Abstract
Early-life adversity increases the risk for psychopathology in later life. The underlying mechanism(s) is unknown, but epigenetic variation represents a plausible candidate. Early-life exposures can disrupt epigenetic programming in the brain, with lasting consequences for gene expression and behavior. This evidence is primarily derived from animal studies, with limited study in humans due to inaccessibility of the target brain tissue. In humans, although there is evidence for DNA methylation changes in the peripheral blood of psychiatric patients, a fundamental question remains as to whether epigenetic markers in the blood can predict epigenetic changes occurring in the brain. We used in utero bisphenol A (BPA) exposure as a model environmental exposure shown to disrupt neurodevelopment and exert long-term effects on behavior in animals and humans. We show that prenatal BPA induces lasting DNA methylation changes in the transcriptionally relevant region of the Bdnf gene in the hippocampus and blood of BALB/c mice and that these changes are consistent with BDNF changes in the cord blood of humans exposed to high maternal BPA levels in utero. Our data suggest that BDNF DNA methylation in the blood may be used as a predictor of brain BDNF DNA methylation and gene expression as well as behavioral vulnerability induced by early-life environmental exposure. Because BDNF expression and DNA methylation are altered in several psychiatric disorders that are associated with early-life adversity, including depression, schizophrenia, bipolar disorder, and autism, BDNF DNA methylation in the blood may represent a novel biomarker for the early detection of psychopathology.
Keywords: epigenetics, biomarker, BDNF, bisphenol A, early life
Early-life adversity is associated with increased risk for the development of many behavioral and psychiatric disorders, including depression, anxiety, and schizophrenia (1). The mechanisms through which early-life exposures contribute to later-life psychopathology are unknown, but epigenetic mechanisms represent a plausible candidate (2). Exposure to stress (3), toxicants (4), and maternal neglect (5) in early life can disrupt epigenetic programming in the brain, with lasting consequences for brain gene expression and behavior. This evidence is primarily derived from animal studies, with limited study in humans (6) due to the inaccessibility of the target brain tissue.
However, emerging evidence suggests that epigenetic biomarkers in peripheral tissues may be used to predict disease phenotypes in humans. Peripheral biomarkers may be of particular relevance for psychiatric disorders because, unlike the brain, tissues such as blood, saliva, or buccal cells are easily accessible in living patients. Several ubiquitous environmental exposures known to affect brain development (e.g., prenatal stress, famine, pollution/toxins) have been associated with epigenetic variation in human peripheral tissues (7–11). In addition, there is evidence for DNA methylation changes in the peripheral blood of psychiatric patients (12–14). However, because epigenetic marks are by definition tissue- and cell type-specific, a fundamental question remains as to whether epigenetic markers in the blood can predict epigenetic changes occurring in the brain and related phenotypic changes.
To address this question, we used in utero bisphenol A (BPA) exposure as a model environmental exposure. BPA is an endocrine disruptor and ubiquitous environmental toxicant (15, 16) that has been shown to disrupt neurodevelopment and exert long-term effects on behavior in animals and humans (17–19). Our previous study showed that environmentally relevant doses of BPA given to female mice during pregnancy induce lasting epigenetic disruption in the prefrontal cortex and hypothalamus of the offspring and that these changes are sex-specific (4). In the current study, we focused on a 200 µg/kg per day dose of BPA that was shown to induce long-term behavioral changes (4). We explored the epigenetic effects of BPA on the gene encoding brain-derived neurotrophic factor (BDNF), which has an essential role in neurodevelopment and has been linked to early-life adversity and psychiatric risk (20). In the animal model, we performed parallel molecular analyses in the hippocampus and blood as well as behavioral assessment of offspring prenatally exposed to BPA. To confirm the translational value of the findings, the animal study was complemented with the DNA methylation analysis of cord blood samples derived from a human cohort with characterized BPA levels during pregnancy.
Results
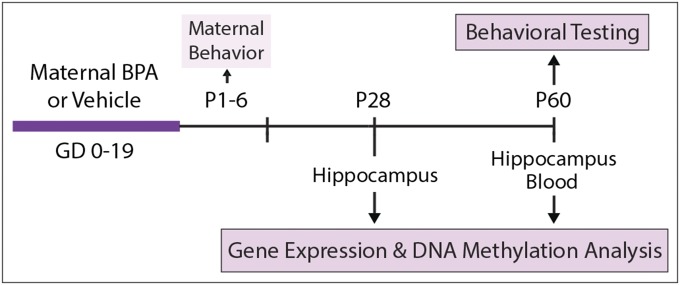
Design of the Animal Study (Fig. 1).
Fig. 1.
Design of the animal study.
Pregnant BALB/c mouse dams were orally treated with either BPA (200 µg/kg per day) or vehicle throughout gestation [gestational days (GD) 0–19]. Mother–infant interactions were characterized from postnatal day (P) 1–6, and maternal behavior (licking/grooming and arched-back nursing) was used as a covariate in all analyses. After weaning at P28, six male and six female offspring per treatment group were killed and examined for hippocampal gene expression and DNA methylation. Remaining offspring were housed under standard conditions (three to four per cage in same-sex, same-treatment groups) and then tested for novel object recognition at P60. After behavioral testing, offspring were killed, and their hippocampi were examined for gene expression and DNA methylation whereas the blood was examined for DNA methylation only.
BPA Effects on Hippocampal Gene Expression and DNA Methylation.
We first assessed whether prenatal BPA treatment induces enduring effects on hippocampal gene expression and DNA methylation at P28 and P60. We examined two genes that have been associated with neuronal plasticity and psychopathology and are known to be epigenetically regulated in response to environmental exposures: brain-derived neurotrophic factor (Bdnf) (21–24) and NMDA receptor 2b subunit (Grin2b) (25, 26). In addition, we also examined expression of the epigenetic regulators implicated in DNA (de)methylation in the brain: growth arrest and DNA damage-inducible beta (Gadd45b) (27) at P28 and P60, and DNA methyltransferases 1 (Dnmt1) and -3a (Dnmt3a) (28) at P60.
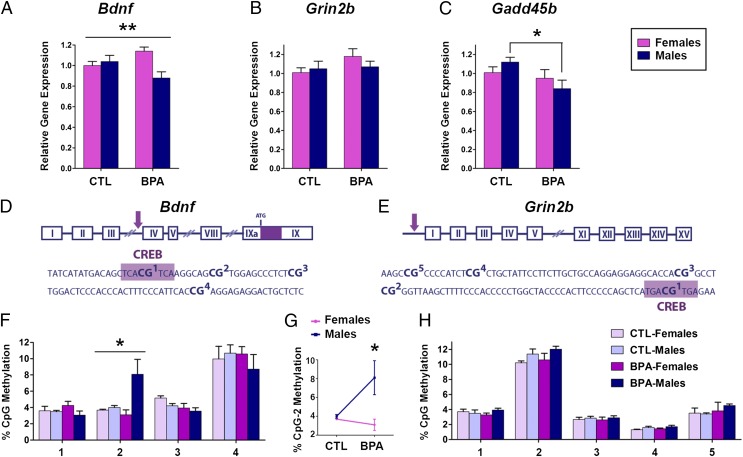
P28 (Fig. 2).
Fig. 2.
Sex-specific changes in hippocampal Bdnf gene expression and DNA methylation at P28 after prenatal BPA exposure. Gene expression analysis of (A) Bdnf, (B) Grin2b, and (C) Gadd45b in the P28 hippocampus of males and females prenatally exposed to BPA (200 µg/kg per day) or vehicle (CTL). Schematic representation of transcriptionally relevant CpG sites in Bdnf IV (D) and Grin2b (E) promoters in the vicinity of CREB-binding sites (purple box). BPA induced a sex-specific effect on DNA methylation of Bdnf IV CpG2 (F) that was primarily driven by hypermethylation induced in males (G). No CpG methylation changes were observed in the Grin2b promoter at P28 (H). *P < 0.05, **P < 0.01.
At P28, prenatal BPA exposure induced significant changes in Bdnf and Gadd45b gene expression whereas there were no changes in Grin2b mRNA levels (Fig. 2 A–C). BPA led to sex-specific changes in Bdnf mRNA (treatment * sex interaction; P < 0.01) (Fig. 2A), with significant decrease in expression observed in males (P < 0.05) and significant increase observed in females (P < 0.05). BPA treatment was also associated with decreased Gadd45b expression primarily in males (P < 0.05) (Fig. 2C). These results suggested that Bdnf down-regulation in males may be partially driven by DNA hypermethylation because Gadd45b has been implicated in active DNA demethylation of the Bdnf gene (27). Our previous analyses indicated no prenatal BPA treatment effect on P28 hippocampal Dnmt1 and Dnmt3a mRNA (4).
To determine the potential role of epigenetic mechanisms in the BPA effect on Bdnf transcription, we examined Bdnf cytosine methylation in two gene regions, promoters IV and IX, which have previously been shown to be regulated by DNA methylation (21, 27, 29). The examined region of the promoter IV included the CREB-binding site (CpG1) and three adjacent CpGs (CpG2 to -4) (Fig. 2D), and DNA methylation levels of this region have been shown to affect CREB binding and consequent transcription (29). Here, we show that BPA induces a sex-specific effect on the cytosine methylation of CpG site 2 (treatment * sex interaction; P < 0.05) (Fig. 2F), which was driven by hypermethylation induced in males (P < 0.05) (Fig. 2G). Importantly, DNA methylation of this site was inversely correlated with Bdnf gene expression in males (Fig. 2A). This effect was specific for Bdnf IV because no significant DNA methylation changes were observed in the Bdnf promoter IX (Fig. S1) or Grin2b gene (Fig. 2H) in either sex. The examined region of Grin2b also contains a CREB-binding site (Fig. 2E) and is sensitive to DNA methylation (25). Therefore, lack of P28 DNA methylation changes in Grin2b was consistent with no change in P28 Grin2b gene expression (Fig. 2B).
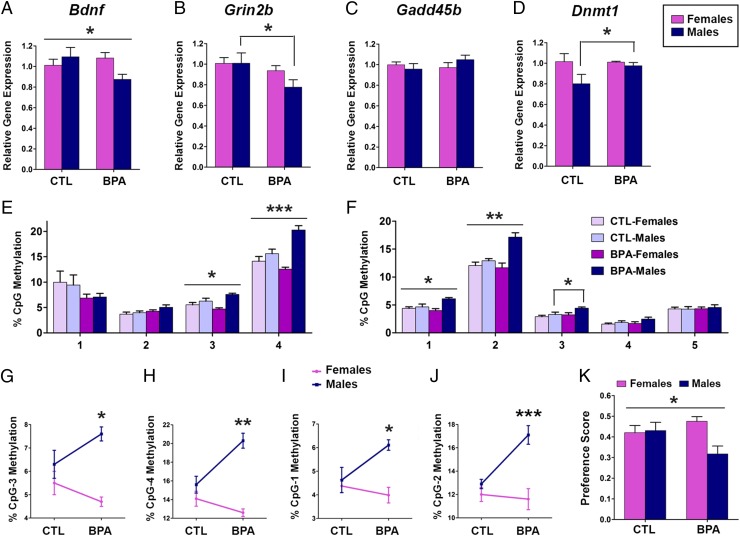
P60 (Fig. 3).
Fig. 3.
Lasting Bdnf gene expression and DNA methylation changes in the P60 hippocampus of BPA-treated animals. Gene expression analysis of (A) Bdnf, (B) Grin2b, (C) Gadd45b, and (D) Dnmt1 in the P60 hippocampus of males and females prenatally exposed to BPA (200 µg/kg per day) or vehicle (CTL). BPA induced sex-specific effect on DNA methylation of Bdnf IV CpG sites 3 and 4 (E) that were primarily driven by hypermethylation induced in males (G and H) as well as on Grin2b sites 1 and 2 (F) that were primarily driven by hypermethylation in males (I and J). Sex-specific changes in gene expression and DNA methylation correspond with BPA-induced changes in the novel object recognition test performed before animals were killed for molecular analyses (K). *P < 0.05, **P < 0.01, ***P < 0.001.
To examine the dynamics of gene expression and DNA methylation after prenatal BPA exposure, we examined the same genes at P60. At this time point, we observed significant BPA-associated changes in Bdnf, Grin2b, and Dnmt1 mRNA whereas there were no changes in Gadd45b and Dnmt3a gene expression (Fig. 3 A–D and Fig. S2). Importantly, BPA effect on Bdnf was persistent and similar to that observed at P28; there was a significant sex-specific effect (treatment * sex interaction; P < 0.05) (Fig. 3A) that was primarily driven by mRNA down-regulation induced in males (P < 0.05). In contrast to P28, at P60, we found significant down-regulation of Grin2b mRNA levels in males (P < 0.05) (Fig. 3B).
We hypothesized that changes in gene expression may be driven by DNA hypermethylation induced by concordant up-regulation of Dnmt1 (28) (Fig. 3D) because no change was noted in the mRNA levels of Gadd45b (Fig. 2C) or Dnmt3a (Fig. S2) at P60. We observed a significant sex-specific effect of BPA at two CpG sites of Bdnf IV-CpG sites 3 and 4 (treatment * sex interaction; P < 0.05 and P < 0.001, respectively) (Fig. 3E), driven by DNA hypermethylation induced at CpG3 and CpG4 in males (P < 0.05 and P < 0.01, respectively) (Fig. 3 G and H). Similarly, BPA induced sex difference in methylation of the CREB-binding site (CpG1) and a site adjacent to it (CpG2) of the Grin2b promoter (treatment * sex interaction; P < 0.05 and P < 0.01, respectively) (Fig. 3F); these BPA effects were primarily driven by CpG hypermethylation in males (P < 0.05 and P < 0.001, respectively) (Fig. 3 I and J). BPA also induced hypermethylation of CpG3 in Grin2b promoter in males (P < 0.05) (Fig. 3F). Therefore, the down-regulation of Bdnf and Grin2b mRNA in males at P60 was associated with the hypermethylation of the corresponding transcriptionally relevant promoter regions.
BPA Effects on Adult Behavior in Mice.
To examine whether there is a behavioral consequence of changes in DNA methylation and gene expression in the hippocampus, we analyzed the results of the novel object recognition test that is largely hippocampus-dependent (30) and was performed at P60 (Fig. 3K). No treatment effects were observed in total duration of exploration during the acquisition phase of the test (i.e., when mice were presented with two identical objects). Importantly, prenatal BPA induced a sex difference in exploration of a novel object (treatment * sex interaction; P < 0.05) (Fig. 3K). Indeed, a significant decrease in the amount of time spent exploring the novel object was seen in males after prenatal BPA treatment (P < 0.05) whereas no significant changes were found in females. These changes were consistent with changes in Bdnf gene expression at P28 and P60 as well as with the down-regulation of Grin2b mRNA observed in males at P60. Although postnatal maternal care has previously been shown to enhance novel object recognition and increase hippocampal Bdnf and Grin2b gene expression in male offspring (31, 32), we did not observe a modulating or mediating effect of maternal behavior on these BPA-associated effects. We have previously shown that BPA induces curvilinear effects on maternal behavior such that, at the 200 µg/kg BPA dose, the frequency of maternal behavior is comparable with that of vehicle-treated dams (4), thus limiting the possibility of detecting maternal care influences on these outcomes.
Bdnf DNA Methylation in the P60 Mouse Blood in Response to in Utero BPA.
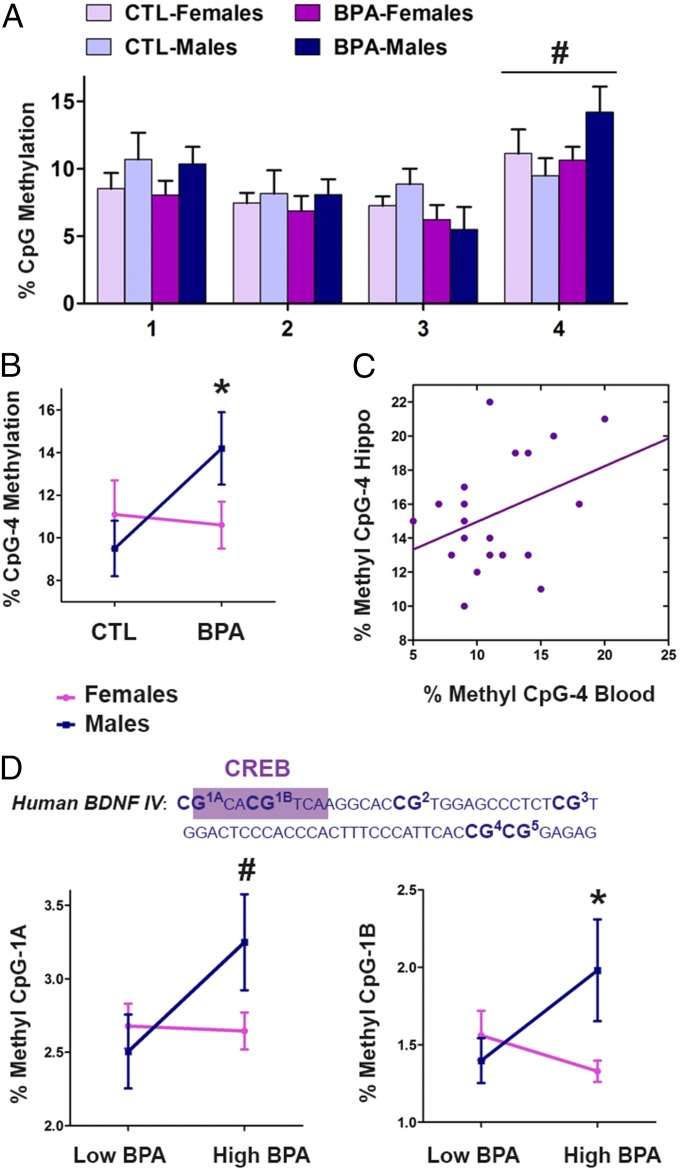
Next, we determined whether Bdnf DNA methylation in the peripheral blood could predict the molecular changes in the brain (hippocampus) and behavioral abnormalities induced by prenatal BPA exposure. To address this question, we analyzed whole-blood samples from P60 mice. Importantly, in our animal model, Bdnf CpG methylation profiles in the blood corresponded with those in the hippocampus (Fig. 4 A–C). BPA induced similar changes in Bdnf IV in the blood as observed in the brain (Fig. 4 A and B); there was a trend for sex-specific effect at CpG site 4 (treatment * sex interaction, P = 0.12) (Fig. 4A), which was driven by hypermethylation induced in males (P < 0.05) (Fig. 4B). In addition, the methylation level of CpG site 4 was significantly correlated between the blood and hippocampus in all animals, independent of BPA exposure (Fig. 4C) (r = 0.53, P < 0.05). Because those same animals were used for hippocampal analysis and behavioral testing, we concluded that Bdnf IV DNA methylation may be used as a predictor of Bdnf methylation and gene expression in the hippocampus as well as of behavioral deficits in male mice.
Fig. 4.
Bdnf DNA methylation patterns in the human and mouse blood mirror Bdnf DNA methylation patterns in the mouse hippocampus. In mice, BPA induced sex-specific effect on DNA methylation of Bdnf IV CpG4 in the blood (A) that was primarily driven by hypermethylation induced in males (B). CpG4 methylation in the blood and CpG4 methylation in the hippocampus are significantly correlated (C). The sequence of the human BDNF promoter IV having 96% homology with the mouse Bdnf IV (CREB-binding site; purple box) (D). In the human cord blood, high maternal BPA levels are associated with altered DNA methylation of two CpG sites in the BDNF IV promoter (D) (low BPA < 1 µg/L; high BPA > 4 µg/L).
BDNF DNA Methylation in Human Cord Blood in Relation to Maternal BPA Levels During Pregnancy.
To explore the translational value of our results, we examined BDNF IV DNA methylation in cord blood samples from the human cohort of the Columbia Center for Children’s Environmental Health (CCCEH) (19). This cohort has characterized maternal BPA levels, which were determined by collecting spot urine samples from the mothers during pregnancy (∼34 weeks). In addition, behavioral analysis of this cohort, conducted on 198 children (87 boys and 111 girls) at 3–5 years of age, showed that high maternal BPA exposure is associated with disturbed emotional regulation and increased aggressive behavior in boys as well as with lower scores for anxiety/depression and reduced aggressive behavior in girls (19). Therefore, the sex-specific behavioral profile of this cohort is consistent with the BPA-induced sex-specific effects in our animal study. To test our hypothesis, we selected cord blood samples of participants that corresponded to approximately the lowest quintile (21 males and 22 females; BPA < 1 µg/L) and highest quintile (19 males and 19 females; BPA > 4 µg/L) of prenatal BPA exposure. The human BDNF IV region that we examined had 96% homology with the sequence of the mouse Bdnf IV region with all four CpG sites being conserved, including the site within the CREB-binding region (Fig. 4D). There were two extra CpG sites in the human region, due to a single nucleotide difference, and we examined all six sites with a single pyrosequencing assay (Fig. 4D and Fig. S3). Importantly, prenatal exposure to high BPA levels was associated with altered DNA methylation of two CpG sites in the human cord blood, in a sex-specific way mimicking the animal data (Fig. 4D). There was a trend for a sex-specific effect of high BPA on CpG1A methylation (treatment * sex interaction, P = 0.09) and a significant sex-specific effect of high BPA exposure on CpG1B methylation levels (treatment * sex interaction, P < 0.05). Importantly, the CpG1B site lies exactly within the CREB-binding site, and our data suggest that the methylation of this site may represent a biomarker in humans predictive of BDNF gene expression in the brain and consequent alterations in brain function and behavior.
Discussion
This study contributes significantly to our current knowledge of epigenetic biomarkers and demonstrates that DNA methylation of BDNF in the blood can be used to predict epigenetic changes in the brain and behavioral vulnerability induced by early-life adversity.
In psychiatry, the identification of biomarkers that predict future risk for psychopathology is of critical importance. Theoretically, epigenetic biomarkers are good candidates because early-life exposures can leave lasting marks on the epigenome of various tissues (2) and these marks could be detected before the disease is fully developed. Although DNA methylation shows significant between-tissue variation, some interindividual differences in DNA methylation are correlated across brain and blood of healthy subjects (33), suggesting that epigenetic profiling of peripheral tissue may be useful for studies of brain disorders. Consistent with this hypothesis, DNA methylation changes have been found both in psychiatric postmortem brain samples (34, 35) and in peripheral blood of the living psychiatric patients (12–14). However, a challenge to this field of study is whether epigenetic markers in peripheral tissues can predict functionally relevant epigenetic changes in the brain and consequent behavioral and psychiatric outcomes.
Here, we provided a direct link between long-term epigenetic effects of an early-life environmental exposure in the brain and blood in relation to a behavioral outcome (Fig. 5). We focused our study on BDNF because it represents a particularly good candidate for early detection of psychiatric risk. BDNF has a well-established role in neurodevelopment and neuronal plasticity, and its deficiency has been linked to several psychiatric disorders that are associated with early-life adversity, including depression (34, 36), schizophrenia (37), bipolar disorder (38), and autism (39). It seems likely that disrupted neural development and neural plasticity, associated with altered BDNF expression emerging in early life, could predispose an individual to behavioral deficits and multiple psychopathologies in adulthood (40).
Fig. 5.
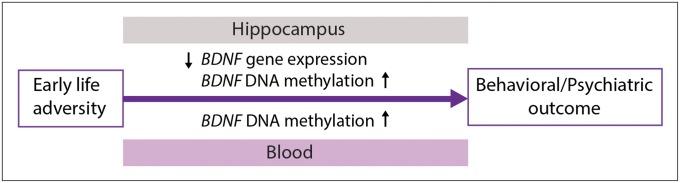
BDNF as biomarker of early-life adversity. Various early-life exposures, such as prenatal stress, maternal toxicological exposures, and maternal neglect, can induce lasting epigenetic changes in the BDNF gene in the brain that contribute to behavioral vulnerability and psychiatric risk. These changes may be reflected in the peripheral blood starting from early life and can be used to predict psychiatric risk.
In particular, our study shows that in utero exposure to a ubiquitous environmental toxicant, BPA (15, 16), induces lasting, sex-specific effects on Bdnf gene expression evident in juvenile (P28) and young adult animals (P60). Importantly, at both ages, we observed the down-regulation of Bdnf expression in males, associated with hypermethylation of CpG sites in the gene regulatory region, and those changes were consistent with behavioral deficits observed in young adult males. In contrast, another gene important for neuronal plasticity in the hippocampus and implicated in the novel object recognition task (41), Grin2b, showed mRNA and DNA methylation changes only in adulthood at the time of observed behavioral changes. Previous studies have likewise reported that prenatal toxicological exposures (e.g., cocaine) can induce either persistent or nonpersistent/delayed effects on gene expression and DNA methylation in the hippocampus depending on a gene locus (42). Our data imply that BPA affects developmental epigenetic programming of Bdnf and induces persistent epigenetic disruption of this gene. In contrast, Grin2b does not seem to be affected during development but is affected in adulthood, likely through an activity-dependent mechanism (26) that may be induced by behavioral testing (exposure to novel environment) (27). These findings strongly suggest that Bdnf (but not Grin2b) DNA methylation status may be an important marker for the early detection of neurodevelopmental abnormalities, with consequences for abnormal behavior in later life. Our study further provides the evidence that Bdnf methylation status in peripheral blood can be used as a predictor of DNA methylation levels in the mouse brain. Our data strongly suggest the translational relevance of these findings to humans because we found that high maternal BPA exposure during pregnancy is associated with altered BDNF methylation in the cord blood.
Interestingly, BDNF DNA methylation changes that we observed in relation to prenatal BPA exposure in both mice and humans are located in a very specific gene regulatory region, promoter IV, which contains the binding site for the transcription factor CREB and is known to be sensitive to DNA methylation as shown by in vitro (29) and in vivo (21, 22) studies. Based on our results, it does not seem to be the case that the same CpG sites remain hypermethylated over time (P28 vs. P60). This finding is perhaps not surprising because, similar to Grin2b, the Bdnf IV promoter in the adult brain is dynamically regulated through DNA methylation in response to neuronal activity (21, 22) and the observed CpG methylation changes (P28 vs. P60) may have been triggered by behavioral testing. Rather than specific CpG sites, a localized region of the Bdnf IV promoter seems to be developmentally targeted for lasting epigenetic disruption. The specific sites within this region that are affected may depend on the specific molecular pathways that are involved in Bdnf regulation and are altered by BPA at each temporal stage (i.e., Gadd45b at P28 vs. Dnmt1 at P60).
Although the sites epigenetically altered in BPA-treated mice that demonstrate strong concordance between brain and blood are adjacent to the CREB-binding site (CpG4 at P60), in human cord blood samples, the BPA-associated epigenetic changes are exactly within the CREB-binding region (CpG1B). This phenomenon may reflect interspecies difference (humans vs. mice) in the regulation and targeting of this genomic region or temporal/between-tissue difference (cord blood vs. adult peripheral blood). However, despite this between-species discordance, it is striking that BDNF DNA methylation differences in humans that may serve as a predictor of behavioral vulnerability could be detected as early as at birth. In addition to promoter IV (34, 43), other BDNF gene regions such as promoters I (13, 14) and VI (10) can also be sensitive to epigenetic effects of early-life adversity and may confer psychiatric risk. Therefore, although our study provides a proof of principle that BDNF DNA methylation in the blood may predict behavioral vulnerability, future more comprehensive epigenetic analyses of the BDNF gene in response to other early-life exposures and in psychiatric patients are warranted.
Our study provides evidence in support of the role of epigenetic mechanisms in the neurodevelopmental toxicity of BPA and reveals BDNF as a target gene of this environmental toxicant. BPA has been detected in more than 90% of the United States population, and, in light of the capacity of BPA to cross the placenta, prenatal exposure to BPA is of great public concern, particularly in relation to neurobehavioral outcomes (18, 44). Although human data linking in utero BPA exposure to psychiatric disorders are as yet unavailable, there is growing evidence that prenatal BPA is associated with increased risk for behavioral changes in young children (17, 19, 45), and animals studies have shown BPA-induced changes in behavior and cognition in early life and adulthood (18). Therefore, our data suggest that lasting epigenetic disruption of BDNF may be an important mechanism that underlies long-term neurobehavioral effects induced by in utero BPA exposure in male offspring.
Males may be generally more vulnerable to prenatal adversity (3, 19), and the sex-specific effect of prenatal BPA on molecular and behavioral outcomes was anticipated because this toxicant is an endocrine disruptor that we have previously shown to induce lasting, sex-specific epigenetic disruption in the brains of offspring (4). Epigenetic regulation seems to be sex-specific throughout life (46), and the differential effect of BPA on male and female epigenetic regulators (such as DNMTs and Gadd45b) may be responsible for sex-specific effects on gene regulation and behavior. It is unclear whether those effects are directly related to BPA action on sex hormone receptors [BPA can act as an estrogenic, antiestrogenic, or antiandrogenic compound (4)]. Exploring the potential role of prenatal testosterone and estradiol levels in conferring vulnerability (males) or resilience (females) in response to in utero exposure to BPA may help to elucidate the mechanisms of these sex-specific effects. For instance, determination of intrauterine position (47), which may expose the fetus to higher vs. lower endogenous levels of testosterone in utero, may help dissociate the mechanism(s) underlying higher male vulnerability in this model.
However, persistent epigenetic changes in BDNF do not seem to be specific to prenatal BPA or male offspring because comparable epigenetic effects have been observed in both sexes in response to other environmental exposures associated with behavioral and psychiatric risk, including prenatal stress (24), early-life maternal neglect (22), and perinatal methylmercury exposure (23). These findings suggest that a BDNF DNA methylation signature may be a hallmark of early-life adversity. Therefore, we propose a model suggesting that early-life adversity may increase the risk for psychopathology through lasting BDNF changes in the brain that can be predicted by examining BDNF DNA methylation in the blood in early life (Fig. 5).
Whether lasting BDNF deficiency will result in a specific psychiatric disorder likely depends on a complex interaction of an individual's genetic make-up, sex, and the life-long environment. Recent studies suggested that concentrations of BDNF protein in the blood (serum or plasma) may represent peripheral manifestation of depression (48) and schizophrenia (49) and could be used as a biomarker for therapeutic efficacy (50). Similarly, BDNF DNA methylation changes have been found in the peripheral blood of patients suffering from depression (13), bipolar disorder (12), schizophrenia (14), and eating disorders (51), and a change in BDNF DNA methylation status correlated with a positive response to psychotherapy in borderline personality disorder (52). In addition, within a psychiatric population, high BDNF DNA methylation was strongly associated with the history of child maltreatment (51, 52). It seems likely that BDNF DNA methylation status may represent a more stable and reliable marker of psychiatric vulnerability compared with the levels of BDNF protein. Therefore, our study offers experimental evidence that BDNF DNA methylation in the blood may represent a novel clinical epigenetic biomarker for the early detection of psychopathology.
Materials and Methods
Animals and Treatments.
BALB/c mice were maintained on a 12:12-h light:dark cycle with ad libitum access to food and water. Before mating, female mice were assigned to two treatment groups (200 μg/kg BPA or vehicle; n = 24 per treatment) and were orally exposed to bisphenol A (BPA) dissolved in tocopherol-stripped corn oil (200 μg/kg per day) or only corn oil (vehicle control, CTL). Mice that became pregnant (around 70%) were placed individually in 10.5 × 19 × 6-inch polysulfone cages and were exposed daily to the assigned treatment during the entire gestational period (GD 0–19). Postpartum maternal behavior was assessed from P1 to P6 as previously described (4). At weaning (P28), six male and six female offspring per treatment [one to two pups per litter from a minimum of five litters per treatment for each sex; total litters, vehicle (n = 8), and 200 μg/kg (n = 7) BPA] were killed, and whole brains were stored at −80 °C to be used for gene expression and DNA methylation analyses. Remaining animals were housed in same-sex/same-treatment cages (n = 3–4 per cage) at the time of weaning and underwent behavioral testing at P60 [male and female offspring from vehicle (n = 11), and 200 μg/kg (n = 8) BPA litters]. After behavioral testing, five to eight male and female offspring per treatment [one to two pups per litter from a minimum of four litters per treatment for each sex; total litters, vehicle (n = 7), and 200 μg/kg (n = 7) BPA] were killed, and whole brains and whole-blood samples were stored at −80 °C to be used for gene expression and/or DNA methylation analyses. All procedures were performed with the approval of the Institutional Animal Care and Use Committee at Columbia University.
RNA and DNA Extraction.
P28 and P60 hippocampal tissues were dissected before nucleic acid extraction, and RNA and DNA were simultaneously isolated using the AllPrep DNA/RNA Mini Kit (Qiagen) according to the manufacturer's instructions. DNA from the mouse P60 whole-blood samples was isolated using the DNeasy Blood & Tissue Kit (Qiagen). DNA from the human cord blood samples was extracted using the standard phenol-chloroform protocol.
Gene Expression Analysis.
Gene expression was assessed using reverse transcription (SuperScript III First-Strand Synthesis System; Invitrogen) followed by quantitative real-time PCR (Fast SYBR Green Master Mix; Applied Biosystems) with a 7500 real-time PCR system (Applied Biosystems) as previously described (4). Primer sequences used to assess the mRNA levels of Bdnf, Grin2b, Gadd45b, Dnmt1, Dnmt3a, and cyclophilin A (CypA) are provided in Table S1. Relative mRNA expression was calculated using the standard 2-ΔΔCT method (53) with female control (CTL) samples as a reference sample and CypA as an endogenous reference gene, based on the following equation: ΔΔCt = (CT,Target − CT,CypA)Group X − (CT,Target − CT,CypA)Female Control Group where Group X is any of the four treatment groups: female controls, BPA-treated females, male controls, or BPA-treated males. Analyses of hippocampal tissue samples indicated no BPA-induced effects on levels of CypA.
DNA Methylation Analysis.
Percent methylation at single CpG sites was determined using the bisulfite-pyrosequencing method as previously described (4, 54). Briefly, 500 ng (hippocampus) or 250 ng (blood) of genomic DNA was used for bisulfite conversion (EpiTect Bisulfite Kit; Qiagen). To amplify gene regions of interest, 20 ng (hippocampus) or 15 ng (blood) of bisulfite converted DNA was run in a PCR using a PyroMark PCR kit (Qiagen) and specific primers, one of which was biotinylated at the 5′ end. Biotinylated PCR products were then processed using the PyroMark Q24 Vacuum Workstation, and subsequent pyrosequencing was performed on a PyroMark Q24 pyrosequencer using PyroMark Gold Q24 Reagents (Qiagen) and a specific sequencing primer. PCR amplification and pyrosequencing primer sequences used to assess CpG methylation levels of the mouse Bdnf (IV and IX) and Grin2b promoter regions as well as the human BDNF IV are presented in Table S2. Average methylation levels of CpG sites were quantified using PyroMark Q24 2.0.4 software (Qiagen).
Novel Object Recognition.
At P60, mice (n = 9–14 per treatment per sex) were assessed in the novel object test (55) that consisted of three phases: habituation, acquisition, and test phase. On the first day, mice were habituated to the novel-object testing arena for 10 min in the absence of any objects. The following day, mice were tested for object recognition using acquisition and test trials separated by a 30-min delay. During the acquisition trial, mice were exposed to two identical objects for 15 min. During the 15-min test trial, mice were exposed to the original object and a new (“novel”) object. After data collection using video recording, all observations were coded using Observer XT software (version 9.0; Noldus Information Technology). The variables of interest were amount of time spent exploring each object and preference for the novel object. Preference score was calculated as the amount of time spent investigating the novel object divided with the total time spent investigating both the novel and familiar objects.
CCCEH Human Cohort.
Sample selection.
Participants for the study were mothers and children enrolled in the Columbia Center for Children’s Environmental Health (CCCEH) New York City (NYC) prospective cohort. These participants were residents of the South Bronx, Washington Heights, or Harlem neighborhoods, recruited during pregnancy between 1998 and 2003 and included women between 18 years and 35 years of age, without a history of diabetes, hypertension, or HIV, who were not tobacco or illicit drug users, that began prenatal care by their 20th week of pregnancy (56). Informed consent was obtained from all participants and the study was approved by the Institutional Review Boards of Columbia University and the Centers for Disease Control and Prevention (CDC).
BPA measures.
BPA measures were based on spot urine samples collected from the mother during pregnancy (range, 24–40 weeks of gestation; mean, 34.0 weeks). Immediately after collection, the urine samples were transported to the CCCEH laboratory, inventoried, and frozen at –80 °C before shipment to CDC for analysis. Total (free and conjugated) urinary concentrations of BPA were measured using online solid-phase extraction coupled with HPLC–isotope dilution–tandem mass spectrometry with peak focusing (57). The limit of detection (LOD) was 0.4 μg/L; concentrations below the LOD were given a value of LOD/2. Urinary dilution was calculated from specific gravity (SG) measurements obtained using a handheld refractometer (Urine-Specific-Gravity-Refractometer-PAL-10-S-P14643C0; TAGO USA, Inc.). All BPA concentrations were corrected for dilution by SG adjustment using a modification of the formula by Hauser et al. (58). In the current study, DNA methylation analyses were conducted on cord blood samples of the participants that corresponded to the lowest (BPA < 1 µg/L) and highest (BPA > 4 µg/L) quintile of maternal urinary BPA levels.
Statistical analysis.
Data were analyzed using two-way ANOVA with BPA treatment and sex as factors, and, where appropriate, a post hoc Student t test was performed to dissociate sex-specific effects. Two measures of maternal behavior (licking/grooming and arched-back nursing) were used as covariates in the analyses. The correlation analysis of Bdnf IV CpG4 methylation in the mouse hippocampus and blood was carried out using Pearson's correlation coefficient. The analyses were performed using IBM SPSS Statistics Software, version 21. Effects were considered significant at P < 0.05. All data are expressed as mean ± SEM.
Supplementary Material
Acknowledgments
We thank Antonia Calafat (the Centers for Disease Control and Prevention) for her ongoing collaboration with the Columbia Center for Children’s Environmental Health and analyses of urinary BPA levels in the human cohort. This research was supported by National Institutes of Health Grant DP2OD001674-01, National Institute of Environmental Health Sciences Grant 5P01ES09600, US Environmental Protection Agency Grant RD834509, Trustees of the Blanchette Hooker Rockefeller Fund, and the Gladys and Roland Harriman Foundation.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Epigenetic Changes in the Developing Brain: Effects on Behavior,” held March 28–29, 2014, at the National Academy of Sciences in Washington, DC. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/Epigenetic_changes.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1408355111/-/DCSupplemental.
References
- 1.Shonkoff JP, Boyce WT, McEwen BS. Neuroscience, molecular biology, and the childhood roots of health disparities: Building a new framework for health promotion and disease prevention. JAMA. 2009;301(21):2252–2259. doi: 10.1001/jama.2009.754. [DOI] [PubMed] [Google Scholar]
- 2.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8(4):253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mueller BR, Bale TL. Sex-specific programming of offspring emotionality after stress early in pregnancy. J Neurosci. 2008;28(36):9055–9065. doi: 10.1523/JNEUROSCI.1424-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kundakovic M, et al. Sex-specific epigenetic disruption and behavioral changes following low-dose in utero bisphenol A exposure. Proc Natl Acad Sci USA. 2013;110(24):9956–9961. doi: 10.1073/pnas.1214056110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weaver IC, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 6.McGowan PO, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12(3):342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heijmans BT, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA. 2008;105(44):17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oberlander TF, et al. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3(2):97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- 9.Radtke KM, et al. Transgenerational impact of intimate partner violence on methylation in the promoter of the glucocorticoid receptor. Transl Psychiatr. 2011;1:e21. doi: 10.1038/tp.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toledo-Rodriguez M, et al. Maternal smoking during pregnancy is associated with epigenetic modifications of the brain-derived neurotrophic factor-6 exon in adolescent offspring. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(7):1350–1354. doi: 10.1002/ajmg.b.31109. [DOI] [PubMed] [Google Scholar]
- 11.Herbstman JB, et al. Prenatal exposure to polycyclic aromatic hydrocarbons, benzo[a]pyrene-DNA adducts, and genomic DNA methylation in cord blood. Environ Health Perspect. 2012;120(5):733–738. doi: 10.1289/ehp.1104056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Addario C, et al. Selective DNA methylation of BDNF promoter in bipolar disorder: Differences among patients with BDI and BDII. Neuropsychopharmacology. 2012;37(7):1647–1655. doi: 10.1038/npp.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchikami M, et al. DNA methylation profiles of the brain-derived neurotrophic factor (BDNF) gene as a potent diagnostic biomarker in major depression. PLoS ONE. 2011;6(8):e23881. doi: 10.1371/journal.pone.0023881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikegame T, et al. DNA methylation analysis of BDNF gene promoters in peripheral blood cells of schizophrenia patients. Neurosci Res. 2013;77(4):208–214. doi: 10.1016/j.neures.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Calafat AM, Ye X, Wong LY, Reidy JA, Needham LL. Exposure of the U.S. population to bisphenol A and 4-tertiary-octylphenol: 2003-2004. Environ Health Perspect. 2008;116(1):39–44. doi: 10.1289/ehp.10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vandenberg LN, et al. Urinary, circulating, and tissue biomonitoring studies indicate widespread exposure to bisphenol A. Environ Health Perspect. 2010;118(8):1055–1070. doi: 10.1289/ehp.0901716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braun JM, et al. Impact of early-life bisphenol A exposure on behavior and executive function in children. Pediatrics. 2011;128(5):873–882. doi: 10.1542/peds.2011-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kundakovic M, Champagne FA. Epigenetic perspective on the developmental effects of bisphenol A. Brain Behav Immun. 2011;25(6):1084–1093. doi: 10.1016/j.bbi.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perera F, et al. Prenatal bisphenol a exposure and child behavior in an inner-city cohort. Environ Health Perspect. 2012;120(8):1190–1194. doi: 10.1289/ehp.1104492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boulle F, et al. Epigenetic regulation of the BDNF gene: Implications for psychiatric disorders. Mol Psychiatry. 2012;17(6):584–596. doi: 10.1038/mp.2011.107. [DOI] [PubMed] [Google Scholar]
- 21.Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28(42):10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65(9):760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onishchenko N, Karpova N, Sabri F, Castrén E, Ceccatelli S. Long-lasting depression-like behavior and epigenetic changes of BDNF gene expression induced by perinatal exposure to methylmercury. J Neurochem. 2008;106(3):1378–1387. doi: 10.1111/j.1471-4159.2008.05484.x. [DOI] [PubMed] [Google Scholar]
- 24.Boersma GJ, et al. Prenatal stress decreases Bdnf expression and increases methylation of Bdnf exon IV in rats. Epigenetics. 2014;9(3):437–447. doi: 10.4161/epi.27558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiang M, et al. The site specific demethylation in the 5′-regulatory area of NMDA receptor 2B subunit gene associated with CIE-induced up-regulation of transcription. PLoS ONE. 2010;5(1):e8798. doi: 10.1371/journal.pone.0008798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S, et al. Activity-dependent NR2B expression is mediated by MeCP2-dependent epigenetic regulation. Biochem Biophys Res Commun. 2008;377(3):930–934. doi: 10.1016/j.bbrc.2008.10.082. [DOI] [PubMed] [Google Scholar]
- 27.Ma DK, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323(5917):1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levenson JM, et al. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281(23):15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- 29.Martinowich K, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302(5646):890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 30.Antunes M, Biala G. The novel object recognition memory: Neurobiology, test procedure, and its modifications. Cogn Process. 2012;13(2):93–110. doi: 10.1007/s10339-011-0430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Champagne FA, Meaney MJ. Transgenerational effects of social environment on variations in maternal care and behavioral response to novelty. Behav Neurosci. 2007;121(6):1353–1363. doi: 10.1037/0735-7044.121.6.1353. [DOI] [PubMed] [Google Scholar]
- 32.Liu D, Diorio J, Day JC, Francis DD, Meaney MJ. Maternal care, hippocampal synaptogenesis and cognitive development in rats. Nat Neurosci. 2000;3(8):799–806. doi: 10.1038/77702. [DOI] [PubMed] [Google Scholar]
- 33.Davies MN, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13(6):R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keller S, et al. Increased BDNF promoter methylation in the Wernicke area of suicide subjects. Arch Gen Psychiatry. 2010;67(3):258–267. doi: 10.1001/archgenpsychiatry.2010.9. [DOI] [PubMed] [Google Scholar]
- 35.Mill J, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82(3):696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dwivedi Y, et al. Altered gene expression of brain-derived neurotrophic factor and receptor tyrosine kinase B in postmortem brain of suicide subjects. Arch Gen Psychiatry. 2003;60(8):804–815. doi: 10.1001/archpsyc.60.8.804. [DOI] [PubMed] [Google Scholar]
- 37.Weickert CS, et al. Reduced brain-derived neurotrophic factor in prefrontal cortex of patients with schizophrenia. Mol Psychiatry. 2003;8(6):592–610. doi: 10.1038/sj.mp.4001308. [DOI] [PubMed] [Google Scholar]
- 38.Kim HW, Rapoport SI, Rao JS. Altered expression of apoptotic factors and synaptic markers in postmortem brain from bipolar disorder patients. Neurobiol Dis. 2010;37(3):596–603. doi: 10.1016/j.nbd.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nickl-Jockschat T, Michel TM. The role of neurotrophic factors in autism. Mol Psychiatry. 2011;16(5):478–490. doi: 10.1038/mp.2010.103. [DOI] [PubMed] [Google Scholar]
- 40.Angelucci F, Brenè S, Mathé AA. BDNF in schizophrenia, depression and corresponding animal models. Mol Psychiatry. 2005;10(4):345–352. doi: 10.1038/sj.mp.4001637. [DOI] [PubMed] [Google Scholar]
- 41.Vedder LC, Smith CC, Flannigan AE, McMahon LL. Estradiol-induced increase in novel object recognition requires hippocampal NR2B-containing NMDA receptors. Hippocampus. 2013;23(1):108–115. doi: 10.1002/hipo.22068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novikova SI, et al. Maternal cocaine administration in mice alters DNA methylation and gene expression in hippocampal neurons of neonatal and prepubertal offspring. PLoS One. 2008;3(4):e1919. doi: 10.1371/journal.pone.0001919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang H-J, et al. BDNF promoter methylation and suicidal behavior in depressive patients. J Affect Disord. 2013;151(2):679–685. doi: 10.1016/j.jad.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Richter CA, et al. In vivo effects of bisphenol A in laboratory rodent studies. Reprod Toxicol. 2007;24(2):199–224. doi: 10.1016/j.reprotox.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Braun JM, et al. Prenatal bisphenol A exposure and early childhood behavior. Environ Health Perspect. 2009;117(12):1945–1952. doi: 10.1289/ehp.0900979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCarthy MM, et al. The epigenetics of sex differences in the brain. J Neurosci. 2009;29(41):12815–12823. doi: 10.1523/JNEUROSCI.3331-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryan BC, Vandenbergh JG. Intrauterine position effects. Neurosci Biobehav Rev. 2002;26(6):665–678. doi: 10.1016/s0149-7634(02)00038-6. [DOI] [PubMed] [Google Scholar]
- 48.Molendijk M, et al. Serum BDNF concentrations as peripheral manifestations of depression: Evidence from a systematic review and meta-analyses on 179 associations (N= 9484) Mol Psychiatry. 2013;19(7):791–800. doi: 10.1038/mp.2013.105. [DOI] [PubMed] [Google Scholar]
- 49.Green MJ, Matheson SL, Shepherd A, Weickert CS, Carr VJ. Brain-derived neurotrophic factor levels in schizophrenia: A systematic review with meta-analysis. Mol Psychiatry. 2011;16(9):960–972. doi: 10.1038/mp.2010.88. [DOI] [PubMed] [Google Scholar]
- 50.Vinogradov S, et al. Is serum brain-derived neurotrophic factor a biomarker for cognitive enhancement in schizophrenia? Biol Psychiatry. 2009;66(6):549–553. doi: 10.1016/j.biopsych.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thaler L, et al. Methylation of BDNF in women with bulimic eating syndromes: Associations with childhood abuse and borderline personality disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2014;54:43–49. doi: 10.1016/j.pnpbp.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 52.Perroud N, et al. Response to psychotherapy in borderline personality disorder and methylation status of the BDNF gene. Transl Psychiatr. 2013;3:e207. doi: 10.1038/tp.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 54.Kundakovic M, Lim S, Gudsnuk K, Champagne FA. Sex-specific and strain-dependent effects of early life adversity on behavioral and epigenetic outcomes. Front Psychiatry. 2013;4:78. doi: 10.3389/fpsyt.2013.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ennaceur A, Delacour J. A new one-trial test for neurobiological studies of memory in rats. 1: Behavioral data. Behav Brain Res. 1988;31(1):47–59. doi: 10.1016/0166-4328(88)90157-x. [DOI] [PubMed] [Google Scholar]
- 56.Perera FP, et al. Effects of transplacental exposure to environmental pollutants on birth outcomes in a multiethnic population. Environ Health Perspect. 2003;111(2):201–205. doi: 10.1289/ehp.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ye X, Kuklenyik Z, Needham LL, Calafat AM. Quantification of urinary conjugates of bisphenol A, 2,5-dichlorophenol, and 2-hydroxy-4-methoxybenzophenone in humans by online solid phase extraction-high performance liquid chromatography-tandem mass spectrometry. Anal Bioanal Chem. 2005;383(4):638–644. doi: 10.1007/s00216-005-0019-4. [DOI] [PubMed] [Google Scholar]
- 58.Hauser R, Meeker JD, Park S, Silva MJ, Calafat AM. Temporal variability of urinary phthalate metabolite levels in men of reproductive age. Environ Health Perspect. 2004;112(17):1734–1740. doi: 10.1289/ehp.7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.