Data from a new in vitro assay measuring inner nuclear membrane (INM) targeting kinetics as well as from mathematical modeling support a diffusion-retention–based mechanism of INM targeting.
Abstract
Newly synthesized membrane proteins are constantly sorted from the endoplasmic reticulum (ER) to various membranous compartments. How proteins specifically enrich at the inner nuclear membrane (INM) is not well understood. We have established a visual in vitro assay to measure kinetics and investigate requirements of protein targeting to the INM. Using human LBR, SUN2, and LAP2β as model substrates, we show that INM targeting is energy-dependent but distinct from import of soluble cargo. Accumulation of proteins at the INM relies on both a highly interconnected ER network, which is affected by energy depletion, and an efficient immobilization step at the INM. Nucleoporin depletions suggest that translocation through nuclear pore complexes (NPCs) is rate-limiting and restricted by the central NPC scaffold. Our experimental data combined with mathematical modeling support a diffusion-retention–based mechanism of INM targeting. We experimentally confirmed the sufficiency of diffusion and retention using an artificial reporter lacking natural sorting signals that recapitulates the energy dependence of the process in vivo.
Introduction
The membranous organelles of eukaryotic cells contain specific sets of proteins that carry out enzymatic reactions or build up structural frameworks. Consequently, an elaborate sorting system is needed for guiding proteins to their correct destination. One of the least understood sorting mechanisms is how membrane proteins specifically enrich at the inner membrane of the nuclear envelope (NE). Dysfunction of an increasing number of these proteins is emerging as a cause of so-called “nuclear envelopathies,” ranging from tissue-specific defects like muscular dystrophies to systemic disorders like progeria (Burke and Stewart, 2014). It is thus crucial to elucidate the mechanism underlying NE biogenesis and homeostasis.
In general, transport into the nucleus occurs through nuclear pore complexes (NPCs). These large assemblies consist of multiples of ∼30 different nucleoporins (Nups; Grossman et al., 2012). The central NPC channel is lined with Nups carrying phenylalanine-glycine (FG)-rich repeats that promote receptor-mediated transport of soluble cargo containing nuclear import signals and limit diffusion of inert macromolecules (Terry and Wente, 2009). Newly synthesized integral inner nuclear membrane (INM) proteins are initially inserted into the ER or the connected outer nuclear membrane (ONM). From the ONM, they translocate to the INM through NPCs as membrane-bound proteins. Peripheral cavities near the pore membrane (Maimon et al., 2012) might allow for passage of membrane proteins.
Four models of INM targeting have been proposed: diffusion-retention, receptor-mediated transport, targeting via INM sorting motifs, and NPC translocation with the help of FG motifs in certain INM proteins (for review see Katta et al., 2014). The diffusion-retention model (Powell and Burke, 1990; Smith and Blobel, 1993; Soullam and Worman, 1993) posits that these proteins distribute by free diffusion within the continuous membranes of the ER, ONM, and INM. Accumulation at the INM is supposed to be driven by interaction with nuclear components. Originally, the diffusion-retention model was based on the observation that an INM protein exchanged between nuclei of interspecies heterokaryons in the presence of an appropriate nuclear retention partner (Powell and Burke, 1990). The concept of retention has been substantiated by the observed reduced mobility of membrane proteins at the NE compared with the ER (Soullam and Worman, 1995; Ellenberg et al., 1997; Östlund et al., 2006; Zuleger et al., 2011). In fact, many INM proteins in metazoans bind to the nuclear lamina and chromatin (Burke and Stewart, 2013), as exemplified by the lamin B receptor (LBR) and LAP2β. Other INM proteins belonging to the conserved SUN protein family are part of multimeric LINC complexes that bridge the NE and connect it to the cytoskeleton (Rothballer et al., 2013).
In contrast, an active step in transport to the INM was proposed based on the energy dependence of a reporter protein in mammalian cells (Ohba et al., 2004). Subsequently, the yeast proteins Heh1 and Heh2 were described to possess bipartite NLSs, conferring receptor-mediated import through the central NPC channel (King et al., 2006; Meinema et al., 2011). Consistently, various metazoan INM proteins, including LBR and SUN2, harbor extralumenal domains that are basic and contain nuclear import signals (Ulbert et al., 2006; Lusk et al., 2007; Ma et al., 2007; Turgay et al., 2010; Tapley et al., 2011). However, the functionality of these NLSs in receptor-mediated translocation of INM proteins has remained elusive. Targeting of INM proteins has also been explained by INM sorting motifs (Saksena et al., 2004, 2006) or intrinsic FG repeats (Zuleger et al., 2011). These internal motifs are proposed to facilitate sorting without the vital need for energy consumption.
To elucidate the mechanisms underlying transport to the INM, we established a visual in vitro assay, which enabled us to measure INM targeting kinetics. We demonstrate that NPC translocation of NLS-containing membrane proteins differs from soluble cargo. Energy depletion retards transport to the INM, accompanied by structural changes and reduced diffusional mobility in the ER. By integrating kinetic targeting and FRAP data into a mathematical model, we propose that INM targeting of our reporters can be explained by a diffusion-retention mechanism.
Results
An in vitro transport assay for INM proteins
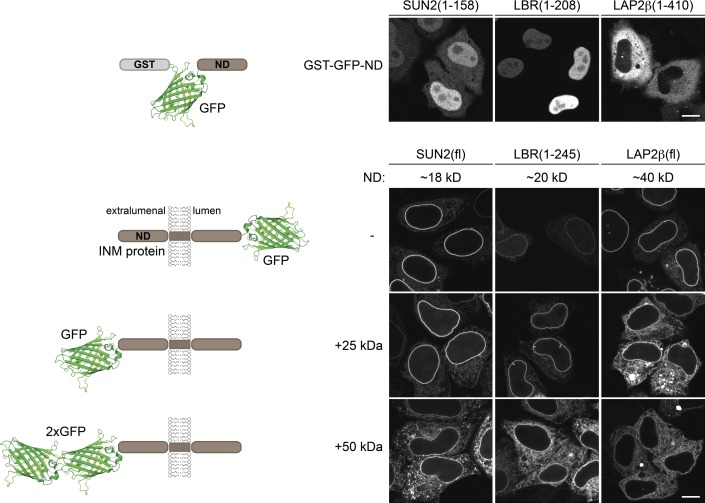
To elucidate the requirements and kinetics of membrane protein transport to the INM, we set out to establish a visual reporter assay in which protein integration into the ER is uncoupled from INM transport. Previous studies had demonstrated that the size of the nucleoplasmic domains (NDs) of INM proteins is restricted to 60 kD (Soullam and Worman, 1995; Ohba et al., 2004; Turgay et al., 2010; Theerthagiri et al., 2010; Zuleger et al., 2011), and proteins with enlarged NDs remain in the ER. First, we tested whether this size limitation applies to INM proteins with and without an NLS. Both SUN2 and LBR contain NLSs (Soullam and Worman, 1993; Ma et al., 2007; Turgay et al., 2010), and their soluble NDs were targeted to the nucleus as fusions with GST-GFP (Fig. 1). In contrast, the ND of LAP2β lacks nuclear import activity, and GST-GFP-LAP2β(ND) remained cytoplasmic.
Figure 1.
INM proteins with NDs >60 kD fail to be efficiently targeted to the INM despite their NLS activity. HeLa cells were transiently transfected with the depicted constructs. Localization was analyzed by confocal microscopy. The NDs of the indicated proteins were expressed as soluble GST-GFP fusions. SUN2(fl), LBR(1–245), and LAP2β(fl) were expressed either fused to a C-terminal, lumenal GFP or as fusions with one or two extralumenal GFP moieties. Bars, 10 µm.
To study how a size increase affected INM targeting of the membrane-integrated proteins, we appended one or two GFPs (25 or 50 kD) to the extralumenal domains of SUN2, LBR(1–245), or LAP2β (Fig. 1). For LAP2β, which has an ND of 40 kD, addition of one GFP was sufficient to restrain the protein from accumulation at the NE. SUN2 and LBR could be trapped in the ER by fusion to two GFPs, resulting in extralumenal domains of 68 and 70 kD, respectively. Thus, the limit of ∼60 kD also applies to INM proteins that harbor NLSs in their NDs.
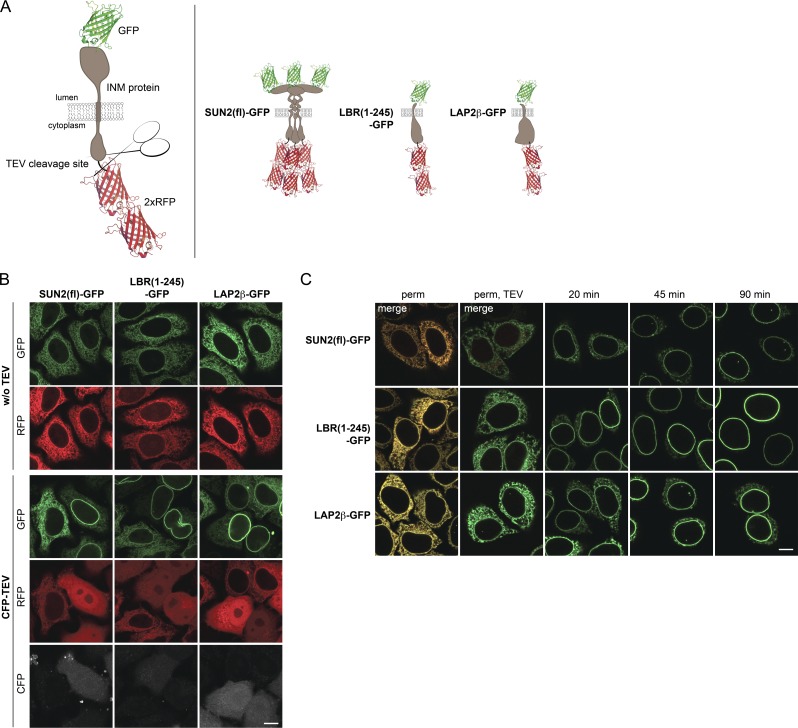
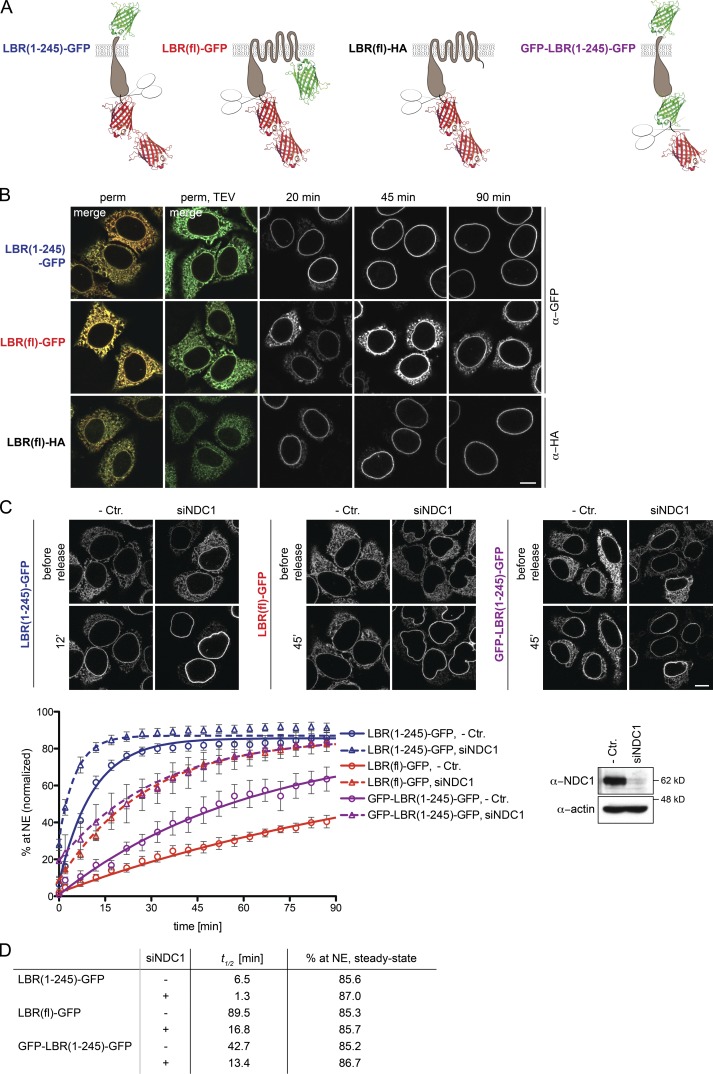
We took advantage of this size restriction for the INM transport assay. We appended two mRFPs to the NDs of selected INM proteins, separated by a tobacco etch virus (TEV) protease site to allow for 2×RFP removal by cleavage (Fig. 2 A). In addition, the reporters were lumenally tagged with GFP for visualization by microscopy. 2×RFP-tagged SUN2(fl)-GFP, LBR(1–245)-GFP, and LAP2β-GFP reporters were successfully trapped in the ER of HeLa cells. Co-expression of CFP-tagged TEV caused 2×RFP release and accumulation of the INM proteins at the NE (Fig. 2 B). This trap/release system was successfully applied to several other INM proteins, including full-length LBR, SUN1, LEM2, Emerin, and NET5 (unpublished data).
Figure 2.
The INM targeting assay. (A) Schematic representation of INM targeting reporters used in this study. (B) Release of INM reporters from the ER to the NE upon TEV cleavage in vivo. Tetracycline-inducible HeLa cell lines expressing 2×RFPtevSUN2(fl)-GFP, 2×RFPtevLBR(1–245)-GFP, or 2×RFPtevLAP2β-GFP were transiently transfected with CFP-TEV protease. Localization of reporters was analyzed after 24 h. (C) Reconstitution of INM targeting in semipermeabilized cells (perm) was initiated by TEV protease cleavage for 10 min at RT (perm, TEV) and started by supplementation with HeLa lysate and an energy regenerating system. At the indicated time points, cells were fixed and analyzed by confocal microscopy. Bars, 10 µm.
For in vitro reconstitution of INM targeting, we used the semipermeabilized cell system that has been instrumental to study the requirements for nuclear import of soluble macromolecules (Adam et al., 1990; Ribbeck and Görlich, 2001). Addition of recombinant TEV to semipermeabilized cells expressing the reporter constructs led to efficient release of 2×RFP (Fig. 2 C and Fig. 3 A). We then reconstituted transport to the INM by addition of a HeLa cell lysate and an energy-regenerating system. All three reporters efficiently targeted to the NE (Fig. 2 C). To confirm that NE accumulation indeed reflects INM targeting of the reporters, INM localization was verified by antibody accessibility of the ND upon differential detergent permeabilization of the NE (Fig. S1).
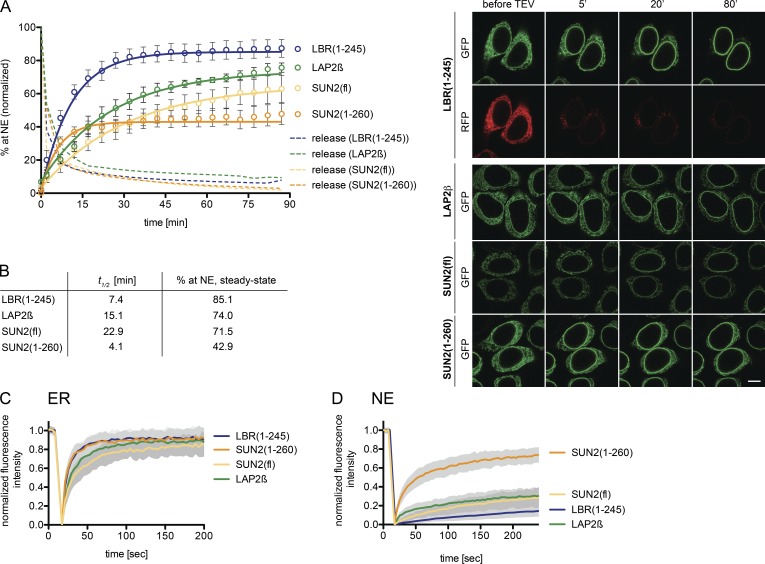
Figure 3.
Kinetics of trafficking of membrane proteins from the ER to the INM. (A) Semipermeabilized reporter cells were supplemented with TEV, HeLa lysate, and an energy mix. Targeting to the NE was followed by confocal time-lapse imaging. Release by TEV cleavage was quantified by the loss of the RFP signal. Mean NE accumulation relative to total fluorescence ± SEM (error bars) is shown; n ≥ 62. Solid lines show least-square fits of the kinetic model. Bar, 10 µm. (B) t1/2 and extrapolated steady-state levels of INM targeting derived from A. (C) FRAP in the ER performed in semipermeabilized cells with HeLa lysate and energy before release by TEV cleavage. Mean ± SD; n ≥ 19. (D) FRAP at the NE after targeting of the reporters. Mean ± SD; n ≥ 15.
Kinetics of translocation to the INM
Next, we measured transport kinetics of SUN2(fl)-GFP, LBR(1–245)-GFP, and LAP2β-GFP by confocal time-lapse microscopy. Half times of 2×RFP release were in the range of 2 min. Accumulation of the reporters at the NE started immediately after addition of TEV protease, HeLa lysate, and energy (Fig. 3 A). LBR accumulated with a half time of ∼7 min and reached a steady-state level of 85%. In contrast, SUN2 and LAP2β were slower (t1/2 of ∼23 min and 15 min), and did not reach steady-state after 90 min (Fig. 3, A and B). To compare targeting kinetics in vitro and in vivo, we also analyzed accumulation of the SUN2 reporter at the NE after microinjection of TEV in living cells. The kinetic curves were nearly superimposable (Fig. S2), suggesting that the in vitro assay adequately recapitulates trafficking to the INM.
The differences in targeting kinetics prompted us to determine the mobility of the reporters in the ER by FRAP. Consistent with its fast targeting kinetics, 2×RFPtevLBR(1–245)-GFP showed fast recovery in the ER (t1/2 = ∼7 s; Fig. 3 C), comparable to published data for LBR (Ellenberg et al., 1997; Zuleger et al., 2011). SUN2 recovered the slowest (t1/2 = ∼16 s), and displayed a lower mobile fraction. The slower diffusional mobility of SUN2 might be due to its homotrimerization (Sosa et al., 2012). In support of this, the ER diffusional mobility of SUN2(1–260), lacking the lumenal coiled-coil and SUN domains, was indeed fast and comparable to LBR (Fig. 3 C). SUN(1–260) also accumulated at the NE with a short t1/2 of ∼4 min. However, it only reached a steady-state level of ∼43% (Fig. 3 B), in agreement with the contribution of LINC complex formation to retention of SUN2 at the NE (Turgay et al., 2010; Sosa et al., 2012).
Considering the simplest model for INM targeting, the diffusion-retention model, the level of accumulation at the INM should be dependent on binding to nuclear interaction partners. As targeting efficiency of the reporters differed, we asked whether this reflects differences in retention strength. We therefore performed FRAP at the NE after in vitro reconstitution of INM targeting (Fig. 3 D). Strikingly, the mobility of LBR at the NE was extremely low, with almost no diffusion-limited recovery and a very slow off-rate limited recovery (t1/2 extrapolated to 26 min). SUN2(fl) and LAP2β displayed a slightly higher binding off-rate (t1/2 = ∼13 and 14 min). In contrast, SUN2(1–260) was highly mobile, even exhibiting a significant diffusion-limited mobile fraction (t1/2 = ∼53 s). These data demonstrate a strong retention for LBR, LAP2β, and SUN2 at the INM. In comparison, SUN2(1–260) is poorly retained. Collectively, the “strength” of retention of the different reporters at the NE correlates by-and-large with their levels at the INM.
The NPC in membrane protein translocation
To dissect the involvement of NPC constituents in INM protein import, we used siRNA-mediated silencing of Nups. We established RNAi conditions for 15 Nups (Fig. 4 A) chosen to cover members of all scaffold subcomplexes, FG Nups, and pore membrane proteins such that depletion could be achieved without diminishing expression of the LBR(1–245) reporter. We also examined whether RNAi-mediated knockdowns led to cooperative loss of whole subcomplexes, reduced NPC numbers, or impaired soluble protein import.
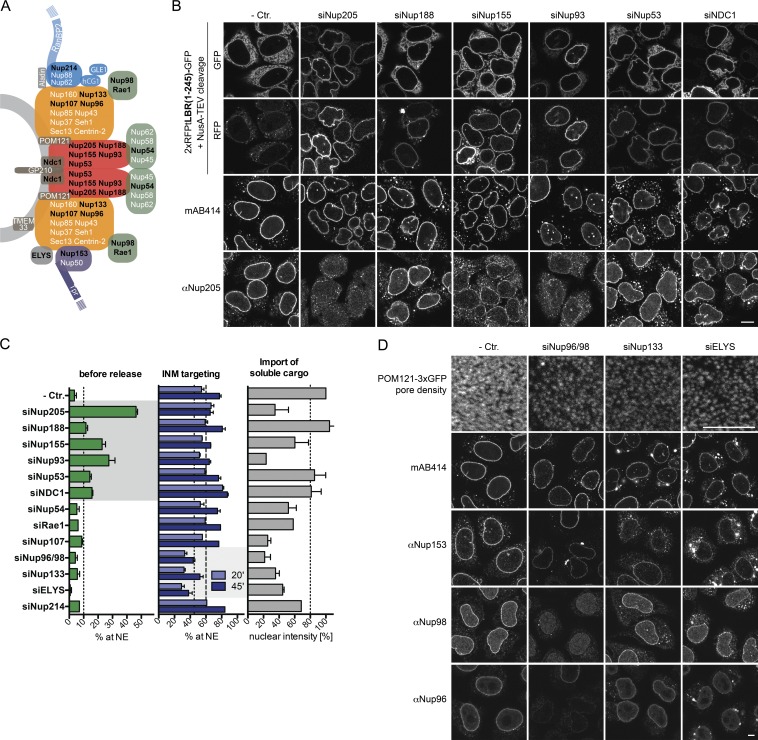
Figure 4.
The Nup93 subcomplex confines passage of INM proteins. (A) Model of NPC organization adapted from Rothballer and Kutay (2012). Nups indicated in black were analyzed. (B) Depletion of Nup93 subcomplex members and NDC1 allow NPC passage of enlarged INM proteins. 2×RFPtevLBR(1–245)-GFP reporter cells were transfected with siRNAs against members of the Nup93 subcomplex and NDC1. 16 h before analysis, expression of the LBR reporter was induced. Reporter cells were semipermeabilized, and protection from NusA-TEV (∼90 kD) served as a readout for INM localization. Immunofluorescence analysis of FG Nups (mAB414) and Nup205 as control for siRNA efficiency was performed. Bar, 10 µm. (C) Quantification of NE accumulation before (as in B) or after reconstitution of INM targeting upon addition of HeLa lysate and energy for 20 or 45 min on cells treated with Nup-specific siRNAs. In parallel, nuclear accumulation of 3×CFP-IBB was assayed after 45 min. Broken lines illustrate cutoffs for significant effects (P ≤ 0.05). Mean ± SEM (error bars); n ≥ 45. (D) Knockdown of Nup96/98, Nup133, and ELYS causes a strong reduction in NPC number visualized on the nuclear surface of a Pom121-3×GFP–expressing HeLa cell line and by immunofluorescence staining with mAB414, Nup153, Nup98, and Nup96 antibodies on cells transfected with control, Nup96/98, Nup133, or ELYS-specific siRNAs. Bars, 5 µm.
First, we analyzed how loss of Nups affected the integrity of the size barrier for membrane proteins using the 2×RFPtevLBR(1–245)-GFP reporter. Depletion of members of the Nup53–93 subcomplex, i.e., Nup205, Nup188, Nup155, Nup93, Nup53, and the transmembrane (TM) Nup NDC1, led to the accumulation of the uncleaved reporter at the NE. At the NE, the reporter was protected from NusA-TEV cleavage, which indicates proper INM localization (Fig. 4, B and C). As depletion of individual Nups of the Nup53–93 subcomplex also led to effects on localization and levels of other members of this subcomplex, e.g., Nup205 (Fig. 4 B), we cannot attribute the phenotype to a single nucleoporin.
Second, we measured INM targeting efficiency in the presence of HeLa lysate and energy 20 or 45 min after release. Knockdown of Nup98/96 (synthesized from one mRNA), Nup133, and ELYS decreased the translocation rate of LBR(1–245) (Fig. 4 C). Importantly, depletion of these Nups caused a strong reduction in NPC number, accompanied by reduced mAB414 staining (Fig. 4 D) and lower import rates for the soluble, importin β–dependent cargo 3×CFP-IBB (Fig. 4 C). However, the effect on translocation of LBR(1–245) did not fully correlate with defects in import of 3×CFP-IBB. For example, knockdown of Nup54 and Rae1 did not affect targeting of LBR, whereas import of 3×CFP-IBB was diminished. As Nup96 and Nup133 are both members of the Nup107–160 complex, and, together with ELYS, are required for NPC biogenesis (for review see Güttinger et al., 2009), their effect on membrane protein import is best explained by a reduction of NPC numbers.
In summary, our data suggest that the Nup53–93 subcomplex and NDC1 define the size exclusion limit for membrane proteins, and that NPC numbers affect targeting kinetics.
NDC1 is an obstacle for membrane protein translocation along the pore membrane
Among the tested Nups, depletion of NDC1 showed an exceptional phenotype. While causing a slight relaxation of the size barrier for 2×RFPtevLBR(1–245)-GFP, it also accelerated translocation of the residual, ER-localized pool to the INM after TEV cleavage (Fig. 4 C). This effect is surprising because NDC1 is crucial for NPC biogenesis and its depletion affects NPC numbers (Mansfeld et al., 2006; Stavru et al., 2006). We reasoned that NDC1, which contains six TM domains and is likely present in 32 copies per NPC (Ori et al., 2013), might restrict translocation of INM proteins within the pore membrane.
To compare the effect of NDC1 depletion on INM proteins with different numbers of TM domains, we measured INM targeting kinetics of LBR(1–245)-GFP and full-length LBR-GFP, which possess one and eight TM segments, respectively. GFP appended to the C terminus of LBR(fl) is, however, extralumenal (Fig. 5 A) and expected to hinder NPC translocation, leading to a slow accumulation at the NE compared with full-length LBR carrying a short hemagglutinin (HA) tag (Fig. 5 B). To account for this, we also constructed an N-terminal GFP fusion of LBR(1–245) to control for changes in transport kinetics that are caused by an extralumenal GFP moiety.
Figure 5.
NDC1 restricts passage of membrane proteins. (A) Schematic depiction of LBR-derived constructs. (B) An extralumenal GFP on LBR retards accumulation at the NE. (C) Representative images and quantification of NE targeting of LBR(1–245)-GFP, LBR(fl)-GFP, or GFP-LBR(1–245)-GFP in the presence of HeLa lysate + energy in cells transfected with control or NDC1-specific siRNAs. Mean ± SEM (error bars); n ≥ 33. Solid lines show least-square fits of the kinetic ODE model. Western blot confirming knockdown of NDC1. Bars, 10 µm. (D) t1/2 and extrapolated steady-state of INM targeting derived from data in C.
GFP-LBR(1–245)-GFP accumulated at the NE slowly, with a t1/2 of 43 min, compared with 7 min for LBR(1–245)-GFP (Fig. 5, C and D), underscoring the size effect of extralumenal domains on translocation kinetics of INM proteins. LBR(fl)-GFP accumulated the slowest, with a t1/2 of 90 min, although its mobility in the ER was comparable to LBR(1–245)-GFP (Zuleger et al., 2011; unpublished data). Importantly, the difference between GFP-LBR(1–245)-GFP and LBR(fl)-GFP can be abrogated by depletion of NDC1, which decreased translocation half times to 13 and 17 min, respectively. Thus, NDC1 appears to present an obstacle within the pore membrane for passage of membrane proteins, especially for those harboring many TM domains.
NPC translocation of membrane protein reporters and soluble cargo differs
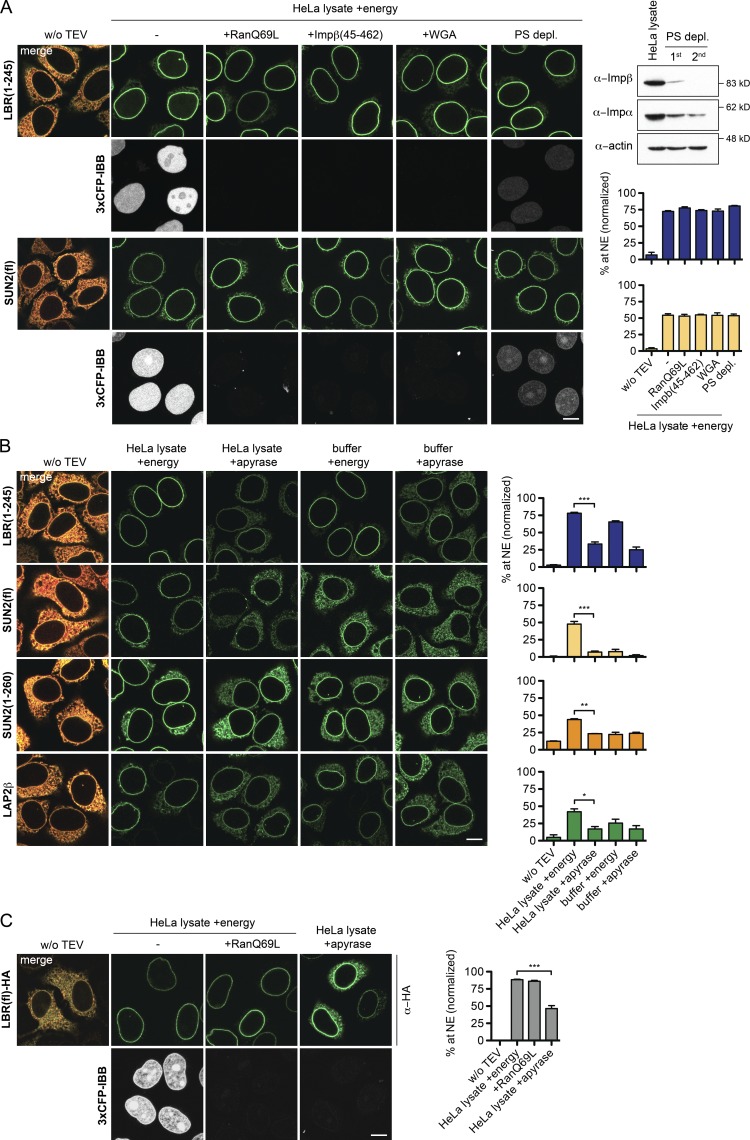
SUN2 and LBR both possess NLSs. Exploiting our in vitro system, we set out to directly test whether reagents that inhibit nuclear import of soluble, NLS-containing cargo would affect their targeting to the INM. To control for the efficacy of inhibition, we simultaneously monitored nuclear uptake of 3×CFP-IBB. As expected, nuclear import of 3×CFP-IBB was impaired when (1) formation of importin–cargo complexes was inhibited by addition of the GTPase-deficient RanQ69L mutant (Klebe et al., 1995), (2) the interaction of importins with FG repeat containing Nups was disturbed by either the lectin wheat germ agglutinin (WGA; Finlay et al., 1987) or a dominant-negative fragment of importin β (Impβ(45–462); Kutay et al., 1997), and (3) upon depletion of importins (Ribbeck and Görlich, 2002). In contrast, translocation of both LBR(1–245) and SUN2 was not affected by any of these treatments (Fig. 6 A). Transport of HA-tagged, full-length LBR was also insensitive to addition of RanQ69L (Fig. 6 C). We conclude that NPC passage of LBR and SUN2 has different requirements than soluble cargo and depends on neither interaction with importins nor FG repeat Nups of the central NPC channel.
Figure 6.
Targeting of LBR and SUN2 in vitro is energy- but not RanGTP-dependent. (A) Targeting of LBR(1–245) and SUN2 is not impaired by inhibition of importin–cargo complex formation (10 µM RanQ69L(GTP)), occlusion of central NPC channels (1 µM Impβ(45–462) and 0.2 mg/ml WGA, 10 min preincubation), or depletion of transport receptors by phenyl-Sepharose (PS-depl.). Efficacy of depletion was controlled by immunoblots for importin α and β. Targeting efficiency of membrane protein reporters and import of the soluble cargo 3×CFP-IBB were analyzed 45 min after release by TEV cleavage. Mean ± SEM (error bars). Blue bars, LBR(1-245); yellow bars, SUN2(fl); n ≥ 27. (B) Targeting to the NE is energy-dependent. Depletion of NTPs by apyrase in reconstitution reactions performed in either HeLa lysate or buffer for 45 min. Mean ± SEM (error bars); n ≥ 109; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (unpaired t-test). (C) Analysis of full-length LBR tagged with a C-terminal HA tag. Mean ± SEM (error bars); n ≥ 32; ***, P ≤ 0.001 (unpaired t test). Bars, 10 µm.
INM targeting of SUN2, LBR, and LAP2β is dependent on the presence of NTPs
One puzzling issue regarding INM protein targeting is its apparent energy dependency in vivo (Ohba et al., 2004), which seems at odds with a diffusion-retention mechanism. To address this point, we compared INM targeting of the different reporters in vitro either in the presence of an energy regenerating system (energy) or upon NTP depletion by apyrase in an endpoint assay (45 min). Targeting of all reporters was significantly reduced by energy depletion (Fig. 6 B). When transport was analyzed in buffer, LBR(1–245) accumulated at the NE efficiently in the presence of energy (65%), demonstrating that accumulation at the INM does not strictly require factors present in the cytosolic extract. In buffer, targeting to the NE was also inhibited by NTP depletion (to ∼25%), but not completely abrogated. This suggests that LBR can in principle translocate to the INM upon energy depletion, albeit inefficiently.
Remarkably, SUN2 was only targeted to the INM efficiently in the presence of both HeLa lysate and energy (Fig. 6 B). In contrast, SUN2(1–260) resembled LBR in its energy dependence, and accumulated at the NE to ∼25% in buffer upon NTP depletion. The behavior of full-length SUN2 might in part be due to its slower targeting kinetics. However, for neither SUN2(1–260) nor SUN2(fl) did addition of NTPs to the buffer restore targeting as efficiently as for LBR, which indicates a specific extract dependency of the SUN2-derived reporters. Interestingly, this extract requirement is not linked to any of the three previously identified NE targeting signals of SUN2, comprising a classical NLS and a Golgi retrieval signal in the ND (both present in SUN2(1–260)), and the lumenal SUN domain (Turgay et al., 2010; Fig. S3). Thus, transport of SUN2 depends on an unknown cytoplasmic component that is not strictly necessary for targeting of LBR. Collectively, these data support a model in which targeting to the INM is in principle possible in the absence of extract and NTPs, perhaps by unassisted diffusion, but the presence of nucleotides increases the efficiency of the process.
Energy depletion affects ER structure and diffusional mobility of ER proteins
As nuclear import of all INM reporters required energy but was insensitive to the addition of RanQ69L, we wondered how the dependency on nucleotide hydrolysis could be explained. We excluded simple possibilities such as protein synthesis or intact microtubules (Fig. S4).
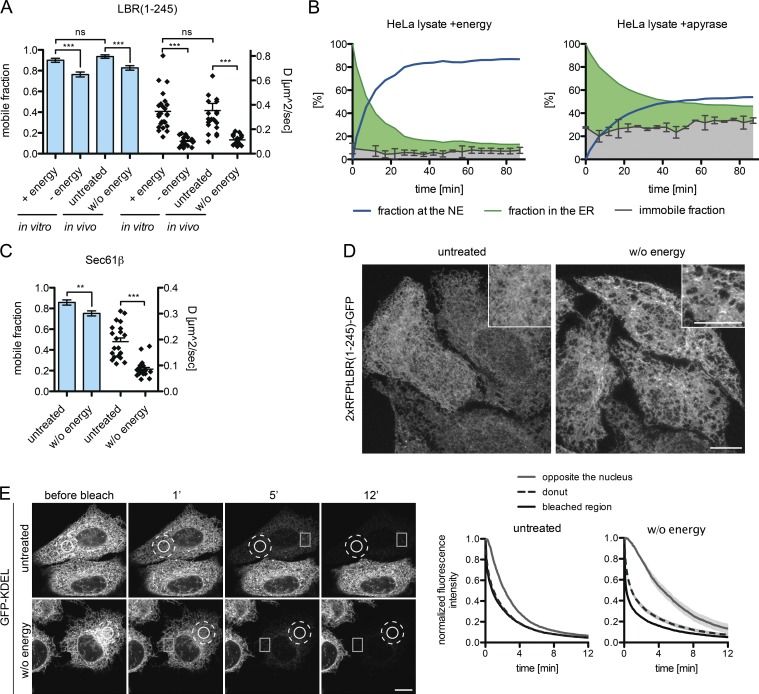
To assess whether NTP levels affect the diffusional properties of our reporters in the ER, we measured the dynamics of the uncleaved LBR(1–245)-GFP reporter in the ER by FRAP in vivo and in vitro upon energy depletion. The apparent diffusion coefficients and the mobile fractions of LBR were very similar in vivo and vitro in untreated controls, indicating that the ER network retains its basic properties in semipermeabilized cells supplemented with energy and HeLa cell lysate (Fig. 7 A). Upon apyrase treatment in vitro, the apparent diffusion coefficient dropped from 0.35 to 0.1 µm2s−1, and the mobile fraction decreased from 91 to 76%. NTP depletion of intact cells by treatment with deoxyglucose and sodium azide attenuated the mobility of LBR in the ER to a comparable extent.
Figure 7.
Energy depletion affects ER structure. (A) Mobile fractions and diffusion coefficient derived from FRAP on 2×RFPtevLBR(1–245)-GFP semipermeabilized reporter cells supplemented with HeLa lysate and either energy or apyrase (in vitro) or intact cells treated with deoxyglucose and NaN3 after 10 min of preincubation (in vivo). Mean ± SEM (error bars); n ≥ 23; ***, P ≤ 0.001 (unpaired t test). (B) Immobile fractions were measured by FRAP in the ER, before (t = 0) and after release of the LBR(1–245) reporter in semipermeabilized cells with HeLa lysate + energy or HeLa lysate + apyrase. INM targeting kinetics were measured as described for Fig. 3 A. The immobile fraction is displayed relative to the whole population. Mean ± SD (error bars). (C) Mobile fractions and diffusion coefficients upon energy depletion derived from FRAP performed on intact cells expressing Sec61β-GFP as in A. Mean ± SEM (error bars); n = 21; **, P ≤ 0.01 (unpaired t test). (D) The ER gets fenestrated upon energy depletion in vivo. Representative images are shown of 2×RFPtevLBR(1–245)-GFP reporter cells treated with deoxyglucose and NaN3 for 20 min before fixation. Maximum intensity projection (1.2 µm) of five confocal slices from the lower cell surface to the NE. (E) FLIP performed on cells transiently transfected with GFP-KDEL, untreated or treated with deoxyglucose and NaN3. Circle, bleached area; dashed circle, “donut” area; square, “opposite the nucleus” area. Mean ± SEM; n ≥ 12. Bars, 10 µm.
The reduced mobile fraction in the ER could explain the decrease in accumulation of LBR at the INM upon energy depletion. We therefore measured the immobile fraction of LBR in the ER by FRAP during the time course of an in vitro transport reaction. The immobile fraction of LBR(1–245)-GFP in the ER relative to the whole population of molecules stayed constant over time, but increased from ∼8% in the presence of energy to ∼30% upon apyrase treatment (Fig. 7 B). The half time of targeting was also prolonged from ∼7 min to ∼15 min, indicating that diminished lateral mobility in the ER impinges on INM targeting kinetics. Thus, lack of NTPs leads to a decreased mobility of the INM reporter in the ER as well as to a larger immobile fraction that fails to become mobilized over time.
When we assessed the dynamics of the ER-resident protein Sec61β upon energy depletion, we observed that its mobile fraction was similarly reduced from 85 to 75%, and also that the apparent diffusion coefficient decreased by a factor of 2 (Fig. 7 C). This indicates that energy depletion affects ER organization more globally.
To test this possibility, we analyzed morphological changes in the ER by high-resolution microscopy of cells expressing the LBR reporter. In untreated cells, the ER appeared as a fine meshwork of interconnected tubules and sheets that was spread evenly throughout the cytoplasm. In contrast, in energy-depleted cells, ER morphology was altered to a more fenestrated structure containing larger sheets that were sparsely connected (Fig. 7 D). To quantitatively assess the influence of these changes on diffusion within the ER, we analyzed the mobility of a soluble, ER-lumenal reporter (EGFP-KDEL) by fluorescence loss in photobleaching (FLIP) upon energy depletion in vivo. Repeated photobleaching of a circular spot led to a nearly simultaneous loss of fluorescence in a surrounding donut in untreated cells, but was delayed by 34 s (t1/2 of bleaching) upon energy depletion (Fig. 7 E). At a distant location, on the opposite side of the nucleus, loss of fluorescence occurred with a delay of ∼70 s in control cells, but only after ∼200 s when energy was depleted. The same tendency was observed in semipermeabilized cells (Fig. S5 B). This deceleration in diffusional exchange of soluble GFP between different parts of the ER upon energy depletion supports the notion that ER morphology changes such that it becomes limiting for rapid exchange.
A kinetic model for INM protein targeting
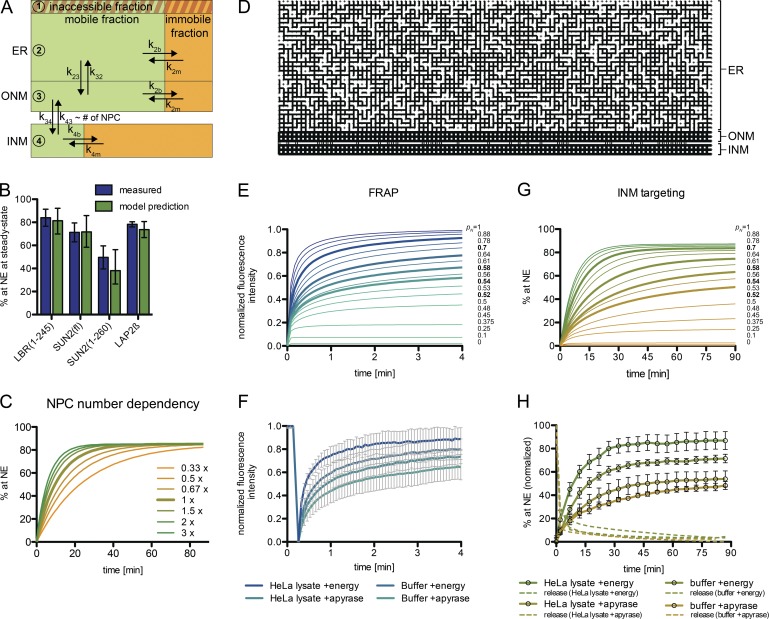
To assess whether a simple diffusion-retention–based mechanism can describe the observed targeting kinetics, we integrated our data into a kinetic model (Fig. 8 A), extending a previous modeling approach (Zuleger et al., 2011). We approximated the size of the membrane compartments (ER, ONM, INM) in which TM proteins reside along their trafficking route. Within these compartments, molecules can belong to a mobile pool, an immobile fraction that is in exchange with the mobile pool, or an inaccessible fraction, as observed for LBR (Fig. 7, A and B). The mobile fraction and apparent diffusion coefficients of molecules in the ER were derived from our FRAP data (Fig. 3 C). Translocation through the NPC was described as a reversible first-order reaction with identical rate constants for both directions. At the INM, we considered immobilization by binding interactions as predicted by the diffusion-retention model. The number of nuclear retention sites was set as unlimited. Mobile fraction and off-rate were derived from a double exponential fit of FRAP recovery curves measured at the NE (Fig. 3 D). Here, we accounted for a diffusive pool at the ONM that influences FRAP measurements at the NE. Using only the mobile fractions and compartment sizes, our model correctly predicted the steady-state of INM targeting for LBR(1–245), SUN2(fl), SUN2(1–260), and LAP2β compared with the experimental data (Fig. 8 B). Thus, a diffusion-retention–based model is able to explain differences in steady-state accumulation at the INM.
Figure 8.
A kinetic model for INM targeting. (A) Flow chart illustrating the kinetic ODE model. (B) Predicted steady-state NE levels are consistent with measured data. Measured values were derived from INM targeting kinetics in Fig. 3 A. Model predictions are based on FRAP data (ER and NE) and spatial assumptions. Mean with 95% CI (error bars). (C) Simulation of INM targeting kinetics of LBR(1–245)-GFP with varied NPC numbers. (D) Spatial ER model. Representation of one realization of a random 2D square network at connection probability pN = 0.7. (E) Simulated FRAP recovery curves in 2D square networks with varied pN. Highlighted in bold are recovery curves resembling measured FRAP curves upon NTP or extract depletion. (F) Measured FRAP in the ER on semipermeabilized 2×RFPtLBR(1–245)-GFP reporter cells revealing lower mobile fractions and higher recovery times upon energy depletion. Mean ± SD (error bars); n ≥ 23. (G) INM targeting simulations in 2D square networks with varied pN. Highlighted in bold are curves resembling the measured kinetics in H. (H) Kinetics of LBR(1–245)-GFP accumulation at the NE measured by time-lapse microscopy in semipermeabilized HeLa cells. Note that targeting kinetics in HeLa lysate + energy were derived from the same measurements as depicted in Fig. 3 A. Mean ± SEM (error bars); n ≥ 85.
The kinetics of INM targeting are mainly determined by three model parameters (Fig. 8 A): the diffusion rates k23/k32 between the ER and ONM, passage through the NPCs described by k34/k43, and immobilization at the INM defined by k4b/k4m. The rate constants k4m and k23/k32 were deduced from FRAP measurements. We were able to confine the residual parameters by fitting the model to the measured targeting kinetics (Fig. 3 A and Fig. 5 C). These fits revealed that NPC translocation is rate-limiting for targeting of all reporters (Table 1). SUN2(fl) was fit to a 4.4-fold lower NPC translocation rate constant k34 than LBR(1–245) and SUN2(1–260), which we attribute to trimerization of SUN2(fl). Notably, we were also able to computationally reproduce targeting kinetics of the different LBR constructs (Fig. 5 C, solid lines), which fitted with differences in k34. For LBR(1–245)-GFP, we can approximate an initial flux of ∼4 molecules/NPC/s at the nonsaturating substrate concentration of ∼107 molecules per cell. Because our model predicts that the NPC translocation rate k34 is rate-limiting for INM targeting, the overall nuclear influx of membrane proteins should in turn be dependent on the number of NPCs. This is consistent with our observation that knockdown of ELYS, Nup133, or Nup96/98 reduced NPC numbers and impaired targeting kinetics of LBR (Fig. 4, C and D). Simulations of INM targeting kinetics for LBR(1–245)-GFP at lower NPC numbers indeed recapitulated the measured retardation in NE accumulation (Fig. 8 C).
Table 1.
Parameters for NE targeting dynamics used for modeling-based analysis
| Model parameters | ER fractions | k2m | k34 (ONM → INM)a | INM fractions | k4b | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mobile | Immobile | Optimum | Optimum | Range (95% CI) | Mobile | Immobile | Value | |||||
| min−1 | min−1 | min−1 | min−1 | |||||||||
| LBR(1-245) | 0.92 | 0.08 | 1.890 | 1.30 | 0.90-1.98 | 0.009 | 0.991 | 2.76 | ||||
| LAP2β | 0.93 | 0.07 | 1.890 | 0.40 | 0.31-0.50 | 0.025 | 0.975 | 2.04 | ||||
| SUN2(fl) | 0.88 | 0.12 | 0.006 | 0.29 | 0.14–1.14 | 0.026 | 0.974 | 1.94 | ||||
| SUN2(1-260) | 0.95 | 0.05 | 4.456 | 1.39 | 0.90-2.20 | 0.112 | 0.888 | 1.46 | ||||
Italics indicate values derived from measurements. CI, confidence interval.
k34 = k43.
To predict how changes in ER connectivity influence INM targeting efficiency, we extended our kinetic model using a spatial ER model represented by a 2D square network with random connectivity. The connectivity between neighboring network points was defined by the probability pN (Fig. 8 D). We then simulated how changes in pN affect molecular dynamics in the ER as well as trafficking from the ER to the INM with parameters for LBR(1–245)-GFP (Fig. 8, E and G; and Fig. S5). FRAP simulations with stepwise reduced connectivity pN resulted in retardation of recovery and a decreased mobile fraction. Importantly, these curves strongly resemble experimental FRAP data on LBR(1–245)-GFP upon energy depletion in vitro (Fig. 8, E and F). A pN value of 0.7, reflecting an ER structure that is on average established by three-way junctions, best represented the experimental FRAP curve with HeLa lysate and energy, whereas pN of 0.54 resembled HeLa lysate plus apyrase. Likewise, simulations of FLIP mirror experimental data measured upon energy depletion (Fig. S5 B).
Next, we simulated INM targeting upon reduction of ER connectivity. Intriguingly, the kinetic curves exhibited a retardation toward lower pN (Fig. 8 G). The same was experimentally observed for INM targeting of LBR upon energy depletion (Fig. 8 H). This reduction in targeting speed at lower pN might be explained by distant parts of the network having fewer direct connections toward the NE, and molecules having to take more “detours” before eventually passing NPCs.
Collectively, our simple model is sufficient to explain targeting of different INM reporters as well as the apparent energy dependence of INM transport. Notably, we did not need to invoke any other contribution than diffusion and retention, with NPC translocation presenting a rate-limiting step.
An artificial reporter (AR) recapitulating diffusion-retention–based INM targeting
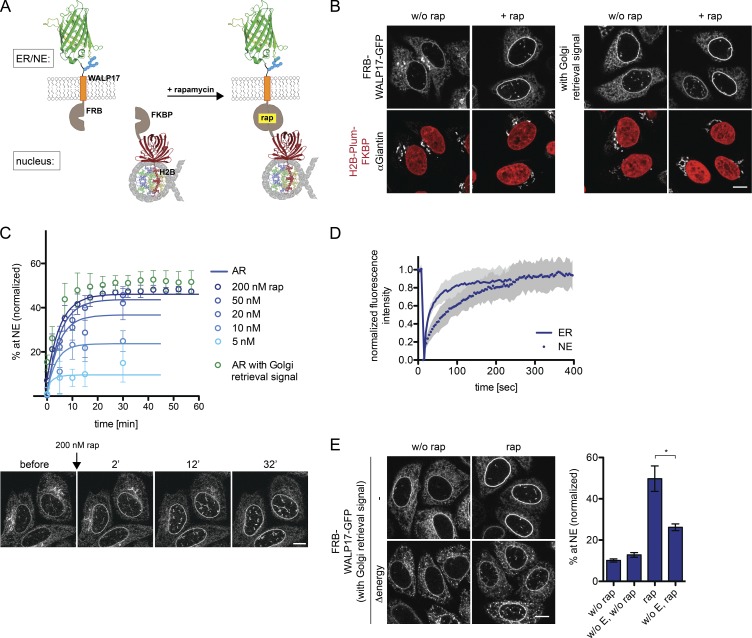
If a diffusion-retention–based mechanism were sufficient for INM targeting, one should be able to create an artificial TM protein that lacks sorting signals present in natural INM proteins, but contains a binding site for a nuclear retention partner. Similar to previously a established reporter-retention system (Ohba et al., 2004), we exploited the high-affinity interaction between FKBP and FRB mediated by rapamycin (Chen et al., 1995).
Our AR is composed of an HA tag, followed by the FRB domain, an artificial TM domain (WALP17; de Planque and Killian, 2003), and GFP (Fig. 9 A). The retention partner FKBP was fused to histone H2B and Plum for visualization. In the absence of rapamycin, the AR equally distributed between ER, NE, and also the Golgi as it lacked a Golgi retrieval signal (Fig. 9 B; Turgay et al., 2010). Insertion of such a signal shifted its localization to the ER. Strikingly, when rapamycin was added to cells, both reporters accumulated at the INM with similar kinetics (Fig. 9, B and C; and Fig. S1). The rapamycin-induced accumulation of the ARs at the INM provides evidence that a retention-driven INM targeting mechanism can operate in the absence of specific sorting signals.
Figure 9.
An AR recapitulates diffusion-retention–based targeting from the ER to the INM in vivo. (A) Schematic representation of AR and retention partner. (B) HeLa cells expressing the AR (with or without the Golgi retrieval signal) and its retention partner were analyzed after incubation with 200 nM rapamycin for 30 min. The Golgi was stained with Giantin antibodies. (C) Kinetics of AR targeting to the NE measured by time-lapse microscopy with decreasing rapamycin concentrations. Mean ± SD (error bars); n ≥ 66. Solid lines are predictions from the kinetic model. (D) FRAP on AR in the ER (without rap) or at the NE, starting 30 min after rapamycin addition. Mean ± SD; n = 23 for ER, n = 14 for NE. (E) INM targeting of the AR is energy-dependent in vivo. Cells expressing the AR were incubated for 30 min in the presence of deoxyglucose and NaN3, before exposure to 200 nM rapamycin for 10 min. Cells were fixed and translocation efficiency was analyzed. Mean ± SEM (error bars); n ≥ 149; *, P ≤ 0.05 (unpaired t test). Bars, 10 µm.
In a retention-dependent trafficking pathway, the binding affinity and number of retention sites determine steady-state accumulation at the destination site. We therefore analyzed how reduction of retention sites would influence INM targeting of the AR by reducing the rapamycin concentration. At 200 and 50 nM, the AR accumulated at similar levels at the INM (Fig. 9 C). However, upon reduction to 20, 10, or 5 nM, the targeting level decreased gradually from ∼37% to 9%, whereas the targeting rates remained similar. When targeting kinetics were predicted by our kinetic model, both under nonlimiting and limiting rapamycin concentrations, targeting dynamics of the AR fitted the measured data well (Fig. 9 C, solid lines). FRAP measurements revealed a high binding off-rate of 0.4 min−1 of the AR at the NE, which is 17-fold faster than for LBR, consistent with the low steady-state accumulation of ∼46% (Fig. 9 D). These data demonstrate the importance of retention in determining the levels of INM proteins at the NE. Importantly, when energy was depleted from cells before the addition of rapamycin, accumulation at the INM was reduced from 46% to 25% (Fig. 9 E). This corroborates our conclusion that energy is not used to power a specific, receptor-mediated translocation pathway.
Discussion
In this study, we established an in vitro assay to investigate requirements for the journey of INM proteins from the ER through NPCs to their nuclear destination. Using LBR, SUN2, and LAP2β as model substrates, we provide evidence that targeting of these membrane-embedded proteins to the INM relies on both a highly interconnected ER network and an efficient retention/immobilization step at the INM. Our observations support a diffusion-retention–based mechanism of INM targeting. Kinetic analysis combined with mathematical modeling identifies NPC translocation as a rate-limiting step. NPC passage of integral membrane proteins strongly responds to the size of the ND of INM proteins, and is restricted by the central NPC scaffold.
Diffusion-retention or active transport?
The membrane system of the ER is continuous with ONM and INM. To explain accumulation of membrane proteins from the ER at the INM against a concentration gradient, one needs to invoke either an energy-requiring step or binding to nuclear partners that remove molecules from the freely diffusive fraction. Similar to previous reports studying INM transport in vivo (Ohba et al., 2004; Zuleger et al., 2011), we observed a decrease in targeting efficiency upon NTP depletion in vitro. However, energy depletion also reduced the diffusional mobility of integral membrane and soluble ER proteins. Thus, the energy requirement of INM transport is not necessarily a contradiction to a diffusion-retention based mechanism.
Using mathematical simulation based on a 2D network model of spatial ER organization, we were able to recapitulate INM targeting kinetics measured in the presence or absence of energy. The agreement between simulated and measured data indicates that decreased ER connectivity in the absence of energy is in principle sufficient to explain the observed targeting defects of our reporters. The energy dependence of an artificial, solely retention-based reporter operating without natural sorting signals further confirmed our interpretation. Of note, our kinetic model recapitulates targeting kinetics of LBR without assuming active transport, i.e., with equal NPC translocation rates toward and from the INM. Nevertheless, we cannot fully dismiss the option that energy-sensitive ER structural changes may mask an additional energy-consuming step.
Both SUN2 and LBR contain signals that confer interaction with importins. However, based on our data, we can exclude a direct contribution of importin-mediated NPC translocation to targeting of SUN2 and LBR because WGA, RanQ69L, and Impβ(45–462) did not affect their import. Notably, many INM proteins contain NLS-like sequences (Lusk et al., 2007). If not used for NPC translocation, these could be of functional importance as nuclear retention motifs, e.g., as part of DNA-binding domains (LaCasse and Lefebvre, 1995; Cokol et al., 2000). In addition, NLSs could be exploited to control reversible chromatin dissociation of INM proteins in cells undergoing open mitosis (Ulbert et al., 2006; Ma et al., 2007). The previously observed Ran dependence of LBR targeting in vivo (Zuleger et al., 2011) could be reconciled with the need for receptor-mediated import of its nuclear binding partners that include histone H3/H4 and HP1 (Olins et al., 2010).
In contrast to the LBR reporter, reconstitution of SUN2 targeting to the INM required the presence of HeLa lysate. The extract (and energy) dependence of SUN2 could not be linked to any of the previously identified targeting signals of SUN2. Rather, the mobile fraction of SUN2 in the ER was strongly reduced without extract (unpublished data). Therefore, we assume that the stimulating activity of the extract might involve factors like chaperones that promote release of SUN2 from transport-inhibitory interactions. It is also possible that mobilization of SUN2 for transport involves binding of a piggyback partner present in the extract, similar to nuclear import of yeast Mps3 along with H2A.Z (Gardner et al., 2011).
Collectively, all our data obtained on SUN2, LBR, LAP2β, and the AR support a diffusion-retention model, and identify maintenance of ER structure as an energy-dependent process required for efficient INM targeting. For generalization of the model, it will, however, be important to analyze additional INM proteins and to identify the nature of the stimulating activity for SUN2 in the cell extract.
Where through the NPC?
The diffusion-retention model posits that targeting of integral membrane proteins from the ONM to the INM occurs by lateral diffusion along the pore membrane. Several studies described lateral openings in the NPC scaffold near the pore membrane. These putative peripheral channels have a diameter of ∼9 nm at the narrowest position (Beck et al., 2007; Maimon et al., 2012), in line with the 60-kD size restriction for the NDs of mammalian INM proteins. Consistently, we confirmed that enlargement of NDs abrogates INM targeting, independently of the presence of NLSs.
Analysis of the translocation capacity after knockdowns of individual Nups revealed that pore passage of INM proteins is confined by the Nup53–93 subcomplex. These Nups constitute the central NPC spoke ring and may form the gateways for INM proteins in proximity to the pore membrane. Which Nups of the Nup53–93 subcomplex exactly encompass these peripheral openings is difficult to deduce from RNAi experiments because most treatments affect the entire subcomplex. A previous nuclear reconstitution study had ascribed a key role to Nup188, a component of the central scaffold (Theerthagiri et al., 2010), in agreement with our data. Interestingly, depletion of NDC1, which provides an anchor point for the Nup53–93 complex at the pore membrane (Mansfeld et al., 2006), accelerated INM targeting, especially for multispanning membrane proteins. We therefore propose that NDC1 presents a steric impediment for diffusion in the pore membrane. Thus, the NPC might form a bottleneck for membrane proteins with respect to both their extralumenal and membrane-embedded domains.
The discovery of receptor-mediated transport of the integral INM proteins Heh1 and Heh2 in yeast shaped an alternative view on NPC translocation. Heh2 contains a strong bipartite NLS and a long unfolded linker between the NLS and the first TM segment (King et al., 2006; Meinema et al., 2011). The linker was proposed to endow Heh proteins with the flexibility to penetrate the NPC scaffold, allowing the NLS bound to Kap60/95 to pass through the central channel of the NPC. In line with NPC passage through the central channel, Heh2-derived tail-anchored reporters are not restricted in size. This raises the question of whether the passageways that we delineate are static channels or transient openings, eventually allowing access to the central channel. Generation of transient gateways would require rearrangements of the NPC that could be either actively induced or reflect intrinsic flexibility. Knockdown of several Nups such as Nup133, Nup96/98, and ELYS as well as Nup205, Nup155, and Nup93 decelerated targeting of LBR. This could hint at an involvement of these Nups in NPC rearrangements. However, considering that NPC passage is rate-limiting, it is more likely that the reduced NPC numbers accompanying these depletions caused retardation of targeting. Because LBR(1–245), SUN2(1–260), and LAP2β reached the INM in the absence of energy, we exclude a strict necessity for energy-dependent NPC rearrangements in human cells.
ER structure and transport to the INM
The ER is a continuous and dynamic membrane network composed of different subdomains shaped into tubules, cisternae, and the NE (Friedman and Voeltz, 2011). In simulations relying on a simplified ER network model, we were able to recapitulate a reduction of diffusional exchange in networks with fewer connections, mimicking the changes that we observed upon energy depletion. The reduction of diffusional exchange within the ER also retarded targeting of membrane proteins to the INM, providing an explanation for the energy dependence of the process.
Notably, our model includes an “inaccessible” fraction; i.e., molecules that are disconnected from the network. Even though we consider it unlikely that parts of the ER are detached at any time, our simulations demonstrate an important principle that is ultimately relevant for the ER network: the larger the distance that molecules have to cover, the stronger a reduction in connectivity impinges on the apparent accessibility. In the time window that we consider, a fraction of proteins can be regarded as temporarily inaccessible. Our work emphasizes the importance of sufficient connections in a network for movement by simple diffusion.
Several ER-resident proteins help to shape the ER by stabilizing membrane curvature or by establishing new connections between neighboring ER tubules and/or sheets. One energy-dependent step important for the maintenance of ER morphology is membrane fusion mediated by GTPases of the atlastin family (Rismanchi et al., 2008; Orso et al., 2009; Liu et al., 2012). A simple explanation for the loss of junctions in the ER network upon NTP depletion might be the loss of activity of atlastin GTPases.
The changes in ER morphology upon energy depletion might indicate that cells constantly need to invest energy to maintain an elaborate ER network. Analogous to the cytoskeleton, this constant investment of energy could confer flexibility for rapid regulation of molecular trafficking or establishment of contact sites to other organelles. Our work on INM protein targeting is exemplary for the far-reaching consequences that an aberrant ER structure might have for cellular homeostasis.
Materials and methods
Molecular cloning
The LBR and LAP2β coding regions were amplified from HeLa cDNA. INM targeting reporter constructs were cloned into pcDNA5 FRT/TO (cytomegalovirus [CMV] promoter; Invitrogen) for generation of stable, tetracycline-inducible cell lines. The respective ORFs encode fusion proteins starting with two monomeric RFPs separated by a linker (SRAQAS), followed by a peptide with a TEV cleavage site (SNSPTTENLYFQ’GRS), the INM protein of choice, and a C-terminal EGFP. After TEV cleavage, the amino acids GRS form the new start of the fusion protein followed by the natural N terminus of the INM protein. Reporter proteins were composed of full-length SUN2 (Turgay et al., 2010), LAP2β, and LBR. Truncated reporters SUN2(1–260) and LBR(1–245), both comprising one TM domain, were generated in the same backbone. In addition, mutant forms of SUN2 (Fig. S3) were derived from constructs described previously (Turgay et al., 2010). The HA-tagged full-length LBR reporter was generated by replacing the C-terminal GFP with the HA tag.
For expression of GST-GFP fusion proteins in HeLa cells, the NDs of SUN2 (aa 1–158), LBR (1–208), and LAP2β (1–410) were subcloned into pK7-GST-GFP (Erkmann et al., 2005), a mammalian expression vector based on pRK7 containing a CMV promoter. A codon usage–optimized sequence of TEV protease for expression in mammalian cells was inserted into pCFPC1 (CMV promoter; modified from Clontech Laboratories; Takara Bio Inc.). EGFP was inserted between an ER signal peptide and KDEL into pCMV/myc/ER (Invitrogen). The coding sequence for human Sec61β was inserted into pEGFPN3 (Takara Bio Inc.). The coding sequence for 3×CFP-IBB, composed of three CFPs and the importin β–binding domain of importin α (RCH1, aa 1–50), was cloned into pQE30 (T5 promoter; QIAGEN) for expression with an N-terminal His6 tag.
For generation of the AR system, the retention partner H2B-Plum-FKBP (FK506 binding protein) was cloned into pIRESpuro2 (CMV promoter; Takara Bio Inc.). The AR contains an N-terminal HA tag, followed by a TEV cleavage site and an FKBP-rapamycin binding domain (FRB; Ho et al., 1996). This extralumenal domain is followed by an artificial TM domain (WALP17; Killian et al., 1996), an N-glycosylation site, and a lumenal GFP. The peptide SRRSRPTT containing the Golgi retrieval signal was inserted between the HA tag and the TEV cleavage site.
The AR was introduced into pEGFPN3 (CMV promoter; Takara Bio Inc.) and subcloned into pcDNA5 FRT/TO for the generation of stable tetracycline-inducible cell lines.
Protein expression and purification
His6-tagged TEV protease fused to NusA, cloned into pET28a (T7 promoter; EMD Millipore), was provided by W. Antonin (Friedrich Miescher Laboratory, Tübingen, Germany), and the protease was purified as described previously (Theerthagiri et al., 2010). For in vitro release of reporters, NusA-TEV was buffer exchanged to permeabilization buffer (PB; 20 mM Hepes, pH 7.5, 110 mM potassium acetate, 5 mM magnesium acetate, 0.5 mM EGTA, and 250 mM sucrose) by dialysis. His6-3×CFP-IBB and Impβ(45–462)-His6 (Kutay et al., 1997) were expressed in E. coli BLR(pREP4) at 25°C by induction with 1 mM IPTG. Cells were lysed by sonication in 20 mM Tris, pH 7.5, 200 mM NaCl, 3 mM MgCl2, and 5% wt/vol glycerol. The lysate was cleared by ultracentrifugation, passed over Ni-NTA Agarose (QIAGEN), and eluted with 400 mM imidazole in lysis buffer. Peak fractions were buffer exchanged to PB. The expression and purification of RanQ69L (Izaurralde et al., 1997) has been described previously.
Antibodies
Rabbit antibodies against Impα2 (RCH1) and Impβ were gifts from D. Görlich (Max-Planck-Institut für Biophysikalische Chemie, Göttingen, Germany), and rabbit antibodies against the N-terminal domain of LBR were provided by R.W. Wozniak (University of Alberta, Edmonton, Alberta, Canada). Commercial antibodies were mAB414 (mouse; BabCo), anti-Nup153 (ab24700, mouse; Abcam), anti-actin (mouse; Sigma-Aldrich), anti-HA (MMS-101P, mouse; Covance), and anti-Giantin (ab24586, rabbit; Abcam). Peptide-specific rabbit antibodies to Nup98 (peptide: CNRDSENLASPSEYPENGER), Nup96 (CSLHHPPDRTSDSTPDPQRV), Nup205 (TPSLSETVNRDGPRQDTQAC), and NDC1 (CQASAEHQKRLQQFLEFKE) have been described previously (Mansfeld et al., 2006; Laurell et al., 2011).
Cell culture, cell lines, transient transfections, and in vivo energy depletion
HeLa cells were maintained in DMEM containing 10% fetal bovine serum and penicillin/streptomycin at 37°C, 5% CO2. Transient transfections were performed using the X-tremeGene transfection reagent (Roche). Reporter constructs were stably integrated into the FRT site of a HeLa cell line containing a tetracycline repressor. Expression of the reporter proteins 2×RFPtevSUN2-GFP, 2×RFPtevSUN2(1–260)-GFP, 2×RFPtevLBR(fl)-GFP, and 2×RFPtevGFP-LBR(1–245)-GFP was induced for 16 h by the addition of 200 ng/ml tetracycline. Expression of 2×RFPtevLBR(1–245)-GFP and 2×RFPtevLAP2β-GFP was induced for 16 h by 20 ng/ml. AR proteins were induced for 8 h with 200 ng/ml tetracycline in HeLa cell lines constitutively expressing H2B-Plum-FKBP. The HeLa cell line (Mitchell et al., 2010) stably expressing rat POM121-3×GFP (Rabut et al., 2004) from the pEGFP backbone (CMV promoter; BD) has been described previously.
For depletion of cellular ATP (and GTP; Schwoebel et al., 2002), cells were treated with 6 mM deoxyglucose (Sigma-Aldrich) and 10 mM NaN3 in glucose-free DMEM containing 0.5% dialyzed FCS. Analysis started 15 min after addition of the energy depletion medium.
RNA interference and immunofluorescence
siRNA transfection was performed using INTERFERin (Polyplus Transfection) at a final siRNA concentration of 10 nM for Nup205 (5′-CTGCGTCAGTTTAAATTTCAAdTdT-3′), Nup93 (5′-AGAGUGAAGUGGCGGACAAdTdT-3′), Nup107 (5′-GAAAGUGUAUUCGCAGUUAdTdT-3′), Nup96/98 (5′-UCUGAAACAAAGACUCAUUdTdT-3′), Nup133 (5′-AACTATAGCCCAGGAAGATAAdTdT-3′), ELYS (5′-AAUAUCUACAUAAUUGCUCUUdTdT-3′), Rae1 (5′-GGGAUACUCGAUCGUCAAAdTdT-3′), and Nup214 (5′-GUCACGGAAACAGUGAAAGdTdT-3′) for 48 h, and for Nup155 (5′-ACAUGCAGGUGUUAGGUUAdTdT-3′) for 72 h. For knockdown of Nup53 (5′-GGAAGUACUCCUAGGAUUUdTdT-3′), NDC1 (5′-CACUGUUCCUGGUUAGAAAdTdT-3′), Nup188 (5′-TACGCTGGGAATACTCCTATAdTdT-3′), and Nup54 (5′-CAGACAGAAGTTGTTATTTATdTdT-3′), RNAi was performed for twice for 48 h with 20 nM of siRNA. AllStars (QIAGEN) was used as a negative control (−ctr.).
For immunofluorescence analysis, cells were fixed in 1% or 4% PFA and washed with PBS. Permeabilization was done for 5 min in 0.1% Triton X-100 at RT. Immunostaining was performed as described previously (Zemp et al., 2009). For preservation of soluble 2×RFP (Fig. 1 C), cells were fixed in 2% PFA, 0.25% glutaraldehyde in PBS for 10 min. Residual unreacted glutaraldehyde was quenched for 10 min in 1 mg/ml NaBH4.
In vitro nuclear import and INM targeting assay
In vitro targeting reactions were performed essentially as described previously (Adam et al., 1990; Görlich et al., 1994). Cells were semipermeabilized with PB containing 0.001% digitonin for 10 min at 4°C, followed by three washing steps in PB buffer without digitonin for 2, 5, and 10 min. Release of the reporter was induced by adding 75 µg/ml NusA-TEV in PB at RT for 10 min. 30 µl import mixture containing a transport-competent cytoplasmic HeLa cell extract and an energy regenerating system (energy mix [1×]: 10 mM creatine phosphate, 0.5 mM GTP, 0.5 mM ATP, 5 mM Hepes, pH 7.5, 0.05 mg/ml creatine kinase, and 12.5 mM sucrose) were added and import/translocation was allowed to proceed at 37°C.
Cytoplasmic extract was prepared by hypotonic lysis (5 mM Tris, pH 7.5, 1 mM magnesium chloride, 0.5 mM EGTA, and 0.5 mM EDTA) of HeLa cells, followed by re-extraction with 220 mM potassium acetate. For depletion of importins, 200 µl of HeLa cell lysate was incubated for 1 h with 30 µl phenyl-Sepharose (low substitution; GE Healthcare) at 4°C. After a second round of depletion, unbound material was used for transport reactions. For NTP depletion, 50 units of apyrase (New England Biolabs, Inc.) were used per reaction. At defined times, coverslips were washed once with PB, cells were fixed with 4% PFA, washed in PBS, and mounted. Confocal microscopy was performed with a microscope (TCS-SP2/AOBS; Leica) using an HCX Plan-Apochromat Lbd Bl 63×, NA 1.4, oil immersion objective lens.
For kinetic measurement of INM targeting, Lab-Tek chambers with semipermeabilized cells were mounted at a microscope (LSM 710-FCS [Carl Zeiss]; 63× 1.4 NA oil DIC Plan-Apochromat immersion objective lens) at 37°C, and supplemented with a HeLa lysate and an energy regenerating system, unless otherwise mentioned. Release from the ER trap, i.e., cleavage of the two RFP moieties by TEV protease, was started after a prerelease image was acquired (0 min time point). Time-lapse imaging was performed at 37°C in 5-min intervals, acquiring z stacks.
FRAP and FLIP
FRAP was performed with a microscope (LSM 710-FCS; Carl Zeiss) using a 63× 1.4 NA oil DIC Plan-Apochromat immersion lens and image recording in 4× zoom at 37°C. Reporter cells were seeded into 8-well Lab-Tek chambers. For in vitro measurements, cells were semipermeabilized and subjected to analysis for the subsequent 45 min. FRAP was recorded on the lumenal EGFP of the reporters. As lateral diffusion of membrane proteins only weakly correlates with molecular size, i.e., an increase from 100 to 500 kD reduces lateral diffusion only 1.2 fold (Sprague and McNally, 2005), mobility in the ER was examined on the uncut, 2×RFP-tagged reporters. Three prebleach images were acquired followed by bleaching of a rectangle of 3.3 × 0.66 µm (2.2 µm2) at full 488-nm laser power and 250 scanning iterations (7.5 s). The bleach area in the ER was enlarged for more reliable detection of ER structural changes to 2 × 10.4 µm (20.8 µm2) for FRAP measurements in Figs. 7, 8, and 9. Fluorescence recovery was measured every 4 s for measurements in the ER and every 10–30 s for FRAP at the NE. Regions of interest of the same size as the bleached region were recorded for correction of bleaching during acquisition and background correction. Data analysis was done using easyFRAP in MATLAB (Rapsomaniki et al., 2012). FRAP data were normalized and fitted by a double exponential. As binding to retention partners retards FRAP recovery compared with a freely diffusing species, a FRAP curve can be subdivided into a first, diffusion-limited recovery phase reflecting unbound proteins, and a second, binding-limited recovery phase determined by the binding off-rate (Sprague and McNally, 2005). Diffusion coefficients were calculated according to Kang et al. (2012) from the first exponent, whereas the binding off rate was derived from the second exponent.
FLIP experiments were performed on transiently transfected ER-targeted EGFP-KDEL on a microscope (LSM 710-FCS; Carl Zeiss) using a 63× 1.4 NA oil DIC Plan-Apochromat immersion objective lens, and images were recorded in 2.5× zoom at 37°C. Two prebleach images were acquired followed by alternating bleaching of a spot (r = 3.16 µm, 50 iterations, 4.5 s) and imaging of the target cell. Fluorescence loss was measured in a ring of 3.16 µm that surrounds the bleached circle (donut) and in a distant region on the opposite side of the nucleus. Bleaching due to acquisition and background were corrected by accounting for measurements of a region in a neighboring cell and the background area, respectively.
Image processing and quantification
Accumulation at the NE was quantified in the mid-section focal plane, as this adequately reflected measurements in stacks over entire cells. For image quantification of endpoint assays, an ImageJ plugin was generated. Basically, first the nuclear contour was detected using DAPI-stained nuclei. From this detection border, the NE was defined as a ring of 230 nm toward the ER and 920 nm toward the nuclear center. The outer border of the ER was defined by thresholding. The integrated fluorescence intensity at the NE was divided by the total fluorescence intensity (in the ER and the NE). Data were normalized relative to the fraction at the NE before release by TEV cleavage (0.205, mean of 670 cells).
Quantification of time-lapse images was done using a MATLAB-based quantification tool. In the selected image set, the measurement background was estimated by calculating the mode (most often appearing background intensity). To achieve an accurate NE contour detection, a Laplacian of Gaussians (log) filter–based edge detector was applied. Contours of smaller or larger than normal nuclear size were removed. Nuclear contours were followed over time based on their spatial location. The NE was defined as an ∼800-nm wide ring and the underlying reporter intensities were collected. Cell borders were delimited by an orthogonal border in the middle of the connecting line between the barycenter of the nuclei. The fraction at the NE was calculated and normalized as described in the preceding paragraph.
Modeling and simulation
We assumed that molecules can be part of four different populations: (1) the inaccessible ER fraction, (2) the accessible ER fraction, (3) the ONM fraction, and (4) the INM fraction. The compartments have the membrane surface areas Ai (i = 1, . . ., 4). Using 3D reconstitution of HeLa cell nuclei, we measured an average ONM and INM surface area of ∼1,050 µm2. The area of the elaborate ER network was more difficult to determine. We estimated 10,300 ± 2,700 µm2 by integrating measurements of the ER volume in semipermeabilized HeLa cells and published average ER surface densities of hepatocytes (Herzfeld et al., 1973).
We denoted the state of molecules in the compartments with j = 0 for the immobile fraction and j = 1 for the mobile fraction. The number of molecules in each compartment, is proportional to the observed cumulative intensity in the compartment. The mobile fraction in each compartment is µi = Ni,0/Ni. Furthermore, we denoted the transport rate constants from compartment i to compartment i′ with the mobilization/dissociation rate constant in compartment i with kim and the corresponding immobilization/binding rate constant with kib. Assuming that transport only applies to the mobile fractions, this results in the following system of ordinary differential equations (ODEs):
to describe INM targeting dynamics (see Fig. 8 A). The observable NE fraction of molecules is:
where λ 1 denotes the bleeding/crosstalk from the ER into the NE due to imperfect NE detection and the limited optical resolution. Further, it was assumed that molecules were evenly distributed in the ER and ONM at the start. The dynamics are obtained by solving the ODE system using MATLAB’s built-in ode15s solver.
The system has eight free parameters: four determining the steady-state and four the kinetics. The steady-state is given by (1) the ratio of the ER to the NE, (2) the ER residual fraction, fit to 7% for all reporters, and (3 and 4) the equilibrium mobile fractions µ2 = µ3 and µ4. The targeting kinetics are further specified by (5 and 6) transport rate constants k23 and k34 (i.e., ER to ONM and ONM to INM), (7) the mobilization rate constants k2m = k3m (we assume equal properties for the ER and the ONM), and (8) the mobilization rate and k4m at the INM. The mobilization rate constant k2m only slightly influences targeting kinetics, as only a minor faction of molecules are immobile in the ER (maximum of 12% for SUN2(fl)). ER and ONM are assumed to be highly connected, resulting in a fast exchange of molecules by diffusion. We fitted k23 to 11.79 min−1 and k32 to 112.59 min−1 for LBR(1–245)-GFP (note that these rates are asymmetric because we are using the total number of molecules in the compartments, and ER and ONM have different areas: k23/k32 = A3/A2). All other kinetic rates or the corresponding equilibrium constants from the fit are given in Table 1. The compartment areas Ai and measured parameters (e.g., from FRAP experiments) were used as initial values and refined together with the unknown parameters in the fitting process using least-square fitting.
Transport between the compartments was assumed to be driven by diffusion. Accordingly, the transport rate constants are determined by the diffusion coefficient Di (we assumed the same D for all compartments) and a structural parameter , which is only determined by the connectivity of the compartments. We assume (i.e., transport from the ER to the ONM is ∼50% slower for Lap2β and full-length SUN2 than for LBR(1–245) and SUN2(1–260) based on the apparent diffusion coefficients derived from FRAP measurements on the ER). Including the diffusion dependency explicitly in the model enables us to fit the structural parameter jointly for all reporters despite their different mobility. Transport through the NPC furthermore depends on the number Np of nuclear pores in the NE. We assumed an average NPC density of 5.2 NPCs/µm2 (Maeshima et al., 2010) and linear mass action kinetics of κ34 ∼ Np. The number of LBR(1–245)-GFP molecules per cell was roughly estimated to 107 by quantitative immunoblotting in comparison to recombinant GFP.
For predictions of INM targeting kinetics of the AR under limiting rapamycin concentrations, we used a Kd for in vitro binding of FRB to FKBP-rap of 12 nM as estimated by previous studies (Banaszynski et al., 2005; Cabantous et al., 2013).
In our spatial ER model, the ER network is modelled as a 2D square lattice. Connections between neighboring points are established randomly with probability pN. At pN = 1, each point of the network is connected to its four neighboring points, whereas at pN = 0, the points are disconnected. The regions of interest are marked on the network and the connectivity between the regions was evaluated using the Network Analysis methods from the MATLAB Bioinformatics Toolbox. The dynamics of FLIP, FRAP, and targeting experiments were computed based on the network structure solving the corresponding PDE with MATLAB. Due to the random nature of the approach, we computed mean and standard deviation for each quantity from 50 realizations of the ER structure.
Online supplemental material
Fig. S1 demonstrates that INM targeting reporters reach the INM. Fig. S2 shows kinetic analysis of SUN2 targeting in vivo. Fig. S3 shows energy and extract dependence of different SUN2 mutants. Fig. S4 demonstrates that nocodazole or cycloheximide do not inhibit INM targeting in vitro. Fig. S5 shows simulation and measurements of FLIP on ER-resident EGFP-KDEL. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201409127/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201409127.dv.
Supplementary Material
Acknowledgments
We thank W. Antonin, D. Görlich, and R. Wozniak for providing reagents, C. Balazs and S. Niederhauser for help with image analysis, C. Larabell for helpful discussion, K. Weis, M. Mayr, and S. Pawar for critical comments on the manuscript, C. Ashiono for excellent assistance, and members of ScopeM for continuous support.
This work was funded by an European Research Council grant (NucEnv) to UK.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AR
- artificial reporter
- CMV
- cytomegalovirus
- FLIP
- fluorescence loss in photobleaching
- HA
- hemagglutinin
- INM
- inner nuclear membrane
- LBR
- lamin B receptor
- ND
- nucleoplasmic domain
- NE
- nuclear envelope
- NPC
- nuclear pore complex
- Nups
- nucleoporins
- ODE
- ordinary differential equation
- ONM
- outer nuclear membrane
- TEV
- tobacco etch virus
- TM
- transmembrane
- WGA
- wheat germ agglutinin
References
- Adam S.A., Marr R.S., and Gerace L.. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111:807–816. 10.1083/jcb.111.3.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski L.A., Liu C.W., and Wandless T.J.. 2005. Characterization of the FKBP.rapamycin.FRB ternary complex. J. Am. Chem. Soc. 127:4715–4721. 10.1021/ja043277y [DOI] [PubMed] [Google Scholar]
- Beck M., Lucić V., Förster F., Baumeister W., and Medalia O.. 2007. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 449:611–615. 10.1038/nature06170 [DOI] [PubMed] [Google Scholar]
- Burke B., and Stewart C.L.. 2013. The nuclear lamins: flexibility in function. Nat. Rev. Mol. Cell Biol. 14:13–24. 10.1038/nrm3488 [DOI] [PubMed] [Google Scholar]
- Burke B., and Stewart C.L.. 2014. Functional architecture of the cell’s nucleus in development, aging, and disease. Curr. Top. Dev. Biol. 109:1–52. [DOI] [PubMed] [Google Scholar]
- Cabantous S., Nguyen H.B., Pedelacq J.D., Koraïchi F., Chaudhary A., Ganguly K., Lockard M.A., Favre G., Terwilliger T.C., and Waldo G.S.. 2013. A new protein-protein interaction sensor based on tripartite split-GFP association. Sci. Rep. 3:2854 10.1038/srep02854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zheng X.F., Brown E.J., and Schreiber S.L.. 1995. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc. Natl. Acad. Sci. USA. 92:4947–4951. 10.1073/pnas.92.11.4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokol M., Nair R., and Rost B.. 2000. Finding nuclear localization signals. EMBO Rep. 1:411–415. 10.1093/embo-reports/kvd092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Planque M.R., and Killian J.A.. 2003. Protein-lipid interactions studied with designed transmembrane peptides: role of hydrophobic matching and interfacial anchoring. Mol. Membr. Biol. 20:271–284. 10.1080/09687680310001605352 [DOI] [PubMed] [Google Scholar]
- Ellenberg J., Siggia E.D., Moreira J.E., Smith C.L., Presley J.F., Worman H.J., and Lippincott-Schwartz J.. 1997. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138:1193–1206. 10.1083/jcb.138.6.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkmann J.A., Wagner E.J., Dong J., Zhang Y., Kutay U., and Marzluff W.F.. 2005. Nuclear import of the stem-loop binding protein and localization during the cell cycle. Mol. Biol. Cell. 16:2960–2971. 10.1091/mbc.E04-11-1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay D.R., Newmeyer D.D., Price T.M., and Forbes D.J.. 1987. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J. Cell Biol. 104:189–200. 10.1083/jcb.104.2.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., and Voeltz G.K.. 2011. The ER in 3D: a multifunctional dynamic membrane network. Trends Cell Biol. 21:709–717. 10.1016/j.tcb.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J.M., Smoyer C.J., Stensrud E.S., Alexander R., Gogol M., Wiegraebe W., and Jaspersen S.L.. 2011. Targeting of the SUN protein Mps3 to the inner nuclear membrane by the histone variant H2A.Z. J. Cell Biol. 193:489–507. 10.1083/jcb.201011017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Prehn S., Laskey R.A., and Hartmann E.. 1994. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 79:767–778. 10.1016/0092-8674(94)90067-1 [DOI] [PubMed] [Google Scholar]
- Grossman E., Medalia O., and Zwerger M.. 2012. Functional architecture of the nuclear pore complex. Annu. Rev. Biophys. 41:557–584. 10.1146/annurev-biophys-050511-102328 [DOI] [PubMed] [Google Scholar]
- Güttinger S., Laurell E., and Kutay U.. 2009. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 10:178–191. 10.1038/nrm2641 [DOI] [PubMed] [Google Scholar]
- Herzfeld A., Federman M., and Greengard O.. 1973. Subcellular morphometric and biochemical analysis of developing rat hepatocytes. J. Cell Biol. 57:475–483. 10.1083/jcb.57.2.475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S.N., Biggar S.R., Spencer D.M., Schreiber S.L., and Crabtree G.R.. 1996. Dimeric ligands define a role for transcriptional activation domains in reinitiation. Nature. 382:822–826. 10.1038/382822a0 [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Kutay U., von Kobbe C., Mattaj I.W., and Görlich D.. 1997. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 16:6535–6547. 10.1093/emboj/16.21.6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M., Day C.A., Kenworthy A.K., and DiBenedetto E.. 2012. Simplified equation to extract diffusion coefficients from confocal FRAP data. Traffic. 13:1589–1600. 10.1111/tra.12008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katta S.S., Smoyer C.J., and Jaspersen S.L.. 2014. Destination: inner nuclear membrane. Trends Cell Biol. 24:221–229. 10.1016/j.tcb.2013.10.006 [DOI] [PubMed] [Google Scholar]
- Killian J.A., Salemink I., de Planque M.R., Lindblom G., Koeppe R.E. II, and Greathouse D.V.. 1996. Induction of nonbilayer structures in diacylphosphatidylcholine model membranes by transmembrane α-helical peptides: importance of hydrophobic mismatch and proposed role of tryptophans. Biochemistry. 35:1037–1045. 10.1021/bi9519258 [DOI] [PubMed] [Google Scholar]
- King M.C., Lusk C.P., and Blobel G.. 2006. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 442:1003–1007. 10.1038/nature05075 [DOI] [PubMed] [Google Scholar]
- Klebe C., Bischoff F.R., Ponstingl H., and Wittinghofer A.. 1995. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry. 34:639–647. 10.1021/bi00002a031 [DOI] [PubMed] [Google Scholar]
- Kutay U., Izaurralde E., Bischoff F.R., Mattaj I.W., and Görlich D.. 1997. Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO J. 16:1153–1163. 10.1093/emboj/16.6.1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCasse E.C., and Lefebvre Y.A.. 1995. Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res. 23:1647–1656. 10.1093/nar/23.10.1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurell E., Beck K., Krupina K., Theerthagiri G., Bodenmiller B., Horvath P., Aebersold R., Antonin W., and Kutay U.. 2011. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell. 144:539–550. 10.1016/j.cell.2011.01.012 [DOI] [PubMed] [Google Scholar]
- Liu T.Y., Bian X., Sun S., Hu X., Klemm R.W., Prinz W.A., Rapoport T.A., and Hu J.. 2012. Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion. Proc. Natl. Acad. Sci. USA. 109:E2146–E2154. 10.1073/pnas.1208385109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk C.P., Blobel G., and King M.C.. 2007. Highway to the inner nuclear membrane: rules for the road. Nat. Rev. Mol. Cell Biol. 8:414–420. 10.1038/nrm2165 [DOI] [PubMed] [Google Scholar]
- Ma Y., Cai S., Lv Q., Jiang Q., Zhang Q., Sodmergen Z., Zhai Z., and Zhang C.. 2007. Lamin B receptor plays a role in stimulating nuclear envelope production and targeting membrane vesicles to chromatin during nuclear envelope assembly through direct interaction with importin β. J. Cell Sci. 120:520–530. 10.1242/jcs.03355 [DOI] [PubMed] [Google Scholar]
- Maeshima K., Iino H., Hihara S., Funakoshi T., Watanabe A., Nishimura M., Nakatomi R., Yahata K., Imamoto F., Hashikawa T., et al. 2010. Nuclear pore formation but not nuclear growth is governed by cyclin-dependent kinases (Cdks) during interphase. Nat. Struct. Mol. Biol. 17:1065–1071. 10.1038/nsmb.1878 [DOI] [PubMed] [Google Scholar]
- Maimon T., Elad N., Dahan I., and Medalia O.. 2012. The human nuclear pore complex as revealed by cryo-electron tomography. Structure. 20:998–1006. 10.1016/j.str.2012.03.025 [DOI] [PubMed] [Google Scholar]
- Mansfeld J., Güttinger S., Hawryluk-Gara L.A., Panté N., Mall M., Galy V., Haselmann U., Mühlhäusser P., Wozniak R.W., Mattaj I.W., et al. 2006. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol. Cell. 22:93–103. 10.1016/j.molcel.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Meinema A.C., Laba J.K., Hapsari R.A., Otten R., Mulder F.A., Kralt A., van den Bogaart G., Lusk C.P., Poolman B., and Veenhoff L.M.. 2011. Long unfolded linkers facilitate membrane protein import through the nuclear pore complex. Science. 333:90–93. 10.1126/science.1205741 [DOI] [PubMed] [Google Scholar]
- Mitchell J.M., Mansfeld J., Capitanio J., Kutay U., and Wozniak R.W.. 2010. Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane. J. Cell Biol. 191:505–521. 10.1083/jcb.201007098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T., Schirmer E.C., Nishimoto T., and Gerace L.. 2004. Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J. Cell Biol. 167:1051–1062. 10.1083/jcb.200409149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins A.L., Rhodes G., Welch D.B., Zwerger M., and Olins D.E.. 2010. Lamin B receptor: multi-tasking at the nuclear envelope. Nucleus. 1:53–70. 10.4161/nucl.1.1.10515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori A., Banterle N., Iskar M., Andrés-Pons A., Escher C., Khanh Bui H., Sparks L., Solis-Mezarino V., Rinner O., Bork P., et al. 2013. Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol. Syst. Biol. 9:648 10.1038/msb.2013.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orso G., Pendin D., Liu S., Tosetto J., Moss T.J., Faust J.E., Micaroni M., Egorova A., Martinuzzi A., McNew J.A., and Daga A.. 2009. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 460:978–983. 10.1038/nature08280 [DOI] [PubMed] [Google Scholar]
- Östlund C., Sullivan T., Stewart C.L., and Worman H.J.. 2006. Dependence of diffusional mobility of integral inner nuclear membrane proteins on A-type lamins. Biochemistry. 45:1374–1382. 10.1021/bi052156n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell L., and Burke B.. 1990. Internuclear exchange of an inner nuclear membrane protein (p55) in heterokaryons: in vivo evidence for the interaction of p55 with the nuclear lamina. J. Cell Biol. 111:2225–2234. 10.1083/jcb.111.6.2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G., Doye V., and Ellenberg J.. 2004. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 6:1114–1121. 10.1038/ncb1184 [DOI] [PubMed] [Google Scholar]
- Rapsomaniki M.A., Kotsantis P., Symeonidou I.E., Giakoumakis N.N., Taraviras S., and Lygerou Z.. 2012. easyFRAP: an interactive, easy-to-use tool for qualitative and quantitative analysis of FRAP data. Bioinformatics. 28:1800–1801. 10.1093/bioinformatics/bts241 [DOI] [PubMed] [Google Scholar]
- Ribbeck K., and Görlich D.. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20:1320–1330. 10.1093/emboj/20.6.1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K., and Görlich D.. 2002. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 21:2664–2671. 10.1093/emboj/21.11.2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rismanchi N., Soderblom C., Stadler J., Zhu P.P., and Blackstone C.. 2008. Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum. Mol. Genet. 17:1591–1604. 10.1093/hmg/ddn046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothballer A., and Kutay U.. 2012. SnapShot: The nuclear envelope I. Cell. 150:868–868: e1. 10.1016/j.cell.2012.07.024 [DOI] [PubMed] [Google Scholar]
- Rothballer A., Schwartz T.U., and Kutay U.. 2013. LINCing complex functions at the nuclear envelope: what the molecular architecture of the LINC complex can reveal about its function. Nucleus. 4:29–36. 10.4161/nucl.23387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S., Shao Y., Braunagel S.C., Summers M.D., and Johnson A.E.. 2004. Cotranslational integration and initial sorting at the endoplasmic reticulum translocon of proteins destined for the inner nuclear membrane. Proc. Natl. Acad. Sci. USA. 101:12537–12542. 10.1073/pnas.0404934101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S., Summers M.D., Burks J.K., Johnson A.E., and Braunagel S.C.. 2006. Importin-α-16 is a translocon-associated protein involved in sorting membrane proteins to the nuclear envelope. Nat. Struct. Mol. Biol. 13:500–508. 10.1038/nsmb1098 [DOI] [PubMed] [Google Scholar]
- Schwoebel E.D., Ho T.H., and Moore M.S.. 2002. The mechanism of inhibition of Ran-dependent nuclear transport by cellular ATP depletion. J. Cell Biol. 157:963–974. 10.1083/jcb.200111077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S., and Blobel G.. 1993. The first membrane spanning region of the lamin B receptor is sufficient for sorting to the inner nuclear membrane. J. Cell Biol. 120:631–637. 10.1083/jcb.120.3.631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa B.A., Rothballer A., Kutay U., and Schwartz T.U.. 2012. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 149:1035–1047. 10.1016/j.cell.2012.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soullam B., and Worman H.J.. 1993. The amino-terminal domain of the lamin B receptor is a nuclear envelope targeting signal. J. Cell Biol. 120:1093–1100. 10.1083/jcb.120.5.1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soullam B., and Worman H.J.. 1995. Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J. Cell Biol. 130:15–27. 10.1083/jcb.130.1.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague B.L., and McNally J.G.. 2005. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 15:84–91. 10.1016/j.tcb.2004.12.001 [DOI] [PubMed] [Google Scholar]
- Stavru F., Hülsmann B.B., Spang A., Hartmann E., Cordes V.C., and Görlich D.. 2006. NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J. Cell Biol. 173:509–519. 10.1083/jcb.200601001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapley E.C., Ly N., and Starr D.A.. 2011. Multiple mechanisms actively target the SUN protein UNC-84 to the inner nuclear membrane. Mol. Biol. Cell. 22:1739–1752. 10.1091/mbc.E10-08-0733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry L.J., and Wente S.R.. 2009. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot. Cell. 8:1814–1827. 10.1128/EC.00225-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theerthagiri G., Eisenhardt N., Schwarz H., and Antonin W.. 2010. The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J. Cell Biol. 189:1129–1142. 10.1083/jcb.200912045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay Y., Ungricht R., Rothballer A., Kiss A., Csucs G., Horvath P., and Kutay U.. 2010. A classical NLS and the SUN domain contribute to the targeting of SUN2 to the inner nuclear membrane. EMBO J. 29:2262–2275. 10.1038/emboj.2010.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbert S., Platani M., Boue S., and Mattaj I.W.. 2006. Direct membrane protein-DNA interactions required early in nuclear envelope assembly. J. Cell Biol. 173:469–476. 10.1083/jcb.200512078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp I., Wild T., O’Donohue M.F., Wandrey F., Widmann B., Gleizes P.E., and Kutay U.. 2009. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J. Cell Biol. 185:1167–1180. 10.1083/jcb.200904048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuleger N., Kelly D.A., Richardson A.C., Kerr A.R., Goldberg M.W., Goryachev A.B., and Schirmer E.C.. 2011. System analysis shows distinct mechanisms and common principles of nuclear envelope protein dynamics. J. Cell Biol. 193:109–123. 10.1083/jcb.201009068 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.