We read with interest the recent report by Londin et al. (1). We reanalyzed the dataset and discovered several challenges.
The first point is related to the cluster analysis of antisense microRNAs (miRNAs). The authors present 13 pairs of miRNAs where precursors on opposite strands overlap with each other (details in dataset S5 of ref. 1). Going through the data we found pairs of miRNAs that have overlapping genomic position and are located on opposite strands, but are not part of the 13 pairs mentioned in the Londin et al. report. One example is the precursor pair:
TJU_CMC_MD2.ID00144.hairpin 1–55784363 55784415
TJU_CMC_MD2.ID00145.hairpin 1 + 55784363 55784417.
We also observed cases in the 13 pairs where we were not able to see a direct overlap between the two miRNA precursors, although Londin et al. (1) describe limiting the search “to only those miRNAs, whose respective precursors directly overlapped with each other on opposite strands”:
TJU_CMC_MD2.ID00833.hairpin 13 + 80583254 80583310
TJU_CMC_MD2.ID00834.hairpin 13–80583317 80583391.
We speculate that this may be because of variations in single samples, although the precursors of “overlapping” miRNAs on different strands in the report are separated by up to 46 bases. Our own analysis of the data identified 63 respective miRNA pairs where precursors on opposite strands showed an overlap. These contain just 5 of the 13 original clusters.
Second, we do not understand the rationale to include cluster 28 as miRNA cluster because the two miRNAs have the same position. Despite whether this miRNA is known or novel, it should be just reported once and not as an miRNA cluster:
8 41517962 41518025 CLUSTER_#28_members = 2_span = 69_averagegap = 23.000000_mingap = 63_maxgap = 64
8 + 41517962 41518025 ID = MI0023622;Alias = MI0023622;Name = hsa-mir-486–2
8 + 41517962 41518025 TJU_CMC_MD2.ID03138.precursor.
Third, we investigated the melting of overlapping novel precursors. Londin et al. (1) state that they “identified hairpins and mature coordinates with end points typically differing by 1–2 base pairs across different samples. To compensate for this effect, overlapping hairpins were merged into a single larger ‘island’, and the mature isomiR with the highest read support across samples was determined to be the mature miRNA.” With this analysis, Londin et al. successfully avoid multiple miRNA precursors that differ by just few bases. However, two precursors with minimal overlap are also merged. This may lead to artificial miRNA precursors that actually look more similar to mirtrons (Fig. 1). Two precursors with a different starting point of 62 bases are predicted, each with two mature forms and showing the typical hairpin structure. As Fig. 1 shows, the 3′ form of the first is almost equal to the 5′ form of the second precursor. The merged precursor has a length of 163 bases and no typical hairpin structure (Fig. 2). The loop between the 3′ and 5′ mature form is 92 bases and the free energy −88.1. Additionally, the GC content of over 88% is remarkable. In the dataset we observed multiple cases where two precursors with substantial shifts are merged.
Fig. 1.
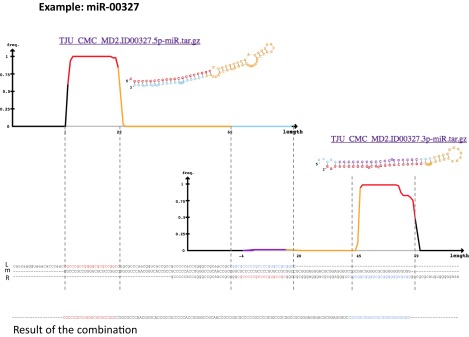
Example of miRNA precursor derived from two initially different precursors shifted by 62 bases. The upper left shows the left precursor with two mature forms and the typical hairpin structure. The right half shows the second precursor also with two mature forms and the typical hairpin structure. The 3′ and 5′ forms of the first and second precursor are almost identical. Below, the image the sequence of the two precursors are highlighted, as well as the merged form.
Fig. 2.
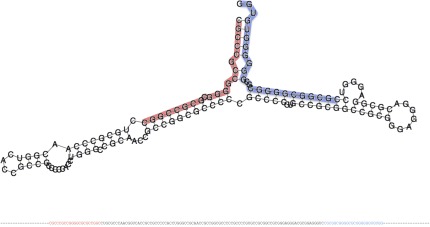
Structure of the merged miRNA precursor from Fig. 1 with the two mature forms colored in red and blue, calculated by RNAFold (rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi).
We appreciate the work of Londin et al. (1) and hope that our insights support the ambition to discover reliable novel miRNAs.
Footnotes
The authors declare no conflict of interest.
References
- 1.Londin E, et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc Natl Acad Sci USA. 2015;112(10):E1106–E1115. doi: 10.1073/pnas.1420955112. [DOI] [PMC free article] [PubMed] [Google Scholar]


