Abstract
Stress plays a substantial role in shaping behavior and brain function, often with lasting effects. How these lasting effects occur in the context of a fixed postmitotic neuronal genome has been an enduring question for the field. Synaptic plasticity and neurogenesis have provided some of the answers to this question, and more recently epigenetic mechanisms have come to the fore. The exploration of epigenetic mechanisms recently led us to discover that a single acute stress can regulate the expression of retrotransposons in the rat hippocampus via an epigenetic mechanism. We propose that this response may represent a genomic stress response aimed at maintaining genomic and transcriptional stability in vulnerable brain regions such as the hippocampus. This finding and those of other researchers have made clear that retrotransposons and the genomic plasticity they permit play a significant role in brain function during stress and disease. These observations also raise the possibility that the transposome might have adaptive functions at the level of both evolution and the individual organism.
Keywords: hippocampus, retrotransposon, histone marks, brain, genomic stress response
The brain is the central organ of stress and adaptation to stressors because it not only perceives what is threatening or potentially threatening and initiates behavioral and physiological responses to those challenges but also is a target of the stressful experiences and the hormones and other mediators of the stress response (1–4). The neural and hormonal mediators of the stress response affect most of the body’s organ systems, and prolonged or severe stressors can have prolonged physiologic and behavioral sequelae that can extend throughout the lifespan and beyond, to leave its imprint on our offspring (2, 5, 6). Short-term activation of stress mediators can be beneficial to cope with challenges, but long-term activation is accompanied by cumulative, potentially detrimental effects referred to, with increasing severity, as “allostatic load and overload” (3, 7, 8) Thus, although the brain is the conductor of this neuroendocrine orchestra, it is also shaped in many ways by its music, with both adaptive and pathogenic results (1, 2, 9).
Stress has a well-established influence on brain structure, function, and behavior; however “stress” is not a unitary phenomenon, nor are its effects upon individuals entirely predictable. The effects of stress upon an individual are dictated by a number of factors: stress duration, severity, controllability, age, and sex have clearly delineated roles in determining the impact of a particular stressor on an individual (10, 11). An individual’s stress history also seems to play an important role in the capacity to resist future stress exposures. Surprisingly, at least from the classical Darwinian perspective, the stress history of parents is a significant factor in the resilience of their offspring (12). The desire to understand how environmental stress transduces its effects into lasting changes on physiology and behavior, which can vary even among genetically identical individuals, has led scientists to hypothesize that epigenetic factors might provide an explanatory mechanism (1, 13, 14). The introduction of next-generation sequencing technologies to the exploration of epigenetics and stress neurobiology has led to greater attention to the possibility that the largely unexplored genomic space represented by retrotransposons might also have functional significance for brain function and stress susceptibility (15–17).
Stress and Brain Plasticity
One of most significant feats of modern neuroscience is the demonstration that the adult mammalian brain demonstrates a capability for reversible structural plasticity in response to experience. Plastic capacity includes alterations in dendritic structure as well as neurogenesis in structures like the hippocampal formation (4). This capacity can be induced by exercise, stimulating environments, or learning (18–22). Stress, both acute and chronic, has long been known to effect brain plasticity, as have steroid hormones, both gonadal and adrenal. Neurotransmitters and trophic factors also influence the molecular and cellular capacity for structural plasticity in the brain (4).
Stress affects plasticity in a number of brain regions, notably the amygdala, hippocampus, and prefrontal cortex. In the medial prefrontal cortex (mPFC) and hippocampus, exposure to chronic stress causes dendritic atrophy and reduced synaptic number, an effect accompanied by functional deficits in cognitive flexibility and memory, a process that seems to be partially reversible in younger brains (23–25). In aged brains, this capacity for recovery of dendritic and synaptic complexity is substantially impaired (26). In the orbitofrontal cortex and basolateral amygdala, the pattern of stress-induced structural plasticity is reversed, with exposure to chronic stress increasing dendritic complexity and synaptic number in parallel with increased vigilance, aggressiveness, and anxiety (27, 28). These findings from animal models fit well with a substantial body of findings linking exposure to stress and elevated levels of adrenal steroids to reduced hippocampal volume (3, 29). These effects also seem to be at least partially reversible with exercise and environmental enrichment (19, 30, 31), which also seems to elevate levels of neurogenesis in man, as it does in model animals (32).
However, youth can also be quite vulnerable to adversity; as the study of adverse childhood experiences in humans and rodent models, such as maternal separation, make plain, stress in early life can alter physiology and behavior across the entire lifespan (1, 33).
Although these stress effects are reversible, at least in youthful brains, it is evident that reversibility is not the same as erasure. For example, whereas recovery from chronic stress results in a return to prestress levels of dendritic length and spine number, these features did not return in the same pattern: distal dendritic retraction was not reversed whereas proximal dendritic length was greater than in stress-naive animals (25). Similar effects have been observed in cortex, hippocampus, and amygdala in response to corticosteroid treatment, suggesting that the network retained a structural memory of the stress exposure (34). This structural stress memory is even more apparent with regard to gene expression in stress-sensitive brain regions like the hippocampus. In a recent study comparing global gene-expression patterns in mice with a variety of stress exposures, it was found that the changes in gene expression induced by an acute forced swim stress in naive animals overlapped by less than 10% with the changes observed in mice exposed to the same swim stress who had a history of chronic stress. Interestingly, this overlap was even smaller when the comparison was with mice that had been chronically stressed and then allowed to recover for 3 wk (35). What these experiments make clear is that individual history is important and that there is a memory of stress history retained by neurons at the cellular level in regions like the hippocampus. Of course these data lead us to the question of how such memories might be created and maintained. One plausible hypothesis to explain neuronal memory of stress experience is via epigenetic mechanisms such as DNA methylation and histone modification.
Epigenetics and the Genomic Stress Response
Epigenetic mechanisms are attractive mechanisms for the transduction of environmental inputs, like stress, into lasting physiological and behavioral changes because many of them show both the malleability and the persistence necessary to explain these outputs. Epigenetics is also attractive to researchers interested in neurodevelopmental and neuropsychiatric disease because many of the most significant disorders show significant “missing heritability” at the genetic level (36–39). Although it is to be expected that rapidly improving sequencing methods will identify rare genetic variants and novel examples of epistasis, it seems unlikely that strong genetic determinism is operative in these disorders; therefore, there needs to be a search for other mechanisms such as epigenetics.
Stress has a number of known effects on epigenetic marks in the brain, producing alterations in DNA methylation and histone modifications in most of the stress-sensitive brain regions examined, including the hippocampus, amygdala, and prefrontal cortex (1, 13). Many of these changes may be maladaptive or contribute to pathologies; however, some of these changes seem to be adaptive. For example, dynamic regulation of DNA methylation is required for the proper formation of fear memories in the mouse hippocampus (40). Our own work has shown that both acute and chronic stress alter methylation of histone H3 in the rat hippocampus. H3 lysine 4 trimethylation is increased modestly by chronic restraint. After acute stress, H3 lysine 27 trimethylation drops by 50% whereas H3 lysine 9 trimethylation (H3K9me3) shows a hippocampus-specific twofold increase (41). H3K9me3 is a heterochromatin mark associated with transcriptional silencing, notably of retrotransposons (42); thus, we hypothesized that it might be a genomic stress response aimed at repressing ectopic overexpression of retrotransposon RNA during stress (16). Chromatin immunoprecipitation (ChIP) sequencing has validated that most of the stress-induced change is in fact targeted at these elements and that some are in fact down-regulated after an acute stress. Further, in the rat hippocampus, the H3K9me3-specific methyltransferase Suv39h2 is bound by the glucocorticoid receptor and shows increased expression after acute stress (15). The glucocorticoid receptor has a number of complex interactions with the epigenome that are still in the process of being mapped out (1, 43). Gonadal steroids have a similar capacity to produce epigenomic reorganization both organizationally and activationally in the context of sex determination and behavior (44–46), suggesting that nuclear hormone receptors in general are significant shapers of chromatin structure, in addition to their long-established role as transcription factors.
Transposons, Stress, and Brain Disorders
Our work demonstrated that an acute environmental stress dynamically regulated the expression of retrotransposon RNA in the brain (15). However, it was not the first time that transposon activity had been linked to stress. In fact, activation of transposons by stress was first proposed by their discoverer, Barbara McClintock (47). Indeed, McClintock’s early observations of transposition also associated the phenomena with heterochromatin and pointed out that mobilization of heterochromatin domains might alter gene expression (48). Thus, epigenetic marks and transposition have been associated since the time of their earliest description. McClintock believed that controlling elements, as she described them, permitted the genome to respond more flexibly to environmental shocks and stresses (47), but this hypothesis has not been the majority view up to the present. Francis Crick and Susumo Ohno lumped all of the transposable and repetitive elements into one heap of “selfish” “junk” (49, 50), and the view that transposons are either useless or parasitic has remained the dominant one.
Our finding, that rodent short interspersed elements (SINEs) are epigenetically repressed in the hippocampus during an acute environmental stress, suggest the capacity of mammalian genomes to regulate retrotransposon RNA expression (15, 16). That heat shock causes an increase in the same RNAs (51) strengthens the support for the idea that expression of SINE RNA at least is regulated in an adaptive fashion. B2 SINE RNA binds to RNA polymerase II and represses transcription (52); thus, increased B2 expression during heat shock could be adaptive in that it would reduce the number of transcripts available for translation and thus reduce the number of misfolded proteins due to hyperthermia. The same function seems to be conserved in the human Alu SINEs (53). On the other hand, protein synthesis is required (54) for memory consolidation in the hippocampus; therefore, suppression of these RNAs during stressful events would be adaptive because it would increase the probability of retaining memories of successful escapes or danger cues.
Transposons have been implicated in pathological processes of course, notably in cancer and autoimmune disorders (55, 56). In the nervous system, dysregulation of transposon expression has been linked to retinal degeneration (57), schizophrenia (58), alcoholism, posttraumatic stress disorder (PTSD) (59, 60), and a number of neurodegenerative disorders (17, 61). Recent work has identified excess transposon expression in the Drosophila melanogaster brain as a factor in age-related neurodegeneration. In the fruit fly brain, dysregulation of the transposon RNA-binding protein TDP-43 results in elevated levels of transposon transcripts and neuronal death (61, 62). Aberrant TDP-43 expression or function has been associated with both amyotrophic lateral sclerosis and fronto-temporal dementia (61, 63). It is notable that these findings demonstrate that control of retrotransposon RNA expression is physiologically important, whether the mechanism is RNA-processing (by dicer1) (57), RNA-binding (TDP-43) (61), or repressive chromatin states (15). Based on these findings, it is certainly plausible to argue that a disorder like schizophrenia, which results in brain atrophy over the course of development and which is characterized by both neuroinflammation and abnormally high retrotransposon activity (58, 64), might result from either environmental insults or genetic susceptibilities that reduce the capacity of the developing brain to control retrotransposon activity.
Transposons: Controlling Elements After All?
Transposon-induced mutagenesis and overexpression are involved in disease and may eventually provide us with an explanation for a variety of complex disorders. The same could be said of the genes themselves, however, and genes are better understood for their normal function, even in those cases where they are discovered and described due to a malfunction or mutation. If parasitism is the only function of transposons, they would be perhaps the only parasites whose hosts expend more energy to replicate than they do on their own genes. Indeed, given how much of the genome is composed of transposons in multicellular organisms, relative to the small fraction present in prokaryotes or even unicellular eukaryotes, it is hard to imagine how multicellular life could have evolved with such a serious competitive disadvantage. Further, that diseases can be caused by aberrant expression of certain genes is certainly well-accepted whereas the idea that genes are parasites cannot even be ascribed to Richard Dawkins in his most extreme moments (65), and the idea itself would render the modern Darwinian synthesis nonsense.
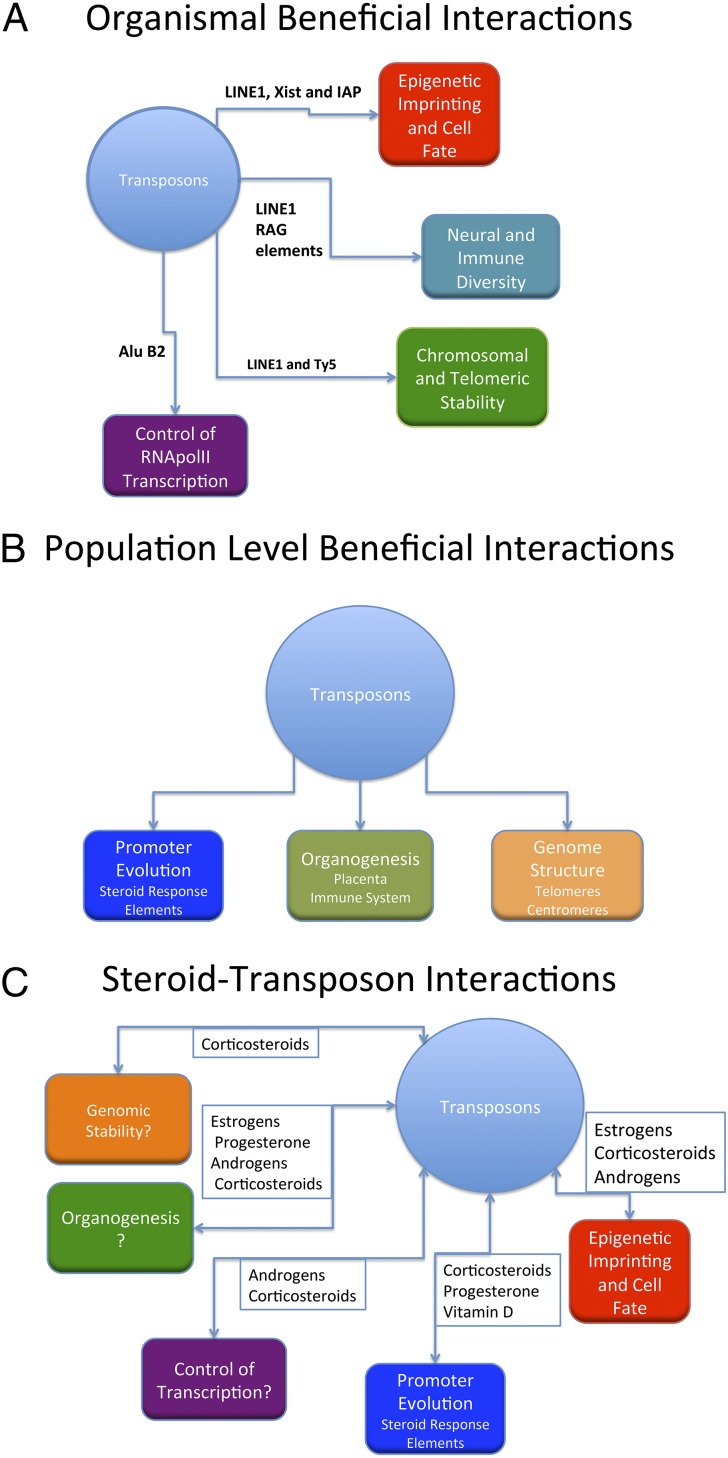
Of course, McClintock did not take the view that controlling elements were parasites; she felt that they helped the organism and the genome respond to changing and stressful environments (66). There is certainly evidence to support this view (Fig. 1); transposons form or can be recruited to telomeres in yeast, plants, insects, and rodents (66, 67). In CHO cells where telomere function is disrupted, L1 retrotransposons preferentially transpose into the region of the telomere, helping preserve telomere function (68). In yeast, the Ty5 LTR-retrotransposon is preferentially directed to the telomeres, save during stress, when a change in phosphorylation status causes it to be directed away from heterochromatin domains like the telomeres and toward actively transcribed genes, allowing a stress-responsive reshuffling of the genome (69). Similar responses have been observed in plants (70). It is now thought that the telomerases that normally maintain the eukaryotic telomere derive from ancient retrotransposon reverse transcriptases (67). Indeed, it has been argued that both the epigenetic machinery that controls transposons and builds telomeres, in concert with transposons, has been a major factor in determining the structure and size of eukaryotic genomes (66). Retrotransposons also play a role in mammalian development from meiosis onward and are thought to be the major drivers behind the evolution of the placenta, which shows the highest level of retrotransposon activity thus far described in a mammalian tissue (71). It is interesting to note that the adrenal cortex, the principal site of glucocorticoid synthesis, expresses the second highest level of endogenous retrovirus (ERV)-type transposons in humans after the placenta (72). Further, retrotransposons have been major factors in the development of epigenetic imprinting (the agouti locus being a notable example) and X-inactivation (Fig. 1A) (73, 74).
Fig. 1.
(A) Description of potentially beneficial actions of transposons that are known to occur at the organismal level. The names next to the connecting lines are those of the elements that have been shown to be involved in the relevant function: e.g., the transposon-derived XIST element governs epigenetically mediated X-chromosome inactivation, and an IAP ERV/LTR retrotransposon created the epigenetically imprinted agouti locus. (B) Description of population-level beneficial effects of transposon endosymbiosis, giving known examples in smaller print: e.g., the evolution of both the mammalian immune system and the placenta have been driven to a large extent by transposon-derived genomic elements. (C) Outline of both known and potential interactions between steroids and transposable elements, with more theoretical interactions followed by a question mark. For instance, those organs that show the highest levels of retrotransposon activity, such as the brain and placenta, also seem to be both steroidogenic and steroid-sensitive although the link remains correlative (see Transposons: Controlling Elements After All).
Transposable elements have also been a significant source of ready-made promoter elements and transcription factor-binding sites during the evolution of the genome (75, 76). This relation with transposable elements seems to be true with regard to steroid receptors in particular (77). Alu SINEs alone are known to form the substrate for response elements for progesterone, glucocorticoids, and vitamin D (78, 79). Steroids and their receptors appear linked in a variety of ways to both chromatin structure and retrotransposons. Steroid receptors themselves are capable of causing interchromosomal interactions up to and including translocations (80), and it is suspected that this capacity is additive or synergistic with the capacity of long interspersed elements (LINEs) to produce chromosomal rearrangements (81). It has been observed that SINE transcription can be induced in rat liver cells by glucocorticoids (82) whereas L1 LINE elements show increased expression when stimulated with androgens (83). The interaction between LINEs and androgen receptors (ARs) seems to be bidirectional in that the LINE-1 ORF1 protein seems to interact directly with AR in prostate-cancer cells and act as a positive coactivator of androgen-regulated genes (84). The observation that steroidogenic organs like the placenta, adrenals, and gonads (85, 86) show the highest levels of retrotransposon activity is worthy of attention, especially given the role of the brain as both an endocrine organ and steroid target.
Steroids drive the development of sexual differentiation (87, 88), and many neuropsychiatric disorders show marked differences in incidence and age of onset between the sexes (4, 89, 90). Therefore, it is reasonable to presume that, to the extent steroids regulate the expression of transposable elements, this regulation could contribute to observed sex differences in mental disorders. To extend the hypothesis about schizophrenia, elaborated above, it could be argued that the earlier age of onset of this disease in males could be due to the underexplored immunosuppressive effects of androgens, which are at peak concentrations in the adolescent males at risk for a first psychotic episode (91, 92). Alternately, it could be due to direct activation of ectopic retrotransposon expression by AR itself. Both of these hypotheses seem plausible and needn’t be mutually exclusive, yet the question remains entirely open. Similar hypotheses could be made with regard to other mental disorders with different incidence between the sexes, such as autism or major depression (93). The interactions between the transposome and both sex and stress steroids (Fig. 1C) seem likely to be fruitful targets for researchers seeking to understand how transposons might contribute usefully to normal function as well as those seeking novel explanations for complex diseases to which sex and stress show clear contributions.
Within the brain there is evidence that transposons may have beneficial roles. Gage and coworkers have demonstrated that L1 line retrotransposition occurs during mammalian brain development and neurogenesis and that the rate of transposition is influenced positively by environmental influences like exercise (94, 95). In parallel to our own observations, L1 activity seems to be under epigenetic control involving DNA methylation and MeCP2 (96). Gage and coworkers have argued that this retrotransposon-induced mosaicism permits increased neuronal diversity in analogy with the retrotransposon-derived V(D)J recombination system that drives antibody diversity in the immune system (97), which is one of the best-described and understood examples of the adaptive value of the transposition machinery (98–100). In this vein, it is interesting to note that there are substantial differences in the regulation of stem-cell L1 elements in humans and apes (101). Although the capacity of these elements to produce neuronal diversity through transposition is clear, it is likely that this genotypic diversity is not the only means by which these elements contribute to brain function. In the human cortex, transposition occurs at a fairly low rate (102), suggesting that diversity produced by transposition may not be as significant a factor there as it is in the hippocampus. We have alluded to the gene regulatory capacity of SINE RNA above, and it is likely that other varieties of retrotransposon RNA could have similar regulatory functions. Transposons are noted for their capacity to induce DNA strand breaks, generally with negative consequences, but recent findings that DNA double-strand breaks are associated with normal exploratory behavior in regions important for memory formation, like the hippocampus (103), suggest that retrotransposons could contribute to memory formation as well although this mechanism is almost completely unexplored.
It remains undetermined whether retrotransposons are truly symbiotic in eukaryotic genomes (104, 105), but it can be hypothesized that this relationship is in fact the case and that they have been coopted so as to be largely beneficial (Fig. 1B). If one compares prokaryotic genomes with eukaryotic genomes, it is immediately plain that the transposon content of the former is orders of magnitude smaller than in the latter whereas, on the other hand, horizontal gene transfer is much more common in prokaryotes than in eukaryotes. In prokaryotes, the energetic and competitive constraints on genome size are quite high, and the common availability of horizontal gene transfer allows bacteria to acquire adaptive genes on an as-needed basis and discard them when the need disappears. Within a multicellular organism, the local genetic diversity is very low (unless an infection has occurred); this low diversity means that opportunities for adaptive horizontal transfer are minimal. As a consequence, multicellular eukaryotes would need to take their genetic diversity along with them if they wished to maintain the flexible genomic response to environmental stress available to prokaryotes via horizontal transfer. If this view of transposons as a portable library of diversity holds true, the transposome can be regarded as a sort of endosymbiont. One prediction of this idea is that eukaryotes with a more prokaryotic lifestyle, with more access to horizontal gene transfer, would have smaller genomes with lower transposon content. This prediction is supported by the observation that unicellular eukaryotic genomes have low (less than 5%) or no detectable transposable elements (106–108). The obverse should also be true: large multicellular eukaryotes should have higher transposon content in their genomes, especially plants, because they cannot rely on behavioral strategies to defeat environmental insults. This prediction too seems to hold, because the human genome is roughly 50% transposon (109) whereas the maize genome, where transposons were originally identified, is 80% transposon (66, 110). It can be seen, then, that a functional, potentially symbiotic role for transposable elements in eukaryotic genomes is entirely plausible.
Conclusions and Outlook for the Future
Transposons are more than junk; they have played a significant role in genome evolution and arguably in the maintenance of genome structure and stability (66). They have been successfully coopted by the mammalian immune system to provide antibody diversity, and a similar process may be at work in generating genomic, and by extension behavioral, diversity in the brain (17, 97, 111). Our work suggests that the brain may also need to regulate retrotransposon RNA expression on a rapid and dynamic basis and that this regulation could have adaptive significance (16). At present, very little other evidence exists to inform our understanding of what the functional role of transposons in the brain might be, but the tools to gather this evidence are readily available in the form of next-generation sequencing and modern bioinformatics. Simple questions, such as how retrotransposon expression differs between brain regions in response to different environmental inputs, remain unanswered. More complex questions, such as how retrotransposons detect stress, how they might promote behavioral diversity, and the mechanisms by which cells choose to regulate their expression, also remain to be answered, both generally and with regard to the nervous system. Epigenetic marks are certainly among the tools cells use to regulate transposon expression, and it seems likely, at least in the context of stress, that steroid hormones will also be involved in orchestrating retrotransposon expression, just as they do with regard to genes. The number of open questions is too great to list here, but it is sufficient to say that, should transposons prove to have a significant functional role in the nervous system, that finding will increase the level of complexity required to understand that system by an order of magnitude.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Epigenetic Changes in the Developing Brain: Effects on Behavior,” held March 28–29, 2014, at the National Academy of Sciences in Washington, DC. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/Epigenetic_changes.
This article is a PNAS Direct Submission. E.B.K. is a guest editor invited by the Editorial Board.
References
- 1.Hunter RG, McEwen BS. Stress and anxiety across the lifespan: Structural plasticity and epigenetic regulation. Epigenomics. 2013;5(2):177–194. doi: 10.2217/epi.13.8. [DOI] [PubMed] [Google Scholar]
- 2.Lupien SJ, McEwen BS, Gunnar MR, Heim C. Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nat Rev Neurosci. 2009;10(6):434–445. doi: 10.1038/nrn2639. [DOI] [PubMed] [Google Scholar]
- 3.McEwen BS, Gianaros PJ. Stress- and allostasis-induced brain plasticity. Annu Rev Med. 2011;62:431–445. doi: 10.1146/annurev-med-052209-100430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McEwen BS. Stress, sex, and neural adaptation to a changing environment: Mechanisms of neuronal remodeling. Ann N Y Acad Sci. 2010;1204(Suppl):E38–E59. doi: 10.1111/j.1749-6632.2010.05568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shonkoff JP, Boyce WT, McEwen BS. Neuroscience, molecular biology, and the childhood roots of health disparities: Building a new framework for health promotion and disease prevention. JAMA. 2009;301(21):2252–2259. doi: 10.1001/jama.2009.754. [DOI] [PubMed] [Google Scholar]
- 6.Bohacek J, Mansuy IM. Epigenetic inheritance of disease and disease risk. Neuropsychopharmacology. 2013;38(1):220–236. doi: 10.1038/npp.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McEwen BS, Wingfield JC. The concept of allostasis in biology and biomedicine. Horm Behav. 2003;43(1):2–15. doi: 10.1016/s0018-506x(02)00024-7. [DOI] [PubMed] [Google Scholar]
- 8.McEwen BS. Protective and damaging effects of stress mediators. N Engl J Med. 1998;338(3):171–179. doi: 10.1056/NEJM199801153380307. [DOI] [PubMed] [Google Scholar]
- 9.McEwen BS, Gianaros PJ. Central role of the brain in stress and adaptation: Links to socioeconomic status, health, and disease. Ann N Y Acad Sci. 2010;1186:190–222. doi: 10.1111/j.1749-6632.2009.05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radley JJ, et al. Stress risk factors and stress-related pathology: Neuroplasticity, epigenetics and endophenotypes. Stress. 2011;14(5):481–497. doi: 10.3109/10253890.2011.604751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karatsoreos IN, McEwen BS. Annual research review: The neurobiology and physiology of resilience and adaptation across the life course. J Child Psychol Psychiatry. 2013;54(4):337–347. doi: 10.1111/jcpp.12054. [DOI] [PubMed] [Google Scholar]
- 12.Yehuda R, Bierer LM. Transgenerational transmission of cortisol and PTSD risk. Prog Brain Res. 2008;167:121–135. doi: 10.1016/S0079-6123(07)67009-5. [DOI] [PubMed] [Google Scholar]
- 13.Hunter RG. Epigenetic effects of stress and corticosteroids in the brain. Front Cell Neurosci. 2012;6:18. doi: 10.3389/fncel.2012.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths BB, Hunter RG. Neuroepigenetics of stress. Neuroscience. 2014;275:420–435. doi: 10.1016/j.neuroscience.2014.06.041. [DOI] [PubMed] [Google Scholar]
- 15.Hunter RG, et al. Acute stress and hippocampal histone H3 lysine 9 trimethylation, a retrotransposon silencing response. Proc Natl Acad Sci USA. 2012;109(43):17657–17662. doi: 10.1073/pnas.1215810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter RG, McEwen BS, Pfaff DW. Environmental stress and transposon transcription in the mammalian brain. Mob Genet Elements. 2013;3(2):e24555. doi: 10.4161/mge.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reilly MT, Faulkner GJ, Dubnau J, Ponomarev I, Gage FH. The role of transposable elements in health and diseases of the central nervous system. J Neurosci. 2013;33(45):17577–17586. doi: 10.1523/JNEUROSCI.3369-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett EL, Diamond MC, Krech D, Rosenzweig MR. Chemical and anatomical plasticity brain. Science. 1964;146(3644):610–619. doi: 10.1126/science.146.3644.610. [DOI] [PubMed] [Google Scholar]
- 19.Erickson KI, et al. Exercise training increases size of hippocampus and improves memory. Proc Natl Acad Sci USA. 2011;108(7):3017–3022. doi: 10.1073/pnas.1015950108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Merzenich MM, Sameshima K, Jenkins WM. Remodelling of hand representation in adult cortex determined by timing of tactile stimulation. Nature. 1995;378(6552):71–75. doi: 10.1038/378071a0. [DOI] [PubMed] [Google Scholar]
- 21.Draganski B, et al. Temporal and spatial dynamics of brain structure changes during extensive learning. J Neurosci. 2006;26(23):6314–6317. doi: 10.1523/JNEUROSCI.4628-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bezzola L, Mérillat S, Gaser C, Jäncke L. Training-induced neural plasticity in golf novices. J Neurosci. 2011;31(35):12444–12448. doi: 10.1523/JNEUROSCI.1996-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McEwen BS. Physiology and neurobiology of stress and adaptation: Central role of the brain. Physiol Rev. 2007;87(3):873–904. doi: 10.1152/physrev.00041.2006. [DOI] [PubMed] [Google Scholar]
- 24.McEwen BS. Stress and hippocampal plasticity. Annu Rev Neurosci. 1999;22:105–122. doi: 10.1146/annurev.neuro.22.1.105. [DOI] [PubMed] [Google Scholar]
- 25.Goldwater DS, et al. Structural and functional alterations to rat medial prefrontal cortex following chronic restraint stress and recovery. Neuroscience. 2009;164(2):798–808. doi: 10.1016/j.neuroscience.2009.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bloss EB, Janssen WG, McEwen BS, Morrison JH. Interactive effects of stress and aging on structural plasticity in the prefrontal cortex. J Neurosci. 2010;30(19):6726–6731. doi: 10.1523/JNEUROSCI.0759-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vyas A, Mitra R, Shankaranarayana Rao BS, Chattarji S. Chronic stress induces contrasting patterns of dendritic remodeling in hippocampal and amygdaloid neurons. J Neurosci. 2002;22(15):6810–6818. doi: 10.1523/JNEUROSCI.22-15-06810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roozendaal B, McEwen BS, Chattarji S. Stress, memory and the amygdala. Nat Rev Neurosci. 2009;10(6):423–433. doi: 10.1038/nrn2651. [DOI] [PubMed] [Google Scholar]
- 29.McEwen BS, Eiland L, Hunter RG, Miller MM. Stress and anxiety: Structural plasticity and epigenetic regulation as a consequence of stress. Neuropharmacology. 2012;62(1):3–12. doi: 10.1016/j.neuropharm.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kempermann G, Kuhn HG, Gage FH. More hippocampal neurons in adult mice living in an enriched environment. Nature. 1997;386(6624):493–495. doi: 10.1038/386493a0. [DOI] [PubMed] [Google Scholar]
- 31.van Praag H, Kempermann G, Gage FH. Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat Neurosci. 1999;2(3):266–270. doi: 10.1038/6368. [DOI] [PubMed] [Google Scholar]
- 32.Pereira AC, et al. An in vivo correlate of exercise-induced neurogenesis in the adult dentate gyrus. Proc Natl Acad Sci USA. 2007;104(13):5638–5643. doi: 10.1073/pnas.0611721104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anda RF, Butchart A, Felitti VJ, Brown DW. Building a framework for global surveillance of the public health implications of adverse childhood experiences. Am J Prev Med. 2010;39(1):93–98. doi: 10.1016/j.amepre.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 34.Gourley SL, Swanson AM, Koleske AJ. Corticosteroid-induced neural remodeling predicts behavioral vulnerability and resilience. J Neurosci. 2013;33(7):3107–3112. doi: 10.1523/JNEUROSCI.2138-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gray JD, Rubin TG, Hunter RG, McEwen BS. Hippocampal gene expression changes underlying stress sensitization and recovery. Mol Psychiatry. 2013 doi: 10.1038/mp.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crow TJ. ‘The missing genes: What happened to the heritability of psychiatric disorders?’. Mol Psychiatry. 2011;16(4):362–364. doi: 10.1038/mp.2010.92. [DOI] [PubMed] [Google Scholar]
- 37.Danchin É, et al. Beyond DNA: Integrating inclusive inheritance into an extended theory of evolution. Nat Rev Genet. 2011;12(7):475–486. doi: 10.1038/nrg3028. [DOI] [PubMed] [Google Scholar]
- 38.Menke A, Klengel T, Binder EB. Epigenetics, depression and antidepressant treatment. Curr Pharm Des. 2012;18(36):5879–5889. doi: 10.2174/138161212803523590. [DOI] [PubMed] [Google Scholar]
- 39.Crow TJ. Schizophrenia as variation in the sapiens-specific epigenetic instruction to the embryo. Clin Genet. 2012;81(4):319–324. doi: 10.1111/j.1399-0004.2012.01830.x. [DOI] [PubMed] [Google Scholar]
- 40.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53(6):857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 41.Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, McEwen BS. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc Natl Acad Sci USA. 2009;106(49):20912–20917. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowe HM, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463(7278):237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 43.Ratman D, et al. How glucocorticoid receptors modulate the activity of other transcription factors: A scope beyond tethering. Mol Cell Endocrinol. 2013;380(1-2):41–54. doi: 10.1016/j.mce.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 44.Gagnidze K, Weil ZM, Faustino LC, Schaafsma SM, Pfaff DW. Early histone modifications in the ventromedial hypothalamus and preoptic area following oestradiol administration. J Neuroendocrinol. 2013;25(10):939–955. doi: 10.1111/jne.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gagnidze K, Weil ZM, Pfaff DW. Histone modifications proposed to regulate sexual differentiation of brain and behavior. BioEssays. 2010;32(11):932–939. doi: 10.1002/bies.201000064. [DOI] [PubMed] [Google Scholar]
- 46.McCarthy MM, Nugent BM. Epigenetic contributions to hormonally-mediated sexual differentiation of the brain. J Neuroendocrinol. 2013;25(11):1133–1140. doi: 10.1111/jne.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226(4676):792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 48.McClintock B. Chromosome organization and genic expression. Cold Spring Harb Symp Quant Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- 49.Orgel LE, Crick FH. Selfish DNA: The ultimate parasite. Nature. 1980;284(5757):604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 50.Ohno S. So much “junk” DNA in our genome. Brookhaven Symp Biol. 1972;23:366–370. [PubMed] [Google Scholar]
- 51.Li T, Spearow J, Rubin CM, Schmid CW. Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene. 1999;239(2):367–372. doi: 10.1016/s0378-1119(99)00384-4. [DOI] [PubMed] [Google Scholar]
- 52.Espinoza CA, Goodrich JA, Kugel JF. Characterization of the structure, function, and mechanism of B2 RNA, an ncRNA repressor of RNA polymerase II transcription. RNA. 2007;13(4):583–596. doi: 10.1261/rna.310307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mariner PD, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29(4):499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 54.Davis HP, Squire LR. Protein synthesis and memory: A review. Psychol Bull. 1984;96(3):518–559. [PubMed] [Google Scholar]
- 55.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 2011;12(9):615–627. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stetson DB. Endogenous retroelements and autoimmune disease. Curr Opin Immunol. 2012;24(6):692–697. doi: 10.1016/j.coi.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tarallo V, et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012;149(4):847–859. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bundo M, et al. Increased l1 retrotransposition in the neuronal genome in schizophrenia. Neuron. 2014;81(2):306–313. doi: 10.1016/j.neuron.2013.10.053. [DOI] [PubMed] [Google Scholar]
- 59.Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32(5):1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ponomarev I, Rau V, Eger EI, Harris RA, Fanselow MS. Amygdala transcriptome and cellular mechanisms underlying stress-enhanced fear learning in a rat model of posttraumatic stress disorder. Neuropsychopharmacology. 2010;35(6):1402–1411. doi: 10.1038/npp.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li W, Jin Y, Prazak L, Hammell M, Dubnau J. Transposable elements in TDP-43-mediated neurodegenerative disorders. PLoS ONE. 2012;7(9):e44099. doi: 10.1371/journal.pone.0044099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li W, et al. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat Neurosci. 2013;16(5):529–531. doi: 10.1038/nn.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Douville R, Liu J, Rothstein J, Nath A. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann Neurol. 2011;69(1):141–151. doi: 10.1002/ana.22149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pasternak O, et al. Excessive extracellular volume reveals a neurodegenerative pattern in schizophrenia onset. J Neurosci. 2012;32(48):17365–17372. doi: 10.1523/JNEUROSCI.2904-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dawkins R. 2006. The Selfish Gene (Oxford Univ Press, Oxford), 30th Anniversary Ed.
- 66.Fedoroff NV. Presidential address: Transposable elements, epigenetics, and genome evolution. Science. 2012;338(6108):758–767. doi: 10.1126/science.338.6108.758. [DOI] [PubMed] [Google Scholar]
- 67.Belfort M, Curcio MJ, Lue NF. Telomerase and retrotransposons: Reverse transcriptases that shaped genomes. Proc Natl Acad Sci USA. 2011;108(51):20304–20310. doi: 10.1073/pnas.1100269109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morrish TA, et al. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446(7132):208–212. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 69.Dai J, Xie W, Brady TL, Gao J, Voytas DF. Phosphorylation regulates integration of the yeast Ty5 retrotransposon into heterochromatin. Mol Cell. 2007;27(2):289–299. doi: 10.1016/j.molcel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 70.Ito H, et al. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature. 2011;472(7341):115–119. doi: 10.1038/nature09861. [DOI] [PubMed] [Google Scholar]
- 71.Dupressoir A, Lavialle C, Heidmann T. From ancestral infectious retroviruses to bona fide cellular genes: Role of the captured syncytins in placentation. Placenta. 2012;33(9):663–671. doi: 10.1016/j.placenta.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 72.Katsumata K, et al. Tissue-specific high-level expression of human endogenous retrovirus-R in the human adrenal cortex. Pathobiology. 1998;66(5):209–215. doi: 10.1159/000028025. [DOI] [PubMed] [Google Scholar]
- 73.Gifford WD, Pfaff SL, Macfarlan TS. Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol. 2013;23(5):218–226. doi: 10.1016/j.tcb.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Keverne EB. Importance of the matriline for genomic imprinting, brain development and behaviour. Philos Trans R Soc Lond B Biol Sci. 2013;368(1609):20110327. doi: 10.1098/rstb.2011.0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rebollo R, Romanish MT, Mager DL. Transposable elements: An abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 2012;46:21–42. doi: 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]
- 76.Faulkner GJ, Carninci P. Altruistic functions for selfish DNA. Cell Cycle. 2009;8(18):2895–2900. doi: 10.4161/cc.8.18.9536. [DOI] [PubMed] [Google Scholar]
- 77.Cotnoir-White D, Laperrière D, Mader S. Evolution of the repertoire of nuclear receptor binding sites in genomes. Mol Cell Endocrinol. 2011;334(1-2):76–82. doi: 10.1016/j.mce.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 78.Jacobsen BM, Jambal P, Schittone SA, Horwitz KB. ALU repeats in promoters are position-dependent co-response elements (coRE) that enhance or repress transcription by dimeric and monomeric progesterone receptors. Mol Endocrinol. 2009;23(7):989–1000. doi: 10.1210/me.2009-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gombart AF, Saito T, Koeffler HP. Exaptation of an ancient Alu short interspersed element provides a highly conserved vitamin D-mediated innate immune response in humans and primates. BMC Genomics. 2009;10:321. doi: 10.1186/1471-2164-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin C, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139(6):1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Holzman DC. Aberrant chromosomes: Not so random after all? J Natl Cancer Inst. 2010;102(6):368–369. doi: 10.1093/jnci/djq082. [DOI] [PubMed] [Google Scholar]
- 82.Sun LH, Frankel FR. The induction of Alu-sequence transcripts by glucocorticoid in rat liver cells. J Steroid Biochem. 1986;25(2):201–207. doi: 10.1016/0022-4731(86)90417-6. [DOI] [PubMed] [Google Scholar]
- 83.Morales JF, Snow ET, Murnane JP. Environmental factors affecting transcription of the human L1 retrotransposon. I. Steroid hormone-like agents. Mutagenesis. 2002;17(3):193–200. doi: 10.1093/mutage/17.3.193. [DOI] [PubMed] [Google Scholar]
- 84.Lu Y, et al. LINE-1 ORF-1p functions as a novel androgen receptor co-activator and promotes the growth of human prostatic carcinoma cells. Cell Signal. 2013;25(2):479–489. doi: 10.1016/j.cellsig.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 85.Pillai RS, Chuma S. piRNAs and their involvement in male germline development in mice. Dev Growth Differ. 2012;54(1):78–92. doi: 10.1111/j.1440-169X.2011.01320.x. [DOI] [PubMed] [Google Scholar]
- 86.Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505(7483):353–359. doi: 10.1038/nature12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zup SL, Forger NG. Hormones and sexual differentiation. In: Ramachandran VS, editor. Encyclopedia of the Human Brain. Elsevier; San Diego: 2002. [Google Scholar]
- 88.McCarthy MM, Arnold AP. Reframing sexual differentiation of the brain. Nat Neurosci. 2011;14(6):677–683. doi: 10.1038/nn.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goldstein JM, Cherkerzian S, Tsuang MT, Petryshen TL. Sex differences in the genetic risk for schizophrenia: History of the evidence for sex-specific and sex-dependent effects. Am J Med Genet B Neuropsychiatr Genet. 2013;162B(7):698–710. doi: 10.1002/ajmg.b.32159. [DOI] [PubMed] [Google Scholar]
- 90.Bao AM, Swaab DF. Sex differences in the brain, behavior, and neuropsychiatric disorders. Neuroscientist. 2010;16(5):550–565. doi: 10.1177/1073858410377005. [DOI] [PubMed] [Google Scholar]
- 91.Angele MK, Pratschke S, Hubbard WJ, Chaudry IH. Gender differences in sepsis: Cardiovascular and immunological aspects. Virulence. 2014;5(1):12–19. doi: 10.4161/viru.26982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bernin H, Lotter H. Sex bias in the outcome of human tropical infectious diseases: Influence of steroid hormones. J Infect Dis. 2014;209(Suppl 3):S107–S113. doi: 10.1093/infdis/jit610. [DOI] [PubMed] [Google Scholar]
- 93.Young LJ, Pfaff DW. Sex differences in neurological and psychiatric disorders. Front Neuroendocrinol. 2014;35(3):253–254. doi: 10.1016/j.yfrne.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 94.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435(7044):903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 95.Muotri AR, Zhao C, Marchetto MC, Gage FH. Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus. 2009;19(10):1002–1007. doi: 10.1002/hipo.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Muotri AR, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468(7322):443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. LINE-1 retrotransposons: Mediators of somatic variation in neuronal genomes? Trends Neurosci. 2010;33(8):345–354. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3(6):e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: A possible source of oncogenic translocations. Cell. 1998;94(4):463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 100.Erwin JA, Marchetto MC, Gage FH. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat Rev Neurosci. 2014;15(8):497–506. doi: 10.1038/nrn3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Marchetto MC, et al. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature. 2013;503(7477):525–529. doi: 10.1038/nature12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Evrony GD, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151(3):483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Suberbielle E, et al. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-β. Nat Neurosci. 2013;16(5):613–621. doi: 10.1038/nn.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Upton KR, Baillie JK, Faulkner GJ. Is somatic retrotransposition a parasitic or symbiotic phenomenon? Mob Genet Elements. 2011;1(4):279–282. doi: 10.4161/mge.18422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ryan FP. Human endogenous retroviruses in health and disease: A symbiotic perspective. J R Soc Med. 2004;97(12):560–565. doi: 10.1258/jrsm.97.12.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: A comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8(5):464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 107.Maumus F, et al. Potential impact of stress activated retrotransposons on genome evolution in a marine diatom. BMC Genomics. 2009;10:624. doi: 10.1186/1471-2164-10-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pritham EJ. Transposable elements and factors influencing their success in eukaryotes. J Hered. 2009;100(5):648–655. doi: 10.1093/jhered/esp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lander ES, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 110.SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL. The paleontology of intergene retrotransposons of maize. Nat Genet. 1998;20(1):43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 111.Baillie JK, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479(7374):534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]