Abstract
pRb and p53 are the two major tumor suppressors. Their inactivation is frequent when cancers develop and their reactivation is rationale of most cancer therapeutics. When pRb and p53 are genetically inactivated, cells irreparably lose the antitumor mechanisms afforded by them. Cancer genome studies document recurrent genetic inactivation of RB1 and TP53, and the inactivation becomes more frequent in more advanced cancers. These findings may explain why more advanced cancers are more likely to resist current therapies. Finding successful treatments for more advanced and multi-therapy resistant cancers will depend on finding antitumor mechanisms that remain effective when pRb and p53 are genetically inactivated. Here, we review studies that have begun to make progress in this direction.
Introduction
Tumorigenesis is genetic and epigenetic evolution to survive and expand when cells acquire tumorigenic mutations. Since our cells have built in tumor suppressing safeguards, tumorigenesis is fundamentally a process of disarming the safeguards. Once a tumor has formed and therapies begin, tumor cells additionally evolve to resist therapies. We will display roles of pRb and p53 in a condensed tumorigenic and tumor suppressive network, and sample a few cancer genome studies for the genetic status of RB1 and TP53 to assess the challenges from cancers with genetically inactivated pRb and p53. We will then discuss publications that have identified mechanisms to block Rb1 (mouse homolog of RB1) knockout (KO), Trp53 (mouse homolog of TP53) KO, or Rb1 and Trp53 doubly knockout (DKO) autochthonous tumors in mouse models. We include studies using xenograft models of human cancer that allow definitive attribution of observed effects to pRb or p53 genetic inactivation. These studies provide insights to the extent of mechanistic, functional, and physiological rigor not feasible with clinical studies. We conclude with a perspective for new approaches to more and better evidence-based treatments and optimism for advanced cancer.
The pRb tumor suppressor safeguard
pRb responds to proliferation signals in the p16-cyclin D1/Cdk4-pRb pathway
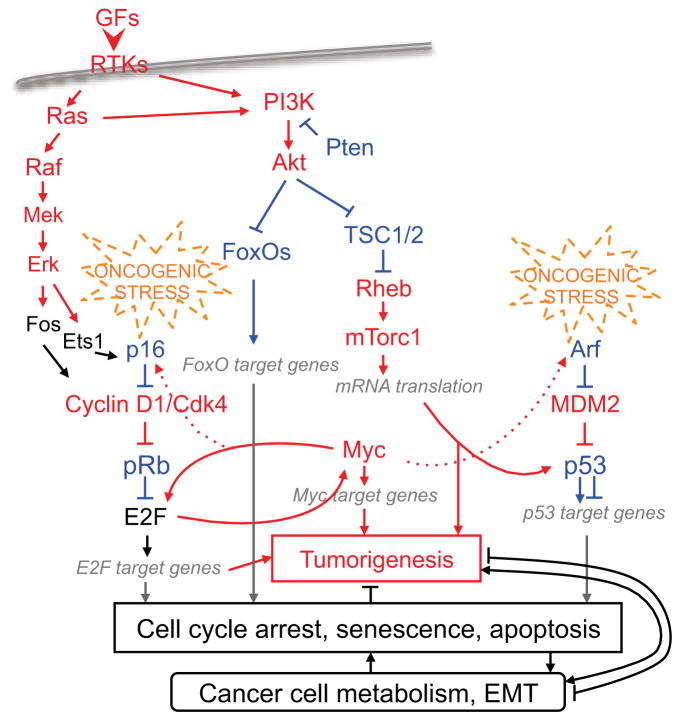
As shown in Figure 1, typical growth factors (GFs) ligate receptor tyrosine kinases (RTKs) to activate Ras-Raf-Mek-Erk and Ras-PI3K-Akt pathways. Since PI3K is also directly activated by RTKs and significantly inhibited by Pten, we will call the latter PI3K(Pten)-Akt. Erk phosphorylates transcription factors, such as Fos, to activate expression of cyclin D1 to phosphorylate and inactivate pRb, or Ets1, to activate expression of p16 (also called p16INK4a) to inhibit cyclin D1/Cdk4 when signaling is aberrant. In the p16-Luc reporter mice, p16 was activated in all 14 tested tumor models (1). A recent study achieved more effective inhibition of PI3K activated breast cancer xenografts when PI3K inhibitors were combined with cyclin D1/Cdk4 inhibitors (2). Overall, pRb is likely activated by most oncogenic signals and subsequently inactivated during tumor progression.
Figure 1. pRb and p53 in a condensed tumorigenic and tumor suppressive network.
Proteins in red are pro-oncogenic; blue ones tumor suppressive. Black ones are neutral in most contexts. Arrows are activating while blunt ends inhibiting. Solid lines demonstrate direct molecular connections, dotted lines show functional connections when molecular steps have not been defined. Pools of target genes are dark gray implying that some target genes of an oncogene such as Myc could also negatively regulate oncogenesis. In their respective positions in the p16-cyclin D1/Cdk4-pRb and Arf-MDM2-p53 pathways, pRb and p53 are activated by most oncogenic events. Cell cycle arrest, senescence, apoptosis are the most fundamental antitumor mechanisms and are mostly implemented by products of E2F and p53 target genes. In parallel, E2F and p53 target genes also directly and indirectly suppress cancer cell metabolism. Tumorigenesis or tumor suppression includes developing compatible cell metabolism. Evidence are emerging that certain aspects of cancer cell metabolism can drive tumorigenesis and disrupting some of them can induce cell cycle arrest, senescence, and apoptosis. Whether these effects may remain effective when pRb and p53 are genetically inactivated have not been tested. Since similar situations also apply to epithelial-mesenchymal transition, EMT is listed in the same box with cancer cell metabolism. General and more focused reviews in the literature provide more expansive and in depth discussions of these topics (14, 98, 142–148).
pRb is best equipped to implement proliferation arrest
pRb binds E2F to bring histone deacetylases and methyltransferases, chromatin remodeling proteins, Polycomb-group proteins, and DNA methyltransferases to the promoters of E2F target genes (3, 4). With these mechanisms, pRb can create local heterochromatic state, sometimes microscopically visible as senescence associated heterochromatic foci (SAHF) (5), to permanently repress expression of DNA synthesis proteins to implement the senescence safeguard.
E2F1 is both proliferative and apoptotic
Then, how could inactivation of pRb benefit tumorigenesis? Following DNA damage, pRb binds more E2F1 on E2F target promoters and brings histone acetylase P/CAF to stimulate expression of apoptotic target genes caspase-7 and p73 but brings histone deacetylase HDAC1 to repress proliferative target gene cyclin A2. Loss of pRb therefore reduced apoptosis in colon epithelium following i.p. injection of doxorubicin (6). In other studies, E2F1 distinguished itself from other E2Fs by forming an additional binding to pRb (7); E2F1’s apoptotic target genes distinguished themselves from proliferative target genes by recruiting FoxO1 and 3 to their promoters (8). Whether pRb uses these mechanisms to activate apoptotic target genes of E2F1 to suppress tumorigenesis requires further study. Other studies attempted to use E2F1’s apoptotic activity to treat pRb deficient tumors (see below).
pRb represses E2F to suppress cancer cell metabolism
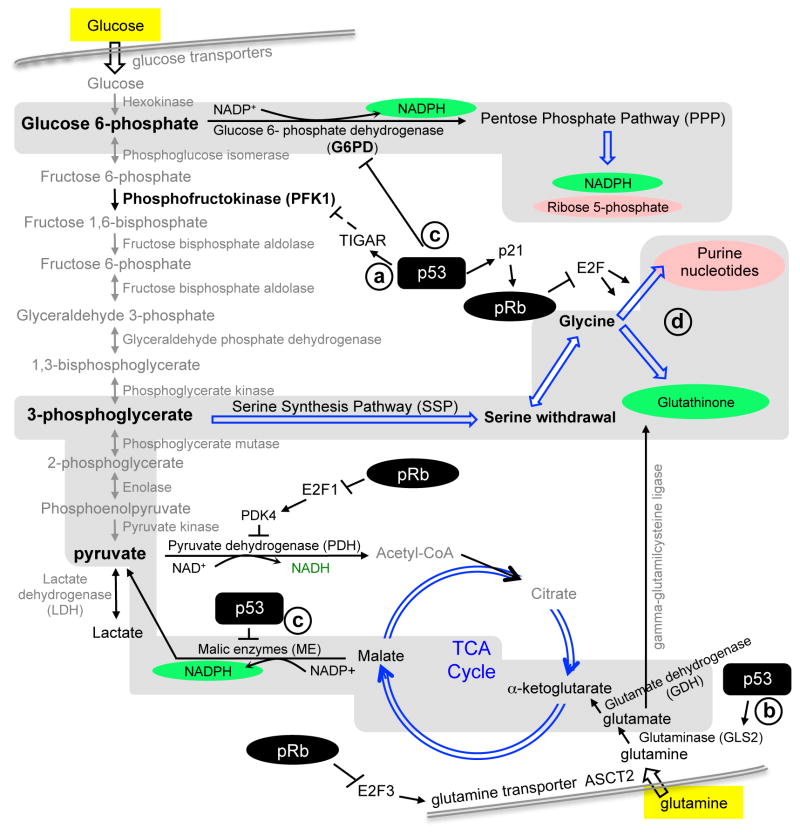
Cancer cells need more free energy to divide and more nucleotides, amino acids, and fatty acids to build new cells. Since a more active metabolism generates more ROS and biosynthesis consumes reductants, cancer cells also rely on stronger supplies of reductive equivalents. As summarized in Figure 2, cancer cells take up more glucose and glutamine and use them to generate more ATP, carbon backbones (light reds), and reductants (greens).
Figure 2. The basics of cancer cell metabolism.
We highlight (yellow) glucose and glutamine as two major fuels for cancer cell metabolism. Their key metabolism steps (in gray fields) produce carbon skeletons (red) for biosynthesis and reductive equivalents (green) for biosynthesis and antioxidant defense. A select few effects of pRb and p53 on cancer cell metabolism are illustrated to accompany the text. Metabolites connected by double blue line-arrows involve more than one steps. Please see text for more details corresponding to (a, b, c, d).
In pRb ChIP experiments with Drosophila third instar larvae (9), genes involved in DNA replication ranked first in numbers as pRb target genes (10). Ranked second were genes involved in pyrimidine metabolism, revealing that pRb coordinates repressing DNA synthesis with reducing nucleotides synthesis. In glycolysis, pyruvate can be converted to acetyl-CoA by pyruvate dehydrogenase (PDH) for oxidative phosphorylation in mitochondria, or to lactate by lactate dehydrogenase (LDH), which is a major feature of cancer cell metabolism. Pyruvate dehydrogenase kinase (PDK) phosphorylates and inactivates PDH; the isoenzyme PDK4 is an E2F1 target gene (11). On the glutamine side, the glutamine transporter ASCT2 is an E2F3 target gene (12). These and other findings suggest that pRb can suppress cancer cell metabolism (13, 14).
The p53 tumor suppressor safeguard: it has become complicated
Classically, p53 safeguard works like this: DNA damage leads to p53 activation and p53 executes apoptosis or cell cycle arrest. As a DNA-binding transcription factor, p53 activates expression of CDKI p21 to arrest cell cycle; and proapoptotic protein Bax, and BH3-only proteins PUMA and NOXA to induce apoptosis. The repressor E2F, E2F7, is a new p53 target gene for cell cycle arrest (15, 16).
Unexpectedly, γ-radiation induced similar tumorigenesis in mice whether or not p53 was present to respond with apoptosis; only after apoptosis subsided, presence of p53 delayed tumorigenesis (17). Oncogenic signaling, but not simple DNA damage, activates Arf (also called p14Arf in humans and p19Arf in mice), which in turn activates p53 in the Arf-MDM2-p53 pathway (17, 18) (Figure 1). Furthermore, while ectopic expression of HRASG112V in MEFs activated Arf-p53 to induce senescence, expression of endogenous KRASG12D stimulated proliferation (19). In the lung, expression of endogenous KRASG12D induced tumors because it did not activate Arf as monitored by the Arf-GFP reporter mice (20, 21). However, KRASG12D strongly activated Arf in the leg muscle and tumorigenesis was therefore blocked (21). Endogenous expression of a different KRAS mutant, KRASG12V, also did not induce Arf-p53 in MEF (22), but induced senescent lesions in the lung and pancreas (23). Tetracycline titrated expression of HRASG12V in mammary glands demonstrated the entire process of tumorigenesis in three steps: initial expression of HRASG12V induced neoplastic lesions, gradually increased expression of HRASG12V triggered Arf-p53 to induce senescence safeguard, and abrogation of this safeguard led to tumorigenesis (24). Dosage dependent activation of Arf-p53 was also observed with graded expression of Myc (25, 26). Thus, activation of Arf depends on oncogenic strength and context. Importantly, when tumors were induced by endogenous KRASG12D in the absence of Arf-p53 activation, restoring p53 induced only mild proliferation inhibition and apoptosis and only in areas of advanced tumor histopathology. In other areas, restored p53 was not activated (27, 28). In contrast, tumors induced by irradiation of newborns (29), viral expression of HRASG12V (30), or Eμ-Myc (31) in the absence of p53 strongly activated restored p53 leading to complete tumor regression. These findings could explain why p53 is more frequently inactivated in more advanced cancer. It will be important to use the p16-Luc mice (1) to learn how p16-pRb responds to differing strengths of oncogenic signals.
The mechanisms by which p53 suppresses tumorigenesis has also become complicated when Li et al generated K-to-R knockin mice to prevent acetylation of K117, K161, and K162 (32). p533KR/3KR MEFs were unable to express p21, PUMA, or NOXA and unable to cell cycle arrest, senesce, or apoptose following DNA damage. p533KR/3KR mice however were still protected from tumorigenesis. From another direction, combined knockout of p21, PUMA, and NOXA disabled proliferation arrest, senescence, or apoptosis in MEFs, but the mice were no more tumor prone than WT mice (33). Thus, the ability of p53 to induce cell cycle arrest, senescence, and apoptosis is dispensable for its tumor suppressor role, leaving us to wonder how p53 suppresses tumorigenesis. Since mutations that completely abolished transcription activity of p53 nullified its tumor suppressor role, p53 likely functions as a transcription factor to suppress tumorigenesis (34). Indeed, p533KR/3KR MEFs can activate expression of TIGAR and GLS2, suggesting that p533KR/3KR may suppress cancer cell metabolism (32) (Figure 2 and see more below). It would be more informative to determine whether p533KR/3KR can activate expression of E2F7 (see above), miR-200c (which suppress EMT (35, 36)), miR-34 (which suppress Snail (37) and stem cell marker CD44 (38)), or can repress expression of CD44 (39), or miR-17~92 (40). p53 also represses hundreds of genes through activating expression of lincRNA-p21 (41).
p53 coordinates inhibition of cell proliferation and growth (42)
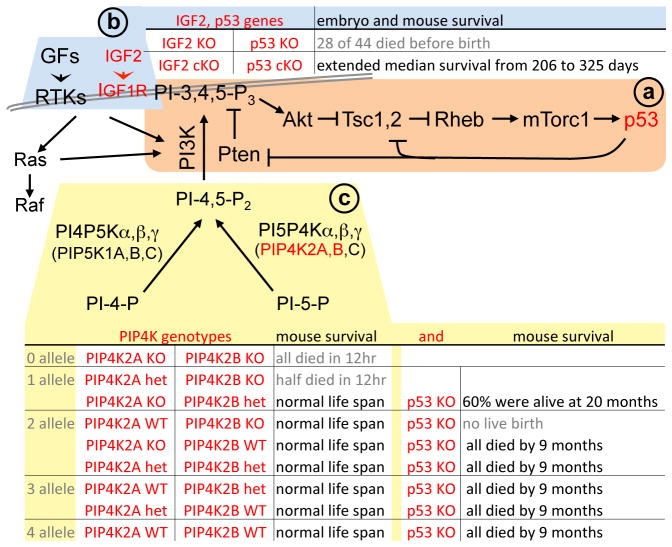
PI3K(Pten)-Akt is a major pathway for growth regulation. Through activating mTORC1, PI3K(Pten)-Akt enhances translation of p53 mRNA to induce senescence independent of Arf (43, 44) (Figure 4a). Reciprocally, p53 negatively regulates cell growth by activating expression of Pten, TSC2, and AMPKβ (45, 46). p53 activates expression of sestrin 1 and 2, which form AMPK-sestrins-TSC2 complex to stimulate AMPK to phosphorylate and activate TSC2 (47). The significance of sestrin and p53 in growth suppression was demonstrable in the liver of sestrin or p53 KO mice following DEN treatment (47). Interestingly, these reciprocal relationships may become synthetic lethal in certain contexts (see below, Figure 4b, c).
Figure 4. Synthetic lethality between IGF2 or PI5P4Ks and p53.
(a) briefly summarizes the reciprocal regulation between IGF2-IGF1R-PI3K(Pten)-Akt and p53 as discussed in the text. (b) displays the synthetic lethal relationship between IGF2 KO and p53 KO in embryogenesis and p53 deficient tumorigenesis reported by Haley et al (87). (c) summarizes the synthetic lethal relationship between specific PIP4K2A,B KO and p53KO (89). In addition to discussions in the text, it is of note that, in the absence of p53, presence of 1 allele of PIP4K2B out of 4 alleles of PIP4K2A and PIP4K2B generated adult mice, but presence of 2 alleles of PIP4K2A without both alleles of PIP4K2B did not produce live birth. This finding demonstrates exquisite specificity distinguishing PIP4K2A from PIP4K2B in substrate specificity, expression dynamics, and relationships with p53, alone or together in various combinations, for further studies.
Cancer genome studies reveal recurrent genetic inactivation of pRb and p53
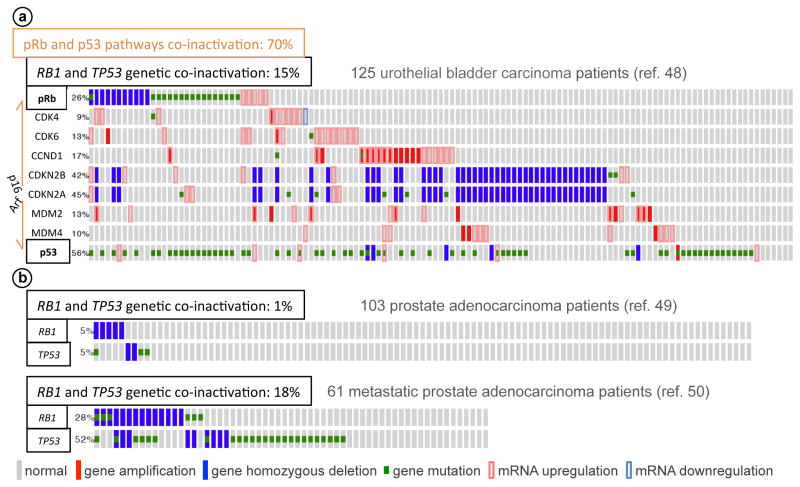
Cancer genome studies have employed large-size collections of primary tumors for multi-platform genome scale characterization. In Figure 3A, we analyzed 125 high-grade bladder carcinomas that yielded gene copy number variations, exome sequencing, and RNA Seq data (48) for components of the p16-cyclin D1/Cdk4-pRb and Arf-MDM2-p53 pathways. Deletion of the CDKN2A locus led to loss of both p16 and Arf. The nearby CDKN2B (encoding p16 family member p15) is generally deleted together. Cyclin D1 and MDM2 are frequently upregulated by gene amplification or overexpression. pRb and p53 are inactivated by cyclin D1/Cdk4 and MDM2/4 at frequencies of 56% and 34%, respectively. Genetically, pRb and p53 is inactivated at 22% and 54%, respectively, with inactivation of both RB1 and TP53 occurred at 15%.
Figure 3. Cancer genome analysis of pRb and p53 status.
We used cBioPortal web-based tool incorporated in TCGA website to display gene copy number alterations, DNA sequence alterations, and mRNA levels, as marked at the bottom of the figure. Cdk6 and MDM4 are included since they function similarly to Cdk4 and MDM2, respectively. Each row represents a query gene, and each column represents a cancer specimen. Only specimens that yielded data on the specified platforms are included.
In Figure 3B, we analyzed data from two prostate cancer studies to assess how the frequencies changed with cancer progression. Taylor et al (49) studied 218 prostate adenocarcinomas samples, 83% of which were obtained at the primary sites; while Grasso et al (50) obtained 61 prostate adenocarcinoma samples mostly (82%) from metastatic sites. In the first study, genetic inactivation of RB1 occurred at 5% and genetic inactivation of TP53 at 5%, with co-occurrence at 1%. In the second study, the corresponding frequencies increased to 28%, 52%, and 18%. Data from low-grade gliomas (TCGA, provisional) and glioblastomas (51) revealed same trend. We therefore suggest that genetic inactivation of pRb, p53 and most certainly both pRb and p53 could explain why more advanced cancer are more likely to resist current therapies.
Antitumor mechanisms for “two hit” pRb deficient tumorigenesis
Two extraordinary characteristics of retinoblastoma are “two hit” RB1 loss and extremely early onset. Retinoblastomas therefore may require few additional mutations. Xu et al reported that cone photoreceptor precursors in normal human retina at mid-gestation express high levels of MDM2 and N-Myc, likely because human MDM2 promoter contains a binding sequence for the cone specific transcription factor RXRγ. Knockdown of RXRγ or another cone specific transcription factor TRβ2 in cultured retinoblastoma cells inhibited their ability to form orthotopic xenografts (52). Laurie et al on the other hand provided evidence that Arf was dramatically activated in retinoblastomas, but MDM4 gene amplification and overexpression prevented the activation of p53 (53). Whole genome sequencing of four retinoblastomas revealed no genetic lesions in known oncogenes or tumor suppressor genes other than RB1 and MYCN (54). Epigenetically, SYK, a proto-oncoprotein, was highly expressed in retinoblastomas. Inhibiting SYK with existing inhibitors significantly delayed orthotopic xenografts by retinoblastoma cells (54). When the genomic studies were expanded to 94 retinoblastomas, MYCN amplification was recurrent at 8.5% with the second most frequently amplified gene OTX2 at 3%. The second most commonly deleted gene following RB1 is BCOR at 4% (55). The significance of these two additional recurrent alterations now needs further study.
“Two hit” pRb deficient tumorigenesis is modeled in melanotrophs in pituitary intermediate lobe (IL) in Rb1+/− mice
Using this model, various pRb targets, including E2F1, showed physiological relevance since IL tumorigenesis was significantly delayed when any one of them was deleted in Rb1+/− mice (56–62). pRb target Skp2 (63, 64) was an exception; its deletion resulted in completely normal IL (65). When POMC-Cre was used to delete Rb1 in melanotrophs, neoplastic transformation occurred across the entire IL in the presence of Skp2, but apoptosis ablated the entire IL in the absence of Skp2. p27T187A knockin (66), which prevents p27 ubiquitination by Skp2, achieved the same effect (65). The accumulation of p27 led to more p27-cyclin A binding to replace E2F1-cyclin A binding on E2F target gene promoters (67). Since cyclin A binding to E2F1 inhibits E2F1 activity (68–71), the cyclin A unbound E2F1 activated E2F target genes to a higher degree than by loss of pRb alone. In this context, deregulated E2F1 became apoptotic, demonstrable by enlarged IL following its deletion (67). These findings suggest strategies to use E2F1’s apoptotic activity to treat pRb deficient cancers.
Modeling retinoblastoma in mice revealed essential roles of p27
Cre mediated Rb1 deletion during eye development and germline KO of Rb family member p107 are required to induce retinoblastoma in mice (72, 73). This could suggest that p107 compensated pRb loss in repressing E2F. However, co-deletion of Rb1 and p27 generated more robust and more penetrant retinoblastoma and a p27 mutation that disrupts cyclin/Cdk bindings was as effective as p27 deletion (74, 75). Therefore, binding and inhibiting cyclin/Cdk by p27, or similarly by p107 (76), likely explains lack of retinoblastoma following Rb1 deletion. Sangwan et al continued to show that i.p. injection of CDK inhibitors (mimicking p27 increase) during pregnancy prevented retinoblastoma in Rb1 and p107 or Rb1 and p27 DKO mice (74).
Co-deletion of Pten and Skp2 induced p53-independent senescence
Tumor suppressor roles of Pten have been amply demonstrated by its absence. However, homozygous deletion of Pten in prostate epithelium (with PB-Cre) induced cellular senescence rather than tumorigenesis (77), likely because oncogenic PI3K(Pten)-Akt activates mTORC1 to activate p53 (see above). Combining Trp53 deletion with Pten deletion avoided senescence and induced tumorigenesis (77). With this system, Lin et al discovered that co-deletion of Pten with Skp2 induced more robust senescence in the prostate (78). Skp2 deletion also inhibited tumorigenesis in Arf−/− mice (78). Detailed studies with MEFs established that co-deletion of Pten and Skp2 induced senescence without activating Arf-p53 and independent of Arf-p53, suggesting that targeting Skp2 could inhibit p53 deficient cancers (78).
Blocking Myc activity inhibited p53 deficient KRASG12D lung tumor
MYC is frequently overexpressed in tumors due to gene amplification (79), fusion, and other oncogenic signaling such RAS (80), and Myc is a potent oncogene (81). To model anti-Myc therapy, Soucek et al generated a dominant negative c-Myc, omomyc, to inhibit C-, N-, and L-Myc together (82). Activation of KRASG12D in the lung for six weeks resulted in multiple neoplastic lesions, systemic expression of omomyc shrank the lesions with increased apoptosis by one week and completely resolved the lesions by one month (82). To determine the role of p53 in the successful treatment, KRASG12D lung tumors were generated in the absence of p53, which increased tumor burden, but omomyc remained effective (83). When lymphomas, liver tumors, or osteosarcomas were induced by c-Myc overexpression, subsequent de-expression regressed tumors with senescence that depended on p53 or pRb (84). Possibly, systemic inhibition of Myc is important since Myc may function in stroma cells to support tumorigenesis (85). Bromodomain inhibitors have shown efficacies targeting Myc (86); it will be interesting to determine whether they could remain effective without p53.
Two synthetic lethal relationships between PI3K(Pten)-Akt and Trp53
As discussed above, activation of PI3K(Pten)-Akt activates p53 (43, 44) and activated p53 inhibit PI3K(Pten)-Akt (42) (Figure 4a). And combined deletion of Pten and Trp53 induces tumorigenesis (77). Interestingly, two alternative scenarios might turn p53 deficiency into therapy advantage (Figure 4b, c). IGFs are typical activators of PI3K(Pten)-Akt. IGF1R ligand IGF2 is often overexpressed in human cancers. Haley et al studied physiological relationship between IGF2 and p53 (87). Mating of IGF2 and Trp53 doubly heterozygous mice produced only 16 live births out of the expected 44 doubly deficient embryos, revealing partial synthetic lethality during embryogenesis. The authors then used conditional deletions to generate doubly deficient mice and found them partially protected from spontaneous tumorigenesis, extending survival from 206 days (p53 cKO mice) to 325 days (cDKO mice), revealing partial synthetic lethality in tumorigenesis as well (88) (Figure 4b).
Emerling et al based their study on frequent co-occurrences of TP53 mutation and PIP4K2B gain in breast cancer (89). As shown in Figure 4c, PIP4K2B encodes for type 2 phosphatidylinositol-5-phosphate 4-kinases β (PI5P4Kβ), which generates PI3K substrate PI-4,5-P2 from PI-5-P. PIP4K2B KO mice grew to adulthood, but PIP4K2B and Trp53 DKO embryos died in uterus. PIP4K2B+/− Trp53−/− mice grew up and showed identical tumor susceptibility as Trp53−/− mice, but additional deletion of PIP4K2A protected PIP4K2A−/− PIP4K2B+/− Trp53−/− mice from tumorigenesis and prolonged survival (P=0.0166). We summarize these findings in Figure 4c. PIP4K2A−/− PIP4K2B+/− seemed a sweet spot: it is just adequate to survive regardless of p53 status; but when p53 is deleted it protected the mice from p53 deficient tumorigenesis. Further reduction in PIP4K2 dosage might produce more dramatic effects if embryo lethality could be avoided by conditional deletions and when the effects of PIP4K2C deletion are tested. Understanding the underlying mechanisms between these synthetic lethal relationships should help identify strategies to most effectively use them.
Deletion of MK2 improved DNA damaging chemotherapy for p53 deficient cancer
DNA damage activates ATM-Chk2, ATR-Chk1, and p38-MK2 kinase cascades to activate p53 to induce apoptosis, which is the rationale of DNA damaging chemotherapies. When p53 is inactivated, Chk2, Chk1, and MK2 can phosphorylate and inhibit Cdc25 to arrest cell cycle for repair, which forms the basis for therapy resistance. If this cell cycle arrest can be prevented, cells with damaged DNA would progress and die by mitotic catastrophe, which forms the rationale to treat p53 deficient cancers (90). In addition to Cdc25 (91), MK2 can phosphorylate hnRNPA0 and PARN to stabilize Gadd45a mRNA (92), induce expression of miR-34c to repress c-Myc (93), and inhibit DNA replication fork movement to stall DNA synthesis (94). MK2 therefore may be most responsible for protecting p53 deficient cells from DNA damaging therapy. When DNA damaging therapy was combined with MK2 inhibition, p53 deficient cancer cells preferentially responded with mitotic catastrophe (95). Inhibition of ATM or Chk2 showed similar effects (96). MK2 studies have now progressed to an autochthonous KRASG12D lung tumor model (97). A new design of Cre mediated recombination generated both MK2+/+ and MK2−/− tumors in the same mice. When the lung tumors were p53+/+, cisplatin treatment showed mild effects on MK2+/+ and MK2−/− tumors. When Trp53 was deleted, cisplatin inhibited MK2−/− tumors with apoptosis much more effectively than MK2+/+ tumors.
p53 deficient cancer could be more vulnerable to metabolic disruptions
A number of metabolic enzymes are encoded by p53 target genes and p53 regulates their expression to improve homeostasis (98). This role of p53 theoretically could benefit tumor cells too. If true, loss of p53 could render tumor cells more vulnerable to metabolism disturbances. We use a few studies to explore this emerging concept.
p53 directly activates expression of TIGAR (99) (Figure 2a). TIGAR indirectly inhibits phosphofructose kinase 1 (PFK1). PFK1 generates fructose 1,6-bisphsphoate in the glycolytic cascade towards pyruvate. Through TIGAR, p53 restricts the flux of glycolysis down from this point and divert the flux towards PPP (100). PPP generates ribose 5-phosphate and reductant NADPH. By activating TIGAR, p53 therefore could support proliferating cells, including tumor cells. Data for these speculations are now available. Deletion of TIGAR impaired intestine mucosa regeneration after injury (normal cell proliferation) and inhibited intestinal tumorigenesis by APC (tumor cell proliferation) (101). On the glutamine side of cancer cell metabolism, p53 activates expression of glutaminase (GLS2) (102, 103) (Figure 2b). Glutamate feeds into the TCA cycle to replenish the anaplerotic carbon skeleton for biosynthesis and generates glutathione for antioxidant defense, again both are advantageous for proliferating cells. Interestingly, when both studies overexpressed GLS2 in lung cancer cell line H1299, cell proliferation in culture was inhibited. While it is not clear how enhanced GLS2 inhibited cell proliferation, these results raised the possibility that highly active p53 might use this mechanism to suppress tumorigenesis.
Interestingly, p53 also functions to inhibit the generation of NADPH. p53 binds and inhibits G6PD in the cytoplasm (104) and represses the expression of malic enzymes (105), both leading to reduction of NADPH (Figure 2c). The consequent buildup of ROS activates p53 via AMPK, establishing a feed-forward loop to senescence (105). However, knockdown of ME1,2 inhibited xenograft growth by HCT116 regardless of their p53 status. Thus, whether p53 uses this mechanism to induce senescence in tumor cells remains unclear. On the other hand, these findings suggest that inhibiting ME1,2 (or otherwise inhibiting NADPH synthesis) could remain effective when p53 is genetically inactivated.
Using the isogenic p53 WT and KO HCT116 cells, Maddocks et al determined the role of p53 in serine withdrawal challenge (106). While withdrawal of the essential amino acid lysine inhibited proliferation of p53 WT and KO cells equally, withdrawal of the non-essential amino acid serine inhibited proliferation of p53 KO cells more significantly than p53 WT cells in culture and in xenografts. In this case, serine withdrawal creates a shortage of purine nucleotides to activate p53. Subsequent increase in p21 stimulated pRb to repress E2F, blocking DNA synthesis to reduce ROS production, and inhibiting nucleotide synthesis to favor glutathione synthesis (Figure 2d). Lack of these two mechanisms in p53 KO cells led to ROS buildup and cell death. These findings are exciting not only because p53 deficiency generated a metabolic vulnerability but also because serine is a non-essential amino acid. Its withdrawal therefore can be tolerated by normally proliferating cells.
Deletion of Skp2 blocked pRb and p53 doubly deficient tumorigenesis co-existent with ongoing DNA synthesis
As the two major tumor suppressors, pRb and p53 together may be responsible for most and best of cells’ antitumor mechanisms, known (Figure 1) and yet to be learned (see above). Genetic inactivation of both pRb and p53 would disable these antitumor mechanisms permanently. Since Skp2 deletion induced synthetic lethality with Rb1 deletion in pituitary melanotrophs (65, 67) and p53 independent senescence in prostate (78), Zhao et al hypothesized that it could block pRb and p53 doubly deficient tumorigenesis in pituitary melanotrophs and prostate. Their study (107) showed that p53 activates expression of Pirh2 and PKC1, two of the three (and Skp2) E3 ubiquitin ligases for p27 (108). Co-deletion of Skp2 and p53 reduced the cellular pool of p27 ubiquitin ligases to allow p27 protein to accumulate in pituitary and prostate. High levels of p27 inhibited cyclin dependent kinases to activate pRb to induce the p53-independent senescence observed by Lin et al (78), but additional deletion of Rb1 rendered DNA replication uninhabitable by p27 and abrogated the senescence. However, inhibition of mitotic cyclin/Cdk by p27 arrested cells in G2/M without pRb. The uninhabitable DNA replication was then maintained as DNA re-replication. E2F1 mediated apoptosis by co-deletion of Rb1 and Skp2 (67) likely could still operate to gradually clear the mitotically blocked tumorigenic cells (please see Figure 5c for more details).
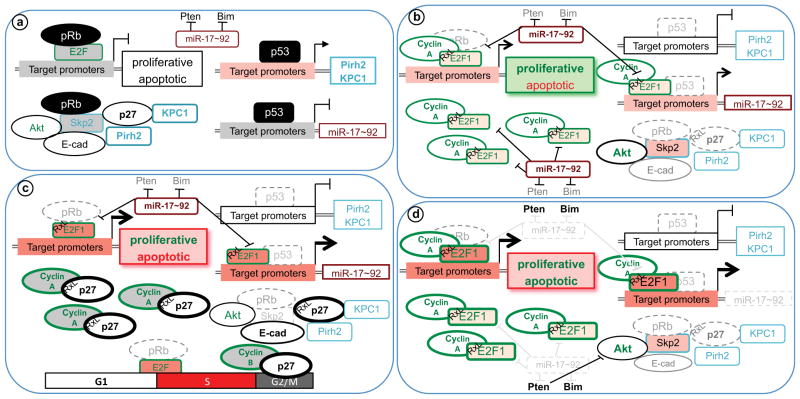
Figure 5. Antitumor mechanisms (experimentally shown and hypothetical) for pRb and p53 doubly deficient tumorigenesis in pituitary, prostate, and retina.
(a) illustrates a normal cell, in which pRb and p53 suppress tumorigenesis using mechanisms involved in studies by Zhao et al and Nittner et al (107, 118). In addition to degrading p27, Skp2 can ubiquitinate and degrade E-cadherin (122), ubiquitinate and activate Akt to promote cancer cell metabolism (121), and many other substrates such as FOXO1 (149). In addition to Pten, Bim and other tumor suppressive regulators, miR-17~92 represses E2F1,2,3 suggesting the importance of restraining these transcription factors in tumorigenesis (109). Molecules and actions are more active when written in bold, drawn in thicker lines, and shaded in bright colors in various combinations; the reverse is used when molecules and actions are repressed. (b) illustrates a tumor cell following loss of both pRb and p53. Prominent tumorigenic features include the activation of E2F1 and loss of p53 safeguard. Importantly, loss of pRb and p53 also enabled intrinsic antitumor mechanisms. An activated E2F1 can induce apoptosis and loss of p53 reduced expression of Pirh2 and KPC1 to render p27 protein more stable. Nevertheless, E2F1 activity is restrained by cyclin A binding through the RxL motif and by miR-17~92 mediated repression and p27 levels are kept low by more active Skp2. The outcome of these activating and restraining effects on E2F1 and p27 is the appropriate balance between proliferation and apoptosis for successful tumorigenesis. (c) displays the effects of combined Skp2 deletion. Lack of Skp2 decreased the pool of p27 ubiquitin ligases (Skp2, Pirh2, and KPC1) further to dramatically increase p27 to form more cyclin A-p27 complexes replacing cyclin A-E2F1 complexes and inhibit more cyclin B kinases. The freed E2F1 is further activated to promote proliferation, such that DNA synthesis became uninhabitable. Since expression of apoptotic target genes is also increased, apoptosis also became more prominent. Inhibition of mitotic cyclin B by p27 induces mitotic arrest in the absence of pRb. Combined, mitotic arrest and ongoing apoptosis blocked tumor progression in the presence of active DNA re-replication. Less active Akt and therefore a weaker cancer cell metabolism and more stable E-cadherin and therefore a stronger epithelial maintenance might also contribute in the life-long block. (d) suggests the effects of combined deletion of miR-17~92. Since miR-17~92 is a potent onco-miR, its absence naturally inhibits tumorigenesis, which could be achieved by elevated Pten and Bim. However, the requirement for loss of pRb to trigger synthetic lethality could suggest the involvement of E2F1’s apoptotic activity when it is not repressed by miR-17~92.
Deletion of miR-17~92 prevented Rb1, p107, and Trp53 TKO mouse retinoblastoma
miR-17~92 cluster is an onco-miR (109). Myc activates its expression (110) and it promotes Eμ-Myc lymphomagenesis (111). miR-17~92 cluster contains six microRNA units (109). Five of them can target Pten and Bim (a BH3 only apoptotic protein), explaining its oncogenic activity (109). Two of the units (miR-17 and miR-20a) however also target E2F1 (110). Interestingly, Myc activates expression of E2F1,2,3 mRNA (112, 113), suggesting a “promoting and dampening” regulation. Furthermore, E2F1,2,3 activate expression of Myc (114) and miR-17~92 (115, 116). While pRb represses E2F1,2,3, p53 represses expression of miR-17~92; and miR-17~92 is frequently overexpressed in human retinoblastoma (117, 118). Thus, miR-17~92 provides another mechanism to dampen E2F1 after loss of pRb (Figure 5a, b). Remarkably, deletion of miR-17~92 completely prevented TKO (Rb1, p107, Trp53) mouse retinoblastoma, resulting in normal retina (118). Using a Cre linked AP reporter, of all the possible genotype combinations, only QKO (miR-17~92 KO and TKO) retina were devoid of AP expression, suggesting that QKO synthetic lethally eliminated retina progenitor cells as soon as they were generated (please see Figure 5d for more details).
Summary and perspective
A number of antitumor mechanisms have been identified that can effectively inhibit tumorigenesis when the genes coding for pRb, p53, or both are deleted in autochthonous mouse tumor models. One of them, combining DNA damaging therapy with MK2 deletion, was rationally developed; most others were based on previous indirect rationales. Another approach to candidate mechanisms is genetic screen with lower model organisms, such as Drosophila (119, 120). Based on the findings so far, we predict continuous progress in these directions.
Over 25 years of studies predict that cancers with genetic inactivation of both pRb and p53 would be severely deficient in antitumor mechanisms
It is encouraging that evidence is now available that pRb and p53 doubly deficient tumorigenesis can be completely blocked at the levels of genetically defined autochthonous mouse tumor models. With Skp2 deletion, the block was nevertheless accompanied with life-long BrdU-labeling lesions, providing a new perspective in cancer treatment and raising concerns to address whether tumor evolution in this context will overcome the block. The lack of Skp2 mediated activation of Akt to promote cancer cell metabolism (121) and Skp2 mediated degradation of E-cadherin (122) might have kept the block durable. Blocking by deleting miR-17~92 involved deletion of p107, which added some uncertainty since p107 is not deleted in human cancer. Since miR-17~92 was deleted locally, the extent of side effects when miR-17~92 is targeted globally needs to be determined. Mice deficient for miR-17~92 died at birth with hypoplastic lung, heart ventricular septal defect, and B cell development failure (123).
Phamacological inhibition of Skp2 and miR-17~92 are rapidlly developing
Skp2 functions as a substrate recruiting subunit of SCFSkp2 ubiquitin ligase. Among the many SCFSkp2 inhibitors (124–126), Compound #25 (C25) has shown inhibitory effects on xenograft tumors by human lung cancer cell line A549 (activating mutation G12S in KRAS) and prostate cancer cell lines PC3 (WT RB1, frameshift mutation in TP53), after systemic administration (i.p. injection) (126). This line of preclinical studies could be directly extended to test pRb and p53 doubly deficient human cancer cell lines.
An important feature of p27 polyubiquitnation by SCFSkp2 is the additional requirement for Cks1 (127, 128), which provided an opportunity to selectively inhibit SCFSkp2-Cks1 (129–131). On the other hand, since combined inactivation of Skp2 and p53 reduced the cellular pool of p27 ubiquitin ligases including Skp2, Pirh2, and PKC1 to accumulate more p27 protein (107), broader inhibition of p27 ubiquitination and degradation might prove beneficial. Remarkably, the proteasome inhibitor Argyrin A inhibited various tumor cell lines by stablizing p27 protein in them (132). Finally, SCFSkp2 mediates K63-linked ubiquitination of Akt for activation and this function of SCFSkp2 promotes tumorigenesis by enhancing Warburg effects (121). C25 mediated inhibition of human cancer xenografts was correlated with both accumulation of p27 protein and prevention of Akt activation and Glut1 expression (126). More studies are needed to determine the relative contributions of Skp2 mediated p27 degradation and Akt activation.
The tiny (8-mer targeting seed sequence) LNA (locked nucleic acid) anti-miR technology can target groups of onco-miRs that share seed sequences, and require only saline-formulation for systemic adminstration (133, 134). The miR-17~92 cluster’s six miRNA components can be grouped into four seed sequences: miR-17,20a, miR-18a, miR-19a,19b-1, and miR-92-1. Tiny LNA miR-17 and tiny LNA miR-19 i.v. administration was well tolerated in adult mice while exhibited inhibitory effects on allograft subcutaneous and orthotopic tumors by Ptch1+/− Trp53−/− medulloblastoma cells (135). This approach could be used to determine the toxic effects of inhibiting all four seed sequences in the miR-17~92 cluster in adult mice and, if tolerated, the inhibitory effects on Rb1, p107, Trp53 TKO retinoblastoma. It is possible that inhibiting a subgroup of these four seed sequences could provide tumor inhibition without significant systemic toxicity.
Context is key
Most biological effects are context-dependent. The finality of gene deletions to inactivate both pRb and p53 may provide certain consistency in the context when one attempts to apply the above knowledge to other cancers and ultimately to clinical studies. In this regard, it is notable that genetic inactivation of TP53 is largely by point mutations in human cancers. For the bladder cancer that contained genetic inactivation of TP53 (67 out of 125 specimens, Figure 3a), p53 is null in 19 specimens by homozygous gene deletion, nonsense mutations, or frame-shift mutations. The rest are missense mutations. Some of them, such as p53R175H (2 specimens), R248Q (7 specimens), R273C (2 specimens), and R280T (3 specimens), gained new functions (GOF) (136). Mice with knockin of specific mutations, p53R172H/- or p53R270H/- (equivalent to R175H and R273H in humans), developed dramatically more carcinomas than p53 null mice. p53R273H and p53R280K can cooperate with SREBP to upregulate expression of mevalonate pathway enzymes to disrupt spheroid formation by mammary epithelial cells (137). When KRASG12D induced pancreatic cancer on the background of p53R172H/-, p53R172H binds p73 to dissociate p73/NF-Y interaction to activate expression of PDGFRB leading to metastasis, which is not observed on p53 WT or p53 null background (138–140). These and many other studies (136, 141) indicate the need to identify antitumor mechanisms that remain effective when p53 incurred GOF mutations and pRb is genetically inactivated.
Acknowledgments
Work in the authors’ laboratory was supported by NIH grants RO1CA127901 and RO1CA131421. HZ is a recipient of DOD PCRP Postdoctoral Fellowship (PC121837), and LZ acknowledges support from the Irma T. Hirschl Career Scientist Award.
Footnotes
The authors declare that no conflict of interest exists.
References
- 1.Burd CE, Sorrentino JA, Clark KS, Darr DB, Krishnamurthy J, Deal AM, et al. Monitoring tumorigenesis and senescence in vivo with a p16(INK4a)-luciferase model. Cell. 2013 Jan 17;152(1–2):340–51. doi: 10.1016/j.cell.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vora SR, Juric D, Kim N, Mino-Kenudson M, Huynh T, Costa C, et al. CDK 4/6 Inhibitors Sensitize PIK3CA Mutant Breast Cancer to PI3K Inhibitors. Cancer Cell. 2014 Jul 14;26(1):136–49. doi: 10.1016/j.ccr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chicas A, Wang X, Zhang C, McCurrach M, Zhao Z, Mert O, et al. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell. 2010 Apr 13;17(4):376–87. doi: 10.1016/j.ccr.2010.01.023. Epub 2010/04/14. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talluri S, Isaac CE, Ahmad M, Henley SA, Francis SM, Martens AL, et al. A G1 checkpoint mediated by the retinoblastoma protein that is dispensable in terminal differentiation but essential for senescence. Mol Cell Biol. 2010 Feb;30(4):948–60. doi: 10.1128/MCB.01168-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narita M, Nunez S, Heard E, Lin AW, Hearn SA, Spector DL, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003 Jun 13;113(6):703–16. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 6.Ianari A, Natale T, Calo E, Ferretti E, Alesse E, Screpanti I, et al. Proapoptotic function of the retinoblastoma tumor suppressor protein. Cancer Cell. 2009 Mar 3;15(3):184–94. doi: 10.1016/j.ccr.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dick FA, Dyson N. pRB contains an E2F1-specific binding domain that allows E2F1-induced apoptosis to be regulated separately from other E2F activities. Mol Cell. 2003 Sep;12(3):639–49. doi: 10.1016/s1097-2765(03)00344-7. [DOI] [PubMed] [Google Scholar]
- 8.Shats I, Gatza ML, Liu B, Angus SP, You L, Nevins JR. FOXO transcription factors control E2F1 transcriptional specificity and apoptotic function. Cancer Res. 2013 Oct 1;73(19):6056–67. doi: 10.1158/0008-5472.CAN-13-0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korenjak M, Anderssen E, Ramaswamy S, Whetstine JR, Dyson NJ. RBF binding to both canonical E2F targets and noncanonical targets depends on functional dE2F/dDP complexes. Mol Cell Biol. 2012 Nov;32(21):4375–87. doi: 10.1128/MCB.00536-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicolay BN, Gameiro PA, Tschop K, Korenjak M, Heilmann AM, Asara JM, et al. Loss of RBF1 changes glutamine catabolism. Genes Dev. 2013 Jan 15;27(2):182–96. doi: 10.1101/gad.206227.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsieh MC, Das D, Sambandam N, Zhang MQ, Nahle Z. Regulation of the PDK4 isozyme by the Rb-E2F1 complex. J Biol Chem. 2008 Oct 10;283(41):27410–7. doi: 10.1074/jbc.M802418200. [DOI] [PubMed] [Google Scholar]
- 12.Reynolds MR, Lane AN, Robertson B, Kemp S, Liu Y, Hill BG, et al. Control of glutamine metabolism by the tumor suppressor Rb. Oncogene. 2014 Jan 30;33(5):556–66. doi: 10.1038/onc.2012.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clem BF, Chesney J. Molecular pathways: regulation of metabolism by RB. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012 Nov 15;18(22):6096–100. doi: 10.1158/1078-0432.CCR-11-3164. [DOI] [PubMed] [Google Scholar]
- 14.Nicolay BN, Dyson NJ. The multiple connections between pRB and cell metabolism. Current opinion in cell biology. 2013 Dec;25(6):735–40. doi: 10.1016/j.ceb.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aksoy O, Chicas A, Zeng T, Zhao Z, McCurrach M, Wang X, et al. The atypical E2F family member E2F7 couples the p53 and RB pathways during cellular senescence. Genes Dev. 2012 Jul 15;26(14):1546–57. doi: 10.1101/gad.196238.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvajal LA, Hamard PJ, Tonnessen C, Manfredi JJ. E2F7, a novel target, is up-regulated by p53 and mediates DNA damage-dependent transcriptional repression. Genes Dev. 2012 Jul 15;26(14):1533–45. doi: 10.1101/gad.184911.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christophorou MA, Ringshausen I, Finch AJ, Swigart LB, Evan GI. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature. 2006 Sep 14;443(7108):214–7. doi: 10.1038/nature05077. [DOI] [PubMed] [Google Scholar]
- 18.Efeyan A, Garcia-Cao I, Herranz D, Velasco-Miguel S, Serrano M. Tumour biology: Policing of oncogene activity by p53. Nature. 2006 Sep 14;443(7108):159. doi: 10.1038/443159a. [DOI] [PubMed] [Google Scholar]
- 19.Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004 Apr;5(4):375–87. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 20.Zindy F, Williams RT, Baudino TA, Rehg JE, Skapek SX, Cleveland JL, et al. Arf tumor suppressor promoter monitors latent oncogenic signals in vivo. Proc Natl Acad Sci U S A. 2003 Dec 23;100(26):15930–5. doi: 10.1073/pnas.2536808100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young NP, Jacks T. Tissue-specific p19Arf regulation dictates the response to oncogenic K-ras. Proc Natl Acad Sci U S A. 2010 Jun 1;107(22):10184–9. doi: 10.1073/pnas.1004796107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guerra C, Mijimolle N, Dhawahir A, Dubus P, Barradas M, Serrano M, et al. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003 Aug;4(2):111–20. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 23.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005 Aug 4;436(7051):642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 24.Sarkisian CJ, Keister BA, Stairs DB, Boxer RB, Moody SE, Chodosh LA. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nat Cell Biol. 2007 May;9(5):493–505. doi: 10.1038/ncb1567. [DOI] [PubMed] [Google Scholar]
- 25.Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, et al. Distinct thresholds govern Myc’s biological output in vivo. Cancer Cell. 2008 Dec 9;14(6):447–57. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen D, Kon N, Zhong J, Zhang P, Yu L, Gu W. Differential effects on ARF stability by normal versus oncogenic levels of c-Myc expression. Molecular cell. 2013 Jul 11;51(1):46–56. doi: 10.1016/j.molcel.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Junttila MR, Karnezis AN, Garcia D, Madriles F, Kortlever RM, Rostker F, et al. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature. 2010 Nov 25;468(7323):567–71. doi: 10.1038/nature09526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feldser DM, Kostova KK, Winslow MM, Taylor SE, Cashman C, Whittaker CA, et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010 Nov 25;468(7323):572–5. doi: 10.1038/nature09535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007 Feb 8;445(7128):661–5. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 30.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007 Feb 8;445(7128):656–60. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006 Dec 29;127(7):1323–34. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012 Jun 8;149(6):1269–83. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valente LJ, Gray DH, Michalak EM, Pinon-Hofbauer J, Egle A, Scott CL, et al. p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell reports. 2013 May 30;3(5):1339–45. doi: 10.1016/j.celrep.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 34.Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011 May 13;145(4):571–83. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol. 2011 Mar;13(3):317–23. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon YJ, Volinia S, et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. The Journal of experimental medicine. 2011 May 9;208(5):875–83. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim NH, Kim HS, Li XY, Lee I, Choi HS, Kang SE, et al. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. The Journal of cell biology. 2011 Oct 31;195(3):417–33. doi: 10.1083/jcb.201103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011 Feb;17(2):211–5. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Godar S, Ince TA, Bell GW, Feldser D, Donaher JL, Bergh J, et al. Growth-inhibitory and tumor- suppressive functions of p53 depend on its repression of CD44 expression. Cell. 2008 Jul 11;134(1):62–73. doi: 10.1016/j.cell.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan HL, Xue G, Mei Q, Wang YZ, Ding FX, Liu MF, et al. Repression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosis. EMBO J. 2009 Sep 16;28(18):2719–32. doi: 10.1038/emboj.2009.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010 Aug 6;142(3):409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends in cell biology. 2010 Jul;20(7):427–34. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alimonti A, Nardella C, Chen Z, Clohessy JG, Carracedo A, Trotman LC, et al. A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J Clin Invest. 2010 Mar;120(3):681–93. doi: 10.1172/JCI40535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Astle MV, Hannan KM, Ng PY, Lee RS, George AJ, Hsu AK, et al. AKT induces senescence in human cells via mTORC1 and p53 in the absence of DNA damage: implications for targeting mTOR during malignancy. Oncogene. 2012 Apr 12;31(15):1949–62. doi: 10.1038/onc.2011.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, et al. Regulation of PTEN transcription by p53. Molecular cell. 2001 Aug;8(2):317–25. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 46.Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007 Apr 1;67(7):3043–53. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 47.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008 Aug 8;134(3):451–60. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014 Mar 20;507(7492):315–22. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010 Jul 13;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012 Jul 12;487(7406):239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008 Oct 23;455(7216):1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu XL, Fang Y, Lee TC, Forrest D, Gregory-Evans C, Almeida D, et al. Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling. Cell. 2009 Jun 12;137(6):1018–31. doi: 10.1016/j.cell.2009.03.051. Epub 2009/06/16. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laurie NA, Donovan SL, Shih CS, Zhang J, Mills N, Fuller C, et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006 Nov 2;444(7115):61–6. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- 54.Zhang J, Benavente CA, McEvoy J, Flores-Otero J, Ding L, Chen X, et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature. 2012 Jan 19;481(7381):329–34. doi: 10.1038/nature10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McEvoy J, Nagahawatte P, Finkelstein D, Richards-Yutz J, Valentine M, Ma J, et al. RB1 gene inactivation by chromothripsis in human retinoblastoma. Oncotarget. 2014 Jan 30;5(2):438–50. doi: 10.18632/oncotarget.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamasaki L, Bronson R, Williams BO, Dyson NJ, Harlow E, Jacks T. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1(+/−)mice. Nat Genet. 1998 Apr;18(4):360–4. doi: 10.1038/ng0498-360. [DOI] [PubMed] [Google Scholar]
- 57.Ziebold U, Lee EY, Bronson RT, Lees JA. E2F3 loss has opposing effects on different pRB-deficient tumors, resulting in suppression of pituitary tumors but metastasis of medullary thyroid carcinomas. Mol Cell Biol. 2003 Sep;23(18):6542–52. doi: 10.1128/MCB.23.18.6542-6552.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee EY, Cam H, Ziebold U, Rayman JB, Lees JA, Dynlacht BD. E2F4 loss suppresses tumorigenesis in Rb mutant mice. Cancer Cell. 2002 Dec;2(6):463–72. doi: 10.1016/s1535-6108(02)00207-6. [DOI] [PubMed] [Google Scholar]
- 59.Lasorella A, Rothschild G, Yokota Y, Russell RG, Iavarone A. Id2 mediates tumor initiation, proliferation, and angiogenesis in Rb mutant mice. Mol Cell Biol. 2005 May;25(9):3563–74. doi: 10.1128/MCB.25.9.3563-3574.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takahashi C, Contreras B, Bronson RT, Loda M, Ewen ME. Genetic interaction between Rb and K-ras in the control of differentiation and tumor suppression. Mol Cell Biol. 2004 Dec;24(23):10406–15. doi: 10.1128/MCB.24.23.10406-10415.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takahashi C, Contreras B, Iwanaga T, Takegami Y, Bakker A, Bronson RT, et al. Nras loss induces metastatic conversion of Rb1-deficient neuroendocrine thyroid tumor. Nat Genet. 2006 Jan;38(1):118–23. doi: 10.1038/ng1703. [DOI] [PubMed] [Google Scholar]
- 62.Lin W, Cao J, Liu J, Beshiri ML, Fujiwara Y, Francis J, et al. Loss of the retinoblastoma binding protein 2 (RBP2) histone demethylase suppresses tumorigenesis in mice lacking Rb1 or Men1. Proc Natl Acad Sci U S A. 2011 Aug 16;108(33):13379–86. doi: 10.1073/pnas.1110104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ji P, Jiang H, Rekhtman K, Bloom J, Ichetovkin M, Pagano M, et al. An Rb-Skp2-p27 pathway mediates acute cell cycle inhibition by Rb and is retained in a partial-penetrance Rb mutant. Mol Cell. 2004 Oct 8;16(1):47–58. doi: 10.1016/j.molcel.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 64.Binne UK, Classon MK, Dick FA, Wei W, Rape M, Kaelin WG, Jr, et al. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat Cell Biol. 2007 Feb;9(2):225–32. doi: 10.1038/ncb1532. [DOI] [PubMed] [Google Scholar]
- 65.Wang H, Bauzon F, Ji P, Xu X, Sun D, Locker J, et al. Skp2 is required for survival of aberrantly proliferating Rb1-deficient cells and for tumorigenesis in Rb1+/− mice. Nat Genet. 2010 Jan;42(1):83–8. doi: 10.1038/ng.498. Epub 2009/12/08. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malek NP, Sundberg H, McGrew S, Nakayama K, Kyriakidis TR, Roberts JM. A mouse knock-in model exposes sequential proteolytic pathways that regulate p27Kip1 in G1 and S phase. Nature. 2001 Sep 20;413(6853):323–7. doi: 10.1038/35095083. [DOI] [PubMed] [Google Scholar]
- 67.Lu Z, Bauzon F, Fu H, Cui J, Zhao H, Nakayama K, et al. Skp2 suppresses apoptosis in Rb1-deficient tumours by limiting E2F1 activity. Nat Commun. 2014;5:3463. doi: 10.1038/ncomms4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krek W, Ewen ME, Shirodkar S, Arany Z, Kaelin WG, Jr, Livingston DM. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell. 1994 Jul 15;78(1):161–72. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 69.Krek W, Xu G, Livingston DM. Cyclin A-kinase regulation of E2F-1 DNA binding function underlies suppression of an S phase checkpoint. Cell. 1995 Dec 29;83(7):1149–58. doi: 10.1016/0092-8674(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 70.Dynlacht BD, Flores O, Lees JA, Harlow E. Differential regulation of E2F trans-activation by cyclin-cdk2 complexes. Genes Dev. 1994;8:1772–86. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- 71.Xu M, Sheppard KA, Peng CY, Yee AS, Piwnica-Worms H. Cyclin A/CDK2 binds directly to E2F-1 and inhibits the DNA-binding activity of E2F-1/DP-1 by phosphorylation. Mol Cell Biol. 1994 Dec;14(12):8420–31. doi: 10.1128/mcb.14.12.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen D, Livne-bar I, Vanderluit JL, Slack RS, Agochiya M, Bremner R. Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell. 2004 Jun;5(6):539–51. doi: 10.1016/j.ccr.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 73.Zhang J, Schweers B, Dyer MA. The first knockout mouse model of retinoblastoma. Cell Cycle. 2004 Jul;3(7):952–9. [PubMed] [Google Scholar]
- 74.Sangwan M, McCurdy SR, Livne-Bar I, Ahmad M, Wrana JL, Chen D, et al. Established and new mouse models reveal E2f1 and Cdk2 dependency of retinoblastoma, and expose effective strategies to block tumor initiation. Oncogene. 2012 Nov 29;31(48):5019–28. doi: 10.1038/onc.2011.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Besson A, Hwang HC, Cicero S, Donovan SL, Gurian-West M, Johnson D, et al. Discovery of an oncogenic activity in p27Kip1 that causes stem cell expansion and a multiple tumor phenotype. Genes Dev. 2007 Jul 15;21(14):1731–46. doi: 10.1101/gad.1556607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu L, Harlow E, Dynlacht BD. p107 uses a p21CIP1-related domain to bind cyclin/cdk2 and regulate interactions with E2F. Genes Dev. 1995;9:1740–52. doi: 10.1101/gad.9.14.1740. [DOI] [PubMed] [Google Scholar]
- 77.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005 Aug 4;436(7051):725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010 Mar 18;464(7287):374–9. doi: 10.1038/nature08815. Epub 2010/03/20. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010 Feb 18;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004 Apr;6(4):308–18. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 81.Dang CV. MYC on the path to cancer. Cell. 2012 Mar 30;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008 Oct 2;455(7213):679–83. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Soucek L, Whitfield JR, Sodir NM, Masso-Valles D, Serrano E, Karnezis AN, et al. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 2013 Mar 1;27(5):504–13. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc Natl Acad Sci U S A. 2007 Aug 7;104(32):13028–33. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sodir NM, Swigart LB, Karnezis AN, Hanahan D, Evan GI, Soucek L. Endogenous Myc maintains the tumor microenvironment. Genes Dev. 2011 May 1;25(9):907–16. doi: 10.1101/gad.2038411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011 Sep 16;146(6):904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haley VL, Barnes DJ, Sandovici I, Constancia M, Graham CF, Pezzella F, et al. Igf2 pathway dependency of the Trp53 developmental and tumour phenotypes. EMBO molecular medicine. 2012 Aug;4(8):705–18. doi: 10.1002/emmm.201101105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Clermont F, Nittner D, Marine JC. IGF2: the Achilles’ heel of p53-deficiency? EMBO molecular medicine. 2012 Aug;4(8):688–90. doi: 10.1002/emmm.201201509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Emerling BM, Hurov JB, Poulogiannis G, Tsukazawa KS, Choo-Wing R, Wulf GM, et al. Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell. 2013 Nov 7;155(4):844–57. doi: 10.1016/j.cell.2013.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reinhardt HC, Jiang H, Hemann MT, Yaffe MB. Exploiting synthetic lethal interactions for targeted cancer therapy. Cell Cycle. 2009 Oct 1;8(19):3112–9. doi: 10.4161/cc.8.19.9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Manke IA, Nguyen A, Lim D, Stewart MQ, Elia AE, Yaffe MB. MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G2/M transition and S phase progression in response to UV irradiation. Molecular cell. 2005 Jan 7;17(1):37–48. doi: 10.1016/j.molcel.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 92.Reinhardt HC, Hasskamp P, Schmedding I, Morandell S, van Vugt MA, Wang X, et al. DNA damage activates a spatially distinct late cytoplasmic cell-cycle checkpoint network controlled by MK2-mediated RNA stabilization. Molecular cell. 2010 Oct 8;40(1):34–49. doi: 10.1016/j.molcel.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cannell IG, Kong YW, Johnston SJ, Chen ML, Collins HM, Dobbyn HC, et al. p38 MAPK/MK2-mediated induction of miR-34c following DNA damage prevents Myc-dependent DNA replication. Proc Natl Acad Sci U S A. 2010 Mar 23;107(12):5375–80. doi: 10.1073/pnas.0910015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kopper F, Bierwirth C, Schon M, Kunze M, Elvers I, Kranz D, et al. Damage-induced DNA replication stalling relies on MAPK-activated protein kinase 2 activity. Proc Natl Acad Sci U S A. 2013 Oct 15;110(42):16856–61. doi: 10.1073/pnas.1304355110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell. 2007 Feb;11(2):175–89. doi: 10.1016/j.ccr.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jiang H, Reinhardt HC, Bartkova J, Tommiska J, Blomqvist C, Nevanlinna H, et al. The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev. 2009 Aug 15;23(16):1895–909. doi: 10.1101/gad.1815309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Morandell S, Reinhardt HC, Cannell IG, Kim JS, Ruf DM, Mitra T, et al. A reversible gene-targeting strategy identifies synthetic lethal interactions between MK2 and p53 in the DNA damage response in vivo. Cell reports. 2013 Nov 27;5(4):868–77. doi: 10.1016/j.celrep.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Berkers CR, Maddocks OD, Cheung EC, Mor I, Vousden KH. Metabolic regulation by p53 family members. Cell Metab. 2013 Nov 5;18(5):617–33. doi: 10.1016/j.cmet.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006 Jul 14;126(1):107–20. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 100.Lee P, Vousden KH, Cheung EC. TIGAR, TIGAR, burning bright. Cancer & metabolism. 2014;2(1):1. doi: 10.1186/2049-3002-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cheung EC, Athineos D, Lee P, Ridgway RA, Lambie W, Nixon C, et al. TIGAR is required for efficient intestinal regeneration and tumorigenesis. Dev Cell. 2013 Jun 10;25(5):463–77. doi: 10.1016/j.devcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci U S A. 2010 Apr 20;107(16):7455–60. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A. 2010 Apr 20;107(16):7461–6. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, et al. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol. 2011 Mar;13(3):310–6. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jiang P, Du W, Mancuso A, Wellen KE, Yang X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature. 2013 Jan 31;493(7434):689–93. doi: 10.1038/nature11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013 Jan 24;493(7433):542–6. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhao H, Bauzon F, Fu H, Lu Z, Cui J, Nakayama K, et al. Skp2 Deletion Unmasks a p27 Safeguard that Blocks Tumorigenesis in the Absence of pRb and p53 Tumor Suppressors. Cancer Cell. 2013 Nov 11;24(5):645–59. doi: 10.1016/j.ccr.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Starostina NG, Kipreos ET. Multiple degradation pathways regulate versatile CIP/KIP CDK inhibitors. Trends in cell biology. 2012 Jan;22(1):33–41. doi: 10.1016/j.tcb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell death and differentiation. 2013 Dec;20(12):1603–14. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005 Jun 9;435(7043):839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 111.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005 Jun 9;435(7043):828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sears R, Ohtani K, Nevins JR. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol Cell Biol. 1997 Sep;17(9):5227–35. doi: 10.1128/mcb.17.9.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Adams MR, Sears R, Nuckolls F, Leone G, Nevins JR. Complex transcriptional regulatory mechanisms control expression of the E2F3 locus. Mol Cell Biol. 2000 May;20(10):3633–9. doi: 10.1128/mcb.20.10.3633-3639.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hiebert SW, Lipp M, Nevins JR. E1A-dependent trans-activation of the human MYC promoter is mediated by the E2F factor. Proc Natl Acad Sci U S A. 1989 May;86(10):3594–8. doi: 10.1073/pnas.86.10.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007 Jan 26;282(4):2135–43. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 116.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007 Jan 26;282(4):2130–4. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 117.Conkrite K, Sundby M, Mukai S, Thomson JM, Mu D, Hammond SM, et al. miR-17~92 cooperates with RB pathway mutations to promote retinoblastoma. Genes Dev. 2011 Aug 15;25(16):1734–45. doi: 10.1101/gad.17027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nittner D, Lambertz I, Clermont F, Mestdagh P, Kohler C, Nielsen SJ, et al. Synthetic lethality between Rb, p53 and Dicer or miR-17-92 in retinal progenitors suppresses retinoblastoma formation. Nat Cell Biol. 2012 Sep;14(9):958–65. doi: 10.1038/ncb2556. [DOI] [PubMed] [Google Scholar]
- 119.Li B, Gordon GM, Du CH, Xu J, Du W. Specific killing of Rb mutant cancer cells by inactivating TSC2. Cancer Cell. 2010 May 18;17(5):469–80. doi: 10.1016/j.ccr.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang T, Liao Y, Hsu FN, Zhang R, Searle JS, Pei X, et al. Hyperactivated Wnt signaling induces synthetic lethal interaction with Rb inactivation by elevating TORC1 activities. PLoS genetics. 2014 May;10(5):e1004357. doi: 10.1371/journal.pgen.1004357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, et al. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012 May 25;149(5):1098–111. doi: 10.1016/j.cell.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Inuzuka H, Gao D, Finley LW, Yang W, Wan L, Fukushima H, et al. Acetylation-dependent regulation of Skp2 function. Cell. 2012 Jul 6;150(1):179–93. doi: 10.1016/j.cell.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008 Mar 7;132(5):875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D, et al. Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. 2008 May 1;111(9):4690–9. doi: 10.1182/blood-2007-09-112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rico-Bautista E, Yang CC, Lu L, Roth GP, Wolf DA. Chemical genetics approach to restoring p27Kip1 reveals novel compounds with antiproliferative activity in prostate cancer cells. BMC biology. 2010;8:153. doi: 10.1186/1741-7007-8-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chan CH, Morrow JK, Li CF, Gao Y, Jin G, Moten A, et al. Pharmacological Inactivation of Skp2 SCF Ubiquitin Ligase Restricts Cancer Stem Cell Traits and Cancer Progression. Cell. 2013 Aug 1;154(3):556–68. doi: 10.1016/j.cell.2013.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Spruck C, Strohmaier H, Watson M, Smith APL, Ryan A, Krek W, et al. A CDK-independent function of mammalian Cks1: targeting of SCFSkp2 to the CDK inhibitor p27Kip1. Mol Cell. 2001;7:639–50. doi: 10.1016/s1097-2765(01)00210-6. [DOI] [PubMed] [Google Scholar]
- 128.Ganoth D, Bornstein G, Ko TK, Larsen B, Tyers M, Pagano M, et al. The cell-cycle regulatory protein Cks1 is required for SCFSkp2-mediated ubiquitinylation of p27. Nat Cell Biol. 2001;3:321–4. doi: 10.1038/35060126. [DOI] [PubMed] [Google Scholar]
- 129.Wu L, Grigoryan AV, Li Y, Hao B, Pagano M, Cardozo TJ. Specific small molecule inhibitors of Skp2-mediated p27 degradation. Chemistry & biology. 2012 Dec 21;19(12):1515–24. doi: 10.1016/j.chembiol.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ooi LC, Watanabe N, Futamura Y, Sulaiman SF, Darah I, Osada H. Identification of small molecule inhibitors of p27(Kip1) ubiquitination by high-throughput screening. Cancer science. 2013 Nov;104(11):1461–7. doi: 10.1111/cas.12246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ungermannova D, Lee J, Zhang G, Dallmann HG, McHenry CS, Liu X. High-throughput screening AlphaScreen assay for identification of small-molecule inhibitors of ubiquitin E3 ligase SCFSkp2-Cks1. Journal of biomolecular screening. 2013 Sep;18(8):910–20. doi: 10.1177/1087057113485789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nickeleit I, Zender S, Sasse F, Geffers R, Brandes G, Sorensen I, et al. Argyrin a reveals a critical role for the tumor suppressor protein p27(kip1) in mediating antitumor activities in response to proteasome inhibition. Cancer Cell. 2008 Jul 8;14(1):23–35. doi: 10.1016/j.ccr.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 133.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011 Apr;43(4):371–8. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.van Rooij E, Kauppinen S. Development of microRNA therapeutics is coming of age. EMBO molecular medicine. 2014 Jul;6(7):851–64. doi: 10.15252/emmm.201100899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Murphy BL, Obad S, Bihannic L, Ayrault O, Zindy F, Kauppinen S, et al. Silencing of the miR-17~92 cluster family inhibits medulloblastoma progression. Cancer Res. 2013 Dec 1;73(23):7068–78. doi: 10.1158/0008-5472.CAN-13-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Muller PA, Trinidad AG, Caswell PT, Norman JC, Vousden KH. Mutant p53 regulates Dicer through p63-dependent and -independent mechanisms to promote an invasive phenotype. J Biol Chem. 2014 Jan 3;289(1):122–32. doi: 10.1074/jbc.M113.502138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, Moon SH, Rodriguez-Barrueco R, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012 Jan 20;148(1–2):244–58. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005 May;7(5):469–83. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 139.Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle B, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci U S A. 2010 Jan 5;107(1):246–51. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Weissmueller S, Manchado E, Saborowski M, Morris JPt, Wagenblast E, Davis CA, et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor beta signaling. Cell. 2014 Apr 10;157(2):382–94. doi: 10.1016/j.cell.2014.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jackson JG, Lozano G. The mutant p53 mouse as a pre-clinical model. Oncogene. 2013 Sep 12;32(37):4325–30. doi: 10.1038/onc.2012.610. [DOI] [PubMed] [Google Scholar]
- 142.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011 Mar 4;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 143.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nature reviews Cancer. 2008 Sep;8(9):671–82. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dick FA, Rubin SM. Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol. 2013 May;14(5):297–306. doi: 10.1038/nrm3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009 May 15;137(4):609–22. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nature reviews Cancer. 2014 May;14(5):359–70. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dang CV. Links between metabolism and cancer. Genes Dev. 2012 May 1;26(9):877–90. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nature reviews Cancer. 2013 Feb;13(2):97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 149.Huang H, Regan KM, Wang F, Wang D, Smith DI, van Deursen JM, et al. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc Natl Acad Sci U S A. 2005 Feb 1;102(5):1649–54. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]