Abstract
The building block of chromatin is nucleosome, which consists of 146 base pairs of DNA wrapped around a histone octamer composed of two copies of histone H2A, H2B, H3, and H4. Significantly, the somatic missense mutations of the histone H3 variant, H3.3, are associated with childhood and young-adult tumors, such as pediatric high-grade astrocytomas, as well as chondroblastoma and giant-cell tumors of the bone. The mechanisms by which these histone mutations cause cancer are by and large unclear. Interestingly, two recent studies identified BS69/ZMYND11, which was proposed to be a candidate tumor suppressor, as a specific reader for a modified form of H3.3 (H3.3K36me3). Importantly, some H3.3 cancer mutations are predicted to abrogate the H3.3K36me3/BS69 interaction, suggesting that this interaction may play an important role in tumor suppression. These new findings also raise the question of whether H3.3 cancer mutations may lead to the disruption and/or gain of interactions of additional cellular factors that contribute to tumorigenesis.
Keywords: histone, H3.3, H3.3K36me3, cancer, BS69
The histone H3 variant, H3.3, differs from the canonical histone H3 (H3.2 and H3.1) by only 4 and 5 amino acids, respectively. With the exception of serine 31 located in the H3.3 tail region, the rest of the amino acid difference resides in the nucleosome core, which dictates the differential recognition of the H3.3 variant versus the canonical histone H3 by different histone chaperones as well as its mode of incorporation into the chromatin. The canonical histone H3 proteins are incorporated into chromatin via the histone chaperone CAF-1 (chromatin assembly factor 1) at S phase in a replication-dependent manner (1). In contrast, H3.3 is incorporated into chromatin by HIRA (histone regulator A) and DAXX (death domain-associated protein)/ATRX (alpha thalassemia/mental retardation syndrome X-linked protein), respectively, in a replication-independent manner (1–3), suggesting that H3.3 may play a regulatory role in many chromatin-templated processes outside of the S phase of the cell cycle, including transcription. Consistently, H3.3 has been found at gene bodies and promoters of transcriptionally active genes as well as gene regulatory elements, such as enhancers (1, 4, 5). More recently, it has been shown that the H3.3-containing nucleosomes are compromised in their ability to form compact structure in vitro and that the H3.3 genomic distribution correlates well with DNase I-sensitive regions, suggesting a preferential association with open chromatin environment in vivo (6). Interestingly, H3.3 has also been found at pericentromeric and telomeric regions, where its deposition is largely dependent on the DAXX/ATRX chaperone system; however, the function of H3.3 at these regions remains unclear (7).
Although H3.3 represents only a small portion of the total cellular histone H3 pool (8), emerging evidence suggests that H3.3 carries biological information that is distinct from its counterparts, H3.1 and H3.2. This notion is best exemplified by the exciting recent findings of recurring heterozygous mutations in H3F3A and H3F3B, the only two genes in mammals that encode H3.3, that are associated with a number of pediatric cancers, including pediatric and young-adult high-grade astrocytomas, chondroblastoma, and giant-cell tumor of bone (9–11). These mutations include K27M, G34R/G34V/G34W/G34L, and K36M. Although both H3F3A and H3F3B encode H3.3 with identical amino acid sequences, the H3.3K36M mutation occurs predominantly in H3F3B whereas the other mutations are almost exclusive to H3F3A (9). Furthermore, these different mutations also appear to segregate with distinct types of tumors. For instance, the K27M mutation has been found only in pediatric diffuse intrinsic pontine glioma (DIPG) and high-grade astrocytomas primarily restricted to midline locations (spinal cord, thalamus, pons, brainstem) in children and younger adults (11–17). The majority of K36M mutation has been found in chondroblastoma, and to a lesser extent, in clear-cell chondrosarcoma (9). The G34R/V mutations predominantly associate with pediatric glioblastoma multiforme (GBM) in the cerebral hemispheres (11, 12, 18, 19), and in some very rare cases, in osteosarcoma (9). Interestingly, two different substitutions at the same amino acid position, G34W and G34L, have been found only in giant-cell tumor of bone (9). It remains to be determined whether differential association of these H3.3 mutations with differential cancer types may imply disparate underlying molecular mechanisms.
Recent studies have begun to address the mechanism by which H3.3 mutations may cause cancer. It has been demonstrated that the H3.3K27M mutation affects not only the methylation potential on the mutated histone tail but also global methylation of H3K27me3 in cell-culture models as well as in the primary tumors (19–21). Supporting this finding, a recent study in Drosophila also found that the H3.3K27M ectopic expression phenocopies PRC2 mutants and causes loss of global H3K27me3 and depression of PRC2 target genes (22). In the study from Lewis et al., H3.3K36M was also shown to cause a global reduction of H3K36me3 (19). Cross talk between these mutations and the nearby modifications has also been observed. For instance, G34R/V mutations have been found to cause a significant loss of H3.3K36me3 only in cis (19), suggesting that these mutations may differentially influence the affected epigenomes. Subsequent studies also showed that K27M and G34R/V mutations are mutually exclusive in tumors and are associated with distinct gene expression and DNA methylation profiles (11, 12). The clinical significance of these findings, however, remains to be determined. Taken together, these recent exciting findings suggest that H3.3 mutations may play an oncogenic driver role by reshaping the epigenomes through alterations of either the local or global histone methylation patterns. Although much remains to be learned regarding the mechanism by which these mutations cause cancer, two recent studies unexpectedly found an H3.3K36me3-specific reader, BS69/ZMYND11, which may provide a new avenue to explore H3.3 biology as well as its cancer connection (23, 24).
BS69 (also Known as ZMYND11)
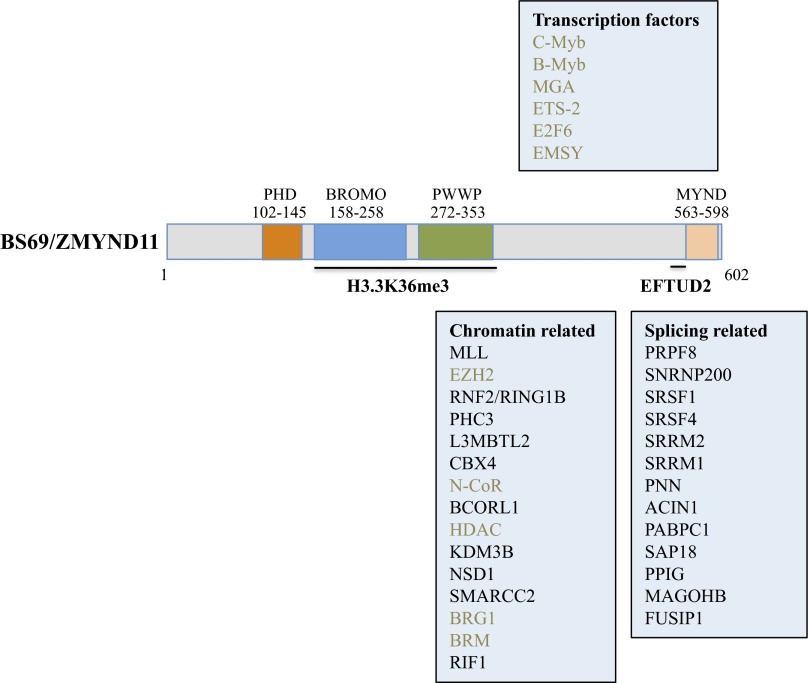
BS69 was originally identified as an interacting protein of the adenoviral E1A oncoprotein and a suppressor of the transactivating function of E1A and has thus been suggested to function as a transcriptional repressor and a candidate tumor suppressor (25–27). The architecture of BS69 is quite interesting; the N-terminal two thirds of BS69 contains three tandemly arranged, putative chromatin recognition modules: namely PHD, BROMO, and PWWP domains (Fig. 1). The C terminus of BS69 contains a zinc-binding motif, the MYND domain, which functions as a protein–protein interaction surface, which mediates interactions of BS69 with transcription factors and chromatin (26–28) (Fig. 1). Compared with the MYND domain, essentially nothing was known about the three N-terminal putative chromatin readers. Previous studies indicated that PHD and BROMO domains are protein modalities that mainly recognize methylated and acetylated lysines located on the histone tails, respectively, whereas the PWWP domain has been suggested to primarily recognize the trimethylated histone H3 lysine 36 (29–31), as well as H4 lysine 20 (32, 33). Therefore, the presence of these reader domains in BS69 suggests that BS69 may also recognize specific chromatin modification patterns, thus playing a bridging role between transcription and chromatin. The two recent publications by Wen et al. (23) and Guo et al. (24) shed light not only on chromatin recognition mediated by these domains (with an interesting and exciting twist) but also how these domains participate in transcription-associated events (see BS69 BROMO-PWWP Domains Read H3.3K36me3).
Fig. 1.
Domain architecture of BS69 protein and its interaction partners. Diagram of the full-length BS69 protein with the various reader domains and the C-terminal MYND domain highlighted in different colors and with their amino acid positions indicated. BROMO and PWWP domains are responsible for H3.3K36me3 recognition (23, 24) whereas the EFTUD2 binding site is located at amino acids 556–562 (24). The other BS69-interacting proteins are grouped into different categories. Interacting proteins reported previously are in brown, and the proteins present in the FLAG-HA-BS69 immunoprecipitate, identified by mass spectrometry reported in the Guo et al. study (24), are in black.
BS69 BROMO-PWWP Domains Read H3.3K36me3
In these two recent studies (23, 24), the BS69 PWWP domain was identified as a specific reader for the K36 trimethylation on histone variant H3.3 (H3.3K36me3) (Fig. 1). In both studies, the authors used an array of methylated and unmethylated histone peptides and demonstrated that, surprisingly, BS69 preferentially recognizes H3.3K36me3. This highly specific recognition is sensitive to both the methyl level on lysine 36: i.e., BS69 binds to H3.3K36me3, but much less to H3.3K36me0/1/2, as well as the adjacent residues, S31 on H3.3 vs. A31 on H3.1/2 (23, 24). Using a cocrystallization approach, the Wen et al. study provided at the atomic level mechanistic understanding of how the BS69 reader modalities discriminate H3.3K36me3 from H3.1K36me3. Interestingly, the recognition of H3.3K36me3 by BS69 involves not only the PWWP but also the adjacent BROMO domain (23). Specifically, the H3.3-dependent recognition is mediated by the encapsulation of the H3.3-specific “S31” residue in a composite pocket formed by the tandem BROMO-PWWP domains of BS69. S31 is a critical contact that is lacking in H3.1/2, thus explaining the preference of BS69 toward H3.3K36me3 (23). The K36me3 contacts involve an aromatic cage formed by F291, W294, and F310 from the PWWP domain, explaining the preference of BS69 for the trimethyl state of H3.3K36 (23). Interestingly, the BS69 BROMO domain does not function as a canonical BROMO domain but rather contributes to the formation of the H3.3K36me3 recognition cavity (23). Consistent with such a role, the lack of a Kac binding pocket and the positive surface charge of the BS69 BROMO domain may impair its ability to function as a histone tail acetyl-lysine binding module (23). In agreement with the structural findings, Guo et al. (24) demonstrated that S31 phosphorylation abrogates BS69 binding to H3.3K36me3 in vitro, suggesting that BS69 recognition of H3.3K36me3 in vivo may be regulated by a signaling pathway that mediates S31 phosphorylation. In addition, Guo et al. also showed that BROMO domain deletion impaired BS69 chromatin association in vivo (24).
Consistent with these in vitro results, BS69 primarily binds gene bodies, which are decorated by H3K36 methylation. Importantly, BS69 gene-body binding is dependent on SETD2 (23, 24), which is the main trimethyltransferase of H3K36 in mammalian cells (34). Both groups also reported a good correlation between genomic occupancy of BS69 and those of H3K36me3 and H3.3 (23, 24). Consistent with the finding that BS69 preferentially binds H3.3K36me3 in vitro, sequential ChIP using H3K36me3 and FLAG antibodies in the H3.3-FLAG–expressing cells revealed that H3.3K36me3 is enriched at BS69 target genes (24). The development of an H3.3K36me3-specific antibody allowing precise mapping of the genome-wide locations of H3.3K36me3 will further inform the biological processes in which H3.3K36me3 is involved.
BS69 Suppresses Transcription Elongation and Promotes Intron Retention
Biochemical fractionation experiments showed that BS69 is mainly associated with chromatin, which is not unexpected for a potential chromatin regulator, and is consistent with the finding that BS69 binds H3.3K36me3 (24). Another interesting observation is that, although BS69 is associated with genes decorated by H3K36me3 and with relatively high expression levels, both groups reported that BS69 knockdown caused only a moderate change in gene expression (23, 24). Thus, it seems that BS69, rather than working as an essential “on/off switch,” may function to “fine-tune” gene expression. Using independent approaches, Wen et al. (23) and Guo et al. (24) reported different, yet not necessarily mutually exclusive, mechanisms of how BS69 regulates gene expression in the context of the H3.3K36me3-decorated chromatin environment.
BS69 Represses Elongation.
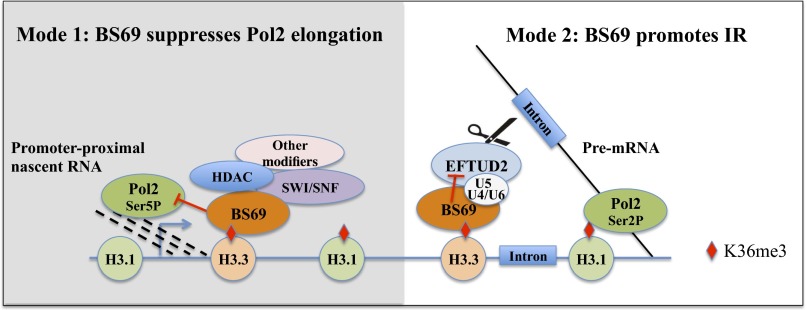
Wen et al. compared the RNA Pol2 gene body occupancies in U2OS cells with BS69 knockdown versus control and observed an enhanced RNA Pol2 elongation rate upon loss of BS69 in the bodies of those genes that are normally suppressed by BS69 (23). In support, the CTD Ser2 phosphorylated form of RNA Pol2, considered as an elongation-specific Pol2, as well as H3K36me3 level, also increases in the BS69 KD cells (23). Based on these results, the authors proposed a model whereby BS69 regulates gene expression by modulating histone modifications and histone exchange in the transcribed regions, thus controlling the release of RNA Pol2 from the paused stage to the elongating stage (35) (Fig. 2). The same study also reported that BS69 knockdown causes up-regulation of c-Myc as a consequence of enhanced elongation and accelerated proliferation (23). This finding provided new insight into how BS69 may function as a tumor suppressor in certain contexts. Interestingly, a previous study identified MYCN, a known driver of glioblastoma in a mouse model (36), as the most highly up-regulated gene in tumors with H3.3G34R/V substitutions (18). As discussed above, the H3.3G34R/V mutations were found to decrease H3.3K36me3 level in cis (19); thus, BS69 binding is predicted to be impaired in these contexts. Whether MYCN deregulation in the tumors with G34R/V mutation is due to the displacement of BS69 from chromatin and has clinical relevance warrant further investigations.
Fig. 2.
Two proposed modes of BS69 action in regulating Pol2 elongation and intron retention. (Left) BS69 binds H3.3K36me3 and regulates local chromatin environment to prevent promoter Pol2 from entering into gene bodies (23). (Right) BS69 is recruited to H3.3K36me3-enriched gene bodies and promotes intron retention by antagonizing the function of EFTUD2 through physical interaction (24). This antagonistic relationship may prevent U4 release, thus blocking the activation of local spliceosome.
In mouse ES cells, RNA Pol2 occupies most of the promoters regardless of the transcriptional status (37). For nonproductive genes, Pol2 stays in a poised state and shows evidence only of initiation but not elongation whereas, for actively transcribed genes, Pol2 signals were seen in both the gene bodies and promoters, indicating that the control of Pol2 pause release is connected to transcriptional activity (37). It is an interesting finding that an H3.3K36me3 binder seems to participate in this process. At the mechanistic level, further work is still required to answer whether BS69 and its associated proteins function to negatively control the Pol2 release from promoters and to reset chromatin after the passage of RNA Pol2, analogous to the better-understood processes in yeast modulated by the Isw1b and Eaf3/Rpd3 protein complexes (35).
BS69 Promotes Intron Retention.
In the second study, Guo et al. purified the BS69 protein complex in a soluble nuclear fraction as well as in an MNase solubilized chromatin fraction (24). In the soluble nuclear fraction, BS69 interacts with a number of DNA binding transcription factors and histone modifiers, such as MLL, NSD1, and KDM3B, as well as ATP-dependent chromatin remodelers (24) (Fig. 1), consistent with a transcriptional role for BS69 suggested by previous studies (25–28). Importantly, these authors found that the chromatin-bound BS69 mainly associates with a different set of proteins that are involved in regulating RNA splicing (24). These splicing-related proteins can be grouped into at least two classes: i.e., the U5 snRNP proteins (EFTUD2, PRPF8, and SNRNP200) and Serine and Arginine-rich (SR) proteins (SRSF1, PNN and ACIN1) (24) (Fig. 1). Consistently, further RNA profiling revealed hundreds of alternatively spliced events in BS69-depleted HeLa cells. Interestingly, the majority of the BS69-regulated alternative splicing (AS) events are intron retention (IR), indicating that BS69 may be a regulator, primarily of IR (24). Loss of BS69 results in a reduced IR whereas knockdown of EFTUD2 results in an increased IR, suggesting that BS69 functions to promote IR by antagonizing EFTUD2 (24). Importantly, the ability of BS69 to promote IR is dependent on its ability to physically interact with EFTUD2 as well as the H3.3K36me3 (24). Collectively, these findings suggest that BS69 functions to connect H3.3K36me3-decorated chromatin to IR regulation (Fig. 2).
The biochemical mechanism by which BS69 regulates IR remains to be fully elucidated. The challenge is that BS69 requires the chromatin context to regulate IR, which demands an in vitro chromatin template-based RNA splicing system, which is currently unavailable. However, some preliminary findings reported by Guo et al. (24) suggested a potential biochemical mechanism. Specifically, in a coimmunoprecipitation experiment, BS69 antibodies seem to bring down EFTUD2 and possibly a subpopulation of U5 snRNP that is surprisingly low in U5 but high in U4 snRNA (24). Given that EFTUD2 is involved in unwinding U4/U6 RNA and eviction of U4 snRNA, which is essential for spliceosome activation (38), the fact that U4 remains high in the BS69-U5 snRNP suggests the tantalizing possibility that BS69 may somehow prevent U4 snRNA release by inhibiting the activity of EFTUD2 through physical interaction.
It is important to note that, from the Guo et al. study, although BS69 binds more than 8,000 genes, only a small set of genes showed altered IR upon depletion of BS69, suggesting that the detection capability may not be optimized enough and/or that there are additional factors involved in this chromatin-templated process (24). Additionally, bioinformatics analyses found that the BS69-regulated introns appear to be significantly longer in length compared with the average size of all IR introns, and with significantly stronger 5′ splice sites (24). However, whether and how these features contribute to the specificity of BS69 regulation remains an interesting question for future investigation. Other factors that can contribute to the regulation of BS69 selectivity may involve additional histone modifications such as acetylation and chromatin remodeling, which are known to be involved in splicing regulation. An example is the histone acetyltransferase Gcn5, which is a member of the yeast SAGA complex involved in cotranscriptional recruitment of the U2 snRNP (39). Similarly, the SWI/SNF chromatin-remodeling complex in higher eukaryotes associates with pre-mRNA and regulates alternative splicing of endogenous genes (40), and treatment with the histone deacetylase inhibitor sodium butyrate (NaB) and loss of HDAC1 affect alternative splicing (41).
Elongation Versus Alternative Splicing Regulation.
A number of previous studies suggested that chromatin plays a role in the regulated pre-mRNA processing (reviewed in ref. 42). Given that BS69 has been shown to regulate both elongation and RNA splicing, are these two functions interconnected? In yeast, the histone H3K36 methyltransferase, Set2, binds to the elongating form of Pol2 (43). In mammals, H3K36me3 occurs primarily in the actively transcribed regions (44). Thus, BS69 is probably recruited as a result of transcription but subsequently negatively regulates promoter Pol2 release for the subsequent rounds of transcription. Mechanistically, BS69 may modify local chromatin through its interaction with HDACs and chromatin remodelers to suppress Pol2 elongation (Fig. 2). Along this line, Wen et al. (23) reported that BS69 knockdown resulted in an increase of gene body H3K36me3, and Guo et al. (24) identified the H3K9 demethylase KDM3B and the H3K36 methyltransferase NSD1, as well as several PRC1 components among the potential BS69 binding proteins (Fig. 1). It will be interesting to examine whether the pattern of other histone modifications, such as acetylation, are altered as a consequence of loss of BS69 and whether these changes have anything to do with Pol2 elongation. On the other hand, it has been documented that activated RNA splicing tends to promote transcription elongation and deposition of H3K36me3 (45, 46). Thus, the increase in elongation and H3K36me3 upon BS69 KD reported by Wen et al. could also be the result of activated splicing, which is normally suppressed by BS69 through inhibiting U5 snRNP (24).
Exons that are surrounded by long introns show a higher level of nucleosome occupancy (47), which in turn results in a slower Pol2 elongation rate (48). In this regard, although the IR introns are in general small in size (median length 548 bp), the BS69-regulated IR introns are significantly longer than the average IR introns (median length 1,025 bp) (24). BS69 and H3K36me3 are found enriched at exons (23, 24); thus, one possibility is that BS69 is recruited to exons with higher nucleosome occupancy through H3.3K36me3 and slows down Pol2 elongation locally. Related to this hypothesis, previous studies found that a reduced elongation rate either by Pol2 mutations or inhibitor treatment causes changes in alternative splicing, specifically the inclusion of alterative exons (49, 50). A more recent study investigating the relationship between IR and Pol2 elongation found that, compared with constitutive introns, the retained introns are more enriched in Pol2 signal, possibly reflecting an increased pausing of Pol2 due to inefficient splicing (51). Thus, the ability of BS69 to promote IR and suppress Pol2 elongation might be interconnected, which needs to be investigated in future studies. Interestingly, the same study also reported that IR is highly prevalent in mammals, and, because IR introns in general have weaker splice sites, they therefore are particularly sensitive to the local availability of splicing factors. This observation is consistent with the finding that BS69 suppresses EFTUD2 function locally through physical interaction in a chromatin context, which is critical for its regulation of IR (24).
Biological Implications: Differentiation and Cancer
As discussed, H3K36me3 marks gene bodies and, when together with H3K4me3, it is associated with active gene expression. In addition, H3.3 has also been suggested to mark active chromatin (4, 6). Interestingly and perhaps somewhat counterintuitively, the H3.3K36me3 reader BS69 primarily suppresses gene expression through the mechanisms discussed in BS69 Suppresses Transcription Elongation and Promotes Intron Retention. One possibility is that this arrangement may represent a fine-tuning mechanism that biological systems tend to use, which involves factors that act in opposite directions. For example, the bivalent domain is marked by both active (H3K4me3) and repressive (H3K27me3) marks, which is an epigenetically poised state found primarily in pluripotent embryonic stem cells. An advantage of such an arrangement is that it offers a quick response to physiological or environmental cues because these genes are already primed for either repression or activation (52). Perhaps, for those genes where BS69 binds and regulates IR, the function of BS69 is to dampen the level of their mRNA expression by promoting IR, thereby allowing a larger magnitude of response to a physiological stimulus. Because BS69 regulates hundreds of IR events, it offers the opportunity to synchronize the regulation of a subgroup of genes through the IR mechanism. Indeed, IR has been demonstrated to coordinate gene expression during neuronal differentiation and granulopoiesis, but whether BS69 or other factors are involved in these processes remains to be determined (53, 54). Interestingly, BS69 was also reported to have the ability to inhibit neuronal and muscle differentiation (55). It will be important to determine whether such an inhibitory activity is due to its ability to regulate IR.
Alterations in IR and ES (exon skipping) have also been identified as significant RNA splicing events in cancer, such as breast and renal-cell carcinomas (56–58). Consistently, an overall up-regulation of splicing factors has been observed in certain cancer models (58). Thus, it is conceivable that BS69 may suppress tumorigenesis through regulation of RNA splicing. This connection has been made even more interesting by the recently identification of H3.3 mutations in cancers (19). Mutations that either directly (K36M) or indirectly (G34R/V) abrogate H3.3K36 trimethylation will likely impair BS69 binding to chromatin in these tumors (Table 1). Furthermore, the main H3K36me3 methyltransferase, SETD2, is also frequently mutated or deleted in cancers, including renal-cell carcinoma, pediatric high-grade glioma, and early T-cell precursor acute lymphoblastic leukemia (59–61), which will also cause global loss of BS69 chromatin association (Table 1). Interestingly, global splicing defects, especially IR, were observed in the SETD2-mutated ccRCCs (57). Furthermore, significant BS69 copy-number loss was also reported in several hematological malignancies, including ALL (acute lymphoblastic leukemia), CML (chronic myeloid leukemia), CLL (chronic lymphoid leukemia), MM (multiple myeloma), and MDS (myelodysplastic syndrome) (62). Thus, we speculate that BS69 may participate in different tumorigenic processes through dynamic regulation of RNA-splicing events such as IR. To explore the connection between BS69, H3.3, SETD2, and IR in cancers, genome-wide RNA profiling data of the related cancer samples should be carefully reexamined for possible alterations in pre-mRNA processing, especially, IR.
Table 1.
Summary of cancer mutations that are predicted to affect BS69 binding to H3.3K36me3
| Cancer mutations | Major cancer association of each mutation |
| H3.3G34R/V | Cerebral hemisphere type pediatric HGA |
| H3.3K36M | Chondroblastoma |
| SETD2 mutation | Cerebral hemisphere HGA, ccRCC, ETP T-ALL |
H3.3K36M and SETD2 deletion are reported to affect global H3K36me3 and therefore are expected to impair overall BS69 binding to chromatin. H3.3G34R/V, on the other hand, affects H3.3K36me3 only in cis and thus will alter BS69 binding only locally. The major cancer association of each mutation is indicated. HGA, high-grade astrocytomas; ccRCC, clear-cell renal carcinoma, ETP T-ALL, early T-cell precursor acute lymphoblastic leukemia.
Finally, Guo et al. also demonstrated that phosphorylation at S31 (H3.3S31PK36me3) significantly reduced the binding of BS69 (24). H3.3S31P has been found to be enriched during mitosis at telomeric ends in mES cells and pericentric heterochromatin after differentiation (63), suggesting that readers such as BS69 may be subjected to regulation by H3.3S31 phosphorylation in vivo. This finding also raises the question of whether S31 may play a role in tumorigenesis and whether its regulators may undergo gain-of–function cancer alterations, which may impair BS69 binding to chromatin. The ongoing efforts of DNA deep sequencing of cancer tissues and identifying the signaling pathway that regulates H3.3S31 phosphorylation will provide insight into these issues.
More Histone Variant Readers?
The identification of BS69 as an H3.3K36me3-specific reader, which is regulated by phosphorylation of the nearby serine at position 31, suggests a possible H3.3-specific pathway. It raises the question whether there are both loss and gain of binding by readers, such as BS69, to the mutant H3.3 tail in the H3.3 mutation-driven cancers. Furthermore, given the proximity of lysine 27 to H3.1A31 or H3.3S31, H3.1K27 and H3.3K27 modifications (such as methylation and acetylation) and their recognition may also be executed by variant-specific sets of enzymes and readers. In support of this idea, a recent study reported a plant H3.1-specific H3K27me3 methyltransferase, ATXR5, which is critical in regulating the integrity of heterochromatin by preventing H3.3K27 from being methylated in the open chromatin (64). It is also worth noting that the majority of previously identified readers and enzymes were discovered using H3.1 histone substrates. For instance, from a focused structure and thermodynamics study, several PWWP domains were reported to bind H3.1K36me3 but with very low affinity (KD values in the mM range) (65). These observations collectively raise the possibility that some of these readers could have a higher affinity for variant histones and are in fact variant histone binders.
Summary
In this article, we have discussed the recent findings of a histone variant H3.3-specific reader, BS69/ZMYND11, and its function in regulating transcriptional elongation and mRNA splicing, especially intron retention. We have also discussed how these discoveries advance the understanding of IR regulation during differentiation and cancers with H3.3 mutations. We envision immediate future efforts to be placed on elucidating the molecular mechanism of how BS69 regulates transcription and transcription-coupled mRNA processing, and whether and how mis-regulation of these processes contributes to the development of cancer and other diseases. Human genome encodes a total of seven H3, namely H3.1 and H3.2, which are the canonical H3, as well as the variant H3.3, H3t, H3.X, H3.Y, and CENP-A. In addition to H3.3, H3t, H3.X, and H3.Y also share significant sequence homology in their tail regions with the canonical H3 (66). We speculate the existence of additional histone variant-specific readers with a role in physiological and pathological processes and expect that the hunt for readers and the investigation of their roles in cancer will prove fruitful. The identification of an H3.3S31-specific kinase and the signaling pathway that regulates H3.3S31 phosphorylation and potential alteration of this pathway in cancer will provide additional insights into the roles of these readers in tumorigenesis.
Acknowledgments
We thank Xiaobing Shi, Yi Xing, Nada Jabado, and Peter W. Lewis for helpful discussions and valuable comments on the manuscript. The Y.S. laboratory at Boston Children’s Hospital (BCH) is supported by NIH Grants CA118487 and MH096066 and by funds from BCH. The work reported in Guo et al. (24) was supported by the “985” Program from the Chinese Ministry of Education and the “973” State Key Development Program of Basic Research of China (Grants 2009CB825602 and 2009CB825603) and in part by BCH funds (to Y.S.). F.L. is a recipient of the China “Thousand Youth Talents” (KHH1340001) and Shanghai “Oriental Scholar” (SHH129002) awards. Y.S. is an American Cancer Society Research Professor.
Footnotes
Conflict of interest statement: Y.S. is a cofounder of Constellation Pharmaceutical, Inc. and a member of its scientific advisory board.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Epigenetic Changes in the Developing Brain: Effects on Behavior,” held March 28–29, 2014, at the National Academy of Sciences in Washington, DC. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/Epigenetic_changes.
This article is a PNAS Direct Submission.
References
- 1.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116(1):51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg AD, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140(5):678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drané P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24(12):1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9(6):1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 5.Ray-Gallet D, et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell. 2011;44(6):928–941. doi: 10.1016/j.molcel.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Chen P, et al. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev. 2013;27(19):2109–2124. doi: 10.1101/gad.222174.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szenker E, Lacoste N, Almouzni G. A developmental requirement for HIRA-dependent H3.3 deposition revealed at gastrulation in Xenopus. Cell Reports. 2012;1(6):730–740. doi: 10.1016/j.celrep.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 8.Bush KM, et al. Endogenous mammalian histone H3.3 exhibits chromatin-related functions during development. Epigenetics & chromatin. 2013;6(1):7. doi: 10.1186/1756-8935-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behjati S, et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet. 2013;45(12):1479–1482. doi: 10.1038/ng.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu G, et al. St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44(3):251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 12.Sturm D, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 13.Buczkowicz P, et al. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat Genet. 2014;46(5):451–456. doi: 10.1038/ng.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontebasso AM, et al. Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat Genet. 2014;46(5):462–466. doi: 10.1038/ng.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu G, et al. St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46(5):444–450. doi: 10.1038/ng.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aihara K, et al. H3F3A K27M mutations in thalamic gliomas from young adult patients. Neuro-oncol. 2014;16(1):140–146. doi: 10.1093/neuonc/not144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor KR, et al. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat Genet. 2014;46(5):457–461. doi: 10.1038/ng.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bjerke L, et al. 2013. Histone H3.3 mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Discov 3(5):512–519.
- 19.Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bender S, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24(5):660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Chan KM, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013;27(9):985–990. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herz HM, et al. Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science. 2014;345(6200):1065–1070. doi: 10.1126/science.1255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen H, et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature. 2014;508(7495):263–268. doi: 10.1038/nature13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo R, et al. BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol Cell. 2014;56(2):298–310. doi: 10.1016/j.molcel.2014.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hateboer G, et al. BS69, a novel adenovirus E1A-associated protein that inhibits E1A transactivation. EMBO J. 1995;14(13):3159–3169. doi: 10.1002/j.1460-2075.1995.tb07318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masselink H, Bernards R. The adenovirus E1A binding protein BS69 is a corepressor of transcription through recruitment of N-CoR. Oncogene. 2000;19(12):1538–1546. doi: 10.1038/sj.onc.1203421. [DOI] [PubMed] [Google Scholar]
- 27.Ladendorff NE, Wu S, Lipsick JS. BS69, an adenovirus E1A-associated protein, inhibits the transcriptional activity of c-Myb. Oncogene. 2001;20(1):125–132. doi: 10.1038/sj.onc.1204048. [DOI] [PubMed] [Google Scholar]
- 28.Velasco G, Grkovic S, Ansieau S. New insights into BS69 functions. J Biol Chem. 2006;281(24):16546–16550. doi: 10.1074/jbc.M600573200. [DOI] [PubMed] [Google Scholar]
- 29.Dhayalan A, et al. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem. 2010;285(34):26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vezzoli A, et al. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat Struct Mol Biol. 2010;17(5):617–619. doi: 10.1038/nsmb.1797. [DOI] [PubMed] [Google Scholar]
- 31.Li F, et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell. 2013;153(3):590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, et al. Regulation of Set9-mediated H4K20 methylation by a PWWP domain protein. Mol Cell. 2009;33(4):428–437. doi: 10.1016/j.molcel.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin S, Min J. 2014. Structure and function of the nucleosome-binding PWWP domain. Trends Biochem Sci 39(11):536–547.
- 34.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27(2):406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen H, Li Y, Li H, Shi X. ZMYND11: An H3.3-specific reader of H3K36me3. Cell Cycle. 2014;13(14):2153–2154. doi: 10.4161/cc.29732. [DOI] [PubMed] [Google Scholar]
- 36.Swartling FJ, et al. Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer Cell. 2012;21(5):601–613. doi: 10.1016/j.ccr.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141(3):432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartels C, Klatt C, Lührmann R, Fabrizio P. The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome. EMBO Rep. 2002;3(9):875–880. doi: 10.1093/embo-reports/kvf172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gunderson FQ, Johnson TL. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet. 2009;5(10):e1000682. doi: 10.1371/journal.pgen.1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Batsché E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13(1):22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 41.Hnilicová J, et al. Histone deacetylase activity modulates alternative splicing. PLoS ONE. 2011;6(2):e16727. doi: 10.1371/journal.pone.0016727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shukla S, Oberdoerffer S. Co-transcriptional regulation of alternative pre-mRNA splicing. Biochim Biophys Acta. 2012;1819(7):673–683. doi: 10.1016/j.bbagrm.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2003;278(11):8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- 44.Huff JT, Plocik AM, Guthrie C, Yamamoto KR. Reciprocal intronic and exonic histone modification regions in humans. Nat Struct Mol Biol. 2010;17(12):1495–1499. doi: 10.1038/nsmb.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim S, Kim H, Fong N, Erickson B, Bentley DL. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci USA. 2011;108(33):13564–13569. doi: 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Almeida SF, et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol. 2011;18(9):977–983. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- 47.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36(2):245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bondarenko VA, et al. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell. 2006;24(3):469–479. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 49.de la Mata M, et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12(2):525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 50.Ip JY, et al. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res. 2011;21(3):390–401. doi: 10.1101/gr.111070.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Braunschweig U, et al. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014;24(11):1774–1786. doi: 10.1101/gr.177790.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 53.Wong JJ, et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154(3):583–595. doi: 10.1016/j.cell.2013.06.052. [DOI] [PubMed] [Google Scholar]
- 54.Yap K, Lim ZQ, Khandelia P, Friedman B, Makeyev EV. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev. 2012;26(11):1209–1223. doi: 10.1101/gad.188037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu B, et al. BS69 undergoes SUMO modification and plays an inhibitory role in muscle and neuronal differentiation. Exp Cell Res. 2009;315(20):3543–3553. doi: 10.1016/j.yexcr.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 56.Eswaran J, et al. RNA sequencing of cancer reveals novel splicing alterations. Scientific reports. 2013;3:1689. doi: 10.1038/srep01689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simon JM, et al. Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res. 2014;24(2):241–250. doi: 10.1101/gr.158253.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shapiro IM, et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011;7(8):e1002218. doi: 10.1371/journal.pgen.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duns G, et al. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 2010;70(11):4287–4291. doi: 10.1158/0008-5472.CAN-10-0120. [DOI] [PubMed] [Google Scholar]
- 60.Fontebasso AM, et al. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol. 2013;125(5):659–669. doi: 10.1007/s00401-013-1095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481(7380):157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang H, et al. Analysis of copy number variations of BS69 in multiple types of hematological malignancies. Ann Hematol. 2010;89(10):959–964. doi: 10.1007/s00277-010-0966-5. [DOI] [PubMed] [Google Scholar]
- 63.Hake SB, et al. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc Natl Acad Sci USA. 2005;102(18):6344–6349. doi: 10.1073/pnas.0502413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jacob Y, et al. Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science. 2014;343(6176):1249–1253. doi: 10.1126/science.1248357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu H, et al. Structural and histone binding ability characterizations of human PWWP domains. PLoS ONE. 2011;6(6):e18919. doi: 10.1371/journal.pone.0018919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Szenker E, Ray-Gallet D, Almouzni G. The double face of the histone variant H3.3. Cell Res. 2011;21(3):421–434. doi: 10.1038/cr.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]