J Biol Chem. 2003;278:29471–29477. doi: 10.1074/jbc.M304545200. [DOI] [PubMed] [Google Scholar]
- Xu M, Hu XT, Cooper DC, Moratalla R, Graybiel AM, White FJ, Tonegawa S. Elimination of cocaine-induced hyperactivity and dopamine-mediated neurophysiological effects in dopamine D1 receptor mutant mice. Cell. 1994;79:945–955. doi: 10.1016/0092-8674(94)90026-4. [DOI] [PubMed] [Google Scholar]
- Yang X, Hegde VL, Rao R, Zhang J, Nagarkatti PS, Nagarkatti M. Histone Modifications Are Associated with Δ9-Tetrahydrocannabinol-mediated Alterations in Antigen-specific T Cell Responses. J Biol Chem. 2014;289:18707–18718. doi: 10.1074/jbc.M113.545210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazigi RA, Odem RR, Polakoski KL. Demonstration of Specific Binding of Cocaine to Human Spermatozoa. Jama. 1991;266:1956–1959. [PubMed] [Google Scholar]
- Yochum C, Doherty-Lyon S, Hoffman C, Hossain MM, Zelikoff JT, Richardson JR. Prenatal cigarette smoke exposure causes hyperactivity and aggressive behavior: Role of altered catecholamines and BDNF. Experimental Neurology. 2014;254:145–152. doi: 10.1016/j.expneurol.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac C, Ivery E, Abel E. Hippocampal morphology in paternally alcohol-exposed offspring of Long-Evans rats. Alcoholism: Clinical and Experimental Research. 1989;13:383. [Google Scholar]
- Zhu J, Lee KP, Spencer TJ, Biederman J, Bhide PG. Transgenerational Transmission of Hyperactivity in a Mouse Model of ADHD. J Neurosci. 2014;34:2768–2773. doi: 10.1523/JNEUROSCI.4402-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhang X, Xu Y, Spencer TJ, Biederman J, Bhide PG. Prenatal nicotine exposure mouse model showing hyperactivity, reduced cingulate cortex volume, reduced dopamine turnover, and responsiveness to oral methylphenidate treatment. J Neurosci. 2012;32:9410–9418. doi: 10.1523/JNEUROSCI.1041-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]