Quantitative analysis relating cell, nuclear, and chromosome size in C. elegans embryos predicts two levels of chromosome length regulation in response to both cell and nuclear size.
Abstract
Multicellular development requires that cells reduce in size as a result of consecutive cell divisions without increase in embryo volume. To maintain cellular integrity, organelle size adapts to cell size throughout development. During mitosis, the longest chromosome arm must be shorter than half of the mitotic spindle for proper chromosome segregation. Using high-resolution time-lapse microscopy of living Caenorhabditis elegans embryos, we have quantified the relation between cell size and chromosome length. In control embryos, chromosome length scaled to cell size. Artificial reduction of cell size resulted in a shortening of chromosome length, following a trend predicted by measurements from control embryos. Disturbing the RAN (Ras-related nuclear protein)-GTP gradient decoupled nuclear size from cell size and resulted in chromosome scaling to nuclear size rather than cell size; smaller nuclei contained shorter chromosomes independent of cell size. In sum, quantitative analysis relating cell, nuclear, and chromosome size predicts two levels of chromosome length regulation: one through cell size and a second in response to nuclear size.
Introduction
During metazoan development, embryonic cells decrease in size by up to two orders of magnitude (from 1.2 mm to 12 µm in Xenopus laevis) as a consequence of multiple rounds of cell division without growth of the embryo. Using Xenopus or Caenorhabditis elegans embryos as model organisms, it has been shown that mitotic structures, including mitotic spindle length, centrosome size, and nuclear size, all scale with cell size (Wühr et al., 2008; Greenan et al., 2010; Levy and Heald, 2010, 2012; Loughlin et al., 2011).
The study of mitotic chromosome scaling has received less attention even though the phenomenon was first reported over 100 yr ago (Conklin, 1912; Schubert and Oud, 1997; Kieserman and Heald, 2011; Neurohr et al., 2011; Hara et al., 2013). The size of an organism’s genome remains constant in all diploid cells in spite of dramatic and rapid changes in cell size during embryonic development. The maximum length of condensed mitotic chromosomes cannot exceed half of the spindle length (Schubert and Oud, 1997). Thus, mitotic chromosomes scale in size in response to decreasing cell size.
Studies in Xenopus have shown that chromosomes reduce in size as the embryo progresses through development. Nuclei isolated from small cells of older embryos incubated in mitotic-arrested egg extract (derived from large cells) condensed their chromosomes to lengths predicted for smaller cells, meaning that the cytoplasm did not dictate chromosome size. Furthermore, allowing small cell–derived nuclei to expand to levels found in large cells (in large cell extract) resulted again in short chromosomes. However, when G2 nuclei were subjected to an entire cell cycle in large cell egg extract, the resulting size of mitotic chromosomes matched those of larger cells. Therefore, it was concluded the nuclear volume had no apparent effect on chromosome size scaling (Kieserman and Heald, 2011). Chromosome length measured in fixed, flattened C. elegans embryos indicated that artificial reduction in nuclear size (through disruption of nuclear import/export trafficking) reduced chromosome size (Hara et al., 2013). In addition, interphase Xenopus nuclei incubated in egg extract and prevented from expanding by inhibiting nuclear import resulted in smaller chromosomes. In sum, studies on chromosome length scaling have not reached a consensus.
To investigate the seemingly disparate results found in C. elegans and Xenopus, we developed a quantitative live cell assay of C. elegans embryos that allowed us to evaluate cell, nuclear, and chromosome size. The C. elegans embryo has a relatively low diploid number (12) and homogeneous chromosome size (varying by <50% in genomic length), providing an excellent system to study chromosome size scaling in vivo. We have correlated cell, nuclear, and chromosome size in measurements derived from intact embryos, allowing statistical analysis of this process. RNAi-based depletion to independently alter cell or nuclei size to decouple developmental program from normal scaling showed that chromosome size scales to cell size as well as nuclear size. We found that depletion of proteins required for establishing the RAN (Ras-related nuclear protein)-GTP gradient decoupled cell size from chromosome size; however, the relation between nuclear size and chromosome length was to some extent maintained. In sum, our results show that chromosome size scales via a predictable, nuclear trafficking–based mechanism in early development.
Results and discussion
Chromosome length regulation during the first four divisions of C. elegans embryos
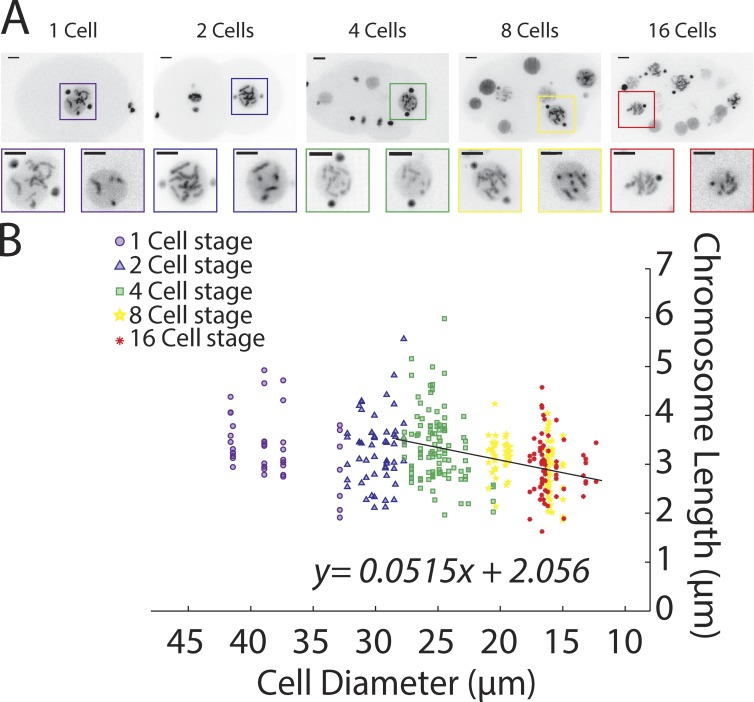
Previous work has not comprehensively analyzed chromosome length scaling relative to cell and nuclear size. To accomplish this goal, we used high-resolution time-lapse microscopy to image mitotic divisions in C. elegans 1- to 16-cell stage embryos expressing H2B-GFP and γ-tubulin–GFP (strain TH32, see Materials and methods). We measured embryo diameter, nuclear size, and chromosome lengths in 3D-rendered images (Fig. 1 A and see Materials and methods for details) in which we could resolve at least two chromosomes clearly. Chromosomes in cells after the 16-cell stage were difficult to resolve, precluding accurate analysis at this time. Our previous analysis showed that chromosome condensation in C. elegans completes to >90% ∼30 s before nuclear envelope breakdown (NEBD); therefore, we measured chromosome length at that time (Maddox et al., 2006). Altogether, this analysis confirmed that condensed chromosomes are shorter in the smaller cells of more developed embryos (Fig. 1 B).
Figure 1.
Chromosome length scales to cell size during early C. elegans embryogenesis. (A) Representative still images of time-lapse movies from embryos expressing H2B::GFP and γ-tubulin::GFP (TH32) at the 1- to 16-cell stage. Insets are an enlargement of the nucleus in prometaphase. Bars, 5 µm. (B) Prometaphase chromosome length measurements at different developmental stages (marks) and in correlation to cell size (up to 28 µm in diameter, linear regression) in control embryos. n = 354 chromosomes.
Our analysis revealed that chromosome size differences between 1- and 4-cell stages were not significant, whereas changes later were (control Bonferroni’s multiple comparisons test [Table S2], following the one-way ANOVA [Table S1]). We interpret the lack of significant difference early in development as indication of a plateau or upper limit to chromosome size (discussed in more detail in "A chromosome length-scaling model" below). A similar concept has been applied to infer upper limits of both spindle and chromosome length during Xenopus early embryonic development (Greenan et al., 2010; Kieserman and Heald, 2011).
Starting after the plateau effect (i.e., cells smaller than 30 µm, ∼4-cell stage and older in controls), a linear regression of chromosome length over cell size (y = ax + b) allowed defining correlation parameters. Specifically, the slope “a” of the linear regression represents the reduction in chromosome length for each 1-µm reduction in cell size. The intercept “b” represents the chromosome length at a theoretical cell size of 0 µm and thus represents the theoretical minimal chromosome length. Applying this rule revealed that, in early C. elegans embryos, chromosome length is reduced predictably by 51.5 ± 10 nm per 1-µm reduction in cell size. At a theoretical cell size of 0 µm, chromosome length would be 2.06 ± 0.22 µm (Table S3). We will use those parameters to determine how different perturbations affect chromosome length regulation.
In early embryos chromosomes shorten in response to decreasing cell size
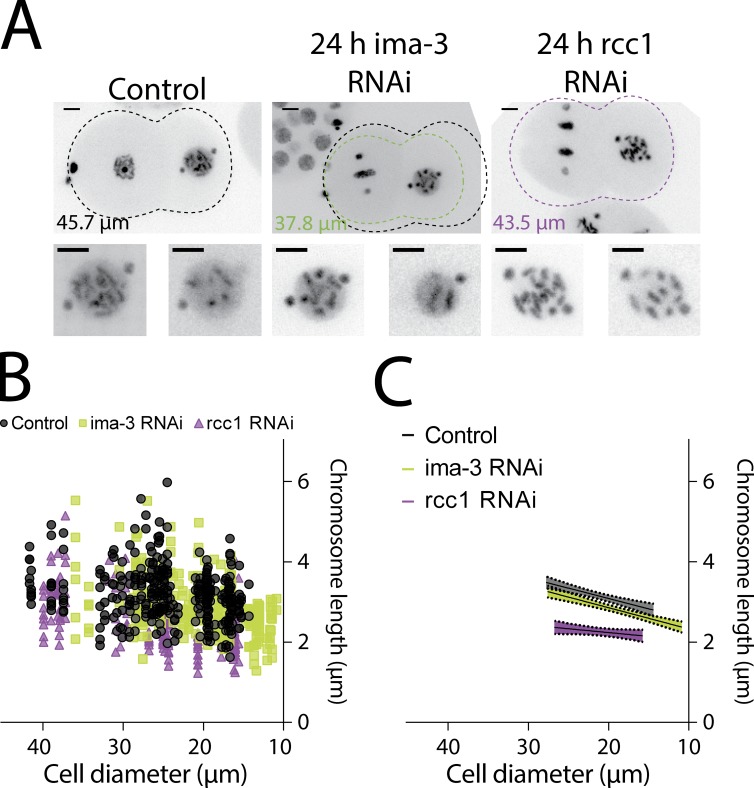
To determine whether we can artificially generate chromosome scaling anomalies as noted in the previous section, we sought to decouple cell size and developmental stage by changing the overall size of the embryo. We successfully reduced embryo size by depleting an Importin-α protein, IMA-3 (Fig. 2 A and Fig. S1 B; Askjaer et al., 2002). Partial depletion (24 h) of IMA-3 resulted in smaller embryos while maintaining chromosome condensation, whereas longer depletion (48 h) resulted in pleiotropic effects including loss of chromosome condensation (Fig. 2 A and Fig. S1 A). The developmental program of the IMA-3 partially depleted embryos was not altered as indicated by the proper nuclear localization of a PIE-1::GFP protein (a germline transcription factor) and proper cell polarity at the 4-cell stage (Fig. S1 C; Reese et al., 2000).
Figure 2.
Chromosome length scales to cell size, not developmental program. (A) Representative still images of time-lapse movies from TH32 after control, IMA-3, and RCC1 depletion at the 2-cell stage. Shown is the outline of the embryo and size of the depicted embryo: black, control; green, IMA-3 RNAi; and purple, RCC1 RNAi. The control outline is overlaid on ima-3 to illustrate differences in size. Bars, 5 µm. (B) Chromosome length correlated to cell size in control and IMA-3– and RCC1-depleted embryos. (C) Graph representing the linear regression of cell size versus chromosome length. The shadows represent the 95% confidence interval. In B, control measurements are duplicated from Fig. 1 to compare the other conditions. IMA-3 RNAi, n = 287 chromosomes; RCC1 RNAi, n = 207 chromosomes.
When comparing chromosome length and cell size for each developmental stage, our analysis confirmed an early plateau in smaller IMA-3–depleted embryos (Table S2). The plateau in IMA-3–depleted embryos spanned only the 1–2-cell embryo stage instead of the 1–4-cell stage seen in controls. As predicted from a cell size–based regulatory mechanism, the rate of chromosome length scaling in relation to cell size after depletion of IMA-3 was not statistically different than in control embryos (53 ± 7 nm per 1-µm reduction of cell size; Table S3 and Fig. 2, B and C). The minimal chromosome size (y intercept), a measure which is cell size and not developmental stage based, was also not statistically different from controls (Table S3). In sum, these results are in agreement with a cell size–based mechanism regulating chromosome length and not a developmental switch hypothesis.
Cell size and nuclear size independently regulate chromosome length scaling
We next examined the possibility that chromosome size scales through nuclear size. It was previously shown in Xenopus embryos that nuclei scaled to cell size through limited nuclear import of structural components (Levy and Heald, 2010). Specifically, increasing the available cytoplasmic fraction of importin-α correlated with increased nuclear size. To test the impact of nuclear import on nuclear size in C. elegans, we correlated cell to nuclear size in the IMA-3–depleted embryos. We found that in addition to having smaller than normal cells, the nuclei were proportionally smaller (Fig. S1 D and Table S3). Therefore, as we observed for chromosome size regulation, nuclei were appropriately sized for the corresponding cell size in IMA-3–depleted embryos. These data raise the hypothesis that chromosome size scales to nuclear size, which in turn scales to cell size.
We depleted other known regulators of nuclear import and looked for a specific effect on nuclear rather than cell size. We found that partial depletion of the RAN–guanine nucleotide exchange factor (GEF; activator) RCC1 (regulator of chromosome condensation 1; RAN-3 in C. elegans, hereafter termed RCC1) fit these criteria (Fig. 2 A). In RCC1-depleted embryos, the nuclear size was smaller than predicted based on both control and IMA-3–depleted embryos (Fig. S1 D and Table S3).
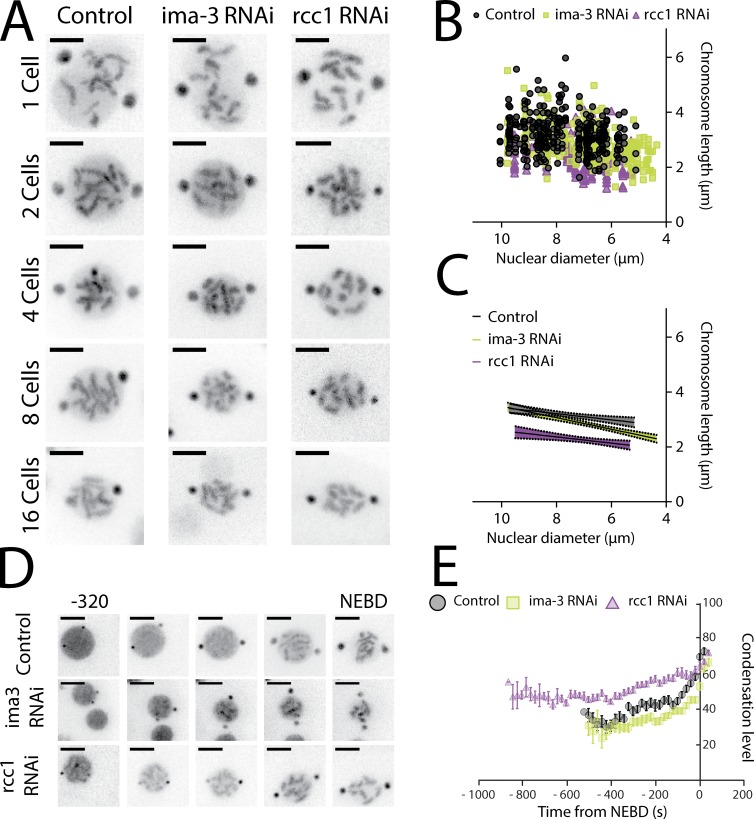
Visual inspection revealed that chromosome length was dramatically reduced after RCC1 depletion (Figs. 2 A and 3 A), an observation confirmed by 3D measurement. Linear regression analysis of chromosome length to cell size in RCC1-depleted embryos revealed a distinct correlation with cell size compared with controls (Fig. 2, B and C). Specifically, chromosome length reduced in relation to cell size with a lower magnitude (19 ± 12 nm/µm) in RCC1-depleted embryos compared with controls (52 ± 10 nm/µm; Table S3). Interestingly, the minimal predicted chromosome size at a cell size of 0 µm was indistinguishable from controls.
Figure 3.
When uncoupling nuclear to cell size, chromosome length scales to nuclear size. (A) Representative still images from time-lapse movies of nuclei from TH32 embryos after RNAi depletion of control, IMA-3, and RCC1 at the 1- to 16-cell stage. (B) Chromosome length correlated to nuclear size in control, IMA-3, and RCC1 RNAi. (C) Graph representing the linear regression of nuclear size versus chromosome length. The shadows represent the 95% confidence interval. (D) Representative still images from time-lapse movies of TH32 at the 8-cell stage from −320 s to NEBD. (A and D) Bars, 5 µm. (E) Kinetic plot of the condensation parameter. Error bars are standard error of the mean. (For more details on the assay, see Materials and methods and refer to Maddox et al., 2006.) Control, n = 39; IMA-3 RNAi, n = 45; and RCC1 RNAi, n = 29. Images are 40-s intervals.
To test the hypothesis that nuclear and chromosome size could be coregulated, we correlated chromosome length to nuclear size rather than cell size in control and RCC1- and IMA-3–depleted embryos (Fig. 3, B and C). In RCC1-depleted embryos, the constant value of chromosome length reduction in relation to nuclear size was indistinguishable from the control condition (130 ± 33 nm in control vs. 120 ± 36 nm in RCC1 RNAi; Table S3). In contrast, in IMA-3–depleted embryos, the constant of chromosome length reduction to nuclear size was significantly different from the control and RCC1-depleted embryos (211 ± 26 nm/µm; Fig. 3, B and C; and Table S3). Surprisingly, at a theoretical nuclear size of 0 µm, chromosome length in RCC1-depleted embryos is predicted to be smaller than control (Table S3). Altogether our results suggest that depletion of RCC1 enhances compaction of each individual chromosome by a fixed magnitude of ∼746 nm (Δ y-intercept between control and RCC1 RNAi). Furthermore, the y-axis intercept is very similar in IMA-3– and RCC1-depleted embryos; thus, removal of either protein reduced chromosome length to the same amount over that limited developmental timescale (Table S3). In sum, our results indicate that IMA-3 is required for proper chromosome to nuclear size but not nuclear to cell size scaling, whereas RCC1 contributes to nuclear to cell size scaling, with the final outcome of RCC1 depletion being chromosomes that are inappropriately short for a given cell size.
RCC1 RNAi embryos exhibit a global increased compaction before mitosis
A hypothesis for decreased chromosome length in RCC1-depleted embryos could be differential condensation dynamics in prophase. To test this hypothesis, we used a live-cell fluorescence-based condensation assay to determine the temporal dynamics of chromatin compaction (Maddox et al., 2006). Our condensation assay revealed that the overall dynamics (shape of the curves) of chromatin compaction did not change in relation to cell size in control embryos, as noted previously (Fig. S3; Hara et al., 2013). However, RCC1-depleted embryos showed distinct chromosome compaction dynamics from those observed in IMA-3 or control depletions (Fig. 3, D and E). In fact, chromosomes in RCC1-depleted embryos are more compacted by this measure at the start of prophase compared with control or IMA-3 RNAi embryos. These results suggest that either prophase (and thus chromosome compaction) takes longer or that chromatin is additionally compacted when entering mitosis in RCC1-depleted embryos. By measuring centrosome growth (using the γ-tubulin–GFP), we confirmed that the time spent in prophase/prometaphase (from start of centrosome growth to NEBD) was not different in any condition (Table S4). Thus, we conclude that chromatin in RCC1-depleted embryos is generally more compacted before mitosis.
Next, we sought to determine whether this additional state of compaction is introduced globally onto the chromatin or more locally at specific areas of the genome. To answer this question, we made use of the inherent chromosome length variance; in any individual cell, all chromosomes will not be the same length. We compared the variance at each developmental stage in control and RCC1 RNAi (Table S5). When we analyzed RCC1 RNAi embryos, we observed that the variance remained similar to control at all developmental stages (except at the 4-cell stage), indicating global rather than local length regulation.
The RAN-GTP gradient contributes to nuclear and chromosome size
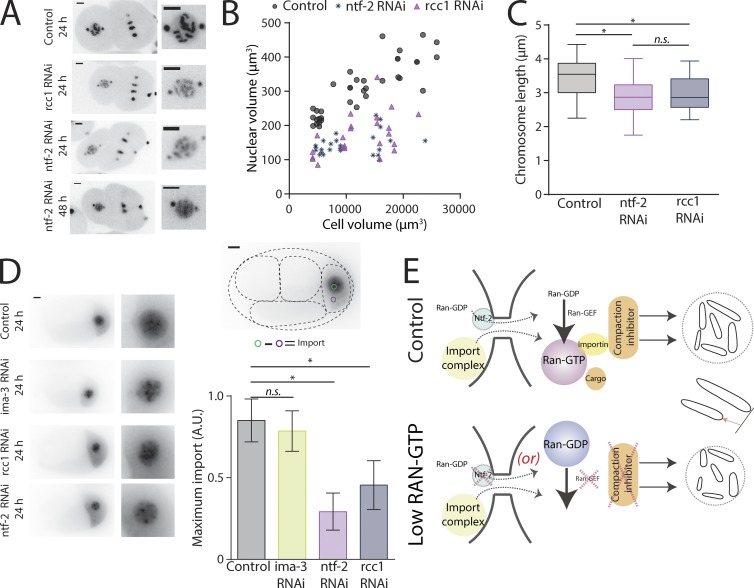
Our experimental design of reducing nuclear and chromosome size through depletion of RCC1 cannot distinguish between a direct effect of RCC1 on chromatin (as it is a known chromatin-binding protein [Makde et al., 2010]) or an effect through disruption of the Ran-GTP gradient across the nuclear membrane. To discriminate between these two possibilities, we depleted other known proteins required to maintain the Ran-GTP gradient. Ntf-2 (nuclear transport factor-2; RAN-4 in C. elegans) is an import factor associated with the nuclear pore that binds Ran-GDP and promotes its nuclear import (Ribbeck et al., 1998), and it should reduce the nuclear pool of RAN-GDP available to be exchange to RAN-GTP by RCC1. After NTF2 depletion, nuclear size was reduced in a manner indistinguishable from RCC1 depletion (Fig. 4, A and B; and Table S3). Partial depletion of NTF2 increases chromosome segregation defects; therefore, we focused on the 2-cell stage, an early time point with fewer accumulated defects and resolvable individual chromosomes. At the 2-cell stage, chromosome length in NTF2-depleted embryos was not statistically different from chromosome length in RCC1-depleted embryos and smaller than controls (Fig. 4 C). Hence, we found that partial depletion of NTF2 phenocopied RCC1 depletion, suggesting that the RAN-GTP gradient is critical to maintain the cell to nuclear size and the cell to chromosome size scaling.
Figure 4.
Ran pathway members regulate chromosome and nuclear size through limited nuclear import. (A) Representative still images from time-lapse movies of nuclei from TH32 embryos after RNAi depletion of control, RCC1, and NTF2 at the 2-cell stage. (B) Nuclear volume correlated to cell volume (in µm3) in control, RCC1, and NTF2 RNAi embryos. See Table S3 for linear regression information. Control, n = 37; RCC1 RNAi, n = 23; and NTF2 RNAi, n = 30. (C) Boxplot of chromosome length at the 2-cell stage. The box represents the 25th and 75th percentiles of the distribution. The line in the middle of the box is the median. The whiskers are the smallest and largest values of the distribution. (D) Representative still images from time-lapse movies of PIE-1::GFP’s nuclear import in P2 cells and mean maximum nuclear import of Pie-1::GFP expressed as a ratio of control’s maximum import. Error bars are standard deviation. (A and D) Bars, 5 µm. (E) Chromosome length scaling model based on a differential level of Ran-GTP in small (low concentration) versus larger cells (high concentration). The low concentration of Ran-GTP schematic represents either (a) depletion of NTF-2, which prevents nuclear import of Ran-GDP, or (b) depletion of the Ran-GEF, which prevents exchange of GDP to GTP on Ran. n.s., not significant; *, P ≤ 0.05.
The depletion of RCC1, NTF2, and IMA-3 did not result in the same phenotype although they are part of the nuclear trafficking pathway. To evaluate the relative contribution of each protein to nuclear import, we measured retention of PIE-1::GFP in the nucleoplasm at the 4-cell stage (see Materials and methods for details). We found that RCC1- and NTF2-depleted embryos have a reduced accumulation of PIE-1::GFP compared with control and IMA-3 RNAi (Fig. 4 D). We did not see any significant differences in nuclear import of PIE-1::GFP after IMA-3 RNAi compared with control. Altogether, these results suggest that RCC1 and NTF-2 depletion reduces global protein import into the nucleus. We hypothesize that factors required to adjust nuclear and chromosome size to cell size fail to import in these conditions. In contrast, IMA-3 perhaps imports a specific factor required for chromosome to nuclear size scaling.
A chromosome length-scaling model
We have shown a connection between cell, nuclear, and chromosome size regulation in developing early embryos. By modifying embryo size, our results suggest that cell size and not developmental program regulates mitotic chromosome size scaling. Cell size–based control is saturated in very large cells present during the first divisions as chromosome length reaches a maximum and does not scale, a phenomenon we term “plateau.” These results lead us to hypothesize a theoretical inhibitor, X, of chromosome compaction in excess in the early embryo. This limiting component model predicts that the amount of X is defined by maternal load and restricts chromosome compaction early during development (Goehring and Hyman, 2012). Its dilution after each round of division results in more compacted chromosomes in smaller cells, independent of developmental stage. This theory is also supported by observations made in tetraploid embryos; although they are twice the volume of diploid embryos, chromosomes are smaller in the first two divisions (Fig. S2). If a developmental program were regulating the plateau, it should be observed irrespective of ploidy. We hypothesize that diploid animals have an excess of X in large cells and the amount of DNA is the limiting component. However, when doubling the amount of DNA, the inhibitor is no longer in excess but is rather the new limiting component, resulting in smaller chromosomes.
Nuclear transport has been proposed to be key for both nuclear to cell size scaling and chromosome to cell size scaling (Levy and Heald, 2010; Kieserman and Heald, 2011; Edens and Levy, 2014; Jevtić and Levy, 2015). In C. elegans, we confirmed that disruption of nuclear import/export resulted in smaller than expect nuclei and chromosomes. Also, as expected, this is influenced by the RAN-GTP nuclear to cytoplasm gradient. We hypothesize that reducing Ran-GTP correspondingly reduces import of inhibitor of compaction in smaller nuclei, decreasing chromosome length (Fig. 4 E). This is in contrast to a model where chromosome condensation enzymes accumulate in nuclei to scale chromosome size.
In conclusion, by using high-resolution time-lapse imaging of living, developing embryos, we determined that mitotic chromosomes scale to cell and nuclear size. Our analysis allowed us to propose two mechanisms regulating chromosome size scaling. The first mechanism regulates chromosome length proportionality to cell and nuclear size through adaptive regulation. The second mechanism reduces chromosome length with a fixed magnitude, perhaps through the incorporation of an unknown epigenetic mark on the chromatin in response to small nuclei size. It is unclear how the two mechanisms are regulated to form chromosomes of specific length during development. In the future, it would be of great interest to determine what are the molecular effectors of this change in chromosome compaction and the nature and dynamics of epigenetic marks introduced during development.
Materials and methods
Worm growth and RNAi experiments
Worm strains N2 (wild type), TH32 (pie-1::bg-1::GFP + unc-119(+), pie-1::GFP::H2B + unc-119(+)), JH2015 (pie-1p::GFP::pie-1 ORF::pie-1 3′utr + unc-119(+)), or SP346 (tetraploid 4A:4X provided by the Caenorhabditis Genetics Center) were grown and maintained at 20°C using standard procedures. Bacterial strains containing a vector expressing dsRNA under the IPTG promoter were obtained from the Ahringer library (from J.-C. Labbé’s laboratory, Institute for Research in Immunology and Cancer, University of Montreal, Montreal, Quebec, Canada). Targets were confirmed by sequencing. Time of protein depletion by bacterial feeding was performed for 24 (low depletion) or 48 h (higher depletion).
Live imaging
All live imaging was performed at RT on a Swept Field confocal mounted on an inverted TE2000 Eclipse microscope or an A1R confocal (Nikon; for chromosome measurement of TH32 expressing H2B::GFP) using the Nis-elements or an inverted DeltaVision microscope (GE Healthcare; for Pie-1::GFP import and DAPI-stained embryos) using SoftWoRx. Time-lapse acquisitions on the Swept Field were executed using a 60×/1.4 NA Plan-Apochromat oil immersion objective at 1.5 magnification and a CoolSNAP HQ2 camera (Photometrics), at 30-s interval and at 0.5-µm spacing. Image acquisition on the A1R confocal was executed with a 60×, GaASP PMT detector (Nikon) using Nis-elements. C. elegans embryos were mounted between a 1.5 coverslip and a microscope slide in M9 buffer on a 5% agarose pad (vs. 2% agarose) and were used to slightly compress the embryo and the chromosomes; this ensures that chromosomes are not completely perpendicular to the z axis, which elongates chromosomes as a result of the point-spread function of the GFP molecules. The coverslip was sealed using Valap (1:1:1 lanolin, petroleum jelly, and parafilm wax).
Image processing and measurements
All experiments were repeated at least three times. The number of cells analyzed is stated in the figure legends. All graphs represent all chromosomes measured in all repeated experiments. TH32 time-lapse videos were visualized in 3D/4D using Imaris (Bitplane). Chromosome lengths were measured in 3D during the prometaphase of C. elegans early embryo mitosis. Measurements were made by visually delimiting chromosomes and assigning 3 points at the extremities and in the middle of individual chromosome. We also delimited the height and the length of the embryo using the fluorescence background seen in the embryo to determine embryo size and cell size. Nuclei size was determined before NEBD by measuring nuclei diameter.
Analysis
Data were exported in MatLab or Prism (GraphPad Software) for analysis. Embryo diameter was obtained by modeling an elliptical form to the embryo. Because of the irregular shape of cells during development, we derived the mean cell diameter from embryo diameter (embryo diameter/cell number). Each individual chromosome was correlated to cell diameter or nuclei diameter using a linear regression of y = ax + b. Evaluating the p-value of the slope being different than 0 determined the presence of correlation between two variables. A difference in slope or intercept between two datasets was determined by nonoverlapping standard error of the slope or the intercept.
We performed Student’s t test to determine the p-value of the null hypothesis in between developmental stage of each condition (Table S2) when the ANOVA test found statistically significant differences (Table S1). The statistical significance of the difference in the variance of each developmental stage for control versus rcc1 RNAi was also tested using Fisher’s F-test.
Nuclear import of Pie-1::GFP in P2 cells was measured using Fiji. For each time point before NEBD, total fluorescent intensity (TFI) of a fixed-size area (after max intensity projection) was measured. For each time point, the TFI value was express as a ratio over control’s maximum intensity. Maximum import was obtained by averaging three time points at corresponding maximum import in control. For each condition, this value was plotted in a bar graph when the maximum import was reached in control embryos.
The condensation assay was performed as described previously (Maddox et al., 2006). Control or RCC1-depleted TH32 worm embryos were imaged on a Swept Field confocal. Using MetaMorph software (Molecular Devices), on the maximum intensity projection of each dividing cell, a square of a given size was placed in the middle of the dividing nuclei during the length of the movie (from start of condensation to NEBD). The fluorescence intensity distribution of the square at each time point is rescaled on a 0–255 gray value scale. The condensation parameter represents the number of pixels with fluorescence intensity above a threshold for each square (example, threshold of 35% of 255 gray values is ∼90 gray values). In designing the assay, multiple thresholds were tested and Fig. S3 shows the graphs for thresholds of 35 or 50%. A modification of the original assay is that for each nuclei size a different area (therefore more pixels in larger nuclei) was used to capture all of the nuclei values at each nuclei size. We found that this modification didn’t change the outcome of the assay.
Online supplemental material
Fig. S1 shows embryo size differences for IMA-1 to -3 RNAi and RCC1 RNAi, PIE-1::GFP localization at the 4-cell stage, and nuclear to cell diameter relation in control, IMA-3, and RCC1 RNAi embryos. Fig. S2 shows chromosome length regulation in diploid versus tetraploid embryos. Fig. S3 shows the graph for the condensation assay using different threshold. Table S1 shows one-way ANOVA comparing developmental stage for each RNAi condition or all RNAi conditions at each developmental stage. Table S2 shows Student’s t test statistical significance of chromosome length differences comparing all developmental stages for each RNAi condition. Table S3 shows correlation between cell size, nuclei size, and chromosome length in control and IMA-3–, RCC1-, and NTF-2–depleted embryos. Table S4 shows time spent in prophase in control and RCC1-depleted embryos. Table S5 shows statistical differences of chromosome length variance at all different stages between control and RCC1-depleted embryos. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201502092/DC1.
Supplementary Material
Acknowledgments
We thank all members the Maddox laboratories and Amy Maddox for helpful conversation. We would like to thank Kerry Bloom, Bob Goldstein, Jean-Claude Labbé, and E.D. Salmon for reading the manuscript. We are grateful to Jean-Claude Labbé for sharing reagents. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440).
P.S. Maddox is supported as a William Burwell Harrison Fellow of Biology. This work was supported in part by the Canadian Institute for Health Research. A.-M. Ladouceur is a predoctoral fellow of the Fonds de Recherche du Québec en Santé.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- NEBD
- nuclear envelope breakdown
- Ran
- Ras-related nuclear protein
References
- Askjaer P., Galy V., Hannak E., and Mattaj I.W.. 2002. Ran GTPase cycle and importins α and β are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos. Mol. Biol. Cell. 13:4355–4370. 10.1091/mbc.E02-06-0346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin E.G. 1912. Cell size and nuclear size. J. Exp. Zool. 12:1–98. 10.1002/jez.1400120102 [DOI] [Google Scholar]
- Edens L.J., and Levy D.L.. 2014. cPKC regulates interphase nuclear size during Xenopus development. J. Cell Biol. 206:473–483. 10.1083/jcb.201406004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring N.W., and Hyman A.A.. 2012. Organelle growth control through limiting pools of cytoplasmic components. Curr. Biol. 22:R330–R339. 10.1016/j.cub.2012.03.046 [DOI] [PubMed] [Google Scholar]
- Greenan G., Brangwynne C.P., Jaensch S., Gharakhani J., Jülicher F., and Hyman A.A.. 2010. Centrosome size sets mitotic spindle length in Caenorhabditis elegans embryos. Curr. Biol. 20:353–358. 10.1016/j.cub.2009.12.050 [DOI] [PubMed] [Google Scholar]
- Hara Y., Iwabuchi M., Ohsumi K., and Kimura A.. 2013. Intranuclear DNA density affects chromosome condensation in metazoans. Mol. Biol. Cell. 24:2442–2453. 10.1091/mbc.E13-01-0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevtić P., and Levy D.L.. 2015. Nuclear size scaling during Xenopus early development contributes to midblastula transition timing. Curr. Biol. 25:45–52. 10.1016/j.cub.2014.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieserman E.K., and Heald R.. 2011. Mitotic chromosome size scaling in Xenopus. Cell Cycle. 10:3863–3870. 10.4161/cc.10.22.17975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D.L., and Heald R.. 2010. Nuclear size is regulated by importin α and Ntf2 in Xenopus. Cell. 143:288–298. 10.1016/j.cell.2010.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D.L., and Heald R.. 2012. Mechanisms of intracellular scaling. Annu. Rev. Cell Dev. Biol. 28:113–135. 10.1146/annurev-cellbio-092910-154158 [DOI] [PubMed] [Google Scholar]
- Loughlin R., Wilbur J.D., McNally F.J., Nédélec F.J., and Heald R.. 2011. Katanin contributes to interspecies spindle length scaling in Xenopus. Cell. 147:1397–1407. 10.1016/j.cell.2011.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P.S., Portier N., Desai A., and Oegema K.. 2006. Molecular analysis of mitotic chromosome condensation using a quantitative time-resolved fluorescence microscopy assay. Proc. Natl. Acad. Sci. USA. 103:15097–15102. 10.1073/pnas.0606993103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makde R.D., England J.R., Yennawar H.P., and Tan S.. 2010. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 467:562–566. 10.1038/nature09321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurohr G., Naegeli A., Titos I., Theler D., Greber B., Díez J., Gabaldón T., Mendoza M., and Barral Y.. 2011. A midzone-based ruler adjusts chromosome compaction to anaphase spindle length. Science. 332:465–468. 10.1126/science.1201578 [DOI] [PubMed] [Google Scholar]
- Reese K.J., Dunn M.A., Waddle J.A., and Seydoux G.. 2000. Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol. Cell. 6:445–455. 10.1016/S1097-2765(00)00043-5 [DOI] [PubMed] [Google Scholar]
- Ribbeck K., Lipowsky G., Kent H.M., Stewart M., and Görlich D.. 1998. NTF2 mediates nuclear import of Ran. EMBO J. 17:6587–6598. 10.1093/emboj/17.22.6587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert I., and Oud J.L.. 1997. There is an upper limit of chromosome size for normal development of an organism. Cell. 88:515–520. 10.1016/S0092-8674(00)81891-7 [DOI] [PubMed] [Google Scholar]
- Wühr M., Chen Y., Dumont S., Groen A.C., Needleman D.J., Salic A., and Mitchison T.J.. 2008. Evidence for an upper limit to mitotic spindle length. Curr. Biol. 18:1256–1261. 10.1016/j.cub.2008.07.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.