STIM2β is a novel STIM2 splice isoform that inhibits Orai channels.
Abstract
Store-operated calcium entry (SOCE) regulates a wide variety of essential cellular functions. SOCE is mediated by STIM1 and STIM2, which sense depletion of ER Ca2+ stores and activate Orai channels in the plasma membrane. Although the amplitude and dynamics of SOCE are considered important determinants of Ca2+-dependent responses, the underlying modulatory mechanisms are unclear. In this paper, we identify STIM2β, a highly conserved alternatively spliced isoform of STIM2, which, in contrast to all known STIM isoforms, is a potent inhibitor of SOCE. Although STIM2β does not by itself strongly bind Orai1, it is recruited to Orai1 channels by forming heterodimers with other STIM isoforms. Analysis of STIM2β mutants and Orai1-STIM2β chimeras suggested that it actively inhibits SOCE through a sequence-specific allosteric interaction with Orai1. Our results reveal a previously unrecognized functional flexibility in the STIM protein family by which alternative splicing creates negative and positive regulators of SOCE to shape the amplitude and dynamics of Ca2+ signals.
Introduction
Store-operated calcium entry (SOCE) generates sustained and oscillatory cytosolic Ca2+ signals that regulate diverse cellular functions such as transcription, differentiation, motility, and secretion (Parekh and Putney, 2005; Hogan et al., 2010; Lewis, 2011). The most well-characterized store-operated channel is the Ca2+ release-activated Ca2+ (CRAC) channel, and defects in its function cause severe combined immunodeficiency (Feske et al., 2006, 2010) as well as deficits in muscle development and function (Stiber et al., 2008; Darbellay et al., 2010; Wei-LaPierre et al., 2013), platelet function (Varga-Szabo et al., 2011), and skin homeostasis (Vandenberghe et al., 2013).
SOCE is activated by the depletion of ER Ca2+ stores, typically upon activation of cell surface receptors. The stromal interaction molecule (STIM) family of ER Ca2+ sensors (STIM1 and STIM2; Liou et al., 2005; Roos et al., 2005; Zhang et al., 2005) and the Orai Ca2+ channels (Orai1, 2, and 3; Feske et al., 2006; Vig et al., 2006) are key molecular mediators of SOCE (Cahalan, 2009; Hogan et al., 2010; Lewis, 2011). Store depletion triggers oligomerization (Stathopulos et al., 2006; Liou et al., 2007; Covington et al., 2010) and conformational rearrangements of STIM proteins (Muik et al., 2011). These rearrangements expose the C-terminal polybasic domain, which interacts with phosphatidylinositol 4,5-bisphosphate in the plasma membrane (PM) and drives STIM accumulation at ER–PM junctions (Wu et al., 2006; Liou et al., 2007; Ercan et al., 2009; Park et al., 2009). Although STIM1 and STIM2 respond similarly to store depletion, STIM2 differs from STIM1 in being partially localized at ER–PM junctions even in store-replete cells, likely as a result of its lower affinity for ER Ca2+ relative to STIM1 (Brandman et al., 2007; Zheng et al., 2008). At ER–PM junctions STIM proteins directly bind to and trap Orai channels (Park et al., 2009; Wu et al., 2014) through their CRAC activation domains (CADs; also known as SOAR [STIM1 Orai1 activation region] or CCb9; Kawasaki et al., 2009; Park et al., 2009; Yuan et al., 2009). STIM binding to Orai opens the channel by a nonlinear process that is highly sensitive to binding stoichiometry (Hoover and Lewis, 2011; Li et al., 2011).
The amplitude and dynamics of SOCE-mediated Ca2+ signals are important factors in shaping Ca2+-dependent responses such as gene expression (Dolmetsch et al., 1997, 1998). Several mechanisms that affect the magnitude of SOCE have been identified, such as transcriptional regulation (Ritchie et al., 2010), posttranslational modifications (Smyth et al., 2009; Hawkins et al., 2010; Pozo-Guisado et al., 2010), and accessory proteins (Srikanth et al., 2010; Palty et al., 2012; Miao et al., 2013). Significantly, all of these mechanisms modulate the activity of STIM proteins without altering their role as activators of SOCE.
A largely unexplored mechanism with the potential to qualitatively alter STIM function is alternative splicing. Recent studies have shown that most, if not all, multiexonal proteins undergo alternative splicing (Kornblihtt et al., 2013). With more than 10 annotated exons, both STIM1 and STIM2 are thus likely to exist as multiple splice isoforms with varying properties. The only characterized splice variant in the STIM family thus far is STIM1L, which includes an actin binding site that prelocalizes it near ER–PM junctions in striated muscle and may thereby facilitate rapid SOCE kinetics (Darbellay et al., 2011; Horinouchi et al., 2012). All presently known STIM isoforms, including STIM1L, serve as activators of Ca2+ influx through Orai channels.
In this study, we describe a novel STIM2 splice isoform, STIM2β, which inhibits Orai function. STIM2β splicing is evolutionarily conserved and developmentally regulated. It contains an eight-residue insert in its CAD that disrupts binding to Orai. However, heterodimerization with other STIM isoforms recruits STIM2β to CRAC channels where it inhibits Ca2+ influx through an allosteric mechanism. Our results establish STIM2β as the first STIM isoform that inhibits Orai channels and introduce alternative splicing as a means of controlling the balance between SOCE activators and inhibitors, thereby tuning the magnitude and time course of calcium entry.
Results
STIM2β is a novel and widely expressed STIM2 splice isoform
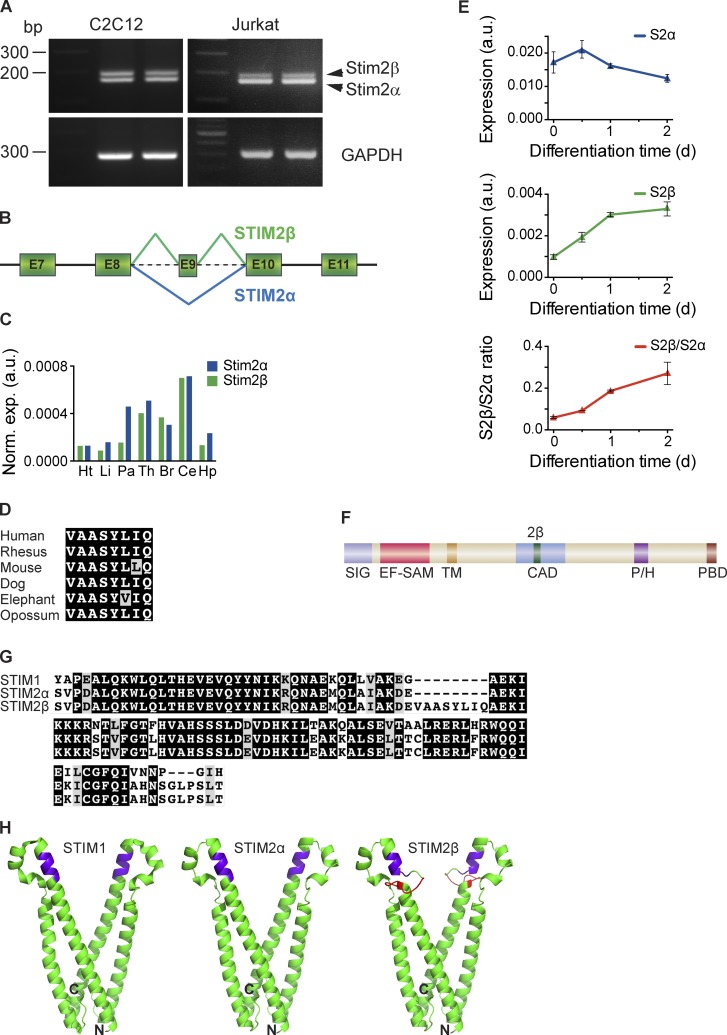
Our attempts to amplify portions of the STIM2 cytosolic domain from cDNA generated from several cell lines unexpectedly produced a doublet of bands when visualized on a standard agarose gel (Fig. 1 A). Sequencing of the higher molecular weight band revealed that it corresponded to a novel splice isoform formed by in-frame splicing of exon 9 of the STIM2 gene (Fig. 1 B). We named the new isoform STIM2β and will refer hereafter to the conventional isoform (without exon 9) as STIM2α. STIM2β is widely expressed across tissues, as shown by analysis of human tissue RNA samples (Fig. 1 C).
Figure 1.
STIM2β is a novel, widely expressed STIM2 splice isoform. (A, top) cDNA from Jurkat and C2C12 cells was amplified using primers targeting the CAD domain of STIM2. The topmost band represents the STIM2β splice isoform. (bottom) Primers targeting GAPDH were used as a positive control. (B) Partial schematic of the STIM2 genomic locus. In-frame inclusion of exon 9 produces STIM2β. (C) GAPDH-normalized expression levels of STIM2α and STIM2β in human tissue RNA samples, measured by quantitative RT-PCR. Means of technical replicates are shown (Ht, heart; Li, liver; Pa, pancreas; Th, thymus; Br, brain; Ce, cerebellum; Hp, hippocampus). (D) Sequence alignment of the 2β insert across six mammalian species (also see Fig. S1 A). Conservative differences are marked in gray. (E) GAPDH-normalized expression levels of STIM2α (top) and STIM2β (middle) mRNA in differentiating cultured C2C12 myoblasts. An approximately fivefold increase in the STIM2β/STIM2α ratio (bottom) occurs during the first 2 d of differentiation. Error bars represent SEM of three independent wells. (F) Domain structure of STIM2. Inclusion of exon 9 leads to an insert (2β, green) in the CAD (SIG, signal peptide; EF-SAM, EF hand/sterile-α motif; TM, transmembrane segment; P/H, proline/histidine-rich domain; PBD, polybasic domain). (G) Alignment of partial CAD sequences from human STIM1, STIM2α, and STIM2β. Sequence identity and similarity are shown in black and gray, respectively. (H) Predicted structures of STIM2α- (center) and STIM2β-CAD (right) derived from the crystal structure of STIM1-CAD (left; Yang et al., 2012) by homology modeling. The stretch of basic residues involved in Orai1 binding is highlighted in purple, and the 2β insert is highlighted in red. N and C termini of the front monomer in each structure are marked for orientation. a.u., arbitrary unit.
STIM2 exon 9 is present in most mammalian species and is highly conserved at the amino acid level (Fig. 1 D and Fig. S1 A), suggesting that its alternative splicing may serve a physiological function. To examine this possibility, we asked whether STIM2β splicing is developmentally regulated, using the serum withdrawal-induced myogenic differentiation of C2C12 myoblasts as a model system (Burattini et al., 2004). Quantitative RT-PCR analysis showed a slight reduction in STIM2α mRNA levels during the first 48 h of differentiation into myotubes; however, STIM2β mRNA levels increased significantly over this period, generating an approximately fivefold increase in the STIM2β/STIM2α ratio (Fig. 1 E). Up-regulation of STIM2β splicing was also observed during neuronal differentiation in vitro (unpublished data). These results show that STIM2β splicing is regulated and support the possibility of a physiological function.
The in-frame splicing of exon 9 inserts eight amino acids (the “2β insert”) into the highly conserved CAD of STIM2 (Fig. 1, F and G). The CAD region is critical for binding and activating Orai1 (Park et al., 2009) as well as stabilizing STIM dimers and oligomers (Covington et al., 2010; Yang et al., 2012). We used homology modeling (Bennett-Lovsey et al., 2008) based on the STIM1-CAD crystal structure (Yang et al., 2012), as well as de novo structure prediction (Lupas et al., 1991), to predict the effect of the 2β insert on CAD structure. While the predicted structure of STIM2α-CAD is quite similar to that of STIM1-CAD, the 2β insert significantly disrupts the helical topology of STIM2β-CAD (Fig. 1 H and Fig. S1 B). In particular, the model predicts that the helical stretch of basic residues (KIKKKR; Fig. 1 H, highlighted in purple) known to play a critical role in binding to Orai1 (Calloway et al., 2009, 2010; Korzeniowski et al., 2010) is likely to be disrupted in STIM2β, whereas the regions responsible for STIM–STIM dimerization (Yang et al., 2012; Stathopulos et al., 2013) may remain intact.
STIM2β inhibits Orai1-mediated Ca2+ influx
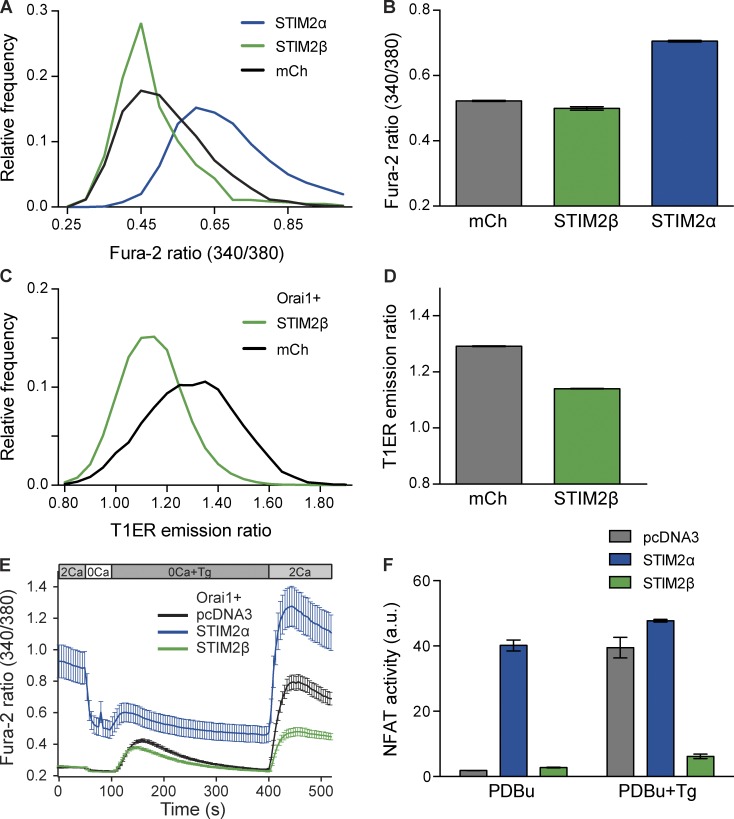
To determine the functional consequences of the 2β insert, we compared the effects of STIM2β and STIM2α on resting cytosolic Ca2+ ([Ca2+]i) and ER Ca2+ ([Ca2+]ER) levels in HEK293T cells. STIM2α overexpression led to a large increase in resting [Ca2+]i as reported previously (Fig. 2, A and B; Brandman et al., 2007). In contrast, overexpression of STIM2β caused a small but significant decrease. In cells expressing the Förster resonance energy transfer (FRET)–based [Ca2+]ER sensor T1ER (Bandara et al., 2013), coexpression of STIM2β with Orai1 caused a significant decrease in [Ca2+]ER (Fig. 2, C and D). In this case, a comparison with STIM2α was not possible, as STIM2α overexpression for the extended period of time required for T1ER coexpression led to large-scale cell death, presumably caused by a prolonged increase in [Ca2+]i (unpublished data). These experiments indicate that, in direct contrast to STIM2α, STIM2β negatively regulates the resting levels of both cytosolic and ER Ca2+.
Figure 2.
STIM2β inhibits Orai1-mediated SOCE. (A and B) Resting cytosolic [Ca2+] in HEK293T cells transfected with STIM2α, STIM2β, or mCherry (mCh). Frequency distribution (A) and means ± SEM (B) of fura-2 ratios are shown. STIM2β overexpression caused a small but significant reduction in the cytosolic fura-2 ratio (n > 800 cells for each condition, P < 0.0001, Mann–Whitney test). (C and D) Resting ER [Ca2+] in HEK293T cells transfected with Orai1 and STIM2β or mCherry. Frequency distribution (C) and means ± SEM (D) of T1ER emission ratios (see Materials and methods) are shown. Higher T1ER ratio signifies higher ER Ca2+ levels. STIM2β overexpression significantly reduced the T1ER ratio (n > 5,000 cells for each condition, P < 0.0001, Mann–Whitney test). (E) Effects of STIM2α and STIM2β on SOCE in HEK293 cells expressing Orai1. Solution changes are indicated, with extracellular Ca2+ concentration in millimolar. STIM2α but not STIM2β elevated resting [Ca2+]i. After depletion of ER Ca2+ stores with 1 µM Tg in Ca2+-free solution, SOCE is shown by the response to 2 mM Ca2+. Compared with the pcDNA3 control, STIM2α increased SOCE, whereas STIM2β strongly inhibited SOCE (n > 30 cells for each condition). (F) NFAT activity in store-replete (PDBu) or store-depleted (PDBu+Tg) HEK293T cells. Overexpression of STIM2α but not STIM2β drives constitutive NFAT activity in store-replete cells, whereas overexpression of STIM2β strongly inhibits NFAT activation by store depletion (n = 3 wells for each condition). Error bars show means ± SEM. a.u., arbitrary unit.
The effect of STIM2α on [Ca2+]i and [Ca2+]ER arises from its ability to activate SOCE through interactions with Orai1 (Brandman et al., 2007). We applied thapsigargin (Tg) to deplete Ca2+ stores and examine the effects of STIM2α and STIM2β on SOCE in HEK293 cells overexpressing Orai1 (Fig. 2 E). With Orai1 expression alone, reintroduction of extracellular Ca2+ after store depletion evoked a large increase in [Ca2+]i reflecting activation of SOCE by endogenous STIMs. Coexpression of STIM2α with Orai1 enhanced the level of SOCE, in addition to causing a large increase in resting [Ca2+]i. In contrast, overexpression of STIM2β with Orai1 strongly inhibited SOCE to levels below those seen with Orai1 overexpression alone (Fig. 2 E).
To further confirm the inhibitory effects of STIM2β, we examined SOCE-activated signaling through the transcription factor nuclear factor of activated T cells (NFAT). NFAT-mediated transcription requires elevated [Ca2+]i as well as PKC activity (Rao et al., 1997). HEK293T cells transfected with an NFAT-luciferase reporter and treated with phorbol 12,13-dibutyrate (PDBu) to activate PKC showed a strong up-regulation of luciferase expression after store depletion with Tg (Fig. 2 F). Overexpression of STIM2α increased NFAT-driven luciferase activity in PDBu-treated cells even in the absence of Tg (Fig. 2 F), as expected from the large increase in basal [Ca2+]i seen in Fig. 2 E. In contrast, overexpression of STIM2β strongly inhibited the Tg-mediated increase in luciferase activity, consistent with its inhibition of SOCE in Fig. 2 E. Collectively, the results shown in Fig. 2 indicate that unlike STIM2α, STIM2β strongly inhibits SOCE generated by endogenous STIM and Orai.
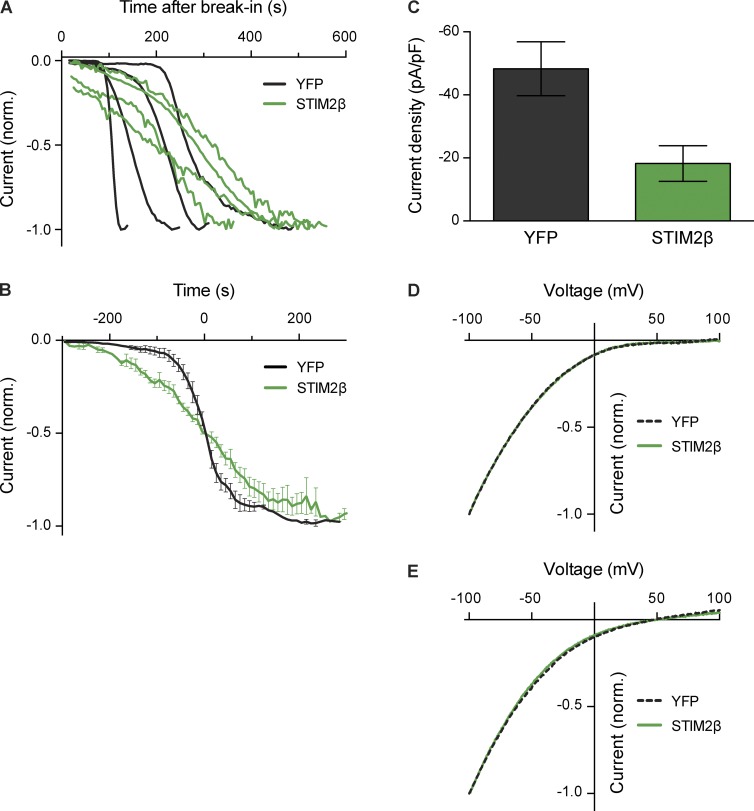
The inhibition of Ca2+ entry by STIM2β could in principle result from a direct effect on the Orai1 channel (inhibition of activity or a loss of Ca2+ selectivity), or an indirect effect such as membrane depolarization (reduction of the driving force for Ca2+ entry). To resolve this question, we used whole-cell recording to measure Orai1-mediated CRAC currents (ICRAC) induced passively by intracellular dialysis with EGTA through the recording pipette. In HEK293 cells stably overexpressing STIM1 and Orai1, coexpression of STIM2β-YFP altered the kinetics of ICRAC induction (Fig. 3, A and B), resulting in a shortened lag phase before current initiation and a reduced maximal rate of current development. Importantly, STIM2β coexpression reduced the ICRAC density at steady state by ∼60% compared with coexpression of YFP only (Fig. 3 C). In contrast, the current–voltage relation for ICRAC in 20 mM Ca2+ was unaffected, showing normal inward rectification with a lack of a well-defined reversal potential up to approximately +80 mV, consistent with the characteristic high selectivity of Orai1 for Ca2+ over monovalent cations (Fig. 3 D). STIM2β coexpression also did not affect the reversal potential measured in the absence of divalent cations (48.6 ± 2.5 mV for YFP and 49.4 ± 4.0 mV for STIM2β, mean ± SEM), indicating that STIM2β does not alter the relative permeability of the channels to Cs+ and Na+ (Fig. 3 E). Together, the results of Figs. 2 and 3 show that STIM2β inhibits SOCE directly by reducing CRAC channel activity without significantly affecting its ion selectivity.
Figure 3.
STIM2β reduces CRAC current amplitude without altering ion selectivity. (A) STIM2β alters the time course of CRAC current induction. HEK293 cells stably expressing STIM1 and Orai1 were transiently transfected with either STIM2β-YFP or YFP only. Currents were measured at −100 mV in 20 mM extracellular Ca2+. Normalized (norm.) current density (I/Imax) after break-in is shown for single cells expressing STIM2β-YFP or YFP (representative of 5–7 cells per condition). (B) Comparison of averaged traces from A. To highlight activation kinetics, traces for each cell were shifted along the time axis before averaging, such that half-maximal activation occurred at t = 0 (n = 5–7 cells per condition). The maximal rate of activation (at 50% of maximal current) was 1.1% s−1 and 0.3% s−1 for YFP and STIM2β, respectively. (C) STIM2β reduces the steady-state amplitude of ICRAC (n = 9 cells for each condition, P = 0.01, Mann–Whitney test). For each cell, Imax was measured as described in A. (D and E) STIM2β expression does not affect the I–V relationship in 20 mM Ca2+ (D) or divalent-free (DVF) Ringer’s solutions (E; mean of 4–6 cells per curve). Error bars show means ± SEM. Error bars in D and E are comparable to the thickness of the curves and are not shown.
The 2β insert disrupts the interaction of STIM2β with Orai1
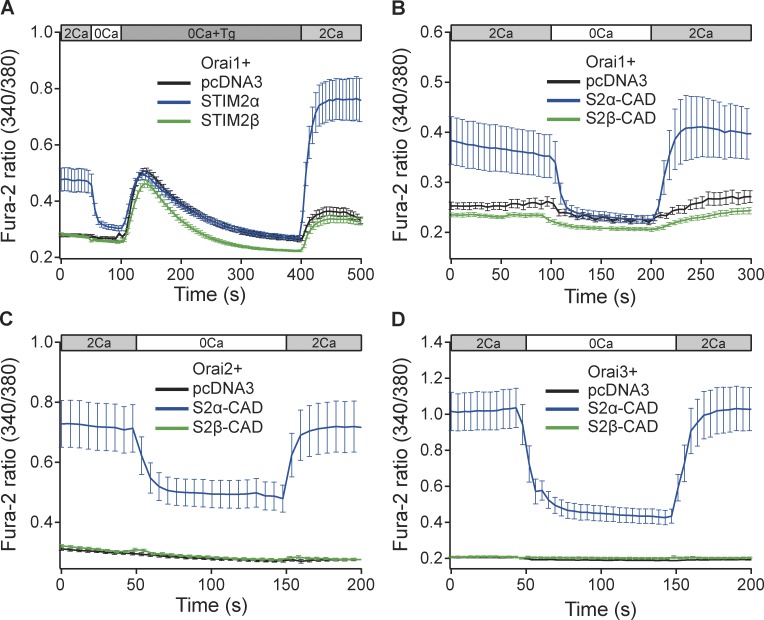
To understand how STIM2β inhibits ICRAC, we studied its interaction with Orai channels. Coexpression of STIM2α and Orai1 fully reconstituted SOCE in Neuro2A neuroblastoma cells, a SOCE-deficient cell line (Fig. 4 A). In contrast, coexpression of STIM2β with Orai1 failed to produce detectable SOCE, indicating that the 2β insert disrupts the functional interaction between STIM2β and Orai1.
Figure 4.
STIM2β cannot activate Orai channels. (A) STIM2α but not STIM2β can reconstitute SOCE in Neuro2A cells when coexpressed with Orai1. (B–D) STIM2α-CAD, but not STIM2β-CAD, elevates basal [Ca2+]i in Neuro2A cells cotransfected with Orai1 (B), Orai2 (C), or Orai3 (D). Expression levels of Orai1, 2, and 3 in B–D are not directly comparable (n ≥ 15 cells for each condition in all panels). Error bars show means ± SEM.
The CAD/SOAR region of STIM proteins is known to be necessary and sufficient to activate Orai channels (Park et al., 2009; Yuan et al., 2009). To confirm that the inability of STIM2β to activate Orai1 resulted from altered function of its CAD, we coexpressed Orai1 with STIM2α- or STIM2β-CAD. As with the full-length STIM2 proteins, coexpression of STIM2α-CAD with Orai1 produced robust increases in [Ca2+]i, whereas STIM2β-CAD with Orai1 failed to do so (Fig. 4 B). STIM2β-CAD also failed to produce increases in [Ca2+]i with Orai2 and Orai3 (Fig. 4, C and D), isoforms that are more tolerant of CAD mutations than Orai1 (Frischauf et al., 2009), further underscoring the complete inability of STIM2β-CAD to activate Orai channels.
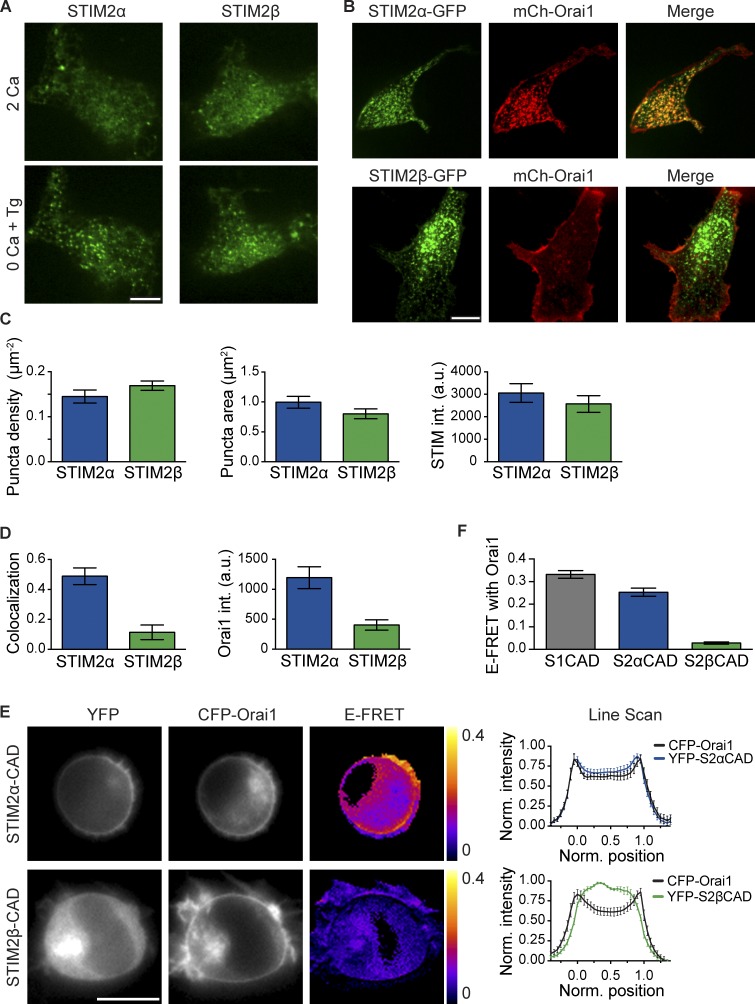
The failure of STIM2β to activate Orai1 could result from a deficient response to Ca2+ store depletion or from a more proximate defect in STIM2β–Orai1 coupling, as the CAD region is involved in both of these processes (Park et al., 2009; Covington et al., 2010; Yang et al., 2012). To test whether STIM2β can respond to store depletion, we expressed fluorescently tagged STIM2 constructs in HEK293 cells and imaged their accumulation at ER–PM junctions as fluorescent puncta. Both STIM2α and STIM2β formed puncta in most resting cells even without store depletion (Fig. 5 A), consistent with the partial activity of STIM2 in resting cells (Fig. 2 E; Brandman et al., 2007). In cells expressing low amounts of either isoform, the brightness and number of puncta were increased after store depletion with Tg (Fig. 5 A). Quantitative analysis showed that STIM2α and STIM2β formed puncta of similar density, area, and intensity (Fig. 5, B and C). Furthermore, in cells coexpressing tagged STIM1 and STIM2β, STIM2β puncta coincided precisely with STIM1 puncta (Fig. S2 A). Thus, STIM2β by itself maintains the ability to redistribute to ER–PM junctions in response to Ca2+ store depletion.
Figure 5.
STIM2β responds normally to store depletion but shows weakened Orai1 binding. (A) STIM2β accumulates at ER–PM junctions upon store depletion. Fluorescent puncta in two representative HEK293 cells expressing either mCherry (mCh)-STIM2α or -STIM2β are shown before (top) and after (bottom) store depletion with 1 µM Tg. (B) STIM2α-GFP, but not STIM2β-GFP, recruits mCherry-Orai1 into puncta after store depletion with Tg. (C) STIM2β forms puncta to a similar extent as STIM2α. Density (left), area (middle), and intensity (right) of puncta in store-depleted cells were quantified from experiments similar to B (n > 15 cells for each bar, P > 0.1 for each comparison, two-tailed t test). (D) Compared with STIM2α, STIM2β shows lower colocalization with Orai1 (left, measured as Pearson correlation; P < 0.0001, two-tailed t test) and elicits a lower Orai1 intensity (int.) in puncta (right, P = 0.0002, two-tailed t test; n > 15 cells for each bar). Data were compiled from experiments similar to B. (E) Binding of STIM2β-CAD to Orai1 is disrupted. FRET in HEK293 cells transfected with CFP-Orai1 and YFP-tagged STIM2α- or STIM2β-CAD. Unlike STIM2α-CAD (top), STIM2β-CAD (bottom) shows neither membrane recruitment nor significant FRET, indicating marginal binding to Orai1. Averaged line scans across cells (right) show high PM colocalization of Orai1 and STIM2α-CAD (n = 8 cells) but poor PM colocalization for Orai1 and STIM2β-CAD (n = 7 cells). Positions 0 and 1 represent the opposite edges of each cell. (F) Comparison of mean E-FRET between Orai1 and STIM1-, STIM2α-, or STIM2β-CAD from experiments like those in E (n > 18 cells for each bar, P < 0.0001, Mann–Whitney test). Error bars show means ± SEM. a.u., arbitrary unit. Bars: (A and B) 10 µm; (E) 5 µm.
STIM proteins activate SOCE by first binding and trapping Orai channels at ER–PM junctions (Park et al., 2009; Wu et al., 2014). To determine whether STIM2β retains the ability to trap Orai1, we coexpressed STIM2α or STIM2β with Orai1 in HEK293 cells. STIM2α robustly recruited Orai1 into puncta as measured by increased Orai fluorescence in puncta and colocalization with STIM2α. In contrast, STIM2β's recruitment of Orai1 was significantly impaired (Fig. 5, B and D). Roughly 60% of STIM2β puncta showed no accumulation of Orai1, while the remainder showed some recruitment (Fig. S2 B), suggesting that STIM2β interacts only weakly with Orai1.
The STIM2β–Orai interaction was quantified by measuring FRET between CFP-Orai1 and YFP-tagged STIM2α- and STIM2β-CADs. When coexpressed with CFP-Orai1, YFP-STIM2α-CAD localized close to the PM (Fig. 5 E) and showed substantial FRET (Fig. 5, E and F), indicating significant binding between STIM2α-CAD and Orai1. In contrast, under similar conditions, STIM2β-CAD maintained a cytosolic distribution and did not generate significant FRET (Fig. 5, E and F; and Fig. S4 B), confirming that its binding to Orai1 is disrupted.
Heterodimerization with STIM1 recruits STIM2β to Orai1 channels
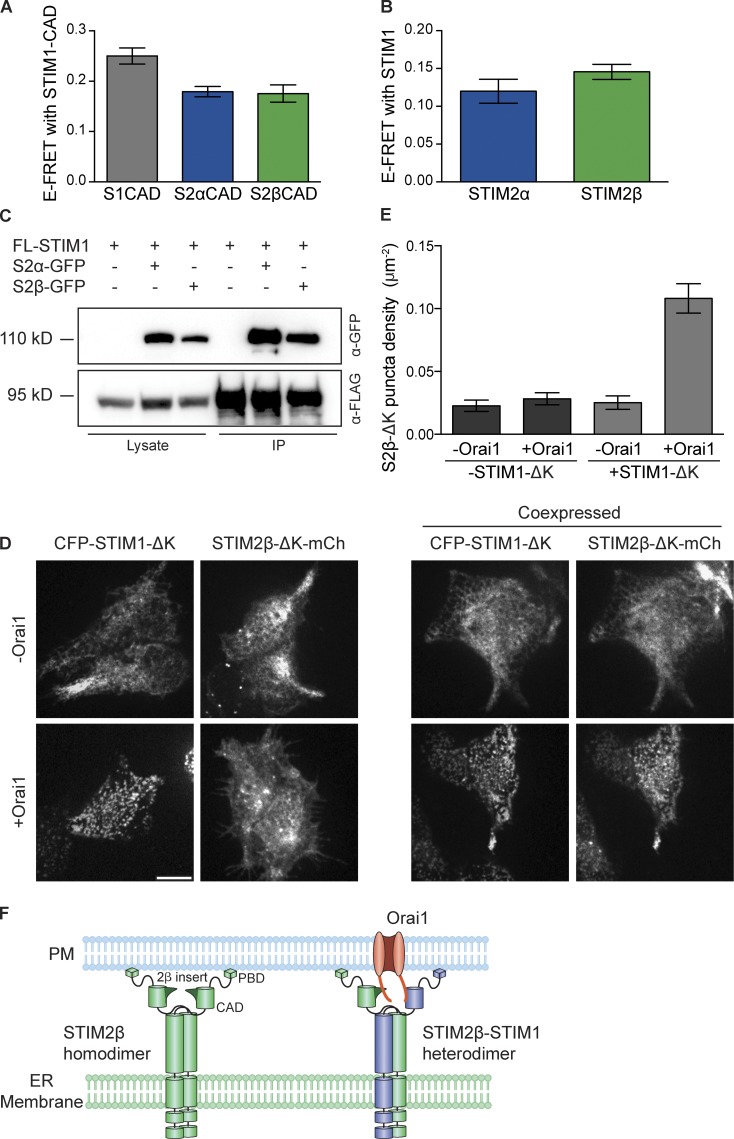
The weakened binding of STIM2β to Orai1 stands in apparent contradiction to its strong inhibition of SOCE. However, STIM1 and STIM2 are known to form heterodimers (Williams et al., 2001; Soboloff et al., 2006; Darbellay et al., 2010), which could provide a mechanism to recruit STIM2β to Orai channels and facilitate inhibition. We used a FRET assay to assess the ability of STIM2β to heterodimerize with STIM1. YFP-labeled STIM2α- and STIM2β-CADs showed similar levels of FRET with CFP-labeled STIM1-CAD (Fig. 6 A), indicating that STIM2β-CAD heterodimerizes normally with STIM1-CAD. Similarly, full-length STIM2α- and STIM2β-CFP generated comparable levels of FRET with full-length STIM1-YFP (Fig. 6 B). Lastly, STIM2α- and STIM2β-GFP coimmunoprecipitated to a similar extent with FLAG-STIM1 (Fig. 6 C). Thus, STIM2β can heterodimerize with STIM1 to a similar degree as STIM2α. Similarly, FRET between STIM2α and STIM2β and between their respective CADs (Fig. S3, A and B) indicated that STIM2β can also heterodimerize with STIM2α.
Figure 6.
Heterodimerization with STIM1 tethers STIM2β to Orai1. (A and B) FRET in cells coexpressing CFP-STIM1-CAD with YFP-STIM2α-CAD or YFP-STIM2β-CAD (A), or STIM1-YFP with STIM2α-CFP or STIM2β-CFP (B). Both STIM2 isoforms as well as their CADs interact with STIM1 (n > 17 cells for each bar, P > 0.1 for each comparison, Mann–Whitney test). (C) STIM2α- or STIM2β-GFP coimmunoprecipitate to a similar extent with FLAG-STIM1 (FL-STIM1). IP, immunoprecipitation. (D) STIM1-ΔK can tether STIM2β-ΔK to Orai1 channels. (left) In store-depleted cells, STIM1-ΔK forms puncta only in the presence of Orai1, whereas STIM2β-ΔK cannot form puncta even when Orai1 is coexpressed. (right) However, coexpression of STIM1-ΔK is sufficient to recruit STIM2β-ΔK to puncta in store-depleted cells expressing Orai1. Bar, 10 µm. (E) Comparison of puncta formation by STIM2β-ΔK from experiments like those in D (n ≥ 10 cells for each bar). Significant puncta above background are formed only upon coexpression of Orai1 and STIM1-ΔK (P < 0.0001, one-way analysis of variance). (F) Model of STIM2β tethering to Orai1 through heterodimerization with STIM1. STIM2β homodimers accumulate at ER–PM junctions through interactions of the polybasic domain with the PM (left) but do not interact strongly with Orai1. STIM2β-STIM1 heterodimers can bring STIM2β into close proximity to Orai1 through STIM1-mediated binding to the Orai1 C terminus. PBD, polybasic domain. Error bars show means ± SEM.
To test whether heterodimerization with STIM1 can recruit STIM2β to Orai1 channels, we expressed STIM constructs lacking the C-terminal polybasic domain (ΔK) in HEK293 cells and depleted Ca2+ stores with Tg. In the absence of the polybasic domain, STIM proteins cannot bind to phosphatidylinositol 4,5-bisphosphate in the PM, and their trapping at ER–PM junctions becomes absolutely dependent on the CAD-mediated interaction with Orai1 (Fig. 6 D, left; Park et al., 2009). As expected from the lack of strong binding between STIM2β and Orai1, STIM2β-ΔK failed to form puncta when expressed with Orai1 alone (Fig. 6 D, left). However, STIM2β-ΔK did form distinct puncta when coexpressed with Orai1 and STIM1-ΔK (Fig. 6 D, right). This STIM1-ΔK–dependent formation of STIM2β-ΔK puncta (Fig. 6 E) suggests that the interaction between STIM1-ΔK and Orai1 is sufficient to recruit STIM2β-ΔK–STIM1-ΔK heterodimers to Orai1. Consistent with this result, coexpression of STIM1 led to significantly increased FRET between STIM2β-YFP and CFP-Orai1 (Fig. S3 C). Thus, heterodimerization with STIM1 (or STIM2α) provides an essential means of recruiting STIM2β to Orai1 and enabling channel inhibition (Fig. 6 F).
The STIM2β insert sequence has a critical role in inhibiting SOCE
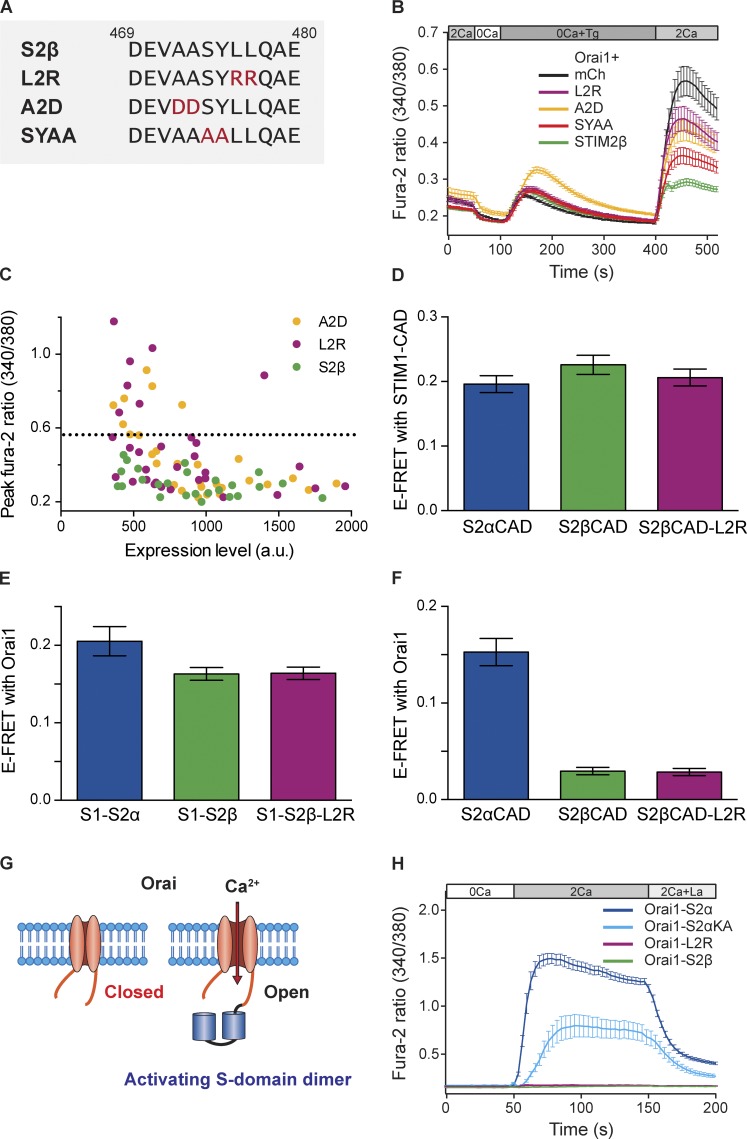
The high evolutionary conservation of the 2β insert suggests that the amino acid sequence itself may be an important functional determinant of STIM2β’s inhibitory activity. To test this idea, we made a series of pairwise mutations of its central amino acids (Fig. 7 A); the A2D and L2R mutants introduce polarity at positions with a strong preference for nonpolar residues (Fig. S1 A), whereas the SYAA mutant removes a pair of highly conserved residues in the central part of the insert. When expressed with Orai1, all three mutants showed diminished inhibition of SOCE compared with wild-type STIM2β, as assessed by the peak fura-2 ratio after Ca2+ readdition (Fig. 7 B). Although the SYAA mutant retained ∼75% of the inhibition seen with wild-type STIM2β, the A2D and L2R mutations produced only ∼40% inhibition (Fig. 7 B). These results are mean values obtained from cells expressing STIM2β proteins at varying levels. A closer look at SOCE in single cells as a function of STIM2β expression revealed that the L2R and A2D mutations were even more effective at diminishing the inhibitory action of STIM2β when expressed at low to moderate levels (Fig. 7 C). These results show that the specific sequence of the 2β insert is critical for enabling the potent inhibition of SOCE.
Figure 7.
The 2β insert sequence is critical for inhibition of SOCE by STIM2β. (A) Sequences of the wild type and mutant 2β inserts. Mutated residues are highlighted in red. Residue numbering is based on the reference sequence in Materials and Methods. (B) STIM2β mutants show reduced inhibition of Orai1-mediated SOCE in HEK293 cells (n > 38 cells for each condition). Wild-type and mutant STIM2β constructs were cotransfected with Orai1 in HEK293 cells, and the response to Ca2+ readdition in mCherry cells indicates the full level of SOCE mediated by endogenous STIM. (C) Peak fura-2 ratios plotted against STIM2β expression levels in single cells from the experiments shown in B. The dashed line indicates the mean peak ratio in mCherry (mCh)-expressing control cells. a.u., arbitrary unit. (D) The L2R mutation does not reduce STIM2β heterodimerization with STIM1. YFP-tagged CADs from the STIM2β-L2R mutant and wild-type STIM2β interact similarly with CFP-STIM1-CAD as shown by FRET (n > 25 cells for each bar, P = 0.1384, Mann–Whitney test). (E) The L2R mutation does not affect the binding of STIM1–STIM2β heterodimers to Orai1. FRET was measured between YFP-tagged tandem S-domain heterodimers and CFP-Orai1. Both wild-type and L2R mutant STIM2β heterodimers show similar levels of Orai1 binding (n ≥ 13 cells per bar, P = 0.64, Mann–Whitney test). (F) The L2R mutation does not restore STIM2β binding to Orai1. YFP-tagged CADs from the L2R mutant or wild-type STIM2β interact poorly if at all with CFP-Orai1 (n > 12 cells for each bar, P = 0.7783, Mann–Whitney test). (G) Chimeric system for detecting activating interactions. A dimer of S domains is covalently tethered to Orai1 channels. As a result of its high local concentration, any activating interactions between the SS dimer and Orai1 are detected as constitutive Ca2+ influx. (H) Fusion of SS dimers from wild-type STIM2α or the STIM2α-KA mutant to Orai1 evokes constitutive Ca2+ influx upon exposure to 2 mM Ca2+. Influx is inhibited by 10 µM La3+, consistent with Orai1 activity. In contrast, fusion of S-domain dimers from wild-type STIM2β or the L2R mutant STIM2β to Orai1 fails to elicit Ca2+ influx (n > 16 cells for each curve). Error bars show means ± SEM.
We considered several hypotheses to explain how mutations in the 2β insert reduce the ability of STIM2β to inhibit SOCE. First, the mutations might lead to misfolding or mislocalization of the protein. However, the L2R mutant showed normal ER localization and puncta formation upon store depletion (Fig. S4 A), making such a defect unlikely. A second possibility is that the mutations inhibit heterodimerization of STIM2β with STIM1, thus reducing the amount of STIM2β tethered to Orai1 channels and freeing more STIM1 homodimers to effectively activate SOCE. However, formation of heterodimers appeared to be unaffected, as judged by FRET between YFP-L2R-CAD and CFP-STIM1-CAD (Fig. 7 D and Fig. S4 B). Similarly, FRET experiments with covalent heterodimers of CAD-containing fragments from STIM1 and STIM2β (referred to as S domains; Li et al., 2011; McNally et al., 2013), indicated that the L2R mutation does not affect the binding of these heterodimers to Orai1 (Fig. 7 E).
A third possibility is that mutations in the 2β insert restore the ability of STIM2β to bind and activate Orai1. However, upon coexpression with CFP-Orai1, YFP-L2R-CAD was neither recruited to the PM nor showed significant levels of FRET (Fig. 7 F and Fig. S4 B). Furthermore, all three STIM2β mutants failed to reconstitute SOCE in Neuro2A cells when coexpressed with Orai1 (Fig. S4 C). Although these experiments rule out the restoration of strong Orai1 binding or activation in the mutants, they do not exclude the possibility that mutations in STIM2β restore weak interactions with Orai1, which are sufficient to activate Orai1 when the mutants are tethered to it as heterodimers with STIM1. To test this possibility, we constructed chimeras of Orai1 with a dimer of S domains (Fig. 7 G). This chimeric system mimics the high local STIM2β concentrations created by the binding of STIM2β–STIM1 heterodimers to Orai1 and also allows STIM2β–Orai1 interaction to be measured directly, i.e., without the interference of STIM1. Fusion of STIM2α S domains to Orai1 produced strong constitutive Ca2+ influx, similar to fusions of STIM1 S domains reported previously (Li et al., 2011; McNally et al., 2013), confirming that the chimeric constructs form functional channels (Fig. 7 H). Chimeras with S domains from a STIM2α-KA mutant (KIKKKR → KIAAAR), which cannot bind or activate Orai1 when expressed in soluble form (Fig. S5, A–C), also showed significant Ca2+ influx (Fig. 7 H), demonstrating that the chimeric system is sufficiently sensitive to detect even weak activating interactions. Significantly, chimeras containing S domains from either wild-type STIM2β or its L2R mutant did not produce any detectable Ca2+ influx (Fig. 7 H), indicating that mutations in the 2β insert did not restore even weak Orai1-activating ability.
These data indicate that mutations in the 2β insert significantly reduce the ability of STIM2β to inhibit SOCE and that this is not caused by defects in folding or trafficking, impaired heterodimerization with STIM1, or restored binding and activation of Orai1. To explain the sequence-specific inhibition of SOCE by STIM2β, we next considered an active inhibition mechanism in which STIM2β delivers an inhibitory signal to Orai1 through a sequence-specific interaction.
STIM2β inhibits Ca2+ influx through a sequence-specific interaction with Orai1
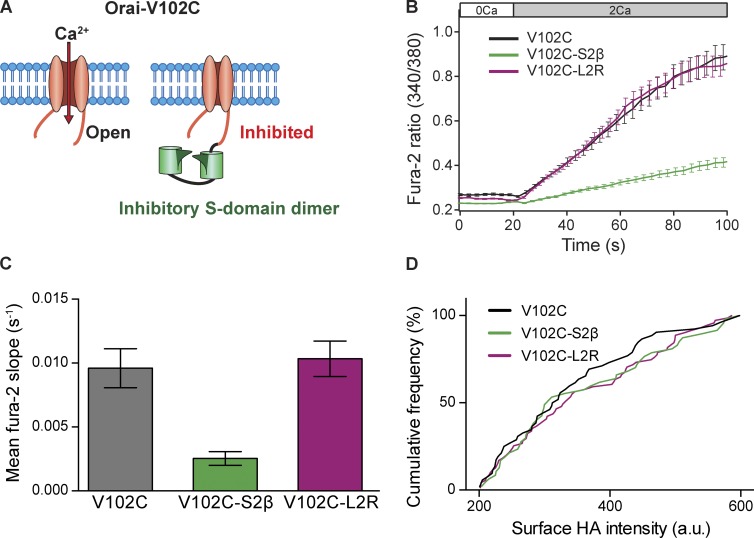
To test for active inhibition of Orai1 by STIM2β, we used Orai1(V102C), a pore mutant that is constitutively active in the absence of STIM1 (McNally et al., 2012). The Orai1(V102C) channel by itself produced constitutive Ca2+ influx when Ca2+ was introduced into the extracellular medium (Fig. 8 B). As expected from the disrupted binding of STIM2β homodimers to Orai1, coexpression with STIM2β did not inhibit Ca2+ flux through Orai1(V102C) channels (Fig. S5 D). To mimic the tethering effect of STIM1–STIM2β heterodimers, we constructed chimeras of the STIM2β S domain with Orai1(V102C) (Fig. 8 A). [Ca2+]i measurements were correlated with surface expression of chimeras at the single-cell level using an extracellular HA tag inserted into the Orai1(V102C) III–IV loop (see Materials and methods). Tethering a dimer of STIM2β S domains to the C terminus of Orai1(V102C) significantly suppressed Ca2+ influx (74% reduction in dRatio/dt slope). This inhibitory effect was absent in chimeras made with L2R mutant S domains, consistent with the reduced inhibition of SOCE by STIM2β bearing this mutation (Fig. 8, B and C). Surface HA staining (Fig. 8 D) indicated that the differences in Ca2+ influx were not caused by altered surface expression levels of the chimeras and thus are likely to reflect an inhibitory interaction between the STIM2β S domain and Orai1(V102C) that is abolished by the L2R mutation. These results indicate that STIM2β delivers an inhibitory signal to Orai1 through a sequence-specific interaction, thus inhibiting SOCE by an active mechanism.
Figure 8.
STIM2β inhibits Orai1 channels through a sequence-specific interaction. (A) Chimeric system for detecting inhibitory interactions. A dimer of S domains is covalently attached to constitutively active Orai1(V102C). Inhibitory interactions between the S domains and Orai are detected by a reduced rate of constitutive Ca2+ influx. (B) Constitutive Ca2+ influx in HEK293 cells expressing Orai1(V102C) or Orai1(V102C)-SS chimeras, measured upon addition of 2 mM Ca2+ (n > 45 cells for each curve). (C) The relative Ca2+ influx rates from the experiments in B, quantified by the initial slopes of the fura-2 ratio. Fusion of wild-type (P < 0.0001), but not L2R mutant (P = 0.67), STIM2β S domains to Orai1(V102C) strongly inhibited constitutive Ca2+ entry (Mann–Whitney test). (D) The cumulative frequency distribution of surface HA intensities for the cells analyzed in B and C shows that surface expression for all three channel constructs was similar. a.u., arbitrary unit. Error bars show means ± SEM.
Discussion
The STIM family of proteins is well recognized as having two primary functions: to sense [Ca2+]ER and to activate store-operated channels. Our studies of STIM2β show how the insertion of an eight-residue sequence in the CAD region converts STIM2α into a potent inhibitor of Orai, thus creating the first inhibitory member of the STIM family. In this way, alternative splicing of STIM2 presents a new type of mechanism for tuning the magnitude of SOCE to match physiological needs.
STIM2β differs in several fundamental ways from STIM2α. Although STIM2α was initially described as a SOCE inhibitor (Soboloff et al., 2006), this was later determined to be an artifact of overexpression (Parvez et al., 2008). STIM2α activates Orai channels less effectively than STIM1 (Bird et al., 2009; Wang et al., 2014); thus, in the presence of STIM1 and limiting amounts of Orai1, overexpressed STIM2α can reduce SOCE by competing with STIM1 for binding to Orai1 channels. When this competition is eliminated, e.g., by the simultaneous overexpression of Orai1, STIM2α robustly activates SOCE (Fig. 2 E; Parvez et al., 2008; Bird et al., 2009). In contrast, STIM2β cannot effectively bind or activate Orai channels by itself, and it inhibits SOCE even when Orai1 is overexpressed, ruling out a simple competitive mechanism as an explanation for its inhibitory effect. The opposing effects of STIM2α and STIM2β on ER and cytosolic Ca2+ levels and NFAT activation further underscore the inhibitory action of STIM2β. Together, our results establish a unique role for STIM2β among all known STIM isoforms as an inhibitor of Ca2+ influx through Orai channels.
How does STIM2β inhibit SOCE? We were initially surprised to find that STIM2β binding to Orai1 is disrupted, making it unlikely that it interacts as a homodimer with Orai channels like the other STIM proteins. However, experiments with STIM2β-ΔK (Fig. 6 D) and full-length STIM2β (Fig. S3 C), as well as S-domain dimers (Fig. 7 E), indicate that heterodimerization with STIM1 or STIM2α can tether STIM2β to Orai1 channels, thus increasing its local concentration to enable it to effectively inhibit Orai1 channels despite its low affinity for them. Once STIM2β is recruited to the channel in heterodimeric form, there are two broad mechanisms by which it could inhibit Orai1. In one case, passive inhibition could result from STIM2β occluding STIM binding sites on the channel or sequestering STIM1 or STIM2α in heterodimers that interact with the channel with a lower affinity (Fig. 7 E). In these scenarios, STIM2β would reduce the number of active CAD domains bound to the channel, which would be expected to limit channel activation (Hoover and Lewis, 2011; Li et al., 2011).
A second possible mechanism is active inhibition in which STIM2β delivers an inhibitory signal through interactions with Orai1 or Orai1-bound STIM1 or STIM2α. Although it is currently impossible to assess inhibition of STIM1/2α activity directly (i.e., in the absence of Orai1), our results with the Orai1(V102C)-STIM2β chimeras strongly support a mechanism in which STIM2β delivers an inhibitory signal to Orai1 (Fig. 8). The inhibition is sequence specific, as it was greatly diminished by mutations in the 2β insert. Interestingly, these mutants were still able to inhibit SOCE when expressed at high levels, apparently in a sequence-independent way (Fig. 7 C). A likely explanation is that at high expression levels, STIM2β mutants bind most of the STIM1/2α in the form of heterodimers, which then reduce Orai1 activation through the passive mechanism. It should be noted that such passive inhibition is unlikely to be significant for wild-type STIM2β under physiological conditions, as STIM2β is generally not expressed at high levels compared with STIM1, and, even if it were, the effects of the stronger active inhibition would likely dominate.
The 2β insert may exert its inhibitory effect by interacting with Orai1 directly or by altering the conformation of STIM2β to generate inhibitory interactions between other regions of STIM2β-CAD and Orai1. In either case, we expect these interactions to exhibit low affinity, as STIM2β by itself interacts poorly with Orai1 in FRET or puncta formation assays (Fig. 5). Our electrophysiology results showed that STIM2β diminishes the total current through Orai1 channels without affecting their ion selectivity (Fig. 3, C–E). In principle, the reduced current could reflect inhibition of channel opening or inhibition of ion flow through the open pore. Interestingly, Orai1(V102C/A) channels are thought to acquire their constitutive activity from the removal of a hydrophobic barrier to ion permeation near V102 (McNally et al., 2012; Dong et al., 2013; Gudlur et al., 2014). Thus, the ability of the tethered STIM2β SS construct to inhibit Ca2+ flux through Orai1(V102C) supports the latter possibility, that STIM2β inhibits Orai1 by imposing a new barrier to conduction, although additional effects on channel opening cannot be ruled out. Further studies of the inhibited state of wild-type Orai1 as well as Orai1(V102C) will be needed to define the structural basis of the STIM2β–Orai interaction and the changes in the pore that underlie the inhibitory allosteric effect.
An intriguing finding from patch–clamp recordings was that STIM2β expression altered the kinetics of ICRAC development in response to passive store depletion. STIM2β significantly shortened the lag phase before current initiation (in some cases, a small number of channels were even active at the time of break-in), and reduced themaximal rate of current development (Fig. 3, A and B). Although multiple factors shape the activation kinetics of CRAC channels, the observed effects of STIM2β may be attributable to the lower ER Ca2+ affinity of STIM2 as compared with STIM1. As a result of this lower affinity, STIM2 can respond to even mild store depletion, whereas STIM1 requires a higher level of store depletion to be reached before it is activated (Brandman et al., 2007; Luik et al., 2008). If STIM2β shifts the overall Ca2+ sensitivity of STIM1–STIM2β heterodimers toward that of STIM2, this may explain the lack of a lag phase and altered kinetics of ICRAC induction observed in these cells. A more detailed explanation of these kinetic effects and their possible relationship to the inhibitory action of STIM2β awaits further study.
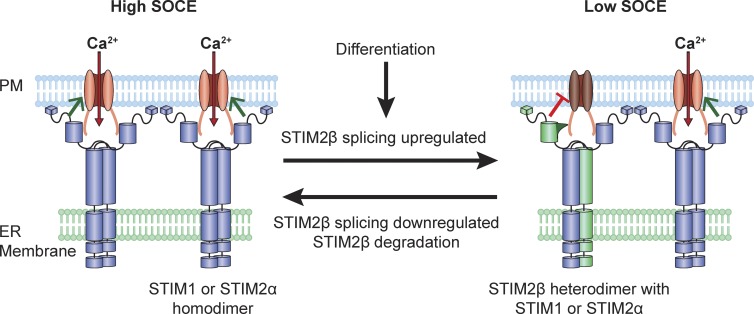
The widespread expression of STIM2β and its high evolutionary conservation suggests that STIM2β is a physiologically important mechanism for modulating SOCE (Fig. 9). The generation of STIM2β through alternative splicing and STIM2β’s ability to actively inhibit Orai have several important implications for its role as an SOCE modulator. As a result of the ability of STIM2β to heterodimerize with STIM2α and actively inhibit Orai, even small increases in the STIM2β/STIM2α ratio may produce large inhibitory effects. Notably, alternative splicing is a particularly effective way of changing this ratio, as it produces simultaneous and opposite changes in STIM2β and STIM2α levels. Increasing STIM2β through splicing may also reduce SOCE levels more rapidly than is possible by down-regulating STIM2α transcription, which would be slowed by the long half-life of STIM2α (>24 h; unpublished data).
Figure 9.
Proposed model for modulation of SOCE by alternative splicing of STIM2. Under conditions of low STIM2β splicing, STIM1 and STIM2α effectively activate Orai1 channels and enable a high level of SOCE. Up-regulation of STIM2β splicing, e.g., during cell differentiation or in response to environmental cues, promotes the formation of STIM2β heterodimers, which inhibit Orai function through a sequence-dependent interaction to reduce the capacity for SOCE. Reversal of this process may occur through down-regulation of STIM2β splicing and degradation of existing STIM2β.
The presence of multiple developmental defects (e.g., in muscle, tooth enamel, and sweat glands) in patients carrying mutations in STIM or Orai genes (Feske, 2010; Nesin et al., 2014) indicates a broad role for SOCE in regulating developmental processes. Among the best studied of these is the role of SOCE in regulating the differentiation of muscle (Stiber et al., 2008; Darbellay et al., 2010) and neural tissue (Somasundaram et al., 2014). We have observed an increase in STIM2β splicing during the in vitro differentiation of both muscle (Fig. 1 E) and neural (unpublished data) progenitors. This change in splicing suggests a possible role for STIM2β in regulating differentiation through modulation of SOCE. Differentiation of these tissues is known to be accompanied by widespread changes in splicing (Hall et al., 2013; Li et al., 2014), including that of key calcium signaling components (Brandt and Vanaman, 1994; Tang et al., 2009). Thus, the up-regulation of STIM2β splicing we have observed may provide an effective way of coordinating the modulation of SOCE with the changes initiated by global splicing programs during development.
Materials and methods
Cell culture, transfections, and solutions
HEK293 and HEK293T cells (ATCC) were cultured in antibiotic-free DMEM (Gibco) with l-glutamine and 10% FBS (Invitrogen). Neuro2A cells (ATCC) were grown in antibiotic-free Eagle’s minimal essential medium (ATCC) with 10% FBS. Jurkat cells (clone E6-1) were cultured in RPMI 1640 with 10% FBS and l-glutamine. C2C12 cells were a gift from H. Blau (Stanford University, Stanford, CA) and were maintained at low confluency in DMEM with GlutaMAX (Invitrogen) and 20% FBS. HEK293, HEK293T, and Neuro2A cells were cultured to 70–80% confluency before transient transfections using Lipofectamine 2000 (Invitrogen). Transfections were performed using the manufacturer’s protocol, except for ER and cytosolic Ca2+ measurements, in which fourfold less Lipofectamine 2000 than recommended was used to minimize perturbations to cell health. For imaging experiments, cells were transferred to polyornithine-coated glass coverslips or 96-well plates and bathed in 2 mM Ca2+ Tyrode’s solution (129 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 30 mM glucose, and 25 mM Hepes, pH 7.4) at 22–25°C unless otherwise specified.
Plasmids and primers
The following primers were used for initial detection of STIM2β (Fig. 1 A): human S2_forward, 5′-ATGCAGCTAGCTATTGCTAAAGATG-3′, and S2_reverse, 5′-TCGTTCTCGTAAACAAGTTGTCAACTC-3′; and mouse mS2_forward, 5′-ATGCAGCTAGCCATCGCTAAGGACG-3′, and mS2_reverse, 5′-CCGTTCTCGCAAGCACGTGGTCAGCTC-3′. These primer sets generate 192- and 168-bp bands corresponding to STIM2β and STIM2α, respectively.
Mouse and human STIM2α and STIM2β cDNAs were amplified using RT-PCR from total mRNA from C2C12 and HEK293 cells, respectively. STIM2α and STIM2β constructs with C-terminal mCherry, GFP, or myc-His tags were generated using Gateway cloning (Life Technologies) into destination vectors based on pGW1, EGFP-C1, and pCDNA3 vectors, respectively. STIM2α and STIM2β CADs were identified by alignment with STIM1-CAD, and constructs with N-terminal mCherry, CFP, or YFP tags were created using Gateway cloning. Gateway destination vectors with GFP, CFP, and YFP tags were derived from pEGFP/ECFP/EYFP-C1/N1 (Takara Bio Inc.) and driven by cytomegalovirus promoters. Destination vectors with mCherry tags were derived from pGW1 vector. QuikChange (Agilent Technologies) mutagenesis was used to generate STIM2α or STIM2β mutants. STIM2β S domain dimers were based on previously published STIM1 S domains (Li et al., 2011) and consisted of residues E427–P472 joined with a 24-residue linker (GGSGGSGGGILQSTGGSGGSGGSG; see primer sequences below; residue numbers based on reference sequence below). Orai1 or Orai1(V102C) was cloned between the NheI and XhoI sites of the pEYFP-N1 vector downstream of a cytomegalovirus promoter, and STIM2α or STIM2β S-domain dimers were then inserted between the Xho1 and BamH1 sites to produce YFP-tagged chimeric constructs with a 13-residue linker (LEGVSTATMGGSG). STIM1 and Orai1 constructs used here have been previously described (Park et al., 2009; Covington et al., 2010). Flag-Myc-Orai1, mCherry-Orai1, Flag-Myc-Orai2, mCherry-Orai3, and FLAG-STIM1 were generated by Gateway cloning into destination vectors based on the pGW1 backbone (New England Biolabs, Inc.).
Primers for Gateway entry clones of STIM2α/β (full length and CAD) were as follows: (human) huSTIM2_forward, 5′-ATGAACGCAGCCGGGATCAGAG-3′, huSTIM2_reverse, 5′-TCACTTAGATTTCTTCTTAAAAAGGCTTTTG-3′, huSTIM2_CAD_forward, 5′-ATGTCTGTTCCAGATGCACTTCAGAAATGG-3′, and huSTIM2_CAD_reverse, 5′-TCAGGTCAGGCTGGGGAGTCC-3′; (mouse) msSTIM2_forward, 5′-GCCACCATGAACGCGGCGGCGAGCCGAGCTTCGCGGGCC-3′, msSTIM2_reverse, 5′-TCACTTAGACTTCTTCTTGAAAAGGCTTTTGATTTTGG-3′, msS2CAD_forward, 5′-ATGTCTGTCCCTGACGCACTACAGAAATGG-3′, and msS2CAD_reverse, 5′-TCAGGTGAGACTGGGGAGCCCAGA-3′.
Primers for cloning STIM2α/β S domains for chimeric constructs were as follows: Xho1_msSTIM2_S_forward, 5′-GAGCTCGAGGGGGTATCAACCGCCACCATGGGTGGTTCCGGCGAACTGAGAAGCAGCTGGTCTGTC-3′; EcoR1_msSTIM2_S_reverse, 5′-GGGGAATTCCACCTCCGCTACCTCCAGAGCCGCCGGGTGTGTCTTCATCGAGGTCATC-3′; Sal1_msSTIM2_S_forward, 5′-GAGGTCGACGGGTGGTTCCGGTGGGTCCGGCGGTTCCGGCGAACTGAGAAGCAGCTGGTCT-3′; and BamHI_msSTIM2_S_reverse, 5′-GGGGGATCCGCACCTCCGCTACCTCCAGAGCCGCCGGGTGTGTCTTCATCGAGGTCATC-3′.
STIM2α residue numbers referred to in various constructs are based on the following reference STIM2α sequence: MNAAGIRAPEAAGADGTRLAPGGSPCLRRRGRPEESPAAVVAPRGAGELQAAGAPLRFHPASPRRLHPASTPGPAWGWLLRRRRWAALLVLGLLVAGAADGCELVPRHLRGRRATGSAATAASSPAAAAGDSPALMTDPCMSLSPPCFTEEDRFSLEALQTIHKQMDDDKDGGIEVEESDEFIREDMKYKDATNKHSHLHREDKHITIEDLWKRWKTSEVHNWTLEDTLQWLIEFVELPQYEKNFRDNNVKGTTLPRIAVHEPSFMISQLKISDRSHRQKLQLKALDVVLFGPLTRPPHNWMKDFILTVSIVIGVGGCWFAYTQNKTSKEHVAKMMKDLESLQTAEQSLMDLQERLEKAQEENRNVAVEKQNLERKMMDEINYAKEEACRLRELREGAECELSRRQYAEQELEQVRMALKKAEKEFELRSSWSVPDALQKWLQLTHEVEVQYYNIKRQNAEMQLAIAKDEAEKIKKKRSTVFGTLHVAHSSSLDEVDHKILEAKKALSELTTCLRERLFRWQQIEKICGFQIAHNSGLPSLTSSLYSDHSWVVMPRVSIPPYPIAGGVDDLDEDTPPIVSQFPGTMAKPPGSLARSSSLCRSRRSIVPSSPQPQRAQLAPHAPHPSHPRHPHHPQHTPHSLPSPDPDILSVSSCPALYRNEEEEEAIYFSAEKQWEVPDTASECDSLNSSIGRKQSPPLSLEIYQTLSPRKISRDEVSLEDSSRGDSPVTVDVSWGSPDCVGLTETKSMIFSPASKVYNGILEKSCSMNQLSSGIPVPKPRHTSCSSAGNDSKPVQEAPSVARISSIPHDLCHNGEKSKKPSKIKSLFKKKSK (833 amino acids).
mRNA expression analysis
For expression analysis of C2C12 cells (Fig. 1 E), differentiation was initiated by growing cells to 70–80% confluency and then transferring them to differentiation media. At the required time points, cells were lysed, and total RNA was extracted using the RNeasy kit (QIAGEN). For expression analysis of human tissues (Fig. 1 C), total RNA was purchased from the BioChain Institute. For Fig. 1 A, total RNA was extracted from Jurkat and C2C12 cells using the TRIzol reagent (Life Technologies).
Total RNA was subjected to DNase digestion and reverse transcription (SuperScript III reverse transcription kit; Life Technologies) to obtain cDNA. Quantitative RT-PCR was performed using the SYBR green system (Roche) on a thermocycler (RealPlex4; Eppendorf). Isoform-specific primers were used to quantify STIM2α and STIM2β, with GAPDH primers used as normalization controls. For C2C12 cells, the differentiation marker MyoG was also quantified to monitor progress of differentiation. The ratio of STIM2β and STIM2α was calculated directly by subtracting cycle threshold values for STIM2α from those of STIM2β for the same sample.
Coimmunoprecipitation
HEK293T cells (500,000 per well) were transfected with FLAG-STIM1 and STIM2α-GFP or STIM2β-GFP. After 24 h, cells were washed twice with PBS and then lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, and protease inhibitor cocktail). Lysates were centrifuged at 12,000 rpm for 10 min, and the supernatant was incubated with anti-FLAG M2 agarose beads (Sigma-Aldrich) for 4–12 h at 4°C to collect immunoprecipitate. Cell lysates and immunoprecipitates were analyzed by Western blotting using anti-FLAG (M2 mouse monoclonal; Sigma-Aldrich) or anti-GFP (rabbit polyclonal; MBL International) antibodies.
Structural modeling
De novo secondary structure predictions for the CAD regions of STIM2α and STIM2β were made using COILS (http://embnet.vital-it.ch/software/COILS_form.html). Homology modeling of the 3D structure of STIM2α/β CAD domains was performed using the Phyre2 server (http://www.sbg.bio.ic.ac.uk/phyre2). The available crystal structure of the STIM1 CAD (PDB accession no. 3TEQ) was selected as the best fit among the models suggested by Phyre2, and structures of the STIM2α and STIM2β CAD were generated accordingly. Structures were displayed in PyMOL (Schrödinger).
Electrophysiology
ICRAC was recorded from tetracycline-inducible HEK293 cells expressing equal amounts of mCherry-STIM1 and myc-Orai1 protein (Sadaghiani et al., 2014). Cells were transiently transfected with either STIM2β-YFP or control cytosolic YFP plasmid 2 d before recording. STIM1 + Orai1 expression were induced with 1 µg/ml tetracycline 1 d before recording.
Whole-cell voltage clamp recordings were made using an amplifier (Axopatch 200B; Molecular Devices) interfaced to an ITC-16 input/output board and a computer running custom Igor routines developed in house. The time course of ICRAC induction in response to passive ER store depletion was monitored in 20 mM Ca2+ Ringer’s using a step-ramp stimulus (100-ms step to −100 mV followed by a 100-ms ramp to 100 mV) applied at 5-s intervals from a holding potential of 30 mV. After ICRAC reached steady-state, stimuli were delivered every 2 s to monitor current in 20 mM Ca2+, divalent-free (DVF), or 2 mM Ca2+ + 100 µM LaCl3 (for leak subtraction). Solutions were perfused locally using a perfusion pencil coupled to an eight-channel electronic valve controller (AutoMate Scientific).
2 mM Ca2+ Ringer’s solution contained (mM): 155 NaCl, 4.5 KCl, 2 CaCl2, 1 MgCl2, 10 d-glucose, and 5 Hepes (pH 7.4 with NaOH). 20 mM Ca2+ Ringer’s was similar to 2 mM Ca2+ Ringer’s but with 130 mM NaCl and 20 mM CaCl2. DVF Ringer’s contained (mM): 150 NaCl, 10 2-hydroxyethyl EDTA, 1 EDTA, 10 tetraethylammonium-Cl, and 10 Hepes (pH 7.4 with NaOH). Internal (pipette) solution contained (mM): 150 Cs aspartate, 8 MgCl2, 10 EGTA, and 10 Hepes (pH 7.2 with CsOH).
To compare I/V curves among cells, ramp currents were leak subtracted and normalized to the peak inward current at −100 mV. ICRAC time courses were normalized to the maximal steady-state current at −100 mV and fit to a sigmoid function in Igor Pro (f(t) = base + max/{1 + exp[(t1/2 − t)/rate])} to obtain t1/2, the time at half-maximal current. The maximal rate of ICRAC induction was determined by fitting a line to a 20-s time segment of the ICRAC trace centered at t1/2. The current was well described by a straight line within this time window.
NFAT luciferase assays
NFAT luciferase assays were performed using the Dual-Luciferase Assay kit (Promega). HEK293T cells (∼250,000 per well) were transfected with NFAT-luciferase reporter (firefly luciferase driven by a 4×-NFAT binding site from the IL-2 promoter) and a control Renilla luciferase plasmid (pRLTK; Renilla expression driven by a thymidine kinase promoter) along with STIM2α or STIM2β. 16 h after transfection, cells were stimulated with 1 µM PDBu (LC Laboratories) with or without 1 µM Tg (LC Laboratories) for 6 h and lysed using passive lysis buffer (Promega). NFAT luciferase activity was quantified as the ratio of firefly to Renilla luciferase activity measured using a 96-well automated luminometer (Turner BioSystems).
Confocal microscopy
HEK293 cells were transfected with 100–150 ng of fluorescently tagged STIM and Orai constructs and imaged 12–15 h after transfection. For imaging store depletion-mediated translocation of STIM2α and STIM2β, live cells were imaged before and 10 min after treatment with 1 µM Tg at 22–25°C. For quantifying maximal puncta formation, cells were fixed with 4% PFA and 8% sucrose in PBS immediately after Tg treatment. Cell footprints were imaged with the confocal microscope (UltraVIEW VoX; PerkinElmer) using a 63× Plan Apochromat (NA 1.4) oil immersion objective (Carl Zeiss).
For quantification of puncta, regions of interest (ROIs) were drawn manually just inside the cell edge and analyzed using the Volocity software package (PerkinElmer). Pearson correlation coefficients between STIM and Orai were calculated for the entire ROI. Puncta were identified as regions with STIM intensity greater than three standard deviations above the cell mean (using Volocity’s “identify objects based on intensity” function). STIM and Orai intensity within each punctum was quantified in Volocity, and mean puncta number, area, and intensities were calculated using MATLAB (MathWorks).
Resting ER and cytosolic Ca2+ measurements
For ER Ca2+ measurements, HEK293T cells were transfected with pcDNA3-T1ER (Bandara et al., 2013) along with mCherry or mCherry-tagged STIM2 constructs. After 40–48 h, cells were plated on 96-well plastic bottom plates (Costar) and imaged using an automated epifluorescence microscope (ImageXpress Micro XL; Molecular Devices) with a 10× objective (NA 0.3) at 37°C. Images were acquired for CFP (excitation = 430 ± 12 nm; emission = 480 ± 20 nm), YFP (excitation = 500 ± 10 nm; emission = 535 ± 15 nm), FRET (excitation = 430 ± 12 nm; emission = 535 ± 15 nm), and mCherry channels. After subtracting background fluorescence, FRET/CFP emission ratios were calculated within single cells identified by the thresholded mCherry image. All image processing was performed in MATLAB.
For cytosolic Ca2+ measurements, HEK293T cells were plated on 96-well glass-bottom plates (In Vitro Scientific) and loaded with 1 µM fura-2/AM (Invitrogen) for 30–45 min at 37°C in serum-free media. Cells were washed with Tyrode’s solution and imaged using an automated epifluorescence microscope (ImageXpress 5000A; Molecular Devices) with a 10× objective (NA 0.3) at 37°C. Images were acquired for fura-2 (excitation = 340 ± 6 nm or 380 ± 6 nm; emission = 510 ± 40 nm) and mCherry (excitation = 565 ± 27; emission = 650 ± 37). Fura-2 340/380 ratios were calculated within single cells identified by the thresholded mCherry image.
Calcium imaging
HEK293 or Neuro2A cells on glass coverslips were loaded with 1 µM fura-2/AM in serum-free media for 30 min at 37°C and then washed with Tyrode’s solution before imaging. For experiments with Orai1–S-domain chimeras, cells were incubated in Ca2+-free Tyrode’s solution (CaCl2 replaced with MgCl2) for 15–30 min to restore [Ca2+]i in all cells to a low baseline before imaging. Coverslips were mounted in perfusion chambers and imaged using 340- and 380-nm excitation with an inverted microscope (Eclipse 2000-U; Nikon) equipped with 40× Nikon Fluor (NA 1.3) oil immersion objective, xenon arc lamp (Sutter Instrument), excitation filter wheel (Lambda-10; Sutter Instruments), and a charge-coupled device camera (Orca; Hamamatsu Photonics). Manual perfusion through syringes was used to exchange extracellular solutions. Transfected cells were identified using mCherry or YFP fluorescence, and ROIs were drawn manually. Mean fura-2 340/380 ratio within each ROI was quantified using custom scripts in Openlab (PerkinElmer). Igor Pro (WaveMetrics) and Prism 6 (GraphPad Software) were used for data analysis and plotting.
Correlated Ca2+ imaging and surface HA staining
HEK293 cells were transfected with Orai1 constructs having a HA tag inserted into the extracellular III–IV loop. After collection of Ca2+ imaging data, the cells were immediately fixed using 4% PFA and 8% sucrose in standard PBS. To measure Orai expression on the cell surface, cells were stained without permeabilization using monoclonal anti-HA antibody (3F10; Roche) followed by Alexa Fluor 594–coupled anti–rat secondary antibody (both at 1:1,000 dilution; Life Technologies), and imaged using the same microscope used for Ca2+ imaging. Fiduciary markings on the coverslips were used to identify the cells that were analyzed by Ca2+ imaging and to correlate HA staining with [Ca2+]i at the single-cell level. Custom scripts in MATLAB were used to calculate slopes from the fura-2 340/380 ratio traces of selected cells.
FRET measurements
FRET measurements were made using the three-cube E-FRET method (Zal and Gascoigne, 2004). HEK293 cells expressing CFP- and YFP-tagged constructs were plated on glass coverslips and imaged 30–48 h after transfection using an inverted epifluorescence microscope (Axiovert 200M; Carl Zeiss) with a 40× Fluar (NA 1.3) oil immersion objective (Carl Zeiss) and a polychrome II excitation source (TILL Photonics). Three channels were acquired (all filters obtained from Chroma Technology Corp.): CFP (440 ± 10 nm excitation, 455 DCLP dichroic, and 485 ± 20 nm emission), YFP (500 ± 10 nm excitation, 515 DCXR dichroic, and 535 ± 15 nm emission), and FRET (440 ± 10 nm excitation, 455 DCLP dichroic, and 535 ± 15 nm emission). For FRET experiments between STIM2α and STIM2β (Fig. S3, A and B), imaging was conducted at 20× magnification on a microscope (ImageXpress Micro XL) described above (see section on Resting ER and cytosolic Ca2+ measurements).
CFP, YFP, and FRET images were analyzed using a custom-written script in MATLAB. For FRET experiments with CFP-Orai1, ROIs were drawn manually along the cell membrane as identified by high CFP-Orai1 fluorescence. For STIM–STIM or CAD–CAD FRET experiments, ROIs were drawn randomly in the cytosolic regions of each cell. To avoid bias, only the CFP and YFP channels were visualized during ROI selection. Analysis was limited to cells showing YFP/CFP fluorescence ratios between 0.4 and 4 for all conditions to ensure accurate comparisons of E-FRET values. E-FRET was calculated as E-FRET = Fc/(Fc + G IDD), where Fc = IDA − a (IAA − c IDD) − d (IDD − b IAA), and IDD, IAA, and IDA are the background-corrected intensities in the CFP, YFP, and FRET channels, respectively. G is the instrument-dependent correction factor, and a, b, c, and d are bleedthrough factors calculated using cells expressing only YFP or CFP constructs (Zal and Gascoigne, 2004). Fixed cells expressing the calibrator construct CFP-RPTPα-SpD2-YFP2.1 (Blanchetot et al., 2002) were used to calculate G to estimate the degree of donor quenching from sensitized emission measurements.
Statistics
All statistical analysis was performed in Prism 6 (GraphPad Software) or the GraphPad QuickCalcs website (http://www.graphpad.com/quickcalcs/). All error bars represent SEM. All pairwise differences were tested for significance using a nonparametric two-tailed Mann–Whitney test or a two-tailed t test. Exact p-values are reported wherever possible.
Online supplemental material
Fig. S1 shows alignment of STIM2β insert sequence across multiple species and predicted effects of the 2β insert on coiled-coil formation in CAD. Fig. S2 shows further analysis of puncta formation and Orai1 recruitment by STIM2α and STIM2β. Fig. S3 shows FRET experiments between STIM2α and STIM2β and the effect of STIM1 on FRET between STIM2β and Orai1. Fig. S4 shows characterization of the STIM2β-L2R mutant. Fig. S5 shows characterization of the STIM2α-KA mutant and the lack of inhibitory effect of independently expressed STIM2β on Orai1(V102C). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201412060/DC1.
Supplementary Material
Acknowledgements
The authors thank G. Panagiotakos for help with quantitative PCR experiments, H. Blau for the generous gift of C2C12 cells, and members of the Lewis Laboratory for technical and scientific advice and comments on the manuscript.
This work was supported by the Lucille P. Markey Stanford Graduate Fellowship and Howard Hughes Medical Institute International Student Research Award (A. Rana), National Science Foundation Graduate Research Fellowship Program and National Institutes of Health training grant 5T32AI007290 to the Stanford Immunology Program (M. Yen), National Research Foundation of Korea grant NRF-2012-R1A1-A1044814 by the Korean Ministry of Education, Science and Technology and Future Challenge Project 1.40091.01 funding from Ulsan National Institute of Science and Technology (C.Y. Park), and National Institutes of Health grant R37GM45374 and the Mathers Charitable Foundation (R.S. Lewis).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CAD
- CRAC activation domain
- CRAC
- Ca2+ release-activated Ca2+
- DVF
- divalent-free
- FRET
- Förster resonance energy transfer
- NFAT
- nuclear factor of activated T cells
- PDBu
- phorbol 12,13-dibutyrate
- PM
- plasma membrane
- ROI
- region of interest
- SOCE
- store-operated calcium entry
- STIM
- stromal interaction molecule
- Tg
- thapsigargin
References
- Bandara S., Malmersjö S., and Meyer T.. 2013. Regulators of calcium homeostasis identified by inference of kinetic model parameters from live single cells perturbed by siRNA. Sci. Signal. 6:ra56 10.1126/scisignal.2003649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett-Lovsey R.M., Herbert A.D., Sternberg M.J., and Kelley L.A.. 2008. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 70:611–625. 10.1002/prot.21688 [DOI] [PubMed] [Google Scholar]
- Bird G.S., Hwang S.Y., Smyth J.T., Fukushima M., Boyles R.R., and Putney J.W. Jr. 2009. STIM1 is a calcium sensor specialized for digital signaling. Curr. Biol. 19:1724–1729. 10.1016/j.cub.2009.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchetot C., Tertoolen L.G., and den Hertog J.. 2002. Regulation of receptor protein-tyrosine phosphatase α by oxidative stress. EMBO J. 21:493–503. 10.1093/emboj/21.4.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O., Liou J., Park W.S., and Meyer T.. 2007. STIM2 is a feedback regulator that stabilizes basal cytosolic and endoplasmic reticulum Ca2+ levels. Cell. 131:1327–1339. 10.1016/j.cell.2007.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt P., and Vanaman T.C.. 1994. Splicing of the muscle-specific plasma membrane Ca2+-ATPase isoforms PMCA1c is associated with cell fusion in C2 myocytes. J. Neurochem. 62:799–802. 10.1046/j.1471-4159.1994.62020799.x [DOI] [PubMed] [Google Scholar]
- Burattini S., Ferri P., Battistelli M., Curci R., Luchetti F., and Falcieri E.. 2004. C2C12 murine myoblasts as a model of skeletal muscle development: morpho-functional characterization. Eur. J. Histochem. 48:223–233. [PubMed] [Google Scholar]
- Cahalan M.D. 2009. STIMulating store-operated Ca2+ entry. Nat. Cell Biol. 11:669–677. 10.1038/ncb0609-669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calloway N., Vig M., Kinet J.P., Holowka D., and Baird B.. 2009. Molecular clustering of STIM1 with Orai1/CRACM1 at the plasma membrane depends dynamically on depletion of Ca2+ stores and on electrostatic interactions. Mol. Biol. Cell. 20:389–399. 10.1091/mbc.E07-11-1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calloway N., Holowka D., and Baird B.. 2010. A basic sequence in STIM1 promotes Ca2+ influx by interacting with the C-terminal acidic coiled coil of Orai1. Biochemistry. 49:1067–1071. 10.1021/bi901936q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington E.D., Wu M.M., and Lewis R.S.. 2010. Essential role for the CRAC activation domain in store-dependent oligomerization of STIM1. Mol. Biol. Cell. 21:1897–1907. 10.1091/mbc.E10-02-0145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbellay B., Arnaudeau S., Ceroni D., Bader C.R., Konig S., and Bernheim L.. 2010. Human muscle economy myoblast differentiation and excitation-contraction coupling use the same molecular partners, STIM1 and STIM2. J. Biol. Chem. 285:22437–22447. 10.1074/jbc.M110.118984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbellay B., Arnaudeau S., Bader C.R., Konig S., and Bernheim L.. 2011. STIM1L is a new actin-binding splice variant involved in fast repetitive Ca2+ release. J. Cell Biol. 194:335–346. 10.1083/jcb.201012157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolmetsch R.E., Lewis R.S., Goodnow C.C., and Healy J.I.. 1997. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature. 386:855–858. 10.1038/386855a0 [DOI] [PubMed] [Google Scholar]
- Dolmetsch R.E., Xu K., and Lewis R.S.. 1998. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 392:933–936. 10.1038/31960 [DOI] [PubMed] [Google Scholar]
- Dong H., Fiorin G., Carnevale V., Treptow W., and Klein M.L.. 2013. Pore waters regulate ion permeation in a calcium release-activated calcium channel. Proc. Natl. Acad. Sci. USA. 110:17332–17337. 10.1073/pnas.1316969110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan E., Momburg F., Engel U., Temmerman K., Nickel W., and Seedorf M.. 2009. A conserved, lipid-mediated sorting mechanism of yeast Ist2 and mammalian STIM proteins to the peripheral ER. Traffic. 10:1802–1818. 10.1111/j.1600-0854.2009.00995.x [DOI] [PubMed] [Google Scholar]
- Feske S. 2010. CRAC channelopathies. Pflugers Arch. 460:417–435. 10.1007/s00424-009-0777-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feske S., Gwack Y., Prakriya M., Srikanth S., Puppel S.H., Tanasa B., Hogan P.G., Lewis R.S., Daly M., and Rao A.. 2006. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 441:179–185. 10.1038/nature04702 [DOI] [PubMed] [Google Scholar]
- Feske S., Picard C., and Fischer A.. 2010. Immunodeficiency due to mutations in ORAI1 and STIM1. Clin. Immunol. 135:169–182. 10.1016/j.clim.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischauf I., Muik M., Derler I., Bergsmann J., Fahrner M., Schindl R., Groschner K., and Romanin C.. 2009. Molecular determinants of the coupling between STIM1 and Orai channels: differential activation of Orai1-3 channels by a STIM1 coiled-coil mutant. J. Biol. Chem. 284:21696–21706. 10.1074/jbc.M109.018408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudlur A., Quintana A., Zhou Y., Hirve N., Mahapatra S., and Hogan P.G.. 2014. STIM1 triggers a gating rearrangement at the extracellular mouth of the ORAI1 channel. Nat. Commun. 5:5164 10.1038/ncomms6164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M.P., Nagel R.J., Fagg W.S., Shiue L., Cline M.S., Perriman R.J., Donohue J.P., and Ares M. Jr. 2013. Quaking and PTB control overlapping splicing regulatory networks during muscle cell differentiation. RNA. 19:627–638. 10.1261/rna.038422.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins B.J., Irrinki K.M., Mallilankaraman K., Lien Y.C., Wang Y., Bhanumathy C.D., Subbiah R., Ritchie M.F., Soboloff J., Baba Y., et al. 2010. S-glutathionylation activates STIM1 and alters mitochondrial homeostasis. J. Cell Biol. 190:391–405. 10.1083/jcb.201004152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan P.G., Lewis R.S., and Rao A.. 2010. Molecular basis of calcium signaling in lymphocytes: STIM and ORAI. Annu. Rev. Immunol. 28:491–533. 10.1146/annurev.immunol.021908.132550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover P.J., and Lewis R.S.. 2011. Stoichiometric requirements for trapping and gating of Ca2+ release-activated Ca2+ (CRAC) channels by stromal interaction molecule 1 (STIM1). Proc. Natl. Acad. Sci. USA. 108:13299–13304. 10.1073/pnas.1101664108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi T., Higashi T., Higa T., Terada K., Mai Y., Aoyagi H., Hatate C., Nepal P., Horiguchi M., Harada T., and Miwa S.. 2012. Different binding property of STIM1 and its novel splice variant STIM1L to Orai1, TRPC3, and TRPC6 channels. Biochem. Biophys. Res. Commun. 428:252–258. 10.1016/j.bbrc.2012.10.034 [DOI] [PubMed] [Google Scholar]
- Kawasaki T., Lange I., and Feske S.. 2009. A minimal regulatory domain in the C terminus of STIM1 binds to and activates ORAI1 CRAC channels. Biochem. Biophys. Res. Commun. 385:49–54. 10.1016/j.bbrc.2009.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt A.R., Schor I.E., Alló M., Dujardin G., Petrillo E., and Muñoz M.J.. 2013. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 14:153–165. 10.1038/nrm3525 [DOI] [PubMed] [Google Scholar]
- Korzeniowski M.K., Manjarrés I.M., Varnai P., and Balla T.. 2010. Activation of STIM1-Orai1 involves an intramolecular switching mechanism. Sci. Signal. 3:ra82 10.1126/scisignal.2001122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis R.S. 2011. Store-operated calcium channels: new perspectives on mechanism and function. Cold Spring Harb. Perspect. Biol. 3:a003970 10.1101/cshperspect.a003970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Liu L., Deng Y., Ji W., Du W., Xu P., Chen L., and Xu T.. 2011. Graded activation of CRAC channel by binding of different numbers of STIM1 to Orai1 subunits. Cell Res. 21:305–315. 10.1038/cr.2010.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Zheng S., Han A., Lin C.H., Stoilov P., Fu X.D., and Black D.L.. 2014. The splicing regulator PTBP2 controls a program of embryonic splicing required for neuronal maturation. eLife. 3:e01201 10.7554/eLife.01201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J., Kim M.L., Heo W.D., Jones J.T., Myers J.W., Ferrell J.E. Jr., and Meyer T.. 2005. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 15:1235–1241. 10.1016/j.cub.2005.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J., Fivaz M., Inoue T., and Meyer T.. 2007. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc. Natl. Acad. Sci. USA. 104:9301–9306. 10.1073/pnas.0702866104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luik R.M., Wang B., Prakriya M., Wu M.M., and Lewis R.S.. 2008. Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature. 454:538–542. 10.1038/nature07065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., and Stock J.. 1991. Predicting coiled coils from protein sequences. Science. 252:1162–1164. 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- McNally B.A., Somasundaram A., Yamashita M., and Prakriya M.. 2012. Gated regulation of CRAC channel ion selectivity by STIM1. Nature. 482:241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally B.A., Somasundaram A., Jairaman A., Yamashita M., and Prakriya M.. 2013. The C- and N-terminal STIM1 binding sites on Orai1 are required for both trapping and gating CRAC channels. J. Physiol. 591:2833–2850. 10.1113/jphysiol.2012.250456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Miner C., Zhang L., Hanson P.I., Dani A., and Vig M.. 2013. An essential and NSF independent role for α-SNAP in store-operated calcium entry. eLife. 2:e00802 10.7554/eLife.00802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik M., Fahrner M., Schindl R., Stathopulos P., Frischauf I., Derler I., Plenk P., Lackner B., Groschner K., Ikura M., and Romanin C.. 2011. STIM1 couples to ORAI1 via an intramolecular transition into an extended conformation. EMBO J. 30:1678–1689. 10.1038/emboj.2011.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesin V., Wiley G., Kousi M., Ong E.C., Lehmann T., Nicholl D.J., Suri M., Shahrizaila N., Katsanis N., Gaffney P.M., et al. 2014. Activating mutations in STIM1 and ORAI1 cause overlapping syndromes of tubular myopathy and congenital miosis. Proc. Natl. Acad. Sci. USA. 111:4197–4202. 10.1073/pnas.1312520111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palty R., Raveh A., Kaminsky I., Meller R., and Reuveny E.. 2012. SARAF inactivates the store operated calcium entry machinery to prevent excess calcium refilling. Cell. 149:425–438. 10.1016/j.cell.2012.01.055 [DOI] [PubMed] [Google Scholar]
- Parekh A.B., and Putney J.W. Jr. 2005. Store-operated calcium channels. Physiol. Rev. 85:757–810. 10.1152/physrev.00057.2003 [DOI] [PubMed] [Google Scholar]
- Park C.Y., Hoover P.J., Mullins F.M., Bachhawat P., Covington E.D., Raunser S., Walz T., Garcia K.C., Dolmetsch R.E., and Lewis R.S.. 2009. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 136:876–890. 10.1016/j.cell.2009.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez S., Beck A., Peinelt C., Soboloff J., Lis A., Monteilh-Zoller M., Gill D.L., Fleig A., and Penner R.. 2008. STIM2 protein mediates distinct store-dependent and store-independent modes of CRAC channel activation. FASEB J. 22:752–761. 10.1096/fj.07-9449com [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo-Guisado E., Campbell D.G., Deak M., Alvarez-Barrientos A., Morrice N.A., Alvarez I.S., Alessi D.R., and Martín-Romero F.J.. 2010. Phosphorylation of STIM1 at ERK1/2 target sites modulates store-operated calcium entry. J. Cell Sci. 123:3084–3093. 10.1242/jcs.067215 [DOI] [PubMed] [Google Scholar]
- Rao A., Luo C., and Hogan P.G.. 1997. Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol. 15:707–747. 10.1146/annurev.immunol.15.1.707 [DOI] [PubMed] [Google Scholar]
- Ritchie M.F., Yue C., Zhou Y., Houghton P.J., and Soboloff J.. 2010. Wilms tumor suppressor 1 (WT1) and early growth response 1 (EGR1) are regulators of STIM1 expression. J. Biol. Chem. 285:10591–10596. 10.1074/jbc.M109.083493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos J., DiGregorio P.J., Yeromin A.V., Ohlsen K., Lioudyno M., Zhang S., Safrina O., Kozak J.A., Wagner S.L., Cahalan M.D., et al. 2005. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 169:435–445. 10.1083/jcb.200502019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaghiani A.M., Lee S.M., Odegaard J.I., Leveson-Gower D.B., McPherson O.M., Novick P., Kim M.R., Koehler A.N., Negrin R., Dolmetsch R.E., and Park C.Y.. 2014. Identification of Orai1 channel inhibitors by using minimal functional domains to screen small molecule microarrays. Chem. Biol. 21:1278–1292. 10.1016/j.chembiol.2014.08.016 [DOI] [PubMed] [Google Scholar]
- Smyth J.T., Petranka J.G., Boyles R.R., DeHaven W.I., Fukushima M., Johnson K.L., Williams J.G., and Putney J.W. Jr. 2009. Phosphorylation of STIM1 underlies suppression of store-operated calcium entry during mitosis. Nat. Cell Biol. 11:1465–1472. 10.1038/ncb1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboloff J., Spassova M.A., Hewavitharana T., He L.P., Xu W., Johnstone L.S., Dziadek M.A., and Gill D.L.. 2006. STIM2 is an inhibitor of STIM1-mediated store-operated Ca2+ Entry. Curr. Biol. 16:1465–1470. 10.1016/j.cub.2006.05.051 [DOI] [PubMed] [Google Scholar]
- Somasundaram A., Shum A.K., McBride H.J., Kessler J.A., Feske S., Miller R.J., and Prakriya M.. 2014. Store-operated CRAC channels regulate gene expression and proliferation in neural progenitor cells. J. Neurosci. 34:9107–9123. 10.1523/JNEUROSCI.0263-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth S., Jung H.J., Kim K.D., Souda P., Whitelegge J., and Gwack Y.. 2010. A novel EF-hand protein, CRACR2A, is a cytosolic Ca2+ sensor that stabilizes CRAC channels in T cells. Nat. Cell Biol. 12:436–446. 10.1038/ncb2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopulos P.B., Li G.Y., Plevin M.J., Ames J.B., and Ikura M.. 2006. Stored Ca2+ depletion-induced oligomerization of stromal interaction molecule 1 (STIM1) via the EF-SAM region: An initiation mechanism for capacitive Ca2+ entry. J. Biol. Chem. 281:35855–35862. 10.1074/jbc.M608247200 [DOI] [PubMed] [Google Scholar]
- Stathopulos P.B., Schindl R., Fahrner M., Zheng L., Gasmi-Seabrook G.M., Muik M., Romanin C., and Ikura M.. 2013. STIM1/Orai1 coiled-coil interplay in the regulation of store-operated calcium entry. Nat. Commun. 4:2963 10.1038/ncomms3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiber J., Hawkins A., Zhang Z.S., Wang S., Burch J., Graham V., Ward C.C., Seth M., Finch E., Malouf N., et al. 2008. STIM1 signalling controls store-operated calcium entry required for development and contractile function in skeletal muscle. Nat. Cell Biol. 10:688–697. 10.1038/ncb1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z.Z., Zheng S., Nikolic J., and Black D.L.. 2009. Developmental control of CaV1.2 L-type calcium channel splicing by Fox proteins. Mol. Cell. Biol. 29:4757–4765. 10.1128/MCB.00608-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberghe M., Raphaël M., Lehen’kyi V., Gordienko D., Hastie R., Oddos T., Rao A., Hogan P.G., Skryma R., and Prevarskaya N.. 2013. ORAI1 calcium channel orchestrates skin homeostasis. Proc. Natl. Acad. Sci. USA. 110:E4839–E4848. 10.1073/pnas.1310394110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Szabo D., Braun A., and Nieswandt B.. 2011. STIM and Orai in platelet function. Cell Calcium. 50:270–278. 10.1016/j.ceca.2011.04.002 [DOI] [PubMed] [Google Scholar]
- Vig M., Peinelt C., Beck A., Koomoa D.L., Rabah D., Koblan-Huberson M., Kraft S., Turner H., Fleig A., Penner R., and Kinet J.P.. 2006. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science. 312:1220–1223. 10.1126/science.1127883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang Y., Zhou Y., Hendron E., Mancarella S., Andrake M.D., Rothberg B.S., Soboloff J., and Gill D.L.. 2014. Distinct Orai-coupling domains in STIM1 and STIM2 define the Orai-activating site. Nat. Commun. 5:3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei-LaPierre L., Carrell E.M., Boncompagni S., Protasi F., and Dirksen R.T.. 2013. Orai1-dependent calcium entry promotes skeletal muscle growth and limits fatigue. Nat. Commun. 4:2805 10.1038/ncomms3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.T., Manji S.S., Parker N.J., Hancock M.S., Van Stekelenburg L., Eid J.P., Senior P.V., Kazenwadel J.S., Shandala T., Saint R., et al. 2001. Identification and characterization of the STIM (stromal interaction molecule) gene family: coding for a novel class of transmembrane proteins. Biochem. J. 357:673–685. 10.1042/0264-6021:3570673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.M., Buchanan J., Luik R.M., and Lewis R.S.. 2006. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J. Cell Biol. 174:803–813. 10.1083/jcb.200604014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.M., Covington E.D., and Lewis R.S.. 2014. Single-molecule analysis of diffusion and trapping of STIM1 and Orai1 at endoplasmic reticulum-plasma membrane junctions. Mol. Biol. Cell. 25:3672–3685. 10.1091/mbc.E14-06-1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Jin H., Cai X., Li S., and Shen Y.. 2012. Structural and mechanistic insights into the activation of Stromal interaction molecule 1 (STIM1). Proc. Natl. Acad. Sci. USA. 109:5657–5662. 10.1073/pnas.1118947109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J.P., Zeng W., Dorwart M.R., Choi Y.J., Worley P.F., and Muallem S.. 2009. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat. Cell Biol. 11:337–343. 10.1038/ncb1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zal T., and Gascoigne N.R.. 2004. Photobleaching-corrected FRET efficiency imaging of live cells. Biophys. J. 86:3923–3939. 10.1529/biophysj.103.022087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.L., Yu Y., Roos J., Kozak J.A., Deerinck T.J., Ellisman M.H., Stauderman K.A., and Cahalan M.D.. 2005. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature. 437:902–905. 10.1038/nature04147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Stathopulos P.B., Li G.Y., and Ikura M.. 2008. Biophysical characterization of the EF-hand and SAM domain containing Ca2+ sensory region of STIM1 and STIM2. Biochem. Biophys. Res. Commun. 369:240–246. 10.1016/j.bbrc.2007.12.129 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.