Abstract
MMS19 is an essential component of the cytoplasmic iron-sulfur (Fe-S) cluster assembly complex in fungi and mammals; the mms19 null mutant alleles are lethal. Our study demonstrates that MMS19/MET18 in Arabidopsis thaliana interacts with the cytoplasmic Fe-S cluster assembly complex but is not an essential component of the complex. We find that MMS19 also interacts with the catalytic subunits of DNA polymerases, which have been demonstrated to be involved in transcriptional gene silencing (TGS), DNA repair, and flowering time regulation. Our results indicate that MMS19 has a similar biological function, suggesting a functional link between MMS19 and DNA polymerases. In the mms19 null mutant, the assembly of Fe-S clusters on the catalytic subunit of DNA polymerase α is reduced but not blocked, which is consistent with the viability of the mutant. Our study suggests that MMS19 assists the assembly of Fe-S clusters on DNA polymerases in the cytosol, thereby facilitating transcriptional gene silencing, DNA repair, and flowering time control.
Introduction
Iron-sulfur (Fe-S) proteins are involved in fundamental cellular processes, such as electron transfer in mitochrondria and plastids, assembly of ribosomes, and DNA replication and repair [1–3]. The assembly of Fe-S clusters on their substrates depends on Fe-S cluster assembly proteins, most of which are evolutionarily conserved from microbes to mammals and plants. Fe-S cluster assembly is initiated in the mitochondria when a cysteine desulfurase complex Nfs1-Isd11 releases the sulfur required for iron-sulfur cluster formation [4]. The sulfur source is transferred to the mitochondrial ISC (iron-sulfur assembly complex) assembly machinery and may be exported to the cytosol by the ABC (ATP-binding cassette) transporter ATM1 for the assembly of cytosolic and nuclear Fe-S proteins [2]. In the cytosol of yeast, a diflavin reductase Tah18 first forms a complex with an Fe-S cluster-containing protein Dre2 for generation of stably inserted Fe-S clusters [5, 6], and subsequently Fe-S clusters are assembled on the scaffold complex CFD1-NBP35 [7]. Fe-S clusters are then released from NBP35 and transferred to target proteins, aided by the Fe-S protein NAR1 and the cytosolic iron-sulfur cluster assembly (CIA) complex [2, 7, 8]. NAR1 is likely to act as an adaptor between the CFD1-NBP35 complex and the CIA complex [7, 9]. A WD40-repeat protein CIA1 and a small acidic protein CIA2 are part of the CIA complex required for the maturation of Fe-S target proteins [10–12]. However, no CFD1 ortholog was found in Arabidopsis, where Fe-S clusters are assembled on the NBP35 homodimer [13].
MMS19 was primarily thought to be a component of the transcription factor TFIIH complex, which is involved in transcription, telomere length maintenance, and nucleotide excision repair in the nucleus [14–16]. Moreover, MMS19 was found in a chromatin silencing complex that contains MMS19, DOS2, RIK1, and CDC20 (the catalytic subunit of DNA polymerase ε) and is required for DNA replication, siRNA production, and heterochromatin assembly [17]. MMS19 is also required for proper chromosome segregation in human cells [18]. These findings suggest that MMS19 is involved in multiple processes in the nucleus. MMS19 was recently identified as a component of the CIA complex in yeast and mammals [3, 19, 20]. Using a yeast-two-hybrid assay, it was demonstrated that the Arabidopsis MMS19/MET18 (AT5G48120) physically interacts with CIA2/AE7 [8]. It is necessary to study how MMS19 regulates Fe-S cluster assembly and diverse nuclear processes in Arabidopsis.
DNA polymerases are central to the replication of double-stranded DNAs and are required for both heterochromatin silencing and DNA repair [21–23]. In Arabidopsis, several proteins involved in DNA replication and repair are required for transcriptional gene silencing (TGS) [23–25]. DNA polymerases α, δ, and ε are the main replicative DNA polymerases required for nuclear DNA replication in eukaryotes. DNA polymerase α is responsible for elongating the RNA primer with ~20 nucleotides. DNA polymerases δ and ε synthesize the lagging- and leading-strand DNAs, respectively [26]. The catalytic subunit of DNA polymerase ε contributes to the silencing of heterochromatin by associating with the silencing factors DOS2 and RIK1 in fission yeast [17]. In Arabidopsis, the null mutants of DNA polymerases α and ε are lethal [27–29], whereas their weak mutant alleles show pleotropic developmental phenotypes and are defective in DNA damage repair and TGS [30–32]. DNA polymerases α and ε are required for flowering repression, and their weak mutant alleles show an early flowering phenotype [29–31]. The DNA polymerases α, δ, and ε and the major DNA mutagenesis enzyme ζ were recently demonstrated to contain Fe-S clusters in the C-terminal domain (CTD) of their catalytic subunits in yeast [33]. Loss of Fe-S clusters in the DNA polymerase δ catalytic subunit Pol3 prevents the interaction of the catalytic subunit with the other subunits Pol31 and Pol32, suggesting that Fe-S clusters are necessary for the functioning of DNA polymerases. However, whether and how Fe-S clusters are assembled on DNA polymerases in plants remain unknown.
In this study, we demonstrate that MMS19 associates with the CIA complex in Arabidopsis, which is consistent with findings obtained with yeast and human cells [3, 19, 20]. Our gel filtration assay indicates, however, that MMS19 exists in a high-molecular-weight complex that differs from the other CIA complex components, suggesting a distinct role of MMS19 in Arabidopsis. Our study suggests that MMS19 in Arabidopsis enhances the assembly of Fe-S clusters on DNA polymerases and thereby contributes to TGS, DNA repair, and flowering time control.
Results
MMS19 associates with iron-sulfur assembly complex components and DNA polymerases
MMS19 was previously demonstrated to facilitate heterochromatin silencing in fission yeast, but the underlying mechanism is controversial [17, 19, 20]. Because MMS19 is conserved in eukaryotic organisms, it will be useful to determine whether and how MMS19 is involved in heterochromatin silencing in Arabidopsis. In Arabidopsis, MMS19 is a single protein and has no other homolog. We generated a construct harboring a native promoter-driven MMS19-Myc fusion sequence and transformed the construct into the mms19 T-DNA mutant (mms19-2: Salk_147068) to obtain MMS19-Myc transgenic plants. We used anti-Myc antibody-conjugated beads to purify MMS19-associating proteins in the MMS19-Myc transgenic plants. Mass spectrometric analysis indicated that many proteins were identified by from both the wild-type control and the MMS19-Myc transgenic plants; these proteins are thought to be contaminants. The proteins shown in Table 1 were identified from the MMS19-Myc transgenic plants but not from the wild-type control. MMS19 is the most abundant protein in affinity purification of MMS19-Myc (Table 1). Moreover, other proteins were co-purified, and these included the cytosolic Fe-S assembly complex components CIA1, CIA2/AE7, and NAR1, as well as the catalytic subunits of DNA polymerases α, δ, and ε (ICU2, AT5G63960, and ABO4), suggesting that MMS19 may associate with these proteins (Table 1). We generated CIA1-Flag and CIA2-Flag constructs and transformed the constructs into MMS19-Myc transgenic plants, thereby obtaining plants harboring both MMS19-Myc and CIA1-Flag or CIA2-Flag. The interactions of MMS19 with CIA1 and CIA2 were confirmed by co-immunoprecipitation (co-IP) in these transgenic plants (Fig 1A). In addition, the interaction of MMS19 with ICU2 was confirmed by co-IP in the transgenic plants harboring both MMS19-Flag and ICU2-Myc transgenes (Fig 1B).
Table 1. Mass spectrometric analyses of MMS19, CIA1, and CIA2 affinity.
| MMS19 in WT | CIA1 in WT | CIA1 in mms19 | CIA2 in WT | CIA2 in mms19 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AGI code | Protein | Mascot score | Unique peptides | Mascot score | Unique peptides | Mascot score | Unique peptides | Mascot score | Unique peptides | Mascot score | Unique peptides |
| AT5G48120 | MMS19 | 5429 | 21 | 1658 | 21 | 0 | 0 | 3455 | 31 | 0 | 0 |
| AT2G26060 | CIA1 | 631 | 12 | 9870 | 65 | 16371 | 89 | 2157 | 16 | 2485 | 20 |
| AT5G67100 | ICU2 | 480 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AT1G68310 | CIA2 | 412 | 5 | 1542 | 10 | 2713 | 16 | 6373 | 15 | 6356 | 23 |
| AT1G08260 | ABO4 | 354 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AT3G63460 | 347 | 7 | 140 | 2 | 0 | 0 | 182 | 3 | 69 | 1 | |
| AT5G63960 | Pol δ | 189 | 4 | 314 | 6 | 0 | 0 | 469 | 11 | 0 | 0 |
| AT2G16950 | TRN1 | 88 | 4 | 139 | 3 | 0 | 0 | 182 | 5 | 0 | 0 |
| AT5G20020 | RAN2 | 64 | 2 | 77 | 2 | 0 | 0 | 71 | 1 | 0 | 0 |
| AT4G16440 | NAR1 | 60 | 1 | 69 | 2 | 109 | 2 | 199 | 3 | 270 | 5 |
| AT3G50845 | 0 | 0 | 416 | 7 | 294 | 5 | 0 | 0 | 0 | 0 | |
| AT3G11830 | 0 | 0 | 361 | 8 | 624 | 12 | 161 | 4 | 147 | 5 | |
| AT5G62600 | MOS14 | 0 | 0 | 229 | 4 | 0 | 0 | 284 | 7 | 0 | 0 |
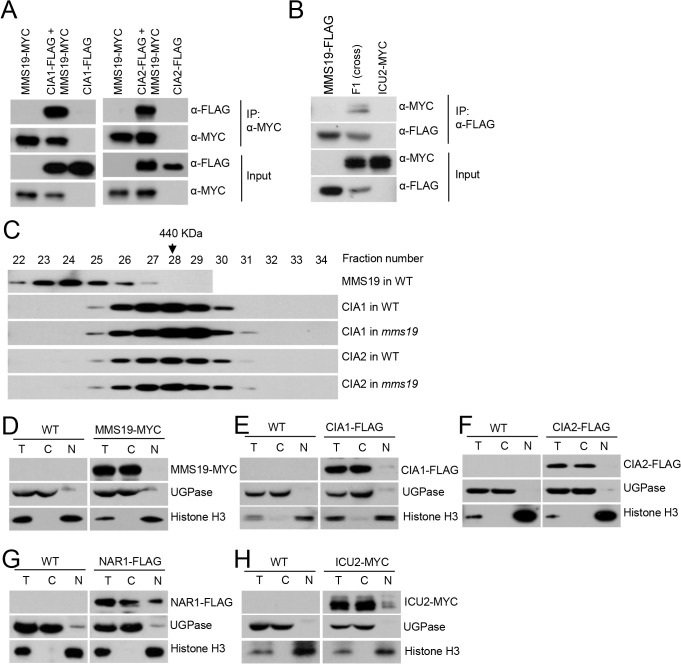
Fig 1. MMS19 interacts with the Fe-S assembly complex components CIA1 and CIA2 and the catalytic subunits of DNA polymerases in the cytoplasm.
(A) MMS19 interacts with CIA1 and CIA2 in vivo as determined by co-IP. (B) MMS19 interacts with ICU2 in vivo. (C) Gel filtration analysis of MMS19, CIA1, and CIA2. The protein extracts were isolated from flowers of transgenic plants (MMS19-Myc, CIA1-Flag, and CIA2-Flag) in the wild-type or mms19 mutant background and were loaded onto a Superose 6 10/300 GL column. The eluted fractions were run on an SDS-PAGE gel and subjected to Western blotting. The fraction numbers and sizes of standard proteins are shown. The subcellular localization of MMS19 (D), CIA1 (E), CIA2 (F), NAR1 (G), and ICU2 (H) as determined by nuclear-cytoplasmic fractionation. T: total extraction proteins, C: cytoplasmic proteins, N: nuclear proteins. MMS19, CIA1, and CIA2 mainly localized in the cytoplasm while NAR1 and ICU2 localized both in the cytoplasm and the nucleus. Histone H3 and UGPase were used as a nuclear marker and a cytoplasmic marker, respectively.
Total proteins were extracted from flowers and used for a gel filtration assay (Fig 1C). The result indicated that CIA1-Flag and CIA2-Flag were co-eluted at ~440 KD (Fig 1C). Previous studies demonstrated that the homologs of CIA1 and CIA2 in fungi and animals exist in a conserved CIA complex [10–12]. Our gel filtration results demonstrated that CIA1 and CIA2 are present in complexes of the same size (Fig 1C), suggesting that the Arabidopsis CIA1 and CIA2 also exist in a conserved CIA complex. We introduced the CIA1-Flag and CIA2-Flag constructs into the mms19 mutant background and found that the size of CIA1- and CIA2-containing complex is not affected in the mms19 mutant as compared to that in the wild type (Fig 1C), suggesting that MMS19 is not an essential component of the CIA complex even though it associates with CIA1 and CIA2. As determined by the gel filtration assay, we found that although the elution of the MMS19-containing complex is partially overlapped with the CIA1- and CIA2-containing complex, the MMS19-containing complex is mostly eluted in high-molecular-weight fractions in which CIA1 and CIA2 are not present (Fig 1C). While MMS19 associates with the CIA complex components CIA1 and CIA2, it may form a high-molecular-weight complex that is different from the CIA complex. As determined by affinity purification of MMS19 (Table 1), the high-molecular-weight MMS19-containing complex may contain DNA polymerases and other uncharacterized proteins. Whether and how these proteins act in Fe-S cluster assembly remain to be studied in future.
DNA polymerases α, δ, and ε are the main replicative DNA polymerases in eukaryotes. The catalytic subunits of the three polymerases in fission yeast have been demonstrated to be Fe-S proteins [33]. In our results, affinity purification of MMS19 co-purified all three polymerase catalytic subunits including ICU2, ABO4/TIL1, and AT5G63960, whereas affinity purification of CIA1 and CIA2 co-purified only AT5G63960 (Table 1). In the mms19 mutant background, however, AT5G63960 was undetectable in the CIA1 and CIA2 affinity-purified proteins (Table 1). MMS19 may facilitate the interaction of CIA1 and CIA2 with the catalytic subunit of DNA polymerase δ.
Subcellular localization of MMS19, CIA1, CIA2, and NAR1
Although MMS19 was recently found to be associated with Fe-S assembly in the cytoplasm [8, 19, 20], MMS19 was previously reported to directly associate with chromatin and act in heterochromatin silencing in the nucleus [17]. Subcellular localization of MMS19 was determined in MMS19-Flag transgenic plants. Nuclear proteins and cytosolic proteins were extracted from the transgenic plants and subjected to Western blotting assay. The antibodies against UGPase and histone H3 were used as markers for cytoplasmic and nuclear proteins, respectively. The results indicated that MMS19 is localized in the cytoplasm but not in the nucleus (Fig 1D). Subcellular localization of CIA1, CIA2, and NAR1 was determined in CIA1-Flag, CIA2-Flag, and NAR1-Flag transgenic plants, respectively. We found that CIA1 and CIA2 are cytoplasmic proteins and that NAR1 exists in both the cytoplasm and the nucleus (Fig 1E, 1F and 1G). The localization of MMS19, CIA1, and CIA2 in the cytoplasm is consistent with the inference that the CIA complex is responsible for Fe-S cluster assembly in the cytoplasm [8, 19, 20]. The yeast NAR1 is predominantly localized in the cytoplasm [9], whereas the Arabidopsis NAR1 is present in both the cytoplasm and the nucleus (Fig 1G). NAR1 was thought to transfer Fe-S clusters from the Fe-S cluster scaffold protein NBP35 to the CIA complex at an early step of the cytosolic Fe-S cluster assembly in yeast [10]. In Arabidopsis, the partially nuclear localization of NAR1 is probably required for Fe-S cluster assembly on nuclear Fe-S proteins.
Among the three DNA polymerases identified by affinity purification of MMS19 (Table 1), ICU2 and ABO4 were previously characterized [32]. However, it is difficult to understand how the cytoplasmic MMS19 interacts with ICU2 and ABO4, which are primarily thought to be located in the nucleus (Table 1 and Fig 1B). To determine the subcellular localization of ICU2 and ABO4, we primarily tried to generate tagged transgenic plants for both ICU2 and ABO4. Because the size of ABO4 is long (~16 Kb), we failed to clone ABO4. Thus, we obtained only ICU2-Myc transgenic plants. The ICU2-Myc signal is predominantly present in the cytoplasm rather than in the nucleus (Fig 1H), suggesting that the interaction of ICU2 with MMS19 occurs in the cytoplasm.
MMS19 is involved in transcriptional gene silencing
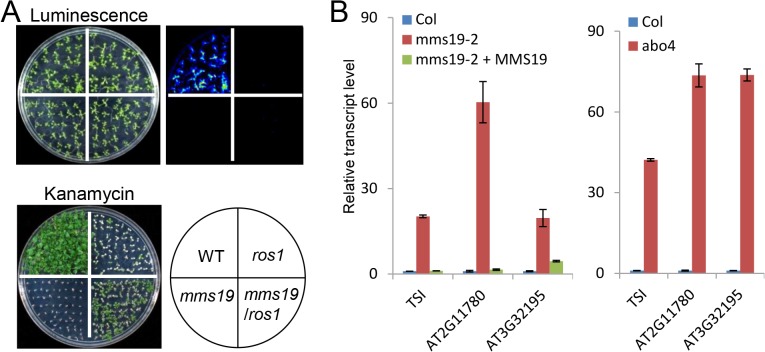
The catalytic subunits of the replicative DNA polymerases α, δ, and ε were co-purified by affinity purification of MMS19 (Table 1 and Fig 1B). Thus, we predicted that MMS19 is functionally associated with the DNA polymerases. Two individual mms19 mutants, mms19-1 (salk_121963) and mms19-2 (salk_147068) were obtained from Arabidopsis Stock Center. The transcript level of MMS19 was not detected in the two mutants by quantitative RT-PCR, indicating that the two mms19 mutants are null allele mutants (S1 Fig). In Arabidopsis, the catalytic subunits of the DNA polymerases α and ε (ICU2 and ABO4) were identified as regulators of TGS [30, 32]. The RD29A promoter-driven luciferase transgene (RD29A-LUC) and the accompanied 35S promoter-driven NPTII transgene (35S-NPTII) are expressed well in the C24 wild-type background but are silenced when the DNA demethylase gene ROS1 is depleted in the ros1 mutant [34]. In the ros1icu2 and ros1abo4 double mutants, the expression of 35S-NPTII but not of RD29A-LUC is reactivated, suggesting that ICU2 and ABO4 are required for the silencing of 35S-NPTII in the ros1 mutant background [30, 32]. To determine whether MMS19 is required for the silencing of RD29A-LUC and 35S-NPTII in the ros1 mutant background, we generated the ros1mms19 double mutant harboring both RD29A-LUC and 35S-NPTII by crossing. We found that although 35S-PTII exists in both the ros1 single mutant and the ros1mms19 double mutant, the ros1mms19 double mutant is much more resistant to kanamycin than the ros1 single mutant, suggesting that mms19 releases the silencing of 35S-NPTII. Meanwhile, the mms19 mutation has no effect on the silencing of RD29A-LUC as determined by luminescence imaging (Fig 2A). The results suggest that MMS19 is required for the DNA polymerase-mediated transgene silencing.
Fig 2. Silencing of 35S-NPTII transgene and endogenous transposable elements and other loci is affected in the mms19 mutant.
(A) The effect of mms19 on the silencing of RD29A-LUC and 35S-NPTII transgenes. Each genotype harboring the RD29A-LUC and 35S-NPTII transgenes was grown on MS medium for 14 days followed by cold treatment for 2 days at 4 °C. The treated seedlings were sprayed with luciferin for luminescence imaging. The seedlings were grown on MS medium supplemented with 150 mg/L kanamycin for 20 days and photographed. (B) The mms19 and abo4 mutants release the silencing of transposable elements. The transcript levels of the transposable element genes TSI, AT2G11780, and AT3G32195 were detected in the wild type, mms19-2 and its complementation line, and abo4 by quantitative RT-PCR. ACT7 was used as an internal control for normalization. Quantitative RT-PCR experiments were biologically repeated three times with similar results. Showing is the result of three technical replicates from one representative experiment.
TSI (transcriptional silencing information) loci are the pericentromeric Athila retrotransposons that are subjected to transcriptional silencing in the wild-type Arabidopsis [35]. The silencing of TSI is significantly released in the icu2 and abo4 mutants [30, 32]. We found that the silencing of TSI is also released in the two individual mms19 mutants and that this effect is rescued by the native promoter-driven MMS19-Myc transgene (Fig 2B and S2 Fig). Moreover, we found that the silencing of the transposon genes AT2G11780 and AT3G32195 is released by mms19 as well as by abo4-1 (Fig 2B and S2 Fig). These results demonstrate that MMS19 is functionally associated with the DNA polymerases α and ε in TGS.
MMS19 and ABO4 have a similar effect on the expression of genes and transposable elements (TEs)
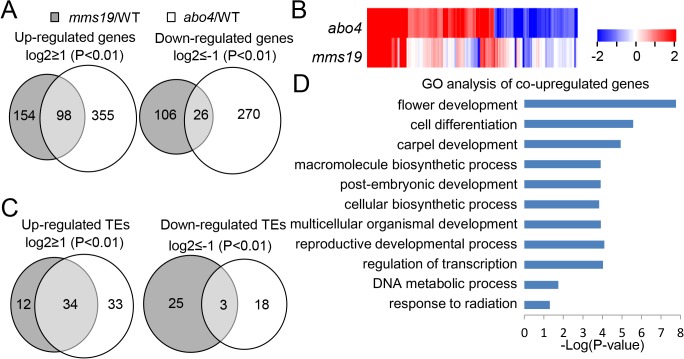
Because of the physical and functional association between MMS19 and ABO4, we performed RNA-sequencing (RNA-seq) analyses to determine whether MMS19 and ABO4 have a similar effect on the expression of genes and TEs. Although null mutant alleles of ABO4 are embryonically lethal [27, 28], the weak missense mutant allele abo4-1 is viable and is defective in TGS [30]. Thus, we used RNA-seq analyses to compare the effect of the null mms19-2 mutation and the weak abo4-1 mutation on the expression of genes and TEs. In total, 5.86x107, 5.37x107, and 5.12x107 reads were obtained from the RNA libraries of the wild type, mms19-1, and abo4-1, respectively, and most of the reads are uniquely matched on the Tair10 Arabidopsis genome. The RNA sequencing data indicated that 252 and 453 genes are significantly upregulated (log2(fold-change)>1; P<0.01) in mms19 and abo4, respectively (Fig 3A, S1 and S2 Tables). Of the 252 genes upregulated in mms19, 98 (98/252 = 38.9%) are also upregulated in abo4 (Fig 3A). The RNA sequencing data also indicated that 132 and 296 genes are significantly downregulated (log2(fold-change)<-1; P<0.01) in mms19 and abo4, respectively (Fig 3A, S1 and S2 Tables). Of the 132 genes downregulated in mms19, 26 (26/132 = 19.7%) are also downregulated in abo4 (Fig 3A, S1 and S2 Tables). The number of target genes shared by MMS19 and ABO4 is significantly greater than expected by random (P<0.01), suggesting that the two components have a similar effect on gene expression. Heat maps further confirmed that differentially expressed genes caused by mms19 and abo4 are positively correlated (Fig 3B), supporting a functional connection between MMS19 and ABO4. We randomly selected seven co-upregulated genes and three co-downregulated genes for validation by quantitative RT-PCR. The results demonstrated that the RNA-seq data are reliable (S3 Fig).
Fig 3. The effect of mms19 on the transcriptome as determined by RNA-seq analyses.
(A) Differentially expressed genes in mms19 and abo4 mutants relative to the wild type are shown by Venn diagrams. (B) Differentially expressed genes in mms19 and abo4 mutants relative to the wild type are shown by heat maps. (C) Differentially expressed TEs in mms19 and abo4 mutants relative to the wild type are shown by Venn diagrams. (D) Gene Ontology (GO) analysis of co-upregulated genes in mms19 and abo4 mutants. The lengths of bars represent statistical values of gene enrichment in the indicated biological processes. The biological processes are listed only when their genes are significantly (P<0.05) enriched.
We compared the effect of mms19 and abo4 on the silencing of TEs based on the RNA-seq data. The results indicated that 46 and 67 TEs are upregulated in mms19 and abo4, respectively (Fig 3C, S3 and S4 Tables). Among the 46 TEs upregulated in mms19, most (34/46 = 73.9%) are also upregulated in abo4 (Fig 3C). We randomly selected three co-upregulated TEs and validated their expression by quantitative RT-PCR; the results confirmed that all three TEs are co-upregulated (S3 Fig). These results suggest that MMS19 and ABO4 have a similar effect on the silencing of TEs. The numbers of downregulated TEs in mms19 and abo4 are 28 and 21, respectively (Fig 3C, S3 and S4 Tables). The downregulation of genes and TEs in mms19 and abo4 is probably due to an indirect effect of mms19 and abo4 on the expression of some uncharacterized transcriptional regulators.
Previous studies reported that icu2 and abo4 are defective in DNA repair and flowering time control [29, 30, 32]. Our RNA-seq data indicated that several DNA repair-related genes and flower development-related genes are upregulated not only in mms19 and but also in abo4 (S1 and S2 Tables). To characterize the common target genes shared by MMS19 and ABO4, we classified the co-upregulated genes by Gene Ontology (GO). The results suggest that flower development-related genes and DNA metabolic process genes are enriched (Fig 3D), which is consistent with the function of MMS19 and ABO4 in flowering time control and DNA metabolism. Additionally, we found that the common target genes of MMS19 and ABO4 are also highly correlated with several other biological processes (Fig 3D). These results will be valuable for understanding additional biological functions of MMS19 and DNA polymerase ε in Arabidopsis.
MMS19 is required for DNA repair
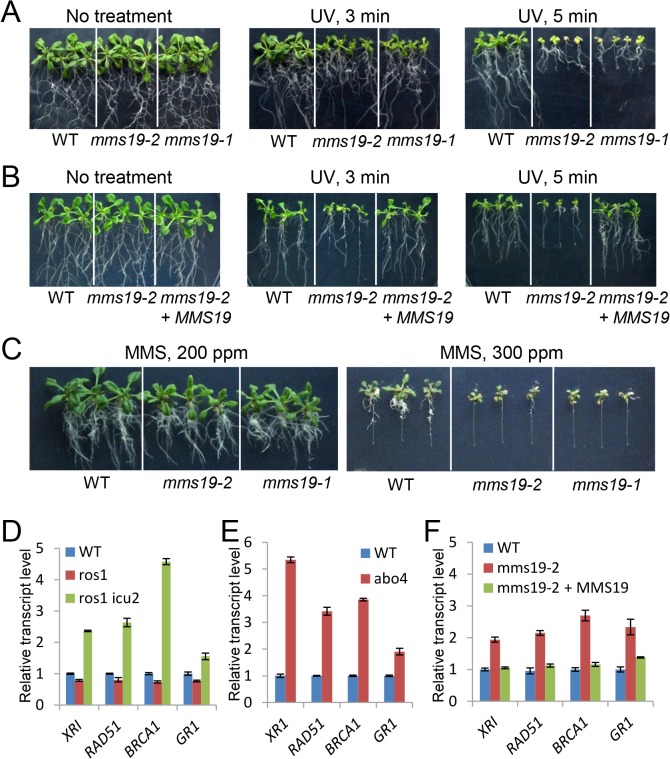
The two catalytic subunits of DNA polymerases, ICU2 and ABO4, are required for DNA repair [23, 30, 32]. That DNA repair-related genes present similar regulation in mms19 and abo4 mutants as determined by our RNA-seq data (S1 and S2 Tables, Fig 3D) suggested that MMS19 may also be required for DNA repair. Five-day-old seedlings of the wild-type control and the two independent mms19 mutants (mms19-1 and mms19-2) were subjected to UV treatment and then grown on vertical MS medium plates for 10 days. Growth was more severely retarded for mms19-1 and mms19-2 than for the wild type (Fig 4A), suggesting that the mms19 mutants are sensitive to UV treatment. Furthermore, the full-length MMS19 transgene restored the UV resistance of the mms19-2 mutant (Fig 4B). To confirm the function of MMS19 in DNA repair, we determined the sensitivity of mms19-1 and mms19-2 to the DNA-damaging regent MMS (methyl methanesulfonate). The result demonstrated that the two mms19 mutants are sensitive to MMS, especially when the concentration of MMS is increased to 300 ppm (Fig 4C). Therefore, like DNA polymerases α and ε, MMS19 is required for DNA repair.
Fig 4. The mms19 mutants are sensitive to DNA damage and constitutively express a high level of DNA repair-related genes.
(A) The mms19 mutants are more sensitive to UV-B treatment than the wild type. Five-day-old seedlings of the wild type and mms19 mutants were treated with UV-B light (1 kJ/m2) for different times and then grown with 16-h-light and 8-h-dark at 22° for 5–7 days before being photographed. (B) The sensitivity of the mms19-2 mutant to UV-B treatment was rescued by the MMS19-Myc transgene in T2 transgenic plants. (C) The mms19 mutants are more sensitive than the wild type to the DNA-damaging reagent methyl methanesulfonate (MMS). Quantitative RT-PCR results indicate that DNA repair-related genes were up-regulated in ros1icu2 (D), abo4 (E), and mms19 (F) mutants compared to their respective control plants. The seedlings were grown on MS medium plates for ~10 days under long-day-condition and then harvested for RNA isolation. Quantitative RT-PCR experiments were biologically repeated with similar results, and three technical replicates of one representative experiment were shown. Error bars represent the standard deviation (SD).
Previous studies reported that several DNA repair-related genes are upregulated by the mutations of the DNA polymerase genes ICU2 and ABO4 [30, 32]. ROS1 is responsible for DNA demethylation and repression of transgene silencing. In the ros1 mutant background, 35S-NPTII transgene is silenced. The mutation of ICU2 in the ros1 mutant background releases the silencing of 35S-NPTII transgene. The function of ICU2 in regulation of DNA repair-related genes was detected not only in the ros1 mutant background but also in the wild-type background [30]. Thus, the ros1icu2 mutant was used to determine the effect of icu2 on the expression of DNA repair-related genes (Fig 4D). Consistent with the previous studies, our quantitative RT-PCR results indicated that the transcript levels of the DNA repair-related genes XRI, RAD51, BRCA1, and GR1 are upregulated by icu2 and abo4 (Fig 4D and 4E). The effect of mms19 on the expression of the DNA repair-related genes was determined by quantitative RT-PCR. The results indicated that the DNA repair-related genes including XRI, RAD51, BRCA1, and GR1 are induced in the mms19-1 and mms19-2 mutants (Fig 4F and S2 Fig). Moreover, the full-length MMS19-Myc transgene restores the transcript levels of these genes in the mms19-2 mutants (Fig 4F). We further determined the effect of the mms19 mutations on the expression of these genes under genotoxic conditions (S4 Fig). The results indicated that XRI, RAD51, BRCA1, and GR1 are induced by MMS treatment not only in the wild type but also in the mms19 mutants (S4 Fig), which is consistent with the notion that these genes are induced by DNA double-strand breaks caused by MMS. Whether or not the seedlings were treated with MMS, the mms19 mutations affect the expression of DNA repair-related genes (S4 Fig). These results suggest that MMS19 is functionally associated with the DNA polymerases α and ε in DNA repair.
MMS19 is involved in flowering time control
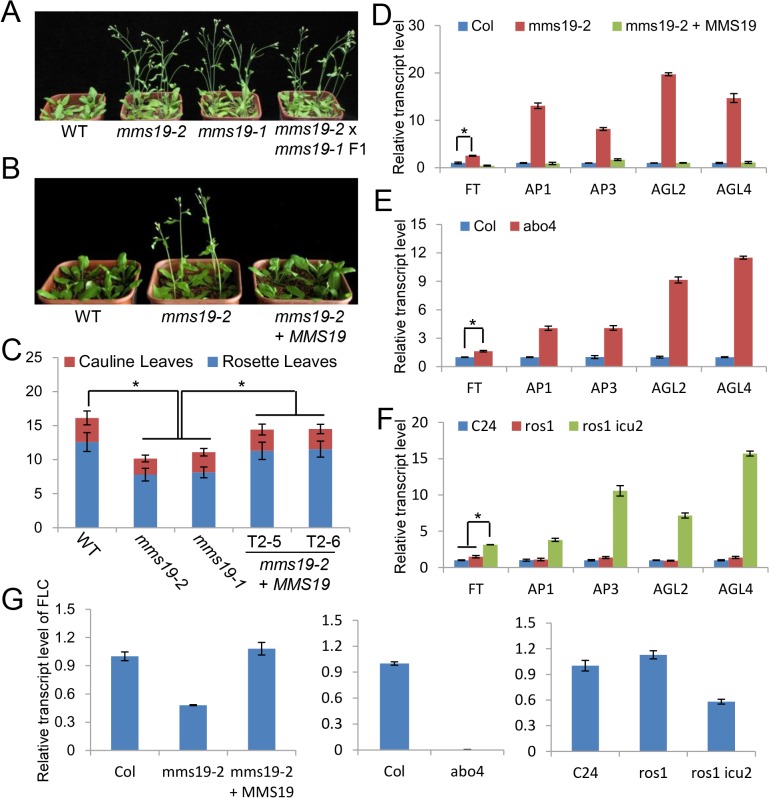
An early flowering phenotype was previously reported in icu2 and abo4 [30, 32]. We found a similar early flowering phenotype in two individual mms19 mutants, mms19-1 and mms19-2 (Fig 5A). An allelic test indicated that the F1 plants obtained from the crossing of mms19-1 and mms19-2 flower as early as their parent plants (Fig 5A), confirming that the early flowering phenotype is caused by the mms19 mutations in mms19-1 and mms19-2. Under long-day conditions (16 h light/8 h dark), the wild-type plants have an average of 12.6 rosette leaves upon flowering, while the two individual mms19 mutants flower with ~8.0 rosette leaves (Fig 5B and 5C). The full-length MMS19-Myc construct was transformed into the mms19-2 mutant for complementation testing. The MMS19-Myc transgene in two individual MMS19-Myc transgenic lines almost restores the flowering time of mms19 to the wild-type level (Fig 5B and 5C). These results suggest that flowering time is regulated not only by icu2 and abo4 but also by mms19.
Fig 5. The mms19 mutants cause an early flowering phenotype by affecting the expression of flowering-related genes.
(A) The early flowering phenotype of mms19 mutants in standard long-day conditions (16-h-light and 8-h-dark at 22°C). (B) The early-flowering phenotype was restored by the MMS19-Myc construction in mms19-2. (C) The statistics of total leaf numbers upon flowering under long-day conditions in the wild type, the mms19 mutants and the complementation lines. T2-5 and T2-6 were two randomly selected individual MMS19-MYC transgenic lines in T2 generation. At least 30 individual plants were counted. Error bars stand for SD. Asterisks indicate significant difference as determined by the t-test (P<0.05). Numbers of rosette and cauline leaves are indicated by blue and red bars, respectively. (D) Leaf numbers under long-day conditions with or without vernalization. (E) Leaf numbers under short-day conditions. (F), (G) and (H) The effect of mms19, abo4, and icu2 on the expression of flowering-related genes as determined by quantitative RT-PCR. ACT7 was amplified as an internal control. The quantitative RT-PCR experiments were biologically repeated for three times and indicated similar results. A representative repetition is shown. Error bars represent SD. Asterisks show significant difference as determined by the t-test (P<0.05).
Flowering time is regulated by various environmental and endogenous signaling pathways, which include vernalization, photoperiod, and autonomous pathways [36, 37]. When a vernalization treatment (4°C for 30 days) was applied, flowering was affected in the mms19 mutants as well as in the wild type (Fig 5D), indicating that the role of MMS19 in flowering time control is independent of the vernalization pathway. When photoperiod was changed from long-day (16-h-light and 8-h-dark) to short-day (8-h-light and 16-h-dark) conditions, the flowering time was delayed in both the wild type and the mms19 mutants (Fig 5E). However, the effect of the photoperiod change on flowering time is much smaller in the mms19 mutants than in the wild type (Fig 5E). The result suggests that the involvement of MMS19 in flowering time is regulated by photoperiod.
FLC acts as a central floral repressor through which the flowering time is regulated by vernalization, photoperiod, and autonomous pathways [37, 38]. FT is a key floral integrator and activates a number of downstream genes required for flower development [36, 39]. The expression of FT is directly repressed by FLC [40, 41]. The early flowering phenotype of icu2 and abo4 is related to the expression of FT [29, 30]. We determined whether the early flowering phenotype of the mms19 mutant is related to the expression of FLC, FT, and other floral integrator genes. Quantitative RT-PCR indicated that the floral integrator genes FT, AP1, AP2, AGL2, and AGL4 are markedly upregulated in the mms19 mutant and that the introduced MMS19-Myc transgene restores the expression of these genes to wild-type levels (Fig 5F). Moreover, the expression of these floral integrator genes is upregulated by both icu2 and abo4 as determined by quantitative RT-PCR (Fig 5G and 5H). Interestingly, the expression of the floral repressor gene FLC is not only downregulated by mms19 but also by icu2 and abo4 (Fig 5F, 5G and 5H). These results suggest that the involvement of MMS19 in flowering time control is likely related to the DNA polymerases α and ε.
MMS19 is involved in Fe-S assembly on DNA polymerase α
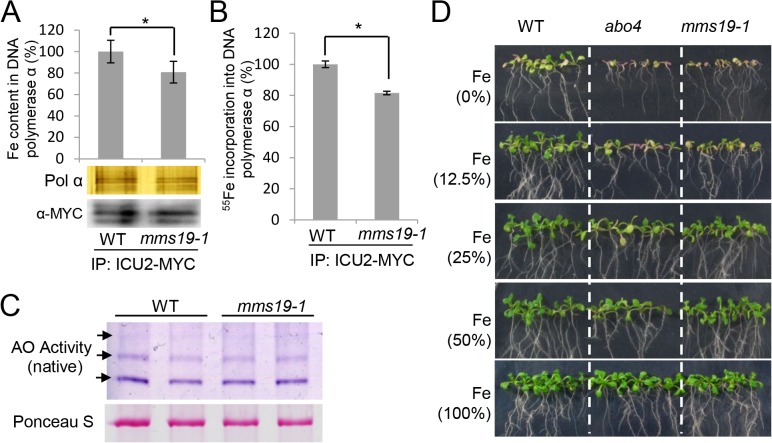
The MMS19 orthologs in yeast and mammals are thought to act as canonical components of the cytosolic Fe-S assembly complexes [19, 20]. Because the catalytic subunits of DNA polymerases α, δ, and ε in yeast contain Fe-S clusters [33], the finding of the interaction between MMS19 and DNA polymerases in Arabidopsis motivated us to determine whether MMS19 is involved in the assembly of Fe-S clusters on the catalytic subunits of DNA polymerases. We transformed an ICU2-Myc construct into the wild type and obtained a transgenic line harboring a single copy of the ICU2-Myc transgene. This ICU2-Myc transgenic line was crossed to the mms19 mutant to introduce the ICU2-Myc transgene into the mms19 mutant background. We performed quantitative PCR to confirm that the same copy of ICU2-Myc transgene is present in the wild type and the mms19 mutant (data not shown). The amount of ICU2-Myc protein was determined by both silver staining and western blotting (Fig 6A). The results suggest that the ICU2-Myc protein level is comparable between the wild type and the mms19 mutant (Fig 6A). By ICP mass spectrometric assay, we found that the Fe content in DNA polymerase α was less in the mms19 mutant than in the wild type (Fig 6A), suggesting that the Fe-S assembly of DNA polymerase α is affected in the mms19 mutant. To confirm the effect of mms19 on the Fe-S assembly of DNA polymerase α, we incubated Arabidopsis seedlings in radioactive 55Fe-labelled liquid MS medium and determined whether the incorporation of 55Fe in the DNA polymerase α is affected in the mms19 mutant. Liquid scintillation counting indicated that 55Fe incorporation is lower in the mms19 mutant than in the wild type (Fig 6B). The two different experiments demonstrated that Fe incorporation into ICU2 is reduced but not blocked in the mms19 mutant, suggesting that MMS19 is not an essential component of the Fe-S assembly complex but acts as an enhancer of the Fe-S assembly of DNA polymerase α. We cannot completely exclude the possibility, however, that the ICU2-Myc protein level is slightly reduced in the mms19 mutant compared to that in the wild type. If it is true, the lower amount of Fe content in the mms19 mutant is at least partially caused by the effect of mms19 on the accumulation of the ICU2-Myc protein.
Fig 6. The mms19 mutant incorporates a reduced level of Fe in ICU2 and is sensitive to Fe absence.
(A) Fe content of ICU2 in the wild type and mms19. The protein extraction was affinity purified by anti-Myc antibody-coupled beads and eluted three times by 0.1 M ammonium hydroxide. The three elutions were combined and subjected to Fe content detection by ICP-MS (Themo ICP-MS XII). The affinity purified ICU2-Myc in the wild type and the mms19 mutant was separated by SDS-PAGE and subjected to silver staining and western blotting as controls. The results of three biological repeats are indicated. Error bars show the SD. Asterisks indicate significant differences as determined by the t-test (P<0.05). (B) 55Fe incorporation into ICU2 in the wild type and the mms19 mutant. The same affinity purification method was used as in (A), and the radioactivity of 55Fe in ICU2 was measured by liquid scintillation counting. Triplicates were performed. Error bars show the SD. Asterisks indicate significant differences as determined by the t-test. (C) The activities of aldehyde oxidase (AO) isozymes in wild-type and mms19 mutant seedlings. The purple bands indicated by arrows represent the activities of AO proteins. The staining by Ponceau S is shown as an equal protein loading control. (D) The mms19 and abo4 seedlings were more sensitive than wild-type seedlings to reduced FeSO4 concentrations. Seedlings were photographed after they had grown for 10 days on MS medium plates with different FeSO4 concentrations. 100%, 50%, 25%, and 12.5% represent 2.78, 1.39, 0.70, and 0.35 g/l of FeSO4.7H2O in MS medium.
We simultaneously tested the effect of mms19 on the activities of aldehyde oxidases (AOs), which carry 2[2Fe-2S] clusters [42]. The enzymes catalyze the conversion of sirtinol into an auxin analog, and the Fe-S cluster assembly is required for their enzymatic activity [42]. In-gel staining indicated that the enzymatic activities of all three AO isoforms are unchanged in the mms19 mutant relative to the wild type (Fig 6C), suggesting that MMS19 is not required for the Fe-S cluster assembly of the three AO isoforms. It is possible that MMS19 is involved in Fe-S cluster assembly on a subset of Fe-S target proteins.
To confirm the involvement of Fe in the function of MMS19 and DNA polymerases, we incubated wild-type, mms19, and abo4 seedlings on MS medium plates containing different concentrations of FeSO4 (Fig 6D). The results indicated that the growth of mms19 and abo4 is retarded compared to that of the wild type when FeSO4 is reduced in the MS medium, whereas both mms19 and abo4 grow as well as the wild type on the control MS medium (Fig 6D). The sensitivity of mms19 and abo4 to Fe demonstrated that Fe is related to the function of MMS19 and ABO4 in Arabidopsis. According to a recent research with yeast, the catalytic subunits of DNA polymerases are Fe-S proteins, and their catalytic functions require Fe-S clusters [33]. Because DNA polymerases are conserved in eukaryotes, MMS19 may contribute to Fe-S cluster assembly on DNA polymerases in Arabidopsis.
Discussion
In Arabidopsis, the components of the cytosolic iron-sulfur cluster assembly complex, including NBP35, NAR1, CIA1, and CIA2, are essential for embryo development [8, 13, 43], which is consistent with the involvement of Fe-S proteins in various essential cellular processes. Although MMS19 was recently identified as a component of the CIA complex, it is nonessential, and two individual mms19 null mutants are viable in Arabidopsis [8]. MMS19 is a single protein without a homolog in Arabidopsis, which excludes the possibility that a homolog is functionally redundant with MMS19. The null mutants of MMS19 are also viable in yeast [16], whereas the mouse MMS19 knockout is embryonically lethal [19]. Although visible defects were reported in the mms19 mutants of yeast and mammals [19, 20], no obvious phenotypic change was observed in the mms19 null mutant of Arabidopsis [8]. We found that the mms19 null mutants are defective in TGS, DNA repair, and flowering time control (Figs 2A and 2B, 4A–4F and 5A–5H), indicating that MMS19 plays an important role in these processes.
Given the absence of obvious phenotypic differences between the mms19 null mutant and the wild type, MMS19 may facilitate Fe-S assembly on a small subset of Fe-S proteins. Fe-S proteins are usually associated with Fe-S cluster assembly components in vivo [19, 20]. By affinity purification of MMS19, we identified the catalytic subunits of DNA polymerases α, δ, and ε but no other known Fe-S proteins (Table 1). The results suggest that the DNA polymerases α, δ, and ε are probably the major Fe-S protein targets of MMS19 in Arabidopsis.
The knockout mutants of the catalytic subunits of the DNA polymerases α and ε are embryonically lethal in Arabidopsis [27–29], suggesting that the two DNA polymerases are necessary for viability. Fe-S cluster assembly is essential for the activity of the DNA polymerases α, δ, ε, and ζ in yeast [33], which is consistent with the essentiality of the Fe-S assembly complex [19, 20]. In Arabidopsis, the null mutants of the Fe-S assembly proteins NBP35, NAR1, CIA1, and CIA2 are embryonically lethal [8, 13, 43], but two independent mms19 null mutants are viable and show no obvious phenotypic defects [8]. We propose that MMS19 is not essential for the Fe-S assembly on DNA polymerases but increases the efficiency of Fe-S cluster assembly. We compared the phenotypes of the mms19 null mutants with those of two previously characterized weak mutant alleles of DNA polymerases α and ε in Arabidopsis [30, 32]. Our results demonstrate that the effect of the mms19 null mutations on TGS, DNA repair, and flowering time (Figs 2A and 2B, 4A–4F and 5A–5H) is comparable to that of the weak mutations in DNA polymerases α and ε. Therefore, our study reveals a functional connection between MMS19 and DNA polymerases in Arabidopsis. The results support the notion that MMS19 increases the efficiency of Fe-S assembly on DNA polymerases.
The yeast MMS19 was originally identified as a component of the transcriptional regulator TFIIH complex [14] and was subsequently also found to directly associate with the chromatin-related proteins DOS2, RIK1, and CDC20 [17]. In yeast, MMS19 is involved in various nuclear processes including transcriptional regulation, telomere length maintenance, DNA replication and repair, and heterochromatin silencing [14–17]. However, the functional mechanism of MMS19 in these nuclear processes has been unclear. MMS19 was recently reported to act as a component of the cytosol Fe-S cluster assembly complex in yeast and mammals [19, 20]. Our results demonstrate that the Arabidopsis MMS19 contributes to various nuclear processes through its effects on the assembly of Fe-S clusters on DNA polymerases in the cytoplasm. Assembly of Fe-S clusters on the catalytic subunits of DNA polymerases is a prerequisite for the formation of active DNA polymerases in mammals [33]. Our study suggests that, during Fe-S cluster assembly on DNA polymerases, MMS19 may increase the efficiency of the Fe-S cluster assembly, thereby affecting nuclear processes including TGS, DNA repair, and flowering time in Arabidopsis.
In Arabidopsis, Fe-S cluster assembly was recently found to participate in genome integrity, embryonic development, and epigenetic regulation [8]. The DNA glycosylase ROS1, a nuclear Fe-S protein, excises 5-methycytosine via the base excision repair pathway and then contributes to DNA demethylation [44]. The demethylation activity of ROS1 was impaired when the Fe-S cluster assembly component AE7/CIA2 was disrupted, suggest that Fe-S cluster assembly facilitates the function of ROS1 in DNA demethylation [8]. DRE2 is a conserved protein required for Fe-S cluster assembly at an early stage of the cytosol Fe-S cluster assembly [5, 6]. In Arabidopsis, AtDRE2 was found to act in DNA demethylation and maternal gene expression mediated by the ROS1 homolog DME [45]. These reports indicate that the Fe-S cluster assembly machinery plays an important role in DNA demethylation, thereby facilitating transcriptional activation. Although MMS19 was found to act as a component of the Fe-S cluster assembly complex, we found that MMS19 is responsible for transcriptional gene silencing. The involvement of MMS19 in transcriptional gene silencing is related to the function of MMS19 in the Fe-S cluster assembly on DNA polymerases. However, we cannot exclude the possibility that MMS19 also takes effect on ROS1 and/or DME to mediate DNA demethylation and transcriptional activation. Further study is required to understand how MMS19 cooperates with other components of the Fe-S cluster assembly complex to act on different target proteins and thereby affects diverse biological processes.
Experimental Procedures
Plant materials and growth conditions
The T-DNA lines Salk_121963 and SALK_147068, which are mms19-1 and mms19-2, respectively, were obtained from the Arabidopsis Stock Center. The native promoter-driven full-length genomic MMS19, CIA1, CIA2, and NAR1 were cloned into the modified pCAMBIA1305 or pRI909 vectors and were tagged with 3xMyc or 3xFlag at their C-terminal ends. For gene transformation, the plant expression constructs were transformed into the agrobacterium strain GV3101 and then introduced into Arabidopsis by the conventional flower-dipping method. The primers used for construction are listed in S5 Table. For plant growth, the seeds were thoroughly washed with sterilized water and sown on MS medium plates. After they were kept for 3 days at 4°C, the plates were transferred to a growth chamber with 16-h-light and 8-h-dark at 22°C. 10-day-old seedlings were transferred to soil and were grown with 16-h-light and 8-h-dark at 22°C to obtain adult plants. For plant crossing, when the plant entered the full-blossom stage, all the six stamens were carefully removed by forceps, and the flowering parent stamen was brought to pollinate the emasculated pistil extensively.
DNA damage treatment
For methyl methanesulfonate (MMS, Sigma, 129925-5G) treatment, 5-day-old seedlings were transferred to upright MS medium plates containing a range of MMS concentrations and were photographed after the seedlings were grown for 10–14 days. For UV-B treatment, 5-day-old seedlings were moved to new MS medium plates and were treated with UV-B light (UV 256 nm, CL-508.G) for 0, 3 and 5 min. After treatment, the seedlings were grown on the MS medium plates for additional 5–7 days and then photographed.
Quantitative RT-PCR
Total RNA was isolated with Trizol reagent (Invitrogen) from Arabidopsis seedlings and was subjected to quantitative RT-PCR. Quantitative RT-PCR was performed according to the operating protocol (TaKaRa). ACT7 was used as an internal control. The primers used for quantitative RT-PCR are listed in the S5 Table. The significant difference was determined by Student's t-test.
RNA-seq analyses
Total RNA was extracted from 14-day-old seedlings of Col-0, mms19-1, and abo4. The total RNA samples were used to produce RNA libraries for deep sequencing (HiSeq 2000, Illumina). The Arabidopsis genome sequences and annotated gene models were downloaded from TAIR10 (www.arabidopsis.org). 45-bp sequences called by the Illumina pipeline were mapped to the TAIR10 Arabidopsis genome. Tophat v2.0.12 was used to align the raw reads to the genome sequences allowing up to two mismatches. Cufflinks (v2.0.1 http://cufflinks.cbcb.umd.edu/) was used to assemble transcripts and calculate transcript abundances. Differences in RNA transcript levels were identified with Cuffdiff. Genes with a significant (p<0.01) and >2-fold change in expression were selected for plotting heat maps using R. The RNA-seq data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE67669).
Affinity purification and mass spectrometric analysis
Samples of flowers or seedlings (6–10 g/sample) from transgenic plants were ground in liquid nitrogen and suspended in lysis buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.1% NP-40, 0.5 mM DTT, 1 mM PMSF, and 1 protease inhibitor cocktail tablet/50 ml [Roche]). The suspension was mixed by slowly rotation on a vertical vortex mixer for 15 min at 4°C. The suspension was then centrifuged, and the supernatant was incubated with anti-Flag or anti-Myc antibody-conjugated beads (Sigma, A2220, A7470) at 4°C; after 2.5–3.0 h, the preparation was centrifuged to precipitate the protein complex. The precipitate was washed once for 6 min with 10 ml of lysis buffer and then five times for 5 min per time with 1 ml of lysis buffer at 4°C. Finally, the protein complex was eluted with 3xFlag peptides (Sigma, F 4799) or 0.1 M ammonium hydroxide at pH 11.3 (for Myc antibody-conjugated beads). The elution samples were run on a 10% SDS-PAGE gel and were silver stained with the ProteoSilver Silver Stain Kit (Sigma PROT-SIL1). The whole proteins were purified from the gel and then subjected to mass spectrometric analysis as previously described [46].
Co-IP and gel filtration
For co-IP, total protein extracts were isolated from about 2–3 g of T2 transgenic plants and incubated with antibody-conjugated beads as described for affinity purification. The beads were washed 5 or 6 times with lysis buffer and boiled in SDS-PAGE sample buffer. The boiled samples were run on SDS-PAGE gels followed by Western blotting assay. For gel filtration, 0.8-g of flowers were ground in liquid nitrogen and suspended in 3 ml of lysis buffer (without NP-40). The samples were centrifuged at 12000 rpm at 4°C. The supernatant was passed through a 0.22-micron filter to remove cell debris and then loaded onto a Superose 6 10/300 GL column (GE Healthcare, 17-5172-01). The elution was collected in a series of fractions (500 μl/fraction). The eluted fractions were run on SDS-PAGE gels for Western blotting assay. A standard prestained protein ladder (Thermo Scientific, 26616) was used for calibration.
Nuclear and cytoplasmic fractionations
Arabidopsis materials (1-g samples) were ground and suspended in protein extraction buffer (20 mM Tris-HCl [pH 7.5], 25% glycerol, 20 mM KCl, 2 mM EDTA, 2.5 mM MgCl2, 250 mM sucrose, 5 mM DTT, 1 protease inhibitor cocktail tablet per 50 ml [Roche]). The suspension was filtered through double layers of Miracloth and centrifuged at 1500 g for 10 min at 4°C. The supernatant was centrifuged at 14000 rpm for 15 min to acquire the cytoplasmic fraction in the supernatant and the nuclear fraction in the precipitate. The precipitate containing the nuclear fraction was washed several times with pre-cooled nuclear resuspension buffer NRBT (20 mM Tris-HCl, pH 7.5, 25% glycerol, 2.5 mM MgCl2, and 0.2% Triton X-100, 5 mM DTT) until the precipitate changed from dark green to white. A 500-μl volume of pre–cooled NRB2 buffer (20 mM Tris-HCl, pH 7.5, 0.25 M sucrose, 10 mM MgCl2, 0.5% Triton X-100, 5 mM DTT, 1 protease inhibitor cocktail tablet per 50 ml [Roche]) was added to the nuclear pellet, which was gently suspended. The suspension was overlaid onto 500 μl of pre-cooled NRB3 buffer (20 mM Tris-HCl [pH 7.5], 1.7 M sucrose, 10 mM MgCl2, 0.5% Triton X-100, 5 mM DTT, 1 protease inhibitor cocktail tablet per 50 ml [Roche]) and centrifuged at 16,000 g for 50 min at 4°C to collect the pellet. Finally, the pellet was resuspended in 250 μl of protein extraction buffer, and then 250 μl of 2xSDS sample buffer was added for denaturation by boiling; the preparation was then subjected to SDS–PAGE and Western blotting. For quality control, UGPase (UDP-glucose pyrophosphorylase, Agrisera, AS05086) and the histone H3 (Abcam, ab1791) were used as a cytoplasmic and nuclear markers, respectively.
Enzyme activity analysis
The in-gel activity assays for AO isoforms were performed as reported previously [42].
Fe content determination and 55Fe incorporation assay
For determination of Fe content, 4 g of 14-day-old transgenic seedlings harboring the homozygous ICU2-Myc transgene in the wild-type Col-0 and mms19 mutant backgrounds was ground into a fine powder and suspended in lysis buffer. After the preparation was centrifuged at 4°C and the supernatant was passed through a 0.22-micron filter, the supernatant was incubated with Myc antibody-conjugated beads (Sigma, A7470) for 2.5 h for binding. The beads were thoroughly washed several times, and the Myc-tag coupled protein complex was eluted three to four times with 100 μl of 0.1 M ammonium hydroxide at pH 11.3. The elutions of each sample were combined for Fe content determination. Briefly, the samples were incubated with HNO3 (Thermofisher, Trace Metal grade) for digestion and then diluted to a certain volume and mixed vigorously for homogeneity. The samples were then subjected to ICP mass spectrometry (Themo ICP-MS Ⅻ) for Fe content determination by the standard curve method.
55Fe incorporation was determined according to a previous method with some modifications [47]. The T3 homozygous ICU2-Myc transgenic plants in the wild-type Col-0 background were crossed to the mms19 mutant to obtain the homozygous ICU2-Myc transgenic plants in the mms19 mutant background from the F2 segregating generation. The homozygous ICU2-Myc transgenic plants in the wild-type and mms19 mutant backgrounds were grown for 1 week on MS medium plates. A 0.5-g quantity of 1-week-old seedlings was transferred to 250-ml conical flasks containing 100 ml of liquid MS medium with one-fourth FeSO4 .7H2O; the preparation was gently shaken on a horizontal shaker for 1 day at 22°C. Thereafter, the seedlings were incubated in newly prepared liquid MS medium without Fe ion for 12 h. A 160-μCi quantity of 55FeCl3 (Perkin Elmer) in 2 ml of 0.1 M sodium ascorbate was then added to the liquid medium, which was incubated for an additional 2 days. The liquid was discarded, and the seedlings were thoroughly washed three times in citrate buffer (50 mM citrate, 1 mM EDTA, pH 7.0) and HEPES buffer (20 mM HEPES, pH 7.4). The seedlings were then collected and stored at -80°C. The target Fe-S protein ICU2-Myc was immunoprecipitated with Myc antibody-conjugated beads (Sigma, A7470). The radioactivity of 55Fe was measured by liquid scintillation counting (scintillation liquid: Ultima Gold, Perkin Elmer, 6013321).
Supporting Information
(A) Diagram of the gene structure of MMS19. Black and white boxes indicate exons and untranslated regions, respectively, and horizontal lines indicate introns. T-DNA insertion positions are marked by open triangles. (B) The transcript level of MMS19 was determined by quantitative RT-PCR. ACT7 was used as an internal control. Error bars indicate SD.
(PDF)
(A) The transposable elements were markedly up-regulated in the mms19 mutant relative to the wild type. (B) The DNA repair-related genes were up-regulated in the mms19 mutant relative to the wild type. ACT7 was an internal control. Three independent experiments were done, and the results from one representative experiment are indicated. Error bars show the SD.
(PDF)
Randomly selected co-upregulated genes (A), co-downregulated genes (B), and co-upregulated TEs (C) in mms19 and abo4 mutants were used for validation. ACT7 was used as an internal control. Error bars represent the SD of three technical replicates.
(PDF)
The seedlings were grown on MS medium plates for ~10 days and then treated with 100 ppm MMS for 0, 12, 24, 48, and 72 h. The transcript levels of XRI (A), BCRA1 (B), RAD51 (C), and GR1 (D) were determined by quantitative RT-PCR. Error bars indicate the SD.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Dr. Zhizhong Gong (China Agricultural University) for providing ros1icu2 and abo4-1 mutant seeds. This work was supported by the National Basic Research Program of China (973 Program) (2012CB910900) and the 973 Program (2011CB812600) from the Chinese Ministry of Science and Technology.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Basic Research Program of China (973 Program) (2012CB910900) and the 973 Program (2011CB812600) from the Chinese Ministry of Science and Technology. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kispal G, Sipos K, Lange H, Fekete Z, Bedekovics T, Janaky T, et al. Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J. 2005;24(3):589–98. 10.1038/sj.emboj.7600541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balk J, Pilon M. Ancient and essential: the assembly of iron-sulfur clusters in plants. Trends Plant Sci. 2011;16(4):218–26. 10.1016/j.tplants.2010.12.006 . [DOI] [PubMed] [Google Scholar]
- 3. Netz DJ, Mascarenhas J, Stehling O, Pierik AJ, Lill R. Maturation of cytosolic and nuclear iron-sulfur proteins. Trends Cell Biol. 2014;24(5):303–12. 10.1016/j.tcb.2013.11.005 . [DOI] [PubMed] [Google Scholar]
- 4. Adam AC, Bornhovd C, Prokisch H, Neupert W, Hell K. The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria. EMBO J. 2006;25(1):174–83. 10.1038/sj.emboj.7600905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Netz DJ, Stumpfig M, Dore C, Muhlenhoff U, Pierik AJ, Lill R. Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nature chemical biology. 2010;6(10):758–65. 10.1038/nchembio.432 . [DOI] [PubMed] [Google Scholar]
- 6. Zhang Y, Lyver ER, Nakamaru-Ogiso E, Yoon H, Amutha B, Lee DW, et al. Dre2, a conserved eukaryotic Fe/S cluster protein, functions in cytosolic Fe/S protein biogenesis. Molecular and cellular biology. 2008;28(18):5569–82. 10.1128/MCB.00642-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Netz DJ, Pierik AJ, Stumpfig M, Muhlenhoff U, Lill R. The Cfd1-Nbp35 complex acts as a scaffold for iron-sulfur protein assembly in the yeast cytosol. Nat Chem Biol. 2007;3(5):278–86. 10.1038/nchembio872 . [DOI] [PubMed] [Google Scholar]
- 8. Luo D, Bernard DG, Balk J, Hai H, Cui X. The DUF59 family gene AE7 acts in the cytosolic iron-sulfur cluster assembly pathway to maintain nuclear genome integrity in Arabidopsis. Plant Cell. 2012;24(10):4135–48. 10.1105/tpc.112.102608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Balk J, Pierik AJ, Netz DJ, Muhlenhoff U, Lill R. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J. 2004;23(10):2105–15. 10.1038/sj.emboj.7600216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Balk J, Aguilar Netz DJ, Tepper K, Pierik AJ, Lill R. The essential WD40 protein Cia1 is involved in a late step of cytosolic and nuclear iron-sulfur protein assembly. Mol Cell Biol. 2005;25(24):10833–41. 10.1128/MCB.25.24.10833-10841.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468(7325):790–5. 10.1038/nature09472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stehling O, Mascarenhas J, Vashisht AA, Sheftel AD, Niggemeyer B, Rosser R, et al. Human CIA2A-FAM96A and CIA2B-FAM96B integrate iron homeostasis and maturation of different subsets of cytosolic-nuclear iron-sulfur proteins. Cell Metab. 2013;18(2):187–98. 10.1016/j.cmet.2013.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bych K, Netz DJ, Vigani G, Bill E, Lill R, Pierik AJ, et al. The essential cytosolic iron-sulfur protein Nbp35 acts without Cfd1 partner in the green lineage. J Biol Chem. 2008;283(51):35797–804. 10.1074/jbc.M807303200 . [DOI] [PubMed] [Google Scholar]
- 14. Lauder S, Bankmann M, Guzder SN, Sung P, Prakash L, Prakash S. Dual requirement for the yeast MMS19 gene in DNA repair and RNA polymerase II transcription. Mol Cell Biol. 1996;16(12):6783–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, et al. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci U S A. 2004;101(23):8658–63. 10.1073/pnas.0401263101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kou H, Zhou Y, Gorospe RM, Wang Z. Mms19 protein functions in nucleotide excision repair by sustaining an adequate cellular concentration of the TFIIH component Rad3. Proc Natl Acad Sci U S A. 2008;105(41):15714–9. 10.1073/pnas.0710736105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li F, Martienssen R, Cande WZ. Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature. 2011;475(7355):244–8. 10.1038/nature10161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ito S, Tan LJ, Andoh D, Narita T, Seki M, Hirano Y, et al. MMXD, a TFIIH-independent XPD-MMS19 protein complex involved in chromosome segregation. Mol Cell. 2010;39(4):632–40. 10.1016/j.molcel.2010.07.029 . [DOI] [PubMed] [Google Scholar]
- 19. Gari K, Leon Ortiz AM, Borel V, Flynn H, Skehel JM, Boulton SJ. MMS19 links cytoplasmic iron-sulfur cluster assembly to DNA metabolism. Science. 2012;337(6091):243–5. 10.1126/science.1219664 . [DOI] [PubMed] [Google Scholar]
- 20. Stehling O, Vashisht AA, Mascarenhas J, Jonsson ZO, Sharma T, Netz DJ, et al. MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science. 2012;337(6091):195–9. 10.1126/science.1219723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feng W, D'Urso G. Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase epsilon are viable but require the DNA damage checkpoint control. Mol Cell Biol. 2001;21(14):4495–504. 10.1128/MCB.21.14.4495-4504.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakayama J, Allshire RC, Klar AJ, Grewal SI. A role for DNA polymerase alpha in epigenetic control of transcriptional silencing in fission yeast. EMBO J. 2001;20(11):2857–66. 10.1093/emboj/20.11.2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu Q, Gong Z. The coupling of epigenome replication with DNA replication. Curr Opin Plant Biol. 2011;14(2):187–94. 10.1016/j.pbi.2010.12.001 . [DOI] [PubMed] [Google Scholar]
- 24. Takeda S, Tadele Z, Hofmann I, Probst AV, Angelis KJ, Kaya H, et al. BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 2004;18(7):782–93. 10.1101/gad.295404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Inagaki S, Suzuki T, Ohto MA, Urawa H, Horiuchi T, Nakamura K, et al. Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell. 2006;18(4):879–92. 10.1105/tpc.105.036798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johansson E, Macneill SA. The eukaryotic replicative DNA polymerases take shape. Trends Biochem Sci. 2010;35(6):339–47. 10.1016/j.tibs.2010.01.004 . [DOI] [PubMed] [Google Scholar]
- 27. Jenik PD, Jurkuta RE, Barton MK. Interactions between the cell cycle and embryonic patterning in Arabidopsis uncovered by a mutation in DNA polymerase epsilon. Plant Cell. 2005;17(12):3362–77. 10.1105/tpc.105.036889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ronceret A, Guilleminot J, Lincker F, Gadea-Vacas J, Delorme V, Bechtold N, et al. Genetic analysis of two Arabidopsis DNA polymerase epsilon subunits during early embryogenesis. Plant J. 2005;44(2):223–36. 10.1111/j.1365-313X.2005.02521.x . [DOI] [PubMed] [Google Scholar]
- 29. Barrero JM, Gonzalez-Bayon R, del Pozo JC, Ponce MR, Micol JL. INCURVATA2 encodes the catalytic subunit of DNA Polymerase alpha and interacts with genes involved in chromatin-mediated cellular memory in Arabidopsis thaliana. Plant Cell. 2007;19(9):2822–38. 10.1105/tpc.107.054130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yin H, Zhang X, Liu J, Wang Y, He J, Yang T, et al. Epigenetic regulation, somatic homologous recombination, and abscisic acid signaling are influenced by DNA polymerase epsilon mutation in Arabidopsis. Plant Cell. 2009;21(2):386–402. 10.1105/tpc.108.061549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. del Olmo I, Lopez-Gonzalez L, Martin-Trillo MM, Martinez-Zapater JM, Pineiro M, Jarillo JA. EARLY IN SHORT DAYS 7 (ESD7) encodes the catalytic subunit of DNA polymerase epsilon and is required for flowering repression through a mechanism involving epigenetic gene silencing. Plant J. 2010;61(4):623–36. 10.1111/j.1365-313X.2009.04093.x . [DOI] [PubMed] [Google Scholar]
- 32. Liu J, Ren X, Yin H, Wang Y, Xia R, Wang Y, et al. Mutation in the catalytic subunit of DNA polymerase alpha influences transcriptional gene silencing and homologous recombination in Arabidopsis. Plant J. 2010;61(1):36–45. 10.1111/j.1365-313X.2009.04026.x . [DOI] [PubMed] [Google Scholar]
- 33. Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, et al. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat Chem Biol. 2012;8(1):125–32. 10.1038/nchembio.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111(6):803–14. . [DOI] [PubMed] [Google Scholar]
- 35. Steimer A, Amedeo P, Afsar K, Fransz P, Mittelsten Scheid O, Paszkowski J. Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell. 2000;12(7):1165–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He Y, Amasino RM. Role of chromatin modification in flowering-time control. Trends Plant Sci. 2005;10(1):30–5. 10.1016/j.tplants.2004.11.003 . [DOI] [PubMed] [Google Scholar]
- 37. Marquardt S, Boss PK, Hadfield J, Dean C. Additional targets of the Arabidopsis autonomous pathway members, FCA and FY. J Exp Bot. 2006;57(13):3379–86. 10.1093/jxb/erl073 . [DOI] [PubMed] [Google Scholar]
- 38. Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11(5):949–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, et al. Activation tagging of the floral inducer FT. Science. 1999;286(5446):1962–5. . [DOI] [PubMed] [Google Scholar]
- 40. Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 2006;46(2):183–92. 10.1111/j.1365-313X.2006.02686.x . [DOI] [PubMed] [Google Scholar]
- 41. Searle I, He Y, Turck F, Vincent C, Fornara F, Krober S, et al. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006;20(7):898–912. 10.1101/gad.373506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bernard DG, Cheng Y, Zhao Y, Balk J. An allelic mutant series of ATM3 reveals its key role in the biogenesis of cytosolic iron-sulfur proteins in Arabidopsis. Plant Physiol. 2009;151(2):590–602. 10.1104/pp.109.143651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakamura M, Buzas DM, Kato A, Fujita M, Kurata N, Kinoshita T. The role of Arabidopsis thaliana NAR1, a cytosolic iron-sulfur cluster assembly component, in gametophytic gene expression and oxidative stress responses in vegetative tissue. New Phytol. 2013;199(4):925–35. 10.1111/nph.12350 . [DOI] [PubMed] [Google Scholar]
- 44. Mok YG, Uzawa R, Lee J, Weiner GM, Eichman BF, Fischer RL, et al. Domain structure of the DEMETER 5-methylcytosine DNA glycosylase. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(45):19225–30. 10.1073/pnas.1014348107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Buzas DM, Nakamura M, Kinoshita T. Epigenetic role for the conserved Fe-S cluster biogenesis protein AtDRE2 in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(37):13565–70. 10.1073/pnas.1404058111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang CJ, Ning YQ, Zhang SW, Chen Q, Shao CR, Guo YW, et al. IDN2 and its paralogs form a complex required for RNA-directed DNA methylation. PLoS Genet. 2012;8(5):e1002693 10.1371/journal.pgen.1002693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pierik AJ, Netz DJ, Lill R. Analysis of iron-sulfur protein maturation in eukaryotes. Nat Protoc. 2009;4(5):753–66. 10.1038/nprot.2009.39 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Diagram of the gene structure of MMS19. Black and white boxes indicate exons and untranslated regions, respectively, and horizontal lines indicate introns. T-DNA insertion positions are marked by open triangles. (B) The transcript level of MMS19 was determined by quantitative RT-PCR. ACT7 was used as an internal control. Error bars indicate SD.
(PDF)
(A) The transposable elements were markedly up-regulated in the mms19 mutant relative to the wild type. (B) The DNA repair-related genes were up-regulated in the mms19 mutant relative to the wild type. ACT7 was an internal control. Three independent experiments were done, and the results from one representative experiment are indicated. Error bars show the SD.
(PDF)
Randomly selected co-upregulated genes (A), co-downregulated genes (B), and co-upregulated TEs (C) in mms19 and abo4 mutants were used for validation. ACT7 was used as an internal control. Error bars represent the SD of three technical replicates.
(PDF)
The seedlings were grown on MS medium plates for ~10 days and then treated with 100 ppm MMS for 0, 12, 24, 48, and 72 h. The transcript levels of XRI (A), BCRA1 (B), RAD51 (C), and GR1 (D) were determined by quantitative RT-PCR. Error bars indicate the SD.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.