Significance
We describe a general strategy that begins to allow us to address the question of whether an expanded genetic code provides an evolutionary advantage to an organism. A large library of β-lactamase variants with distinct noncanonical amino acids substituted randomly at single sites throughout the protein was generated and then subjected to an antibiotic growth-based screen to identify mutants with enhanced catalytic activity. We show that a unique noncanonical mutation in the enzyme β-lactamase significantly increases catalytic activity by unexpected mechanisms. These effects cannot be recapitulated by other canonical amino acids at this site, suggesting that an expanded set of building blocks beyond the canonical 20 may offer unique solutions to organisms in the evolution of new functions.
Keywords: noncanonical amino acid, beta-lactamase, catalytic activity, evolutionary advantage, conformational effects
Abstract
With few exceptions, all living organisms encode the same 20 canonical amino acids; however, it remains an open question whether organisms with additional amino acids beyond the common 20 might have an evolutionary advantage. Here, we begin to test that notion by making a large library of mutant enzymes in which 10 structurally distinct noncanonical amino acids were substituted at single sites randomly throughout TEM-1 β-lactamase. A screen for growth on the β-lactam antibiotic cephalexin afforded a unique p-acrylamido-phenylalanine (AcrF) mutation at Val-216 that leads to an increase in catalytic efficiency by increasing kcat, but not significantly affecting KM. To understand the structural basis for this enhanced activity, we solved the X-ray crystal structures of the ligand-free mutant enzyme and of the deacylation-defective wild-type and mutant cephalexin acyl-enzyme intermediates. These structures show that the Val-216–AcrF mutation leads to conformational changes in key active site residues—both in the free enzyme and upon formation of the acyl-enzyme intermediate—that lower the free energy of activation of the substrate transacylation reaction. The functional changes induced by this mutation could not be reproduced by substitution of any of the 20 canonical amino acids for Val-216, indicating that an expanded genetic code may offer novel solutions to proteins as they evolve new activities.
With the rare exceptions of pyrrolysine and selenocysteine, all known organisms use the same 20 canonical amino acids to carry out their life functions. However, additional regulatory, catalytic, and structural elements have evolved that include organic/inorganic cofactors and posttranslational modifications that expand the structural and functional repertoire of protein sequence space (1, 2). The question arises as to why the genetic code has remained virtually constant over evolution, given that relatively few genes are required to add additional building blocks to the code. All that is necessary is an additional cognate tRNA/aminoacyl-tRNA synthetase (aaRS) pair that is orthogonal to the existing pairs in the host (i.e., does not cross-react with endogenous tRNA/aaRS pairs), selectively recognizes a noncanonical amino acid (ncAA), and uniquely inserts that amino acid into the growing polypeptide chain in response to a nonsense or frameshift codon (3, 4). Indeed, pyrrolysine itself is genetically encoded by an orthogonal amber suppressor tRNA/aaRS pair, and we and others have shown that a large number of ncAAs with unique chemical and physical properties can be added to the genetic codes of prokaryotes with little effect on growth rates under normal laboratory growth conditions (3–6). This expanded genetic code now allows one to begin to explore the potential impact of additional building blocks on protein structure and function.
A number of experimental systems have been reported that begin to explore this question. For example, an in vitro antibody phage display system was developed in which sulfo-tyrosine–containing antibodies outcompeted others in affinity-based panning for binding to gp120, an HIV protein that naturally binds the sulfated cytokine receptor CCR5 (7). A similar phage display system was used to select a Zn(II) finger transcription factor in which the Zn(II) site was replaced with a unique 4-bipyridylalanine·Fe(II) site with no impact on DNA affinity and selectivity (8). A selection system based on survival of Escherichia coli in response to proteolytic stress afforded ribosomal cyclic peptides that contain a keto amino acid and covalently bind and inactivate HIV protease (9). Recently, it was shown that substitution of 3-iodo-tyrosine in the type II holin protein of bacteriophage T7 leads to increased phage fitness (10). Here we describe an alternative approach to explore the possible effects of additional building blocks on protein structure and function in which we generated a large library of β-lactamase variants with distinct ncAAs substituted randomly at single sites throughout the protein (Fig. 1A). We then used this system to identify an enzyme with increased catalytic efficiency that stems from conformational effects on key active site residues resulting from mutation of Val-216 to p-acrylamido-phenylalanine (AcrF).
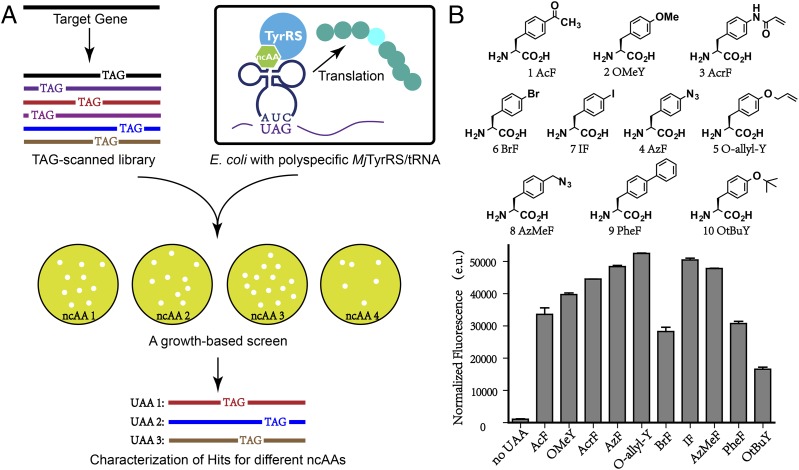
Fig. 1.
ncAA mutagenesis and a growth-based screen. (A) The TAG-scanned library was transformed into cells expressing a polyspecific amber suppressor tRNA/aaRS pair. The growth-based screen was carried out with various ncAAs, and the mutated residues within the target gene were determined by sequence analysis. (B) The structures of ncAAs used in this study. Suppression efficiencies of tRNACUATyr/PolyRS pair in the absence and presence of various amino acids were evaluated by using a GFP-based assay.
Results
Generation of a β-Lactamase Library Containing ncAAs.
Bacterial resistance to expanded-spectrum β-lactam antibiotics has attracted considerable interest, and multidrug-resistant bacteria have resulted in an increasing number of hospital infections. A major mechanism for this resistance is the enzyme β-lactamase. To date, >200 β-lactamase mutants have been found to arise by mutations from class-A β-lactamases, such as TEM and SHV (11). In an effort to determine whether an expanded genetic code might provide an evolutionary advantage to β-lactamases in adapting to new β-lactam antibiotics, we generated a large library of point mutants in which a number of different ncAAs were randomly substituted at each of 144 of the 286 residues in TEM-1 β-lactamase. These residues were chosen based on the X-ray crystal structure to include residues located in the active-site region and positions previously reported to be critical for enzyme function (12).
To simplify construction of a β-lactamase library containing a variety of structurally distinct ncAAs, a polyspecific aminoacyl-tRNA synthetase (PolyRS) was used that is orthogonal to the endogenous translational machinery of E. coli and that selectively incorporates ncAAs and not any of the canonical 20 amino acids in response to the amber nonsense codon. A fluorescence-based screen was first used to determine the suppression efficiency and polyspecificity of this aaRS for a set of substituted aromatic amino acid side chains, (because PolyRS is derived from a Methanococcus jannaschii tyrosyl-tRNA synthetase) (Fig. 1B). This screen was accomplished by growing DH10B cells containing the amber suppressor tRNA/PolyRS pair (encoded on the plasmid pUltra-Poly) and a superfolder green fluorescent protein (sfGFP) with an amber mutation at the permissive site Tyr-151 (encoded on plasmid pET22b-T5-sfGFP*) in LB medium supplemented with various ncAAs at a concentration of 1 mM (Fig. 1B) (13, 14). The highest GFP expression levels were observed in the presence of 10 derivatives of tyrosine or phenylalanine, including p-acetyl-phenylalanine (AcF), O-methyl-tyrosine, AcrF, p-azido-phenylalanine (AzF), O-allyl-tyrosine (O-allyl-Y), p-bromo-phenylalanine (BrF), p-iodo-phenylalanine (IF), p-azidomethyl-phenylalanine (AzMeF), p-biphenylalanine, and O-tert-butyl-tyrosine (Fig. 1B). These 10 amino acids were subsequently used in growth-based screens with the initial β-lactamase library.
A Growth-Based Screen for β-Lactamase Variants with Enhanced Activity.
To generate TEM-1 β-lactamase mutants containing these ncAAs, a TAG nonsense codon was introduced independently at each of 144 sites by site-directed mutagenesis and verified by sequencing. This TAG-scanned library was then introduced into E. coli DH10B cells containing the amber suppressor tRNA/PolyRS pair (Fig. 1A), and the transformed cells were subjected to selective pressure in the presence and absence of the 10 ncAAs with the expanded spectrum β-lactam antibiotics cephalexin or ceftazidime (Fig. 2A). To our delight, hundreds of colonies were observed on the plate with 3 μg⋅mL−1 ceftazidime in the presence of AzMeF, AcF, AzF, O-allyl-Y, BrF, or IF after 2 d. Notably, no colonies were observed on the plates with 3 μg⋅mL−1 ceftazidime in the absence of the ncAAs. Sequencing of 10 individual colonies on each plate revealed a unique mutation (Asp-179–TAG). The presence of the Asp-179–AzMeF mutation resulted in a significantly higher ceftazidime minimal inhibitory concentration (MIC) (14 μg⋅mL−1), compared with the wild-type β-lactamase (0.25 μg⋅mL−1) (Table S1). The X-ray crystal structure of wild-type β-lactamase shows that the guanidinium side chain of Arg-164 is hydrogen-bonded to Asp-179 in the omega loop near the active site (15). Previous studies have demonstrated that loss of this hydrogen bond by point mutation (Asp-179–Asn) may weaken the omega loop, which makes it possible for the enzyme to accommodate bulkier substrates (16, 17).
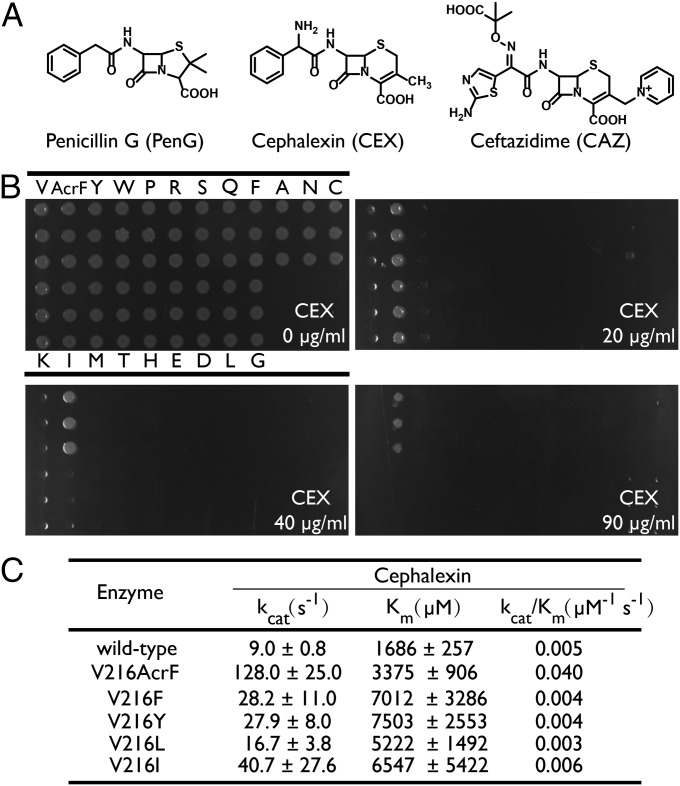
Fig. 2.
Characterization of wild-type and mutant β-lactamases. (A) Structures of β-lactam antibiotics used in this study. (B) MICs of cephalexin for wild-type and V216X mutant enzymes. (C) Kinetic parameters for hydrolysis of cephalexin by wild-type and V216X mutant enzymes.
A Novel β-Lactamase Mutant with Increased Catalytic Efficiency.
Although the above result provided validation for this experimental approach, both canonical and ncAAs led to increased activity when substituted for Asp-179 (Table S1). We therefore carried out additional experiments to identify enzymes with improved activity that results only from mutation to a ncAA. Using a similar growth-based screen with the β-lactam antibiotic cephalexin, we identified a Val-216 to AcrF mutant with a significantly higher MIC (90 μg⋅mL−1) than the wild-type enzyme (10 μg⋅mL−1). This substitution was shown to be AcrF-dependent, because β-lactamases with Val-216 mutated to the other nine ncAAs have MICs for cephalexin that are less than or equal that of the wild-type enzyme (Table S2). To determine whether the enhanced activity resulting from the Val-216–AcrF substitution could be recapitulated by any of the canonical amino acids, we independently mutated Val-216 to the other 19 canonical amino acids. The most active variant (Val-216–Ile) within this pool had at most a twofold higher MIC (20 μg⋅mL−1) than the wild-type enzyme (10 μg⋅mL−1); the MICs of the other mutants were comparable to or lower than the wild-type enzyme (Fig. 2B).
To further assess the effect of the Val-216–AcrF mutation on enzymic activity against cephalexin, the kinetic parameters were determined for the wild-type and mutant enzymes (Fig. 2C) (18). Wild-type β-lactamase and β-lactamases bearing canonical mutations were prepared as described in the previous study (19). For the Val-216–AcrF variant, pET30b-T5-TEM1-216TAG containing a TEM-1 variant with a C-terminal His6 tag and an amber codon at Val-216 was cotransformed with the tRNA/PolyRS pair (in the pUltra expression vector) into E. coli DH10B cells. The expression of the Val-216–AcrF enzyme was carried out in rich medium in the presence of 1 mM AcrF, and protein was purified by Ni-NTA and ion-exchange chromatography. Electrospray ionization (ESI)-MS analysis of proteins confirmed the site-specific incorporation of mutations and the homogeneity of purified enzymes (Fig. S1). The Val-216–AcrF enzyme exhibited a significantly increased kcat (14-fold) and a slightly increased KM (twofold) for cephalexin hydrolysis compared with those values observed for the wild-type enzyme (Fig. 2C). Among the canonical amino acids, phenylalanine and tyrosine are most closely related to AcrF. The Val-216–Tyr and –Phe mutants exhibited a slight decrease in kcat/KM compared with the wild-type enzyme; the most active canonical amino acid mutant (Val-216–Ile) showed a comparable kcat/KM compared with the wild-type enzyme (Fig. 2C). Thus, none of the Val-216–X canonical or noncanonical mutants could recapitulate the functional change imparted by the AcrF mutation.
Structural Analysis of β-Lactamase Variants.
Val-216 lies near the active site, but has not been implicated in playing a role in catalysis. To explore the mechanistic basis for the increase in catalytic activity resulting from the Val-216–AcrF substitution, the crystal structure of the Val-216–AcrF mutant enzyme was obtained at 1.54-Å resolution by molecular replacement (Fig. 3D). Alignment of the Val-216–AcrF crystal structure with the published ligand-free TEM-1 β-lactamase structure [Protein Data Bank (PDB) ID code 1BTL] showed a number of conformational changes in some surface residues (e.g., Glu-140, Glu-240, and Arg-241), which are observed in other structures. More relevant is a side-chain reorientation of Tyr-105 in the free Val-216–AcrF mutant to a minus (m-85°/m-30°) rotamer (Fig. 3D), relative to its conformation in the free wild-type enzyme and the benzylpenicillin acyl-enzyme intermediate, where Tyr-105 adopts a trans (t-80°) rotamer (Fig. 3 A and B) (12, 20, 21). This conformational change likely relieves possible steric interactions between Tyr-105 and the acrylamido moiety of AcrF. In the class A β-lactamases, Tyr-105 is a conserved residue that lies at the entrance to the active site. Saturation mutagenesis studies have suggested that Tyr-105 may play a role in substrate discrimination and binding (22). Interestingly, the minus (m-85°/m-30°) rotamer orientation of Tyr-105 is observed in the crystal structures of the wild-type enzyme complexes with boronic acid transition state inhibitors or the natural protein inhibitors BLIP I and BLIP II. This observation suggests that the minus conformation of Tyr-105 may also stabilize the transition state for the transacylation of cephalexin, resulting in an increased kcat (20, 23, 24). In contrast, in the wild-type enzyme, Tyr-105 is fixed in a trans configuration in the free enzyme (Fig. 3 A and B), and there would be a free energy cost associated with movement of this side chain to the minus conformer upon formation of the enzyme–cephalexin transition state complex (Fig. 3 A and C).
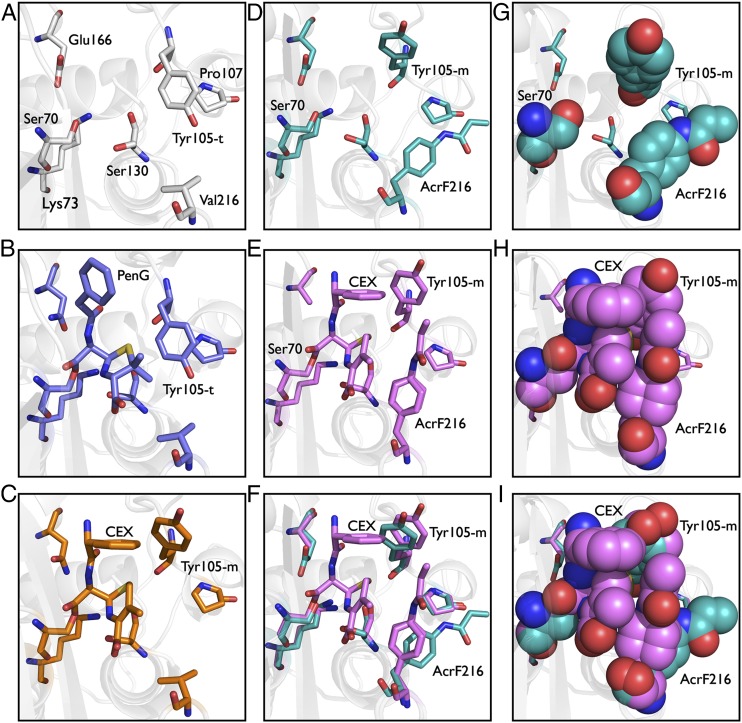
Fig. 3.
Crystal structures of wild-type and Val-216–AcrF mutant β-lactamases. (A) X-ray crystal structure of ligand-free wild-type enzyme (PDB ID code 1BTL). (B) X-ray crystal structure of benzylpenicillin acyl-enzyme intermediate for wild-type enzyme (PDB ID code 1FQG). (C) X-ray crystal structure of cephalexin acyl-enzyme intermediate for wild-type enzyme. (D) X-ray crystal structure of ligand-free Val-216–AcrF mutant enzyme. (E) X-ray crystal structure of cephalexin acyl-enzyme intermediate for the Val-216–AcrF mutant enzyme. (F) Overlay of active-site residues of cephalexin-bound and ligand-free Val-216–AcrF mutant enzymes. (G) X-ray crystal structure of ligand-free mutant enzyme with Val-216–AcrF substitution displayed as cyan spheres. (H) Packing interactions between cephalexin and side chains of mutant enzyme with Val-216–AcrF substitution. (I) Overlay of active site residues as spheres of cephalexin-bound and ligand-free mutant enzymes with Val-216–AcrF substitution.
To support this hypothesis, we crystallized the acyl-enzyme intermediate of a deacylation-defective β-lactamase in which cephalexin is covalently bound to the catalytic serine residue of the wild-type or Val-216–AcrF enzyme. Previously, the crystal structure of a Glu-166–Asn deacylation-defective TEM-1 β-lactamase mutant was solved with a benzylpenicillin bound acyl-enzyme intermediate (12). Using a similar approach, we were able to cocrystallize and solve the structures of a Glu-166–Asn mutant of wild-type β-lactamase (1.80-Å resolution) and a Glu-166–Ala mutant of the Val-216–AcrF mutant (1.70-Å resolution), both with cephalexin covalently bound to Ser-70 (12, 15). Indeed, in both structures, Tyr-105 adopts the m rotamer, which results in a favorable end-on stacking interaction with the benzyl moiety of the cephalexin acyl-enzyme intermediate (Fig. 3 C and E). In addition the m rotamer likely relieves possible unfavorable steric interactions of Tyr-105 with the bulkier dihydrothiazine ring of the antibiotic (Fig. 3 C and E). Thus, the wild-type enzyme must pay an energetic price to reorient Tyr-105 for hydrolysis of cephalexin, whereas in the Val-216–AcrF mutant the same m rotamer of Tyr-105 is observed in both the ligand-free enzyme and the cephalexin acyl-enzyme intermediate (Fig. 3 D–F). In addition, a comparison of the structures of the free and cephalexin acyl-enzyme mutant enzymes shows that the side chain of AcrF–216 undergoes a significant conformational change during the catalytic reaction that leads to improve packing interactions with cephalexin in the acyl-enzyme intermediate (Fig. 3 F and I). This change in the conformation of AcrF between the free enzyme and acyl-enzyme intermediate appears to occur by an induced-fit mechanism, although it is not clear whether this conformational change occurs upon substrate binding or upon formation of the covalent adduct (Fig. 3 G–I).
Discussion
Proteins are innately flexible, undergoing dynamic conformational changes over a wide range of time regimes and amplitudes. Previous studies have shown that changes in the energetics of specific conformational states of proteins cannot affect only binding affinity, but also catalytic efficiency (25–30). Early work demonstrated that some enzymes bind their substrates by an induced-fit mechanism in which active residues adopt distinct conformations upon substrate binding (25, 31, 32). More recent structural and spectroscopic studies have also shown that reduction in the conformational plasticity of germ-line antibodies by somatic mutation results in increased affinity for the cognate antigen (29, 33, 34). In the enzyme cyclophilin A, it has been shown that mutations, which alter the conformational equilibrium of catalytically active and inactive states, affect catalytic efficiency (35). In addition, structural and dynamic studies of dihydrofolate reductase have shown that mutations that affect millisecond-time-scale fluctuations lead to decreased rates of hydride transfer (36). Similarly, crystallographic analysis and microsecond-scale molecular-dynamics simulations have suggested that remote mutations in simvastatin synthase alter catalytic efficiency through conformational effects (37). Other examples include the demonstration of altered catalytic activity in conformational isomers of a catalytic antibody and the effects of distal mutations on HIV protease active-site conformation and catalytic activity (38).
Herein we show that a unique noncanonical mutation (Val-216–AcrF) in the enzyme TEM-1 β-lactamase significantly increases catalytic activity by affecting the conformational energetics of key active-site residues. Hydrolysis of β-lactam antibiotics by the class-A β-lactamases involves a two-step pathway: transacylation to form an acyl-enzyme intermediate and deacylation by attack of water (12). Fluorescence and mass spectral kinetic studies have suggested that the enzyme–substrate transacylation reaction is the rate-limiting step for hydrolysis of first-generation cephalosporins (39, 40). In all previously reported free enzyme structures, Tyr-105 which is in the active site but has not been implicated to play a role in catalysis, has been assigned to the trans rotamer. Our results show that the noncanonical mutation Val-216–AcrF induces a conformational change in Tyr-105 to the minus rotamer in both the free enzyme and acyl-enzyme intermediate, which results in improved packing interactions with the acyl-enzyme intermediate. Furthermore, the cephalexin acyl-enzyme intermediate reveals additional favorable packing interactions between the AcrF side chain and the thiazine ring and benzyl group of cephalexin, which occur by a change in the side-chain conformation of AcrF relative to the free enzyme. Together, these results support the notion that the Val-216–AcrF mutation increases kcat for hydrolysis of cephalexin through conformational changes in both Tyr-105 and AcrF itself that may lower the activation energy of the cephalexin transacylation reaction. Whether these interactions lower the free energy of activation by stabilizing the tetrahedral transition state or by introducing strain into the ground-state complex cannot be determined at present. However, the X-ray crystal structure of boronic acid transition-state analogs bound to the enzyme do show Tyr-105 in the same minus conformation that is present in the cephalexin acyl-enzyme intermediate of the Val-216–AcrF mutant, and the mutation does not show any significant effect on substrate KM (23, 41, 42). A comparison of the structures of the mutant and WT enzyme reveals no significant change in the position of the catalytic water molecule, supporting the notion that the Val-216–AcrF mutation primarily affects the first step of the enzymatic hydrolysis reaction.
Although one cannot exclude the possibility that canonical mutations at other sites might have a similar effect to the Val-216–AcrF mutation, substitution of any canonical amino acids at residue 216 does not significantly affect the catalytic efficiency (kcat/KM) of the enzyme. Thus, we demonstrate that an expanded set of building blocks beyond the canonical 20 can lead to enhanced activity by unexpected mechanisms, which may be distinct from those possible through conventional mutagenesis schemes. It is likely that ncAAs will also provide the opportunity to enhance other properties of proteins—for example, electrophilic keto-containing amino acids may increase the stability of a protein through the formation of covalent cross-links to lysine or act as an electron sink in catalysis, and redox active amino acids might facilitate electron transfer reactions. However, it remains to be determined whether genetically encoded ncAAs can provide a unique evolutionary advantage that cannot be achieved by either single or multiple canonical amino acid mutations at other sites in a protein or the incorporation of nonproteinogenic cofactors. We expect that further advances in site-specific ncAA incorporation technology in combination with other new mutagenesis techniques, such as TAG codon scanning, multiplex automated genomic engineering, and multiplex iterative plasmid engineering, may provide the opportunity to address this question at a more global whole-organism level (43–46).
Materials and Methods
For cloning and propagation of plasmid DNA, E. coli DH10B was used. PBS and Tris/borate/EDTA buffers were obtained from Cellgro. LB agar and 2× yeast–tryptone (YT) were purchased from Fisher Scientific. Isopropyl-β-d-thiogalactoside (IPTG) was purchased from Anatrace, and 4–12% (wt/vol) Bis-Tris gels for SDS/PAGE were purchased from Invitrogen. DNA polymerase was obtained from Agilent Technologies, dNTPs were obtained from New England Biolabs, and oligonucleotides were purchased from Integrated DNA Technologies (Tables S3 and S4 list the oligonucleotides used in this report). Restriction enzymes and T4 DNA ligase were purchased from NEB. Ceftazidime, cephalexin, cefaclor, and cefradine were obtained from Research Products International Corp., Alfa Aesar, Enzo Life Sciences, and Alfa Aesar, respectively. Plasmid DNA preparation was carried out with the QIAprep Spin Miniprep Kit (Qiagen). Unless otherwise mentioned, all other chemicals were purchased from Sigma-Aldrich and used without further purification. AcrF was synthesized as described (47). Absorbance spectra were measured with a Hewlett-Packard 8453 UV-visible spectrophotometer.
Construction of TAG-Scanned Library.
To construct the TAG-scanned library, the WT TEM-1 gene cassette was generated by PCR using primers HX0975 and HX096, and the pBK vector was amplified by using primers HX0976 and HX0977. The WT TEM-1 gene was inserted into the pBK vector by Gibson assembly cloning to provide pBK-wtTEM1. From this plasmid, TEM-1 gene variants bearing TAG mutation were obtained by site-directed mutagenesis using the QuikChange Site Directed mutagenesis kit (Agilent Technologies) and the primers listed in Table S3. These individual mutants were sequenced and combined in a pool to yield the TAG-scanned library (Table S3).
Construction of Expression Vectors.
The WT TEM-1 gene cassette was generated by PCR using primers HX1034 and HX1035 and inserted into pET30b by using NdeI and KpnI sites to provide pET30b-T5-wtTEM1. From this plasmid, pET30b-T5-TEM1-216TAG was generated by site-directed mutagenesis using the QuikChange Site Directed mutagenesis kit (Agilent Technologies) and the following primers: HX1057 and HX1058. pET30b-T5-TEM1-216Phe, pET30b-T5-TEM1-216Tyr, pET30b-T5-TEM1-216Leu, and pET30b-T5-TEM1-216Ile were prepared by site-directed mutagenesis using primers HX1065 and HX1066, HX1063 and HX1064, HX1067 and HX1068, HX1069, and HX1070, respectively (Table S4).
Determination of PolyRS Polyspecificity.
The pET22b-T5-sfGFP* and pUltra-PolyRS plasmids were transformed into E. coli DH10B. Cells were grown in glycerol minimal medium with leucine or 2× YT medium, supplemented with ampicillin (50 μg/mL), spectinomycin (25 μg/mL), and various ncAAs as a concentration of 1 mM at 37 °C to an OD600 of 0.6, at which point IPTG was added to a final concentration of 1 mM. After 16 h at 30 °C, the cells were harvested by centrifugation at 4,700 × g for 10 min and washed three times with PBS. Cells were resuspended in PBS and transferred to a clear-bottom 96-well plate, and GFP-fluorescence was measured by using a plate reader (485 nm excitation and 510 nm emission).
Determination of the MIC.
The MICs of different antibiotics for various TEM-1 β-lactamase mutants were determined as described (48, 49). A single colony of E. coli DH10B harboring plasmids pUltra-PolyRS and pBK (encoding the TEM-1 β-lactamase variants) was used to inoculate 5 mL of LB medium supplemented with spectinomycin (25 μg/mL) and kanamycin (25 μg/mL) at 37 °C. The overnight culture was then diluted to 1:400, and 4 µL of the bacterial culture was placed on an LB agar plate supplemented with spectinomycin (25 μg/mL), kanamycin (25 μg/mL), 1 mM ncAA, and 1 mM IPTG. After 48 h of incubation at 37 °C, the MIC was recorded as the lowest concentration of antibiotics on which no colonies were observed. The MICs were calculated from triplicate experiments.
Expression and Purification of TEM β-Lactamases.
The pUltra-PolyRS and pET30b-T5-TEM1-216TAG plasmids were transformed into the E. coli DH10B strain. Cells were grown in TB medium and supplemented with spectinomycin (25 μg/mL), kanamycin (25 μg/mL), and 1 mM ncAA at 37 °C to an OD600 of 0.6, at which point IPTG was added to a final concentration of 0.5 mM. After 20 h of incubation at 27 °C, the cells were harvested by centrifugation at 4,700 × g for 10 min. To purify the protein, cell pellets were resuspended in loading buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0) and lysed using a microfluidizer at 20,000 psi chamber pressure. The resulting cell lysate was clarified by centrifugation at 18,000 × g for 30 min, and the proteins were purified on Ni2+-NTA resin following the manufacturer’s (Qiagen) instructions. The resulting protein was further concentrated with an Amicon ultrafiltration device (membrane molecular mass cutoff, 10,000 Da), reconstituted in 0.01 M Tris⋅HCl buffer (pH 7.5), and loaded onto a MonoQ anion-exchange column. The protein was eluted with a gradient of NaCl in 0.01 M Tris⋅HCl buffer (pH 7.5) at a flow rate of 2 mL/min. The protein fractions with right molecular mass determined by Coomassie brilliant blue staining were collected and pooled.
Determination of Kinetic Properties of β-Lactamase Variants.
TEM-1 β-lactamases activities against cephalexin was determined with a Hewlett-Packard 8453 UV-visible spectrophotometer in 100 mM phosphate buffer (pH 7.0) at room temperature by monitoring changes in absorption upon β-lactam ring opening, using the following extinction coefficient: cephalexin, Δε266 = 7,800 M−1⋅cm−1. The initial velocities were measured and data were fitted to the Michaelis–Menten equation by using GraphPad Prism5 to determine kcat and KM.
Crystallization of β-Lactamase Variants.
Crystallization screening was performed with Mosquito Robot (TTP Labtech Ltd.) by using the vapor-diffusion method in a sitting-drop manner at room temperature, and the initial crystals were obtained from the Morpheus screen (Molecular Dimension Ltd.). Crystal growth condition was optimized by using vapor diffusion in a hanging-drop manner with a 1:1 ratio of 1 µL of reservoir solution and 1 µL of protein solution at a concentration of 12 mg/mL in 10 mM Tris, pH 7.8, 100 mM NaCl, and 10% glycerol. Single crystals appeared after 24 h and grew to their maximum size within a week. The best crystals grew in 0.1 M 2-(N-morpholino)ethanesulfonic acid (MES), pH 6.5, 15% PEG 20.000, and 15% PEG 550MME at 22 °C. The crystals were flash-frozen in liquid nitrogen before they were mounted for data collection. All crystals grew in similar conditions, but streak seeding or macro seeding from the wild-type protein crystals was essential to initiate the crystal growth.
Crystals were soaked with Cephalexin by transferring a single crystal to a pre-equilibrated drop of reservoir (0.1 M MES, pH 6.5, 17% PEG20.000, and 17% PEG550MME) and 40 mM cephalexin solution (10 mM Tris, pH 7.8, 100 mM NaCl, and 10% glycerol) at a 1:1 ratio. The crystals were transferred to a new drop every 3 h and were incubated overnight in the fifth drop. Crystals were frozen in liquid nitrogen and stored the next day.
X-Ray Diffraction Data Collection and Structure Determination.
The X-ray diffraction data were collected either at an in-house X-ray source (Rigaku FR-E SuperBright) with Rigaku RAXIS-HTC detector or at Stanford Synchrotron Radiation Lightsource using beamlines BL 11-1 and BL 7-1 with PILATUS 6M PAD and ADSC QUANTUM 315R detector, respectively. The collected data were indexed and integrated with imosflm or XDS and scaled by using Scala, a part of the CCP4 suite (50–53). Initial phase information was obtained by molecular replacement using PHASER with the previously solved structure of TEM1 β-lactamase as a search model (PDB ID code 1BTL) (54). During the initial round of the refinement, waters were added by using ArpWarp (55). The structure was improved by iterative rounds of model building and refinement with the programs Coot and Refmac5 (56, 57). The crystals belonged to space group P212121, with one molecule in the asymmetric unit. Crystallographic details and statistics are listed in Table S5.
Supplementary Material
Acknowledgments
We thank Kristen Williams for her assistance in manuscript preparation; Bernhard C. Lechtenberg for his technical assistance with the in-house data collection; and Prof. Sergei Vakulenko for helpful discussions. This work was supported by Division of Materials Sciences, Department of Energy, Grant DE-SC0011787 (to P.G.S.). This is Paper 29058 of The Scripps Research Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4ZJ1, 4ZJ2, and 4ZJ3).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507741112/-/DCSupplemental.
References
- 1.Silverman RB. The Organic Chemistry of Enzyme-Catalyzed Reactions. Academic; San Diego: 2000. [Google Scholar]
- 2.Walsh C. Posttranslational Modification of Proteins: Expanding Nature's Inventory. Roberts; Eaglewood, CO: 2006. [Google Scholar]
- 3.Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu Rev Biophys Biomol Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 4.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 5.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: Charging of a UAG-decoding specialized tRNA. Science. 2002;296(5572):1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 6.Krzycki JA. The direct genetic encoding of pyrrolysine. Curr Opin Microbiol. 2005;8(6):706–712. doi: 10.1016/j.mib.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Liu CC, et al. Protein evolution with an expanded genetic code. Proc Natl Acad Sci USA. 2008;105(46):17688–17693. doi: 10.1073/pnas.0809543105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang M, et al. Evolution of iron(II)-finger peptides by using a bipyridyl amino acid. ChemBioChem. 2014;15(6):822–825. doi: 10.1002/cbic.201300727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young TS, et al. Evolution of cyclic peptide protease inhibitors. Proc Natl Acad Sci USA. 2011;108(27):11052–11056. doi: 10.1073/pnas.1108045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammerling MJ, et al. Bacteriophages use an expanded genetic code on evolutionary paths to higher fitness. Nat Chem Biol. 2014;10(3):178–180. doi: 10.1038/nchembio.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacoby GA, Medeiros AA. More extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1991;35(9):1697–1704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strynadka NC, et al. Molecular structure of the acyl-enzyme intermediate in beta-lactam hydrolysis at 1.7 A resolution. Nature. 1992;359(6397):700–705. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- 13.Chatterjee A, Sun SB, Furman JL, Xiao H, Schultz PG. A versatile platform for single- and multiple-unnatural amino acid mutagenesis in Escherichia coli. Biochemistry. 2013;52(10):1828–1837. doi: 10.1021/bi4000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao H, et al. Genetic incorporation of histidine derivatives using an engineered pyrrolysyl-tRNA synthetase. ACS Chem Biol. 2014;9(5):1092–1096. doi: 10.1021/cb500032c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jelsch C, Mourey L, Masson JM, Samama JP. Crystal structure of Escherichia coli TEM1 beta-lactamase at 1.8 A resolution. Proteins. 1993;16(4):364–383. doi: 10.1002/prot.340160406. [DOI] [PubMed] [Google Scholar]
- 16.Vakulenko SB, Tóth M, Taibi P, Mobashery S, Lerner SA. Effects of Asp-179 mutations in TEMpUC19 beta-lactamase on susceptibility to beta-lactams. Antimicrob Agents Chemother. 1995;39(8):1878–1880. doi: 10.1128/aac.39.8.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levitt PS, et al. Exploring the role of a conserved class A residue in the Ω-Loop of KPC-2 β-lactamase: A mechanism for ceftazidime hydrolysis. J Biol Chem. 2012;287(38):31783–31793. doi: 10.1074/jbc.M112.348540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown NG, Shanker S, Prasad BV, Palzkill T. Structural and biochemical evidence that a TEM-1 beta-lactamase N170G active site mutant acts via substrate-assisted catalysis. J Biol Chem. 2009;284(48):33703–33712. doi: 10.1074/jbc.M109.053819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bae IK, et al. A novel ceftazidime-hydrolysing extended-spectrum beta-lactamase, CTX-M-54, with a single amino acid substitution at position 167 in the omega loop. J Antimicrob Chemother. 2006;58(2):315–319. doi: 10.1093/jac/dkl252. [DOI] [PubMed] [Google Scholar]
- 20.Lim D, et al. Crystal structure and kinetic analysis of beta-lactamase inhibitor protein-II in complex with TEM-1 beta-lactamase. Nat Struct Biol. 2001;8(10):848–852. doi: 10.1038/nsb1001-848. [DOI] [PubMed] [Google Scholar]
- 21.Strynadka NC, Jensen SE, Alzari PM, James MN. A potent new mode of beta-lactamase inhibition revealed by the 1.7 A X-ray crystallographic structure of the TEM-1-BLIP complex. Nat Struct Biol. 1996;3(3):290–297. doi: 10.1038/nsb0396-290. [DOI] [PubMed] [Google Scholar]
- 22.Doucet N, De Wals PY, Pelletier JN. Site-saturation mutagenesis of Tyr-105 reveals its importance in substrate stabilization and discrimination in TEM-1 beta-lactamase. J Biol Chem. 2004;279(44):46295–46303. doi: 10.1074/jbc.M407606200. [DOI] [PubMed] [Google Scholar]
- 23.Strynadka NC, Martin R, Jensen SE, Gold M, Jones JB. Structure-based design of a potent transition state analogue for TEM-1 beta-lactamase. Nat Struct Biol. 1996;3(8):688–695. doi: 10.1038/nsb0896-688. [DOI] [PubMed] [Google Scholar]
- 24.Ferraro MJ. Performance Standards for Antimicrobial Susceptibility Testing: Twelfth Informational Supplement. NCCLS; Wayne, PA: 2002. [Google Scholar]
- 25.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5(11):789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu Rev Biochem. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 27.Schulenburg C, Hilvert D. Protein conformational disorder and enzyme catalysis. Top Curr Chem. 2013;337:41–67. doi: 10.1007/128_2012_411. [DOI] [PubMed] [Google Scholar]
- 28.Ulrich HD, et al. The interplay between binding energy and catalysis in the evolution of a catalytic antibody. Nature. 1997;389(6648):271–275. doi: 10.1038/38470. [DOI] [PubMed] [Google Scholar]
- 29.Wedemayer GJ, Patten PA, Wang LH, Schultz PG, Stevens RC. Structural insights into the evolution of an antibody combining site. Science. 1997;276(5319):1665–1669. doi: 10.1126/science.276.5319.1665. [DOI] [PubMed] [Google Scholar]
- 30.Romesberg FE, Spiller B, Schultz PG, Stevens RC. Immunological origins of binding and catalysis in a Diels-Alderase antibody. Science. 1998;279(5358):1929–1933. doi: 10.1126/science.279.5358.1929. [DOI] [PubMed] [Google Scholar]
- 31.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254(5038):1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 32.Tsai CJ, Kumar S, Ma B, Nussinov R. Folding funnels, binding funnels, and protein function. Protein Sci. 1999;8(6):1181–1190. doi: 10.1110/ps.8.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin J, et al. A comparative analysis of the immunological evolution of antibody 28B4. Biochemistry. 2001;40(36):10764–10773. doi: 10.1021/bi010536c. [DOI] [PubMed] [Google Scholar]
- 34.Schultz PG, Yin J, Lerner RA. The chemistry of the antibody molecule. Angew Chem Int Ed Engl. 2002;41(23):4427–4437. doi: 10.1002/1521-3773(20021202)41:23<4427::AID-ANIE4427>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 35.Fraser JS, et al. Hidden alternative structures of proline isomerase essential for catalysis. Nature. 2009;462(7273):669–673. doi: 10.1038/nature08615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhabha G, et al. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science. 2011;332(6026):234–238. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiménez-Osés G, et al. The role of distant mutations and allosteric regulation on LovD active site dynamics. Nat Chem Biol. 2014;10(6):431–436. doi: 10.1038/nchembio.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weikl TR, Hemmateenejad B. How conformational changes can affect catalysis, inhibition and drug resistance of enzymes with induced-fit binding mechanism such as the HIV-1 protease. Biochim Biophys Acta. 2013;1834(5):867–873. doi: 10.1016/j.bbapap.2013.01.027. [DOI] [PubMed] [Google Scholar]
- 39.Saves I, et al. Mass spectral kinetic study of acylation and deacylation during the hydrolysis of penicillins and cefotaxime by beta-lactamase TEM-1 and the G238S mutant. Biochemistry. 1995;34(37):11660–11667. doi: 10.1021/bi00037a003. [DOI] [PubMed] [Google Scholar]
- 40.Anderson EG, Pratt RF. Pre-steady state beta-lactamase kinetics. The trapping of a covalent intermediate and the interpretation of pH rate profiles. J Biol Chem. 1983;258(21):13120–13126. [PubMed] [Google Scholar]
- 41.Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science. 1994;264(5157):375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- 42.Maveyraud L, et al. Structural basis for clinical longevity of carbapenem antibiotics in the face of challenge by the common class A beta-lactamases from the antibiotic-resistant bacteria. J Am Chem Soc. 1998;120(38):9748–9752. [Google Scholar]
- 43.Daggett KA, Layer M, Cropp TA. A general method for scanning unnatural amino acid mutagenesis. ACS Chem Biol. 2009;4(2):109–113. doi: 10.1021/cb800271f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460(7257):894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y, et al. Multiplex iterative plasmid engineering for combinatorial optimization of metabolic pathways and diversification of protein coding sequences. ACS Synth Biol. 2013;2(11):651–661. doi: 10.1021/sb400051t. [DOI] [PubMed] [Google Scholar]
- 46.Malyshev DA, et al. A semi-synthetic organism with an expanded genetic alphabet. Nature. 2014;509(7500):385–388. doi: 10.1038/nature13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Furman JL, et al. A genetically encoded aza-Michael acceptor for covalent cross-linking of protein-receptor complexes. J Am Chem Soc. 2014;136(23):8411–8417. doi: 10.1021/ja502851h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang W, Palzkill T. A natural polymorphism in beta-lactamase is a global suppressor. Proc Natl Acad Sci USA. 1997;94(16):8801–8806. doi: 10.1073/pnas.94.16.8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3(2):163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 50.Read RJ, Sussman J. Evolving Methods for Macromolecular Crystallography: The Structural Path to the Understanding of the Mechanisms of Action of CBRN Agents. Springer; Dordrecht, The Netherlands: 2007. p. viii. [Google Scholar]
- 51.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 52.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 1):72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 53.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3(7):1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 57.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.