Significance
Ageing is dramatically accelerated in Cockayne syndrome (CS), but the impairments that lead to this phenotype have not been elucidated. The DNA repair proteins CSA or CSB are mutated in CS, but premature ageing is not caused by the DNA repair defect. CSB also affects mitochondrial turnover. Our data reveal a novel pathway that is affected in CS cells. We show that CSB deregulates the expression of a serine protease, which degrades mitochondrial DNA polymerase gamma and impairs mitochondrial function. We rescue this defect, by two independent strategies, in primary cells from patients. Our findings open novel possibilities for developing treatments, which are presently missing for CS patients. Abnormalities revealed here might occur at a slower rate during normal physiological ageing.
Keywords: Cockayne syndrome, premature ageing, serine protease, DNA polymerase gamma, mtSSB
Abstract
UV-sensitive syndrome (UVSS) and Cockayne syndrome (CS) are human disorders caused by CSA or CSB gene mutations; both conditions cause defective transcription-coupled repair and photosensitivity. Patients with CS also display neurological and developmental abnormalities and dramatic premature aging, and their cells are hypersensitive to oxidative stress. We report CSA/CSB-dependent depletion of the mitochondrial DNA polymerase-γ catalytic subunit (POLG1), due to HTRA3 serine protease accumulation in CS, but not in UVsS or control fibroblasts. Inhibition of serine proteases restored physiological POLG1 levels in either CS fibroblasts and in CSB-silenced cells. Moreover, patient-derived CS cells displayed greater nitroso-redox imbalance than UVSS cells. Scavengers of reactive oxygen species and peroxynitrite normalized HTRA3 and POLG1 levels in CS cells, and notably, increased mitochondrial oxidative phosphorylation, which was altered in CS cells. These data reveal critical deregulation of proteases potentially linked to progeroid phenotypes in CS, and our results suggest rescue strategies as a therapeutic option.
Cockayne syndrome (CS) is a rare genetic disorder characterized by photosensitivity, severe neurological and developmental defects, and dramatically precocious aging (1). CS is clinically classified into three subtypes: type I (CS-I), the classical form with symptom onset in early childhood; type II (CS-II), an early-onset form with severe symptoms that appear in utero (in this study, CS-II patients have also been clinically diagnosed as cerebro-oculo-facio-skeletal syndrome, COFS); and type III (CS-III), with mild symptoms and onset in late childhood. CS is caused by mutations in the Cockayne syndrome B (CSB) (∼70% of cases) or A (CSA) gene (∼30% of cases) (2, 3). CSA and CSB proteins are implicated in transcription-coupled nucleotide excision repair, which removes bulky adducts, including UV-induced damage, in actively transcribed DNA regions (4, 5). However, it is difficult to explain the dramatic CS phenotype based solely on transcription repair deficiency (6).
Different mutations in CSA or CSB can give rise to another transcription-repair deficient disease, the UV-sensitive syndrome (UVSS), characterized by photosensitivity and mild skin abnormalities, but normal growth and lifespan, and no premature aging (7). A patient with UVSS (UVSS1VI) carrying a CSA mutation that conferred the hallmark UVSS symptoms, without CS characteristics of premature aging, oxidative stress sensitivity, or neuro-degeneration, has been characterized (8). Comparison of these two types of patient' cells indicates that the impaired UV response was uncoupled from oxidative stress and premature aging. In agreement with this finding, higher levels of reactive oxygen species (ROS) were recently reported in mitochondria from CS compared with UVSS and healthy cells (9). CSA and CSB also act as transcription factors, and CSB has been proposed to remodel chromatin (10–12). Moreover, both CSA and CSB have been detected in the nucleus and mitochondria (13), where they are involved in removing oxidized bases through base excision repair (14). Altered mitochondrial transcription and autophagy have been reported in immortalized CS cells, suggesting that mitochondrial impairments play a key role in CS (15–17).
Mitochondria are essential for many cellular functions, but they are primarily involved in ATP production. They also produce ROS during oxidative phosphorylation (OXPHOS). ROS are considered causative factors of aging (18), but the picture has not been fully elucidated in vivo, as mitochondrial ROS also have signaling function (19), and may also increase the lifespan (20). Mitochondria carry many copies of their own genome, a circular, double-stranded DNA of 16.6 kbp in humans, which is replicated in the organelle by a nuclear-encoded DNA polymerase (POLG), a heterotrimer composed of the catalytic subunit, POLG1, and two copies of the accessory protein, POLG2 (21). The replication machinery also includes the helicase Twinkle. mtDNA replication is stimulated by the mitochondrial single-stranded DNA binding protein (mtSSB). Defects in mitochondrial DNA (mtDNA) maintenance are associated with age-related phenotypes (22).
Here, we investigated primary diploid fibroblasts derived from patients with CS and UVSS, CSB-silenced, CSB-deleted, and CSB-overexpressing cells. Altogether, these complementary models revealed impaired mitochondrial function and reduced respiration due to a depletion of the mitochondrial DNA polymerase (POLG1), which is specific to CS cells and depends on the CSA/CSB deficiency. We also show that POLG1 is degraded due to dramatic increases in the levels of the high temperature requirement A (HTRA) 3 protease, which in turn depends on altered nitroso-redox balance. We rescued this defect, by two independent strategies, in primary cells from patients.
Results
Mitochondrial POLG1 Depletion in Fibroblasts from CS Patients.
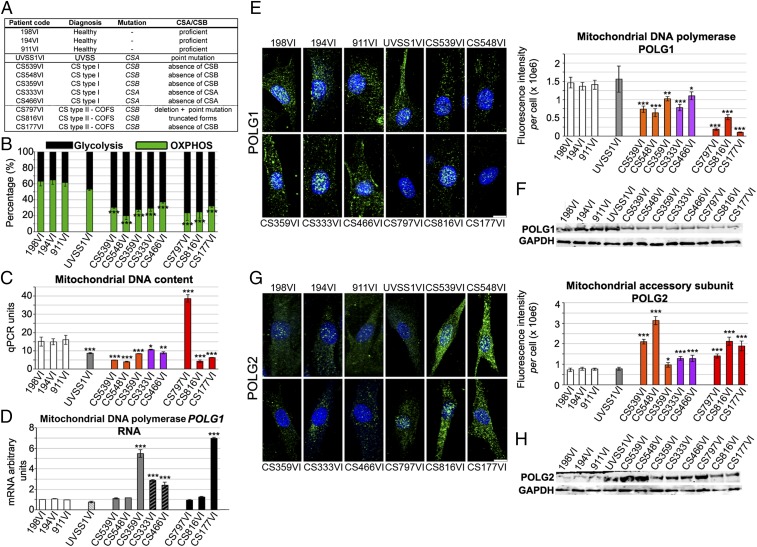
We assessed critical mitochondrial parameters in primary skin fibroblasts derived from control individuals and from patients with UVSS1, CS-I, or CS-II (described in Fig. 1A). The mitochondrial membrane potential, measured with fluorescent Tetramethylrhodamine Ethylamine (TMRE), and which expresses the capacity for generation of ATP, exhibited similar levels in control, UVSS1VI, and most CS cells (Fig. S1 A and B). In agreement with TMRE experiments, total ATP levels per cell were relatively stable (Fig. S1C). Conversely, treatment with oligomycin, an inhibitor of the mitochondrial respiratory chain, revealed significant glycolytic shifts (increased glycolysis/OXPHOS ratio) in all CS cells compared with controls, consistent with a previous report (23), but not in UVSS1VI cells, suggesting that the metabolic shift was CS-specific (Fig. 1B).
Fig. 1.
Glycolytic shift and altered mitochondrial DNA polymerase in fibroblasts from CS patients. (A) Characteristics of primary skin fibroblasts from three individuals without CS (wild type, healthy), and nine individuals with UVSS or CS. (B) Fractions of glycolysis (oligomycin-insensitive, black) and OXPHOS (oligomycin-sensitive, green) activities. (C) Quantitative PCR measurements of mtDNA content (12S region). (D) RT-qPCR shows mitochondrial POLG1 mRNA expression. (E) (Left) 3D reconstructions of cells immunostained for POLG1 (green), counterstained with Hoechst (blue). (Right) Quantification of POLG1 fluorescence intensity per cell. (F) Immunoblot of POLG1 and reference protein GAPDH. (G) (Left) 3D reconstructions of cells immunostained for POLG2 (green), counterstained with Hoechst, (blue). (Right) Quantification of POLG2 fluorescence intensity per cell. (H) Immunoblot of POLG2 and GAPDH. Results are representative of three independent experiments. (Scale bar: 10 μm.) Immunofluorescence, n = 30 cells from three independent experiments, mean ± SEM. Immunoblots and RT-qPCR, n = 3 independent experiments mean ± SD *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 versus control 198VI cells, based on the unpaired t test.
Quantitative PCR [qPCR, evaluated in a late replicated region (12S), Fig. 1C, and in an early replicated region of the mtDNA, Fig. S2C, black columns], and Picogreen staining (Fig. S2A) showed that the mtDNA content was altered in CS cells, but it was not correlated with the metabolic shift. Indeed, although copy number was low in many cases where there was a metabolic shift, the CS797VI cells had elevated mtDNA copy number despite the shift, and UVSS1VI cells had a decrease in copy number with no shift.
mtDNA is unique as it consists of an essentially double-stranded molecule of 16.6 kbp in humans, and a ∼650 nt DNA, the 7S (Fig. S2B), which forms a triple-stranded structure downstream of the main origin of replication, the D-loop (24). The content of 7S DNA has been reported to be highly variable compared with full-length mtDNA and it appears to be linked to mtDNA replication and maintenance. CS cells exhibited an imbalance between the 7S/mtDNA ratio (25) and mtDNA levels compared with controls (elevated 7S/mtDNA ratio when the mtDNA content was low and vice versa for the CS797VI line, Fig. S2C). The d-loop has been proposed to play a role in recruiting POLG2 (26), the accessory protein of the mtDNA replication complex (27), and in turn the 7S DNA might be stabilized by POLG2 after the protein is recruited to the D-loop.
We next examined POLG1 and POLG2 levels and found that despite variable mRNA levels (Fig. 1D), the POLG1 protein content was reduced (range: 22–93% reductions) in all CS cells compared with controls and UVSS cells (Fig. 1 E and F). In particular, the lowest POLG1 protein levels were detected in CS-II cells, which suggested an association between POLG1 amount and disease severity. CS cells, but not UVSS1VI cells, displayed increased levels of POLG2 (range: 28–400% above control levels; Fig. 1 G and H). Thus, the POLG1/POLG2 stoichiometry was severely altered in CS cells (Fig. S2D). These data showed quantitative modifications in the replicative polymerase complex in CS cells, but not in the UVSS1VI fibroblasts.
POLG1 Depletion Depended on CSB Impairment.
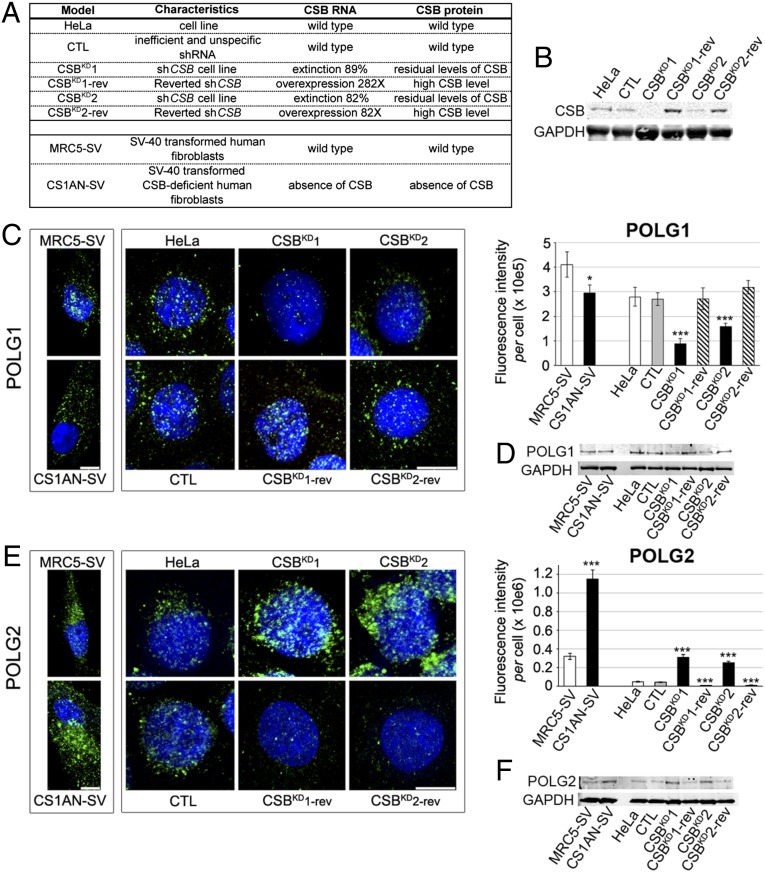
We next determined whether the observed decreased POLG1 content and increased POLG2 levels were consequences of CSB impairment. We used two cellular models: one was a stable, CSB knock-down (two independent knock-downs: CSBKD1 and CSBKD2) in a HeLa cell line, which were compared with a control counterpart (CTL), generated as described (28, 29), and their revertants (CSBKD1-rev and CSBKD2-rev, which expressed higher CSB levels than controls); the second was a CSB-deficient, SV40-transformed cell line (CS1AN-SV), which was compared with normal (CSB-proficient) fibroblasts transformed with control SV40 (MRC5-SV) (Fig. 2 A and B).
Fig. 2.
Alterations in the POLG complex are CSB-dependent. (A) Characteristics of the cellular models used in this study, including CSB status. (B) Immunoblot of CSB and GAPDH loading control in CSBKD and CSBKD-rev HeLa cells. The CSB-silenced clones (CSBKD1 and CSBKD2) display silencing efficiencies of 89% and 82%, respectively, and 3–15% of residual protein, compared with control cells. Controls carried a plasmid with an inefficient, nonspecific short hairpin RNA (shRNA) sequence (29). Due to their episomal features, the silencing plasmids (pEBVsiRNA) disappeared after selection pressure withdrawal. Under these conditions, revertant cells, CSBKD1-rev and CSBKD2-rev, expressed remarkably higher CSB mRNA and protein compared with control cells. (C) (Left) 3D reconstructions of cells immunostained for POLG1 (green), and counterstained with Hoechst (blue). (Right) Quantification of POLG1 fluorescence intensity per cell. (D) Immunoblots of POLG1 and GAPDH loading control. (E) (Left) 3D reconstructions of cells immunostained for POLG2 (green), and counterstained with Hoechst (nuclei, blue). (Right) Quantification of POLG2 fluorescence intensity per cell. (F) Immunoblot of POLG2 and GAPDH loading control. In C and E (Left), immortalized fibroblasts and HeLa cell models are distinctly framed. (Scale bar: 10 μm.) Immunofluorescence, n = 30 cells from three independent experiments, mean ± SEM. Immunoblots, n = 3 independent experiments. ***P ≤ 0.001, based on unpaired t test comparisons of CS1AN-SV versus MRC5-SV or CTL, CSBKD1, CSBKD2, and revertants versus HeLa.
CSB silencing resulted in 68% and 43% reductions in POLG1 immunofluorescence (CSBKD1 and CSBKD2 cells, respectively; Fig. 2C), confirmed by Western blots (Fig. 2D), to a similar extent as that observed in patient-derived CS-I and CS-II fibroblasts. Moreover, the CSB-deficient line (CS1AN-SV) displayed a 28% reduction in POLG1 compared with MRC5-SV controls. Consistent with patient-derived CS cells, both CSB-silencing and CSB-deficient cells also resulted in five- to sixfold increases in POLG2 protein expression (Fig. 2 E and F). Revertant cells that expressed high levels of CSB showed POLG1 and POLG2 contents similar to control levels, and they sometimes exceeded control values. Thus, dramatic alterations observed in the mitochondrial DNA replication complex in patient-derived CS cells were directly linked to CSB deficiency.
Mitochondrial Serine Proteases Accumulated with CSB Deficiency.
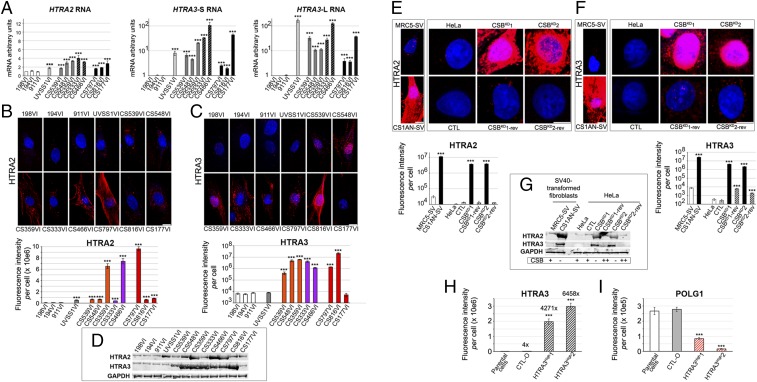
POLG1 depletion in the presence of regular or high levels of POLG1 mRNA suggested that the homeostasis of this protein was altered in CS cells. The expression of genes coding for relevant mitochondrial proteases (LONP1, AGF3L2, and SPG7, which code for Lon protease, an AAA-protease, and paraplegin, respectively) remained unchanged or increased by twofold in CS cells compared with controls and UVSS1VI cells (Fig. S3A). However, the mRNA levels of a mitochondrial stress factor, HTRA2, which codes for a serine protease located in the organelle intermembrane space (30), were 2- to 4-fold higher in patient-derived cells compared with controls (Fig. 3A). Another member of the HTRA serine protease family (31), HTRA3, is a pregnancy-related mitochondrial stress-response factor (32). HTRA3 has multiple potential targets, including mitochondrial POLG1, according to Cytoscape data integration [Prion Disease Database (33), based on ref. 34 for HTRA3; Fig. S3B]. Remarkably, in patient-derived cells the mRNA levels of HTRA3-S/L (Short/Long variants; ref. 35) were 2- to 165-fold higher compared with controls (Fig. 3A). Immunofluorescence confirmed dramatically higher levels of HTRA2 (5- to 122-fold) and HTRA3 (>60-fold) in most CS cells compared with controls (Fig. 3 B and C); these results were corroborated by Western blots (Fig. 3D). In UVSS1VI cells, HTRA2 levels were a few-fold higher than in controls, whereas HTRA3 levels were not elevated, which suggested a tight link between HTRA3 and CS phenotype. It was noteworthy that POLG1 and POLG2 levels were not altered in UVSS1VI cells, resulting in an inverse correlation between HTRA3 and POLG1 levels (with the exception of CS177VI cells).
Fig. 3.
CSB-dependent accumulation of serine proteases in patient-derived CS cells. (A) Quantitative RT-qPCR of serine protease mRNAs: HTRA2 (also called OMI), HTRA3-S (y = log scale), and HTRA3-L (y= log scale) in patient-derived cells. (B and C) (Upper) 3D reconstructions of cells immunostained for HTRA2 (red; B) or HTRA3 (red; C), and counterstained with Hoechst (blue). (Lower) Quantifications of HTRA2 or HTRA3 (y= log scale) fluorescence intensity per cell. (D) Immunoblot of HTRA2, HTRA3, and GAPDH loading control from whole-cell extracts. The anti-HTRA3 antibody recognizes both S- and L- forms. Note that large HTRA2 and HTRA3 protein elevations were not necessarily correlated with elevated mRNA levels, which suggested the protein was stabilized posttranslationally in patient-derived cells, which may be linked to the regulation of HTRA2 and HTRA3 auto-proteolysis (31). Serine protease dynamics might explain the apparently unaffected HTRA3 levels in CS177VI cells, despite the high mRNA levels and strongly reduced POLG1. In UVSS1VI cells, HTRA2, but not HTRA3, levels were a few-fold higher than in controls. (E and F) (Upper) 3D reconstructions of cells stained for HTRA2 (red; E) or HTRA3 (red; F), and counterstained with Hoechst (blue). (Lower) Quantification of HTRA2 (y = log scale) or HTRA3 (y = log scale) fluorescence intensity per cell. Immortalized fibroblasts and HeLa cell models are distinctly framed. (G) Immunoblots of HTRA2, HTRA3, and GAPDH from whole-cell extracts. (H–I) Quantification of immunofluorescence in HeLa parental cells, or cells transfected with an empty vector (CTL-O) or a vector coding for HTRA3 (HTRA3High1 for pBD3188, and HTRA3High2 for pBD3189). (H and I) HTRA3 (fold increase compared with HeLa is shown on top of each column; H), and POLG1 (I). (A–D) Primary fibroblasts. (E–I) CSB cellular models. (Scale bar: 10 μm.) Immunofluorescence, n = 30 cells from three independent experiments, mean ± SEM. Immunoblots and RT-qPCR tests, n = 3 independent experiments, mean ± SD ***P ≤ 0.001; based on unpaired t test comparisons to respective controls. Immunoblots are representative of three independent experiments.
We further tested the idea that reduced POLG1 levels in CS cells were due to elevated expression of HTRA3, and possibly HTRA2, as a consequence of CSB impairment. For this, we shut down CSB expression, and examined the HTRA2/3 protein levels in CSB cellular models. We observed that, in CSBKD and transformed CS1AN-SV cells, HTRA2 and HTRA3 immunofluorescence intensities were ≥103-fold higher than in the respective CSB-proficient controls (Fig. 3 E and F and Fig. S3C). Elevated HTRA2 and HTRA3 protein levels were confirmed by Western blots (Fig. 3G). Moreover, in CSBKD-rev cells, we observed a dramatic drop in both HTRA2 and HTRA3 protein contents compared with their CSBKD counterparts (Fig. 3 E–G). These data suggested a tight link between CSB and HTRA2/3 expression.
The effect of HTRA2 and HTRA3 proteases on POLG1 appeared selective (see below). Moreover, overexpression of HTRA3 in transfected HeLa cells (Fig. 3H) resulted in severe depletion of POLG1 immunofluorescence signal (Fig. 3I). The selectivity of POLG1 depletion in CS cells was confirmed by the observation that the levels of other proteins such as cellular F-actin (Fig. S4A), mitochondrial POLG2 (see above, Fig. 1 G and H), DRP1, a mitochondrial fission protein located in the membrane of the organelle, TFAM, the mtDNA transcription and maintenance factor, and Twinkle helicase located in nucleoids (21) did not systematically decrease in CS cells (Fig. S4 B–D). We nevertheless observed that the nucleoid protein mtSSB decreased by 87–98% in CS and also in UVSS cells (Fig. S4E), confirmed by Western blots (Fig. S4F). Thus, differently from the reduction of POLG1 content, depletion of mtSSB was not specific of Cockayne syndrome cells, suggesting that this alteration is not exclusively linked to CS.
Serine Protease Inhibition Restored POLG1 Levels in CS Fibroblasts.
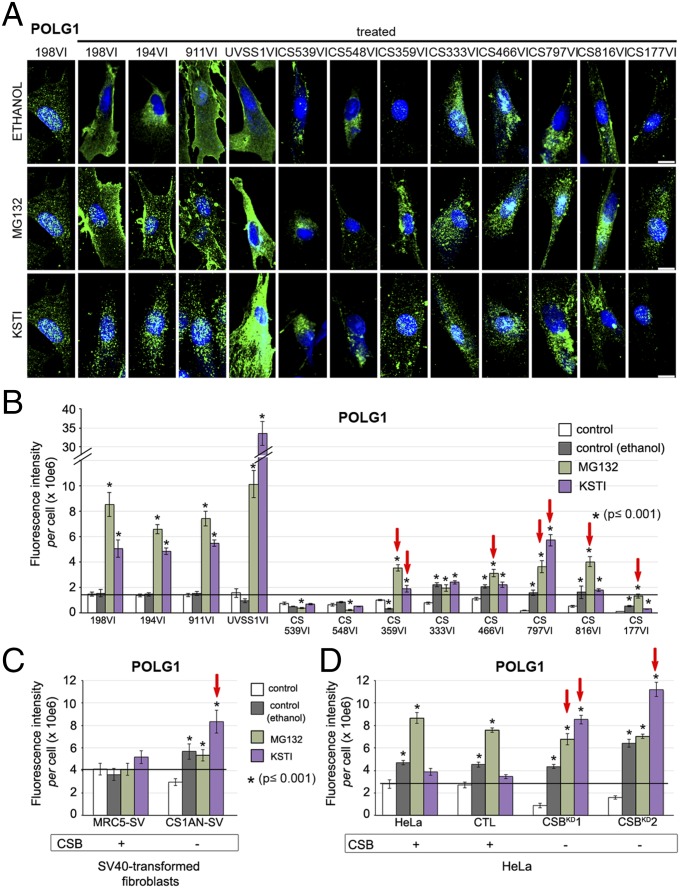
We then determined whether treatment with serine protease inhibitors would increase POLG1 protein levels in patient-derived CS cells. We found that POLG1 levels were controlled by serine proteases in normal cells. Indeed, in control fibroblasts MG132, a proteasome inhibitor which targets cysteine and serine proteases (36) and serpin Kunitz soybean trypsin inhibitor (KSTI), which specifically targets serine proteases (37) increased POLG1 immunofluorescence intensities by 5-fold and 3.6-fold, respectively, compared with fibroblasts grown in the presence of the inhibitor diluent alone (ethanol) (Fig. 4 A and B). Similarly, UVSS1VI cells showed 6- to 34-fold increases in POLG1 signal intensities in the presence of the protease inhibitors. The results were more diverse among CS mutants: two of eight failed to show any significant response (CS539VI and CS548VI), two responded to ethanol, an inducer of autophagy, which requires the activity of proteases (38), (CS333VI and CS466VI), and four displayed a protease inhibitor-based elevation of POLG1. The two cases with no response suggested that either HTRA2/3 targeting by the inhibitor was incomplete or additional factors were involved in maintaining POLG1 levels. Thus, protease inhibitors promoted a large increase of POLG1 in controls, whereas in CS cells that exhibit huge amounts of HTRA2/3 restored, or exceeded by a few-fold, control POLG1 levels, at the doses tested. The strong increase of POLG1 content upon treatment with protease inhibitors in UVSS cells, which express high levels of HTRA2 but not HTRA3 (see above, Fig. 3 B and C), suggests that POLG1 levels are largely under control of HTRA3, as aforementioned (Fig. 3I).
Fig. 4.
Serine protease inhibitors restore POLG1 levels in patient-derived cells. (A) 3D reconstructions of primary fibroblasts immunostained for POLG1 (green), and counterstained with Hoechst (blue). Cells were grown in the presence of ethanol (vehicle control), MG132, or KSTI; sample 198VI is an untreated control (the labeling of the other wild type cells are shown in Fig. 1E). (B) Quantification of POLG1 fluorescence intensity per cell. The horizontal bar indicates the level of untreated controls. Red arrows indicate increase of POLG1 at level of at least healthy controls, and exceeding ethanol treatment. Quantifications of POLG1 fluorescence intensity per cell, in CSB-deficient (CS1ANSV; C) and CSB-silenced (CSBKD; D) cellular models and respective controls. The corresponding 3D reconstructions of immunostained cells are shown in Fig. S3D. The horizontal bars indicate the level of untreated controls. Red arrows indicate increase of POLG1 at level of at least controls, and exceeding ethanol treatment. (Scale bar: 10 μm.) Immunofluorescence, n = 30 cells from three independent experiments, mean ± SEM; *P ≤ 0.001; based on unpaired t test for comparisons to respective controls.
Consistent with these findings, POLG1 increased in response to KSTI in all CSB-impaired cell lines (CS1AN immortalized fibroblasts, Fig. 4C; CSBKD1 and CSBKD2, Fig. 4D, and Fig. S3D). CSBKD1 was also sensitive to the MG132 treatment, whereas CSBKD2 and CS1AN were not more sensitive than ethanol alone. Conversely, MG132 did not appear to affect MRC5-SV and CS1AN-SV transformed cells, as it did in primary cells, whereas it affected HeLa cells, independently of CSB.
Taken together, these data suggested that inhibiting serine proteases increases POLG1 levels, and CSB-dependent POLG1 depletion can be rescued, at least in some cases. These data were consistent with the notion that CSB depletion promoted accumulation of serine proteases HTRA2 and HTRA3, which affected POLG1 protein levels.
Nitroso-Redox Imbalance in CS Fibroblasts.
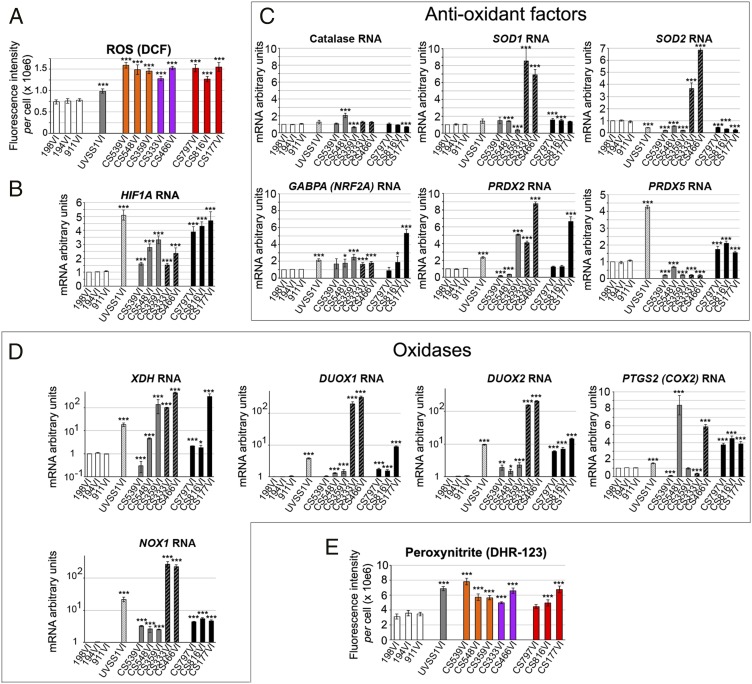
Next, we elucidated the mechanism(s) that gave rise to CSA/CSB effects on HTRA2 and HTRA3. HTRA2 expression increases in tissues undergoing oxidative stress, and CS cells are hypersensitive to oxidative stress (8, 23). All CS cells exhibited higher levels (1.6- to 2-fold) of ROS, evaluated with the fluorescent probe DCFDA, than those observed in controls and UVSS1VI cells (Fig. 5A and Fig. S5A), compatibly with a previous report (9). Moreover, all patient-derived cells displayed very high hypoxia-inducible factor-1 (HIF1α) mRNA levels, which are induced by high ROS content (39), compared with controls (Fig. 5B).
Fig. 5.
Nitroso-redox stress increases in CS cells. (A) Quantification of DCF fluorescence intensity per cell. Cells were treated with DCFDA to detect ROS levels. Representative immunofluorescence labelings are shown in Fig. S5A. (B–D) Quantitative RT-qPCR show mRNA expression levels of HIF1α (B); antioxidant factors catalase and SOD1, primarily mitochondrial antioxidant factors SOD2, GABPA/NRF2A, and peroxynitrite reductases PRDX2 and PRDX5 (C); and ROS-producing oxidases NOX1, XDH, DUOX1, DUOX2, and COX2, (y = log scales with the exception of COX2) (D). (E) Quantification of DHR123 fluorescence intensity per cell. Cells were treated with DHR123 to detect peroxynitrite levels. Representative immunofluorescence labelings are shown in Fig. S5D. Immunofluorescence, n = 30 cells from three independent experiments, mean ± SEM. RT-qPCR and immunoblots, n = 3 independent experiments, mean ± SD *P ≤ 0.05, ***P ≤ 0.001 versus 198VI, based on the unpaired t test.
In turn, HIF1α is reported to stimulate the expression of several stress-related factors (40). Because HIF1α is a transcriptional complex that affects RNA levels of target proteins, we first conducted RT-qPCR analyses of several oxidant and anti-oxidant factors, rather than assessment of protein turnover, as it was the case for POLG1. Compared with controls, UVSS1VI cells displayed similar or lower mRNA levels of the antioxidant enzymes catalase and superoxide dismutases [SOD1 (cytoplasmic) and SOD2 (mitochondrial)] (Fig. 5C). UVSS1VI cells also showed increased levels of primarily mitochondrial antioxidant factors including GABPA/NRF2A, peroxynitrite reductases peroxiredoxin-2 (PRDX2) and particularly, peroxiredoxin-5 (PRDX5) but higher levels of ROS-producing oxidases, namely NADPH oxidase 1 (NOX1), xanthine oxidase/dehydrogenase (XDH), dual oxidase 1 (DUOX1) and 2 (DUOX2) and, to a lesser extent, cyclooxygenase 2 (COX2), compared with controls (Fig. 5D). Altogether, the expression levels of pro- and anti-oxidant factors suggested a robust antioxidant response to counteract constitutively high levels of ROS-producing factors, which might account for the moderate ROS increase in CSA-mutated UVSS cells compared with controls (summarized in Fig. S5B).
Type-I CSA cells, which also displayed high levels of both oxidases (NOX1, XDH, DUOX1, DUOX2, and occasionally COX2, Fig. 5D) and antioxidants (SOD1, SOD2, PRDX2, and to a minor extent, GABPA, Fig. 5C), had particularly low levels of PRDX5, suggesting that PRDX5 plays an important role in the control of ROS levels. Type-I CSB cells, which displayed very high levels of one or more of the NOX1, XDH, DUOX1, DUOX2, and COX2 oxidases, expressed normal or low levels of the main anti-oxidant factors PRDX5, SOD2, and PRDX2 (for the latter, except in the CS539VI cells). Thus, imbalance in redox homeostasis may be responsible for the strong constitutive oxidative stress detected in type-I CSB cells. Importantly, although type-II CSB cells may have a more robust antioxidant profile than type-I CSB cells (particularly because they have higher levels of PRDX5), they exhibited very high levels of all tested oxidases; this resulted in high oxidative stress, as shown above. These data have revealed distinct oxidant/antioxidant profiles in CSA, CBS-I, and CSB-II cells (Fig. S5B), which regulated ROS levels, suggesting that a PRDX5-dependent antioxidant response plays a relevant role in counteracting extensive oxidase activity.
High levels of ROS can react with nitric oxide (NO), which promotes the formation of peroxynitrite, a powerful oxidant and nitration agent (41). UVSS1VI and CS cells expressed high levels of a key NO-producing enzyme, the inducible NO synthase (iNOS), and the genes involved in the biosynthesis of the NO-cofactor BH4 (dihydrofolate reductase DHFR, methylenetetrahydrofolate reductase MTHFR, GTP cyclohydrolase GCH1) (Fig. S5C). Consistent with this finding, the fluorescent probe dihydrorhodamine 123 (DHR123) showed accumulation of peroxynitrite in the UVSS1VI and CS cells compared with normal fibroblasts (Fig. 5E and Fig. S5D).
ROS and peroxynitrite also accumulated in CSBKD and CS1AN-SV cells, and the levels were lowered with CSB restoration in CSBKD-rev cells (Fig. S6), indicating that they are linked to CSB.
ROS and Peroxynitrite Scavenging Rescues Mitochondrial Dysfunction in CS Fibroblasts.
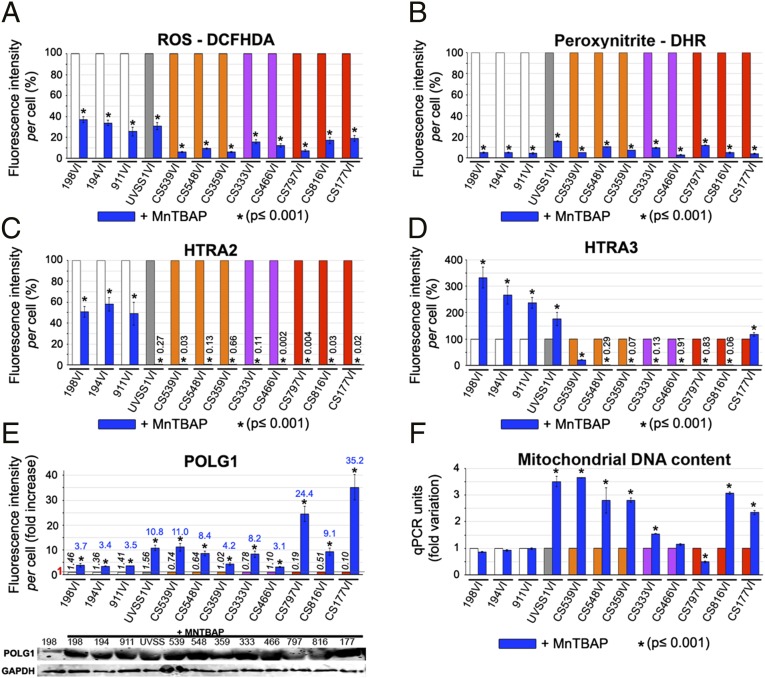
We reasoned that, if ROS and peroxynitrite induced serine protease accumulation, which resulted in POLG1 depletion, then the original parameters might be restored by treating CS fibroblasts with ROS and peroxynitrite scavengers. Manganese(III)tetrakis (4-benzoic acid) porphyrin (MnTBAP) is a synthetic metalloporphyrin that mimics superoxide dismutase and scavenges ROS and peroxynitrite. MnTBAP treatment reduced ROS to a greater extent (80–95%) in CS cells than in controls and UVSS1VI fibroblasts (reduction by two thirds) (Fig. 6A and Fig. S7). Similarly, MnTBAP greatly reduced the levels of peroxynitrite in all control and patient-derived cells (Fig. 6B and Fig. S7). Importantly, a reduction in nitro-oxidative levels following MnTBAP treatment was accompanied by a reduction of HTRA2 expression in all samples, but the reduction was most dramatic in UVSS and in particular CS cells (Fig. 6C and Fig. S7), which originally exhibited elevated levels of this serine protease.
Fig. 6.
Scavenging nitroso-redox stress normalizes mitochondrial parameters in patient-derived cells. Primary fibroblasts were treated with 5 μM MnTBAP for 24h (blue columns) or untreated [white columns = controls, light gray column = UVSS, orange columns = CS-I (CSB), purple columns = CS-I (CSA), red columns = CS-II]. Values for cells treated with MnTBAP are expressed as percent of the respective untreated control in A–D. (A–D) Fluorescence intensities per cell for DCFHDA (A), DHR123 (B), HTRA2 (C), and HTRA3 (D) (low percent values are indicated at the position of the column); see also Fig. S8A. (E) (Upper) Fluorescence intensities per cell for POLG1 (expressed as fold increase, with value indicated in blue on top of the MnTBAP column); the mean absolute value (×106) is indicated in black italic on top of each control column; value 1 is indicated in red on the y axis, and a straight line corresponding to this value is drawn in the graph. (Lower) Immunoblots of POLG1 and GADPH, representative of three independent experiments. (F) qPCR results show mtDNA content (12S); see also Fig. S8B. Representative immunostainings of A–E are shown in Fig. S7.
Strikingly, MnTBAP increased the levels of HTRA3 in control and in UVSS1VI cells, but dramatically reduced the levels in CS cells (Fig. 6D and Fig. S7), with the exception of the CS177VI cell line, thereby restoring HTRA3 to the levels observed in untreated controls (Fig. S8A). These data showed that overexpression of HTRA3 in CSA- or CSB-impaired cells was promoted by high ROS and peroxynitrite levels. Conversely, the nitro-oxidative levels observed in CSA- and CSB-proficient fibroblasts limited HTRA3 expression.
Furthermore, MnTBAP treatment increased POLG1 levels in normal fibroblasts and, to a greater extent, in UVSS1VI and CS cells (Fig. 6E and Fig. S7). The difference indicated that the patient-derived cells were particularly sensitive to ROS and peroxynitrite levels in the context of POLG1 regulation. This finding confirmed our proposed model. Interestingly, in the presence of MnTBAP, the mtDNA content was strongly altered in patient-derived CS cells (Fig. 6F), reaching the level observed in untreated controls (Fig. S8B). This was the case independently of the original alteration, whereas the treatment did not change the mtDNA content in control fibroblasts.
We also observed that the glycolytic shift reported in CS cells was attenuated after treatment with MnTBAP (Fig. S8C). Thus, MnTBAP increased the fraction of ATP produced by mitochondria, and partially restored the OXPHOS values. Conversely, the treatment essentially did not alter the glycolytic/OXPHOS ratio in controls or UVSS1VI cells. In the presence of MnTBAP, CS cells maintained relatively high levels of ATP, between 20% and 50% of untreated cells; in contrast, ATP levels in UVSS1VI and control fibroblasts were about 75% lower than in untreated cells (Fig. S8D). These data showed that scavenging nitro-oxidative molecules had a restoring effect in cells with altered nitroso-redox balance, but may severely alter key parameters in cells with a normal nitroso-redox balance.
Discussion
There is growing evidence that the CSA and CSB-dependent DNA repair defect, which also characterizes the nonprogeroid UVSS syndrome, is not primarily responsible for the precocious aging phenotype in Cockayne syndrome. CSA and CSB are also active in mitochondria, and their impairment leads to defective autophagy and mitophagy (16, 17, 42), although it is not clear whether this is also the case in UVSS cells.
Using primary CS, UVSS and healthy control cells, this study showed that CS-specific defects rely on the overexpression of HTRA3, a serine protease known for its role in pregnancy-related mitochondrial stress and cancer (31). HTRA3 selectively reduced the levels of POLG1, which in turn affected mitochondrial function. Inhibition of serine proteases rescued HTRA3 and POLG1 levels in several primary patient-derived CS cells. Overexpression of HTRA3 was promoted by unbalanced nitroso-oxidative stress in CS cells, and scavenging of the reactive molecules rescued HTRA3 and POLG1 levels, and mitochondrial OXPHOS in CS cells.
POLG1 and POLG2 are essential for mtDNA replication. Mutations in these proteins have been associated with mitochondrial diseases, which largely occur in childhood, and result in reduced lifespan (43, 44). Moreover, alterations in POLG1 proof-reading activity have been associated with precocious aging (45). We showed that the mtDNA replication machinery was dramatically altered in CS cells, with decreased levels of POLG1 and increased levels of POLG2. The effect on POLG2 could be due to a perturbation of the system in POLG1-depleted cells. Moreover other proteins involved in mtDNA replication were reduced in most (Twinkle) or all (mtSSB) CS cells, as well as in UVSS cells. Although these alterations were not CS-specific, in CS cells the depletion of Twinkle and mtSSB cumulated with the imbalance of the POLG1/POLG2 complex, possibly enhancing the DNA replication defect in these cells. Interestingly, these data suggest that mtSSB is far less needed for mtDNA maintenance than hitherto believed (46, 47), and this point may need to be developed in future studies. We showed that mtDNA maintenance was altered in CS cells: mtDNA content was either dramatically reduced or increased in these cells, and affected the 7S/mtDNA ratio. The function of 7S DNA has not been clearly established, although it seems implicated in the organization of mtDNA in nucleoids (26), and it may be linked to age-related mtDNA rearrangements (24). Taken together, our data indicated that the mtDNA replication machinery was affected by CSB deficiencies. Thus, both POLG1 mutations (known effect) and altered POLG1/POLG2 levels (this study) affected mitochondrial function, and may lead to premature aging.
POLG1 is a potential target for serine proteases. In patient-derived CS fibroblasts, but not in normal and UVSS cells, low POLG1 content was associated with dramatic increases in HTRA3 serine protease levels, which depended on impaired CSB or CSA. It is possible that either HTRA3 may act alone, or HTRA3 and HTRA2 (also increased by CSA/CSB deficiency) may act additively in lowering POLG1 expression as in the case for CS177VI cells. A direct effect of HTRA3 on POLG1 levels was also observed after stable overexpression of this serine protease in HeLa cells. HTRA2 is a powerful regulator of autophagy, which may explain the increased autophagy reported in CS-immortalized cells (48).
We do not exclude that HTRA3 targets other proteins than POLG1, and that these proteins may also affect the CS phenotype. In this context, a broader analysis of HTRA3 targets and their relevance to the CS phenotype should be performed in the future.
How do CSA/CSB mutations affect the expression of serine proteases (and possibly other targets), which eventually alter POLG1 and POLG2 levels, and finally lead to mitochondrial dysfunction? The HTRA family of serine proteases is implicated in the cellular response to stresses, like heat shock, inflammation, and cancer (49). These stresses up-regulate and/or affect the mitochondrial localization of HTRA2 and HTRA3. Mitochondria produce ROS and reactive nitrogen species (RNS) which, at high levels, can generate cell-damaging oxidative and nitrosative stress (50). We reported a CSB-dependent increase in ROS/RNS, associated with an imbalance in the expression of oxidases versus antioxidant factors in CS cells, and in particular low levels of mitochondrial PRDX5, which conversely, appeared to protect UVSS cells. Thus, we showed that a CSA/CSB-dependent nitroso-redox imbalance led to increase serine proteases and depletion of POLG1. Treatment with the ROS/RNS scavenger MnTBAP reduced ROS to the greatest extent in patient-derived CS cells, which ultimately displayed lower ROS levels than healthy controls. MnTBAP also dramatically reduced RNS, HTRA3, and HTRA2 in patient-derived cells to levels comparable to those in healthy controls, and consequently, increased the POLG1 content and the mitochondrial respiration fraction in all CS primary fibroblasts.
CSA-mutated cells were indistinguishable from CSB-mutated cells, suggesting that CSA and CSB are components of the same complex that affect serine protease levels. This hypothesis is consistent with clinical observations, because patients with CSA-mutated type-I CS are clinically indistinguishable from those with CSB-mutated type-I CS (2).
In CS cells, but not in UVSS cells, altered mtDNA replication machinery was associated with reduced OXPHOS, despite constant ATP production. Impaired mitochondrial function and alterations in metabolic pathways can maintain regular ATP levels, but this adaptation may result in pathology (51) and premature aging (52).
In summary, by altering the expression of oxidases and antioxidants, CSB/CSA impairments promoted overproduction of serine proteases, which in turn, depleted POLG1, altered the mtDNA replication machinery, and reduced mitochondrial OXPHOS (Fig. 7). CSA and CSB are involved in transcription and possibly chromatin remodeling (10, 12); thus, impairments in these processes may affect the expression of several other factors. Indeed, proteins other than POLG1 could be affected by serine protease overexpression, and factors other than serine proteases may be affected by CSB impairments.
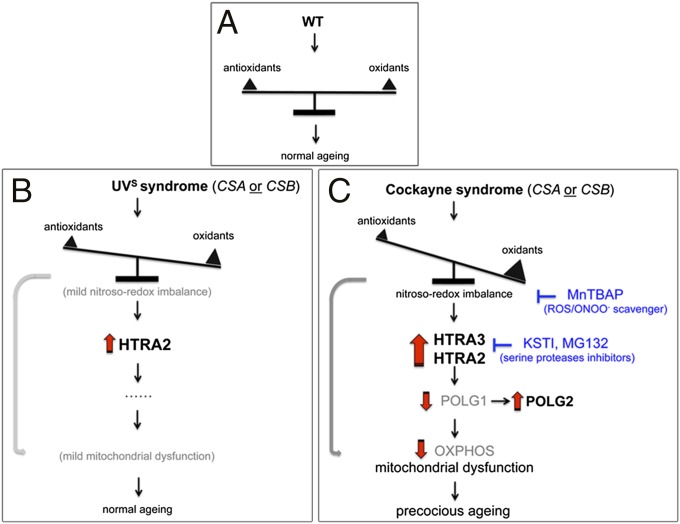
Fig. 7.
Model for alterations in CS cells. (A) Regular nitroso-redox, HTRA3 and POLG1 levels, and normal mitochondrial function in control (WT) cells. (B) UVSS cells display only a moderately altered nitroso-redox imbalance and thereby increased expression of HTRA2, but not HTRA3. Therefore, these cells show physiological values for POLG1, POLG2, and OXPHOS. Only limited mitochondrial dysfunctions occurred, due to presence of some oxidative and nitrosative stress (gray arrow), which result in mild clinical features. (C) In CS cells, CSA/CSB-deficiency causes deregulation of antioxidant defense, oxidases, and NOS. These changes result in ROS and peroxynitrite (ONOO−) stress, which promotes accumulation of mitochondrial serine proteases, HTRA2 and HTRA3, which leads to POLG1 depletion. In relation to the impaired replicative complex, POLG2 expression increases. These alterations lead to a glycolysis shift, and additional mitochondrial alterations (e.g., mtDNA content, 7S/mtDNA ratio). Moreover, the altered nitroso-redox balance also affects mitochondrial function. Dysfunctional mitochondria functioning, in turn, accentuates the oxidative stress, which may cause more mitochondrial and cellular damage (gray arrow). Acting at different levels of this pathway, serine protease inhibitors and ROS/RNS scavengers block the cascade of events, restoring POLG1/POLG2 and mitochondrial function to normal levels. These effects are specific to CS cells. Protein overexpression is indicated in bold, protein depletion or reduced activity in light gray.
Our data revealed a novel pathway that altered mitochondrial, and thus cellular function in progeroid-like CS. The finding that these effects could be reversed in patient-derived cells by targeting two independent pathways has opened novel possibilities for treatments, which are presently lacking for patients with CS. These data also suggested the possibility that the abnormalities described here occurred at a slower pace in normal cells; consequently, they may be linked to physiological aging. Thus, further exploration may open a window to prevention therapy.
Methods
Patient Cells.
Patient fibroblasts were derived from skin biopsies excised from unexposed body sites by dermatologists. Patients provided informed consent to receive diagnosis and genetic analysis. The French Agency of Biomedicine (Paris) (Arrêté n°2001/904 and Ref: AG08-0321 GEN of 27/09/2008; www.agence-biomedecine.fr/Genetique and the European Commission “Geneskin: Genetics of human genodermatosis” (Brussels, 2008) approved this study.
Cells and Culture Conditions.
Human primary skin fibroblasts from normal individuals, and UVSS and CS patients, as indicated in Fig. 1A, MRC5-SV and CS1AN-SV (∆CSB) SV40-transformed cell lines (a generous gift from A. R. Lehmann, Sussex University, Sussex, United Kingdom), HeLa, silenced CSBKD, and revertant CSBKD-rev, or stably overexpressing HTRA3 cells were grown in DMEM supplemented with 10% FBS and Pen-Strep, in 20% O2/5% CO2 at 37 °C. The medium was supplemented with 125 µg/mL hygromycin B for HeLa CSBKD cells, and 0.4 µg/mL puromycin for HTRA3High cells. Experiments with revertant cells, which displayed a phenotype similar to control cells, were completed within 7 d after recovery of CSB levels (selection pressure withdrawal for at least 21 d). When indicated, cells were treated for 5 h with 5 μM MG132 (Z-Leu-Leu-Leu-al dissolved in ethanol, Sigma-Aldrich) or 100 μg/mL KSTI (Kunitz Soybean Trypsin Inhibitor dissolved in water, Sigma-Aldrich) or ethanol, or for 24 h with 100 μM MnTBAP (Merck Millipore).
Generation of CSB Silenced and Revertant HeLa Cells.
Design and cloning in pEBVsiRNA vectors carrying a hygromycin B resistance cassette as well as establishment of stable knockdown (KD) and control HeLa clones were carried out as described (28). RNAi targeted sequences for CSB (NM_000124) stretched over nucleotides 1253–1271 (pBD920) or 2655–2673 (pBD1282). HeLa cells carrying the pBD650 plasmid that expressed an inefficient shRNA sequence were used as control. HeLa cells stably overexpressing HTRA3 were constructed using two pEBVCAG-HTRA3-puro plasmids carrying the HTRA3 cDNA (Thermo Scientific, Clone ID: 5588608) with or without an endogenous 3′UTR sequence (pBD3189 and pBD3188, respectively).
Immunofluorescence, Reagents, and Antibodies.
Cells plated on glass slides were fixed with 2% (wt/vol) paraformaldehyde (PFA) and permeabilized with 0.5% Triton X-100. The glass slides were incubated in blocking buffer [BSA 5% (wt/vol) in PBS] overnight at 4 °C then 1 h at room temperature with the primary antibody, and finally with the secondary antibody and 10 μg/mL Hoechst for 1 h at room temperature. POLG1, POLG2, HTRA2, HTRA3 antibodies and Hoechst 33342 were from Sigma-Aldrich; goat anti-mouse and goat anti-rabbit Alexa Fluor 555- or Alexa Fluor 488-conjugated secondary antibodies from Life Technologies.
Detection of Peroxynitrite and ROS.
Detection and quantitation of peroxynitrite were based on oxidation of the cell-permeant nonfluorescent DHR123 (Dihydrorhodamine 123, Sigma-Aldrich) to green fluorescent rhodamine, which acts as prevalent marker of peroxynitrite. Detection and quantitation of ROS (mostly O2− and H2O2) were obtained with the cell-permeable nonfluorescent DCFH-DA (2’,7’-Dichlorodihydrofluorescin diacetate, Sigma-Aldrich) that is oxidized by ROS to form green fluorescent DCF (2’,7’-dichlorofluorescein). Detection of peroxynitrite and ROS levels was performed incubating cells with 10 μM of DHR123 (Sigma-Aldrich) or DCFH-DA (Sigma-Aldrich), respectively, for 1 h at 37 °C, in 20% O2/5% CO2.
Confocal Acquisition, 3D Reconstruction, and Quantification.
Confocal acquisitions were performed using a spinning-disk Perkin-Elmer Ultraview RS Nipkow Disk, an inverted laser-scanning confocal microscope Zeiss Axiovert 200M with an Apochromat 63×/1.4 oil objective and a Hamamatsu ORCA II ER camera (Imagopole, PFID, Institut Pasteur, Paris). Optical slices were taken each 200-nm interval along the z axis covering the whole depth of the cell, at resolution of 1.024/1.024 pixels (53). 3D reconstruction was achieved using the Imaris software (Bitplane). Scale bar = 10 μm. Fluorescence quantification was done using a single-imaging frame collection and ImageJ 1.47v software (postacquisition analysis). For each condition, 30 cells were analyzed from three independent experiments.
Mitochondrial Membrane Potential, Total ATP Steady-State Levels, and Glycolysis/OXPHOS Fraction.
Mitochondrial membrane potential was measured by treating cells with 200 nM Tetramethylrhodamine ethylamine (TMRE; Sigma) for 1 h at 37 °C. TMRE is a lipophilic cation that accumulates into mitochondria in direct proportion to the mitochondrial membrane potential. For ATP levels, 10,000 cells were treated with 10 µM oligomycin for 1 h (for glycolytic ATP) or untreated (for total ATP levels); then, cells were tested with the CellTiter-Glo Luminescent assay (Promega), according to supplier instructions.
Total RNA Extraction and RT-qPCR.
Total RNA was isolated from cells using the RNAeasy Micro kit (Qiagen), treated with DNaseI (Qiagen), then reverse-transcribed with SuperScriptIII Reverse transcriptase (Invitrogen). Real-time quantitative PCR was performed using Power Sybr Green PCR Master Mix (Applied Biosystems) and the rate of dye incorporation was monitored using the StepOne Plus RealTime PCR system (Applied Biosystems). Three biological replicates were used for each condition. Data were analyzed by StepOne Plus RT PCR software v2.1 and Microsoft excel. TBP transcript levels were used for normalization of each target (=∆CT). Real-time PCR CT values were analyzed using the 2-∆∆Ct method to calculate the fold expression (54). qPCR Primers used are listed in Table S1, which includes the relative reference.
mtDNA Content Analysis by qPCR.
The quantification of mtDNA was performed as described (55). Real-time PCR amplification was performed on 200 pg of total DNA using the StepOne Plus RealTime PCR system (Applied Biosystems) and Power Sybr Green PCR Master mix (ABI) following the supplier instruction. The region tested on mtDNA was included in the 12S gene (Human mitochondrial DNA accession number, www.mitomap.org/bin/view.pl/MITOMAP/HumanMitoSeq). When indicated, a region within 7S, and a region downstream of 7S (in the direction of replication) were also amplified, as described (25). The nuclear encoded 18S rRNA gene was used as endogenous reference. The level of mtDNA was calculated using the ΔCT of average CT of mtDNA and nDNA (ΔCT = CT nDNA - CT mtDNA) as 2∆CT (56). Primers used are listed in Table S1.
Protein Extraction and Western Blot.
Cells were lysed with lysis buffer (50 mM Tris⋅HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1 mM EDTA, and protease inhibitor mixture). Lysed cells were not centrifuged, and the whole extract was collected. The protein content was determined with the Bradford reagent (Sigma-Aldrich), and 20 µg of protein were loaded for SDS/PAGE. After blotting, Hybond ECL nitrocellulose filters were probed with primary antibodies, then with IRDye secondary antibodies. Detection was performed using Odyssey Infrared Imaging system scanner and Odyssey application software v3.0 (LI-COR Biosciences). Protein bands were quantified, normalized to GAPDH, and then normalized to control cells. Experiments were done in triplicate, and a representative Western Blot was shown.
Statistical Analysis.
The significance of differences between data were determined using Student’s t test for unpaired observations; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Supplementary Material
Acknowledgments
We thank the ‘Imagopole’ (PFID) of Institut Pasteur. This work was supported by grants from the DARRI Institut Pasteur, Agence Nationale de la Recherche (ANR 11 BSV2 025 02) (to M.R.), and from the Association Française des Myopathies (AFM, no. 2008.0547, Evry, France) (to A.S.).
Footnotes
Conflict of interest statement: A European Patent application (EP 14 305 203), based on the findings of the present manuscript, has been submitted for the therapeutic methods against progeroid-like syndromes or symptoms.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1422264112/-/DCSupplemental.
References
- 1.Laugel V, et al. Deletion of 5′ sequences of the CSB gene provides insight into the pathophysiology of Cockayne syndrome. Eur J Hum Genet. 2008;16(3):320–327. doi: 10.1038/sj.ejhg.5201991. [DOI] [PubMed] [Google Scholar]
- 2.Laugel V, et al. Mutation update for the CSB/ERCC6 and CSA/ERCC8 genes involved in Cockayne syndrome. Hum Mutat. 2010;31(2):113–126. doi: 10.1002/humu.21154. [DOI] [PubMed] [Google Scholar]
- 3.Mallery DL, et al. Molecular analysis of mutations in the CSB (ERCC6) gene in patients with Cockayne syndrome. Am J Hum Genet. 1998;62(1):77–85. doi: 10.1086/301686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9(12):958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 5.Mayne LV, Lehmann AR, Waters R. Excision repair in Cockayne syndrome. Mutat Res. 1982;106(1):179–189. doi: 10.1016/0027-5107(82)90200-7. [DOI] [PubMed] [Google Scholar]
- 6.Vélez-Cruz R, Egly JM. Cockayne syndrome group B (CSB) protein: At the crossroads of transcriptional networks. Mech Ageing Dev. 2013;134(5-6):234–242. doi: 10.1016/j.mad.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Sarasin A. UVSSA and USP7: New players regulating transcription-coupled nucleotide excision repair in human cells. Genome Med. 2012;4(5):44. doi: 10.1186/gm343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nardo T, et al. A UV-sensitive syndrome patient with a specific CSA mutation reveals separable roles for CSA in response to UV and oxidative DNA damage. Proc Natl Acad Sci USA. 2009;106(15):6209–6214. doi: 10.1073/pnas.0902113106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cleaver JE, et al. Mitochondrial reactive oxygen species are scavenged by Cockayne syndrome B protein in human fibroblasts without nuclear DNA damage. Proc Natl Acad Sci USA. 2014;111(37):13487–13492. doi: 10.1073/pnas.1414135111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Citterio E, et al. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol Cell Biol. 2000;20(20):7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le May N, et al. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol Cell. 2010;38(1):54–66. doi: 10.1016/j.molcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, et al. Regulation of the Rhp26ERCC6/CSB chromatin remodeler by a novel conserved leucine latch motif. Proc Natl Acad Sci USA. 2014;111(52):18566–18571. doi: 10.1073/pnas.1420227112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aamann MD, et al. Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J. 2010;24(7):2334–2346. doi: 10.1096/fj.09-147991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khobta A, Epe B. Repair of oxidatively generated DNA damage in Cockayne syndrome. Mech Ageing Dev. 2013;134(5-6):253–260. doi: 10.1016/j.mad.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Berquist BR, Canugovi C, Sykora P, Wilson DM, 3rd, Bohr VA. Human Cockayne syndrome B protein reciprocally communicates with mitochondrial proteins and promotes transcriptional elongation. Nucleic Acids Res. 2012;40(17):8392–8405. doi: 10.1093/nar/gks565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamenisch Y, Berneburg M. Mitochondrial CSA and CSB: Protein interactions and protection from ageing associated DNA mutations. Mech Ageing Dev. 2013;134(5-6):270–274. doi: 10.1016/j.mad.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Scheibye-Knudsen M, et al. Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. J Exp Med. 2012;209(4):855–869. doi: 10.1084/jem.20111721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harman D. Aging: A theory based on free radical and radiation chemistry. J Gerontol. 1956;11(3):298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 19.Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48(2):158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaar CE, et al. Mitochondrial and cytoplasmic ROS have opposing effects on lifespan. PLoS Genet. 2015;11(2):e1004972. doi: 10.1371/journal.pgen.1004972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 22.Park CB, Larsson NG. Mitochondrial DNA mutations in disease and aging. J Cell Biol. 2011;193(5):809–818. doi: 10.1083/jcb.201010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pascucci B, et al. An altered redox balance mediates the hypersensitivity of Cockayne syndrome primary fibroblasts to oxidative stress. Aging Cell. 2012;11(3):520–529. doi: 10.1111/j.1474-9726.2012.00815.x. [DOI] [PubMed] [Google Scholar]
- 24.Nicholls TJ, Minczuk M. In D-loop: 40 years of mitochondrial 7S DNA. Exp Gerontol. 2014;56:175–181. doi: 10.1016/j.exger.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 25.Chatre L, Ricchetti M. Prevalent coordination of mitochondrial DNA transcription and initiation of replication with the cell cycle. Nucleic Acids Res. 2013;41(5):3068–3078. doi: 10.1093/nar/gkt015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Re M, et al. The accessory subunit of mitochondrial DNA polymerase gamma determines the DNA content of mitochondrial nucleoids in human cultured cells. Nucleic Acids Res. 2009;37(17):5701–5713. doi: 10.1093/nar/gkp614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seo AY, et al. New insights into the role of mitochondria in aging: Mitochondrial dynamics and more. J Cell Sci. 2010;123(Pt 15):2533–2542. doi: 10.1242/jcs.070490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biard DS. Untangling the relationships between DNA repair pathways by silencing more than 20 DNA repair genes in human stable clones. Nucleic Acids Res. 2007;35(11):3535–3550. doi: 10.1093/nar/gkm195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biard DS, Despras E, Sarasin A, Angulo JF. Development of new EBV-based vectors for stable expression of small interfering RNA to mimick human syndromes: Application to NER gene silencing. Mol Cancer Res. 2005;3(9):519–529. doi: 10.1158/1541-7786.MCR-05-0044. [DOI] [PubMed] [Google Scholar]
- 30.Kang S, Fernandes-Alnemri T, Alnemri ES. A novel role for the mitochondrial HTRA2/OMI protease in aging. Autophagy. 2013;9(3):420–421. doi: 10.4161/auto.22920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clausen T, Kaiser M, Huber R, Ehrmann M. HTRA proteases: Regulated proteolysis in protein quality control. Nat Rev Mol Cell Biol. 2011;12(3):152–162. doi: 10.1038/nrm3065. [DOI] [PubMed] [Google Scholar]
- 32.Bowden MA, et al. High-temperature requirement factor A3 (Htra3): A novel serine protease and its potential role in ovarian function and ovarian cancers. Mol Cell Endocrinol. 2010;327(1-2):13–18. doi: 10.1016/j.mce.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 33. Gehlenborg N, et al. (2009) The Prion Disease Database: A comprehensive transcriptome resource for systems biology research in prion diseases. Database 2009:bap011. [DOI] [PMC free article] [PubMed]
- 34.Lehner B, Fraser AG. A first-draft human protein-interaction map. Genome Biol. 2004;5(9):R63. doi: 10.1186/gb-2004-5-9-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nie GY, Hampton A, Li Y, Findlay JK, Salamonsen LA. Identification and cloning of two isoforms of human high-temperature requirement factor A3 (HtrA3), characterization of its genomic structure and comparison of its tissue distribution with HtrA1 and HtrA2. Biochem J. 2003;371(Pt 1):39–48. doi: 10.1042/BJ20021569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kisselev AF, Goldberg AL. Proteasome inhibitors: From research tools to drug candidates. Chem Biol. 2001;8(8):739–758. doi: 10.1016/s1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi H. Prevention of cancer and inflammation by soybean protease inhibitors. Front Biosci (Elite Ed) 2013;5:966–973. doi: 10.2741/e676. [DOI] [PubMed] [Google Scholar]
- 38.Chen G, et al. Autophagy is a protective response to ethanol neurotoxicity. Autophagy. 2012;8(11):1577–1589. doi: 10.4161/auto.21376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pamplona R, Costantini D. Molecular and structural antioxidant defenses against oxidative stress in animals. Am J Physiol Regul Integr Comp Physiol. 2011;301(4):R843–R863. doi: 10.1152/ajpregu.00034.2011. [DOI] [PubMed] [Google Scholar]
- 40.Hellwig-Burgel T, Stiehl DP, Wagner AE, Metzen E, Jelkmann W. Review: Hypoxia-inducible factor-1 (HIF-1): A novel transcription factor in immune reactions. J Interferon Cytokine Res. 2005;25(6):297–310. doi: 10.1089/jir.2005.25.297. [DOI] [PubMed] [Google Scholar]
- 41.Poyton RO, Ball KA, Castello PR. Mitochondrial generation of free radicals and hypoxic signaling. Trends Endocrinol Metab. 2009;20(7):332–340. doi: 10.1016/j.tem.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 42.Fang EF, et al. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell. 2014;157(4):882–896. doi: 10.1016/j.cell.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stumpf JD, Saneto RP, Copeland WC. Clinical and molecular features of POLG-related mitochondrial disease. Cold Spring Harb Perspect Biol. 2013;5(4):a011395. doi: 10.1101/cshperspect.a011395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young MJ, et al. Biochemical analysis of human POLG2 variants associated with mitochondrial disease. Hum Mol Genet. 2011;20(15):3052–3066. doi: 10.1093/hmg/ddr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trifunovic A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 46.Miralles Fusté J, et al. In vivo occupancy of mitochondrial single-stranded DNA binding protein supports the strand displacement mode of DNA replication. PLoS Genet. 2014;10(12):e1004832. doi: 10.1371/journal.pgen.1004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oliveira MT, Kaguni LS. Reduced stimulation of recombinant DNA polymerase γ and mitochondrial DNA (mtDNA) helicase by variants of mitochondrial single-stranded DNA-binding protein (mtSSB) correlates with defects in mtDNA replication in animal cells. J Biol Chem. 2011;286(47):40649–40658. doi: 10.1074/jbc.M111.289983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li B, et al. Omi/HtrA2 is a positive regulator of autophagy that facilitates the degradation of mutant proteins involved in neurodegenerative diseases. Cell Death Differ. 2010;17(11):1773–1784. doi: 10.1038/cdd.2010.55. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki Y, et al. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol Cell. 2001;8(3):613–621. doi: 10.1016/s1097-2765(01)00341-0. [DOI] [PubMed] [Google Scholar]
- 50.Noctor G, De Paepe R, Foyer CH. Mitochondrial redox biology and homeostasis in plants. Trends Plant Sci. 2007;12(3):125–134. doi: 10.1016/j.tplants.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 51.Jose C, Melser S, Benard G, Rossignol R. Mitoplasticity: Adaptation biology of the mitochondrion to the cellular redox state in physiology and carcinogenesis. Antioxid Redox Signal. 2013;18(7):808–849. doi: 10.1089/ars.2011.4357. [DOI] [PubMed] [Google Scholar]
- 52.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chatre L, Ricchetti M. Large heterogeneity of mitochondrial DNA transcription and initiation of replication exposed by single-cell imaging. J Cell Sci. 2013;126(Pt 4):914–926. doi: 10.1242/jcs.114322. [DOI] [PubMed] [Google Scholar]
- 54.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 55.Parone PA, et al. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS ONE. 2008;3(9):e3257. doi: 10.1371/journal.pone.0003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fan AX, et al. Mitochondrial DNA content in paired normal and cancerous breast tissue samples from patients with breast cancer. J Cancer Res Clin Oncol. 2009;135(8):983–989. doi: 10.1007/s00432-008-0533-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.