Significance
This report reveals the influence of sialylation on the activation of epidermal growth factor receptor (EGFR) and sensitivity to tyrosine kinase inhibitors (TKIs) against EGFR phosphorylation. By utilizing biochemical approaches, we demonstrated that EGFR sialylation suppresses EGFR phosphorylation by inhibiting EGF binding and EGFR dimerization. In the TKI-resistant lung cancer cell line with L858R/T790M mutations on EGFR, the levels of phosphorylation at Y1068, Y1086, and Y1173 are upregulated, and sialylation can partially suppress the phosphorylation of EGFR at these sites and enhance EGFR sensitivity to TKI. These findings suggest that sialylation has an important role in tumorigenesis and sensitivity to TKIs by modulating EGFR phosphorylation and the associated signaling network and provide insights for therapeutic intervention.
Keywords: glycosylation, dimerization, lung cancer, gefitinib, tyrosin kinase inhibitor
Abstract
Epidermal growth factor receptor (EGFR) is a heavily glycosylated transmembrane receptor tyrosine kinase. Upon EGF-binding, EGFR undergoes conformational changes to dimerize, resulting in kinase activation and autophosphorylation and downstream signaling. Tyrosine kinase inhibitors (TKIs) have been used to treat lung cancer by inhibiting EGFR phosphorylation. Previously, we demonstrated that EGFR sialylation suppresses its dimerization and phosphorylation. In this report, we further investigated the effect of sialylation on the phosphorylation profile of EGFR in TKI-sensitive and TKI-resistant cells. Sialylation was induced in cancer progression to inhibit the association of EGFR with EGF and the subsequent autophosphorylation. In the absence of EGF the TKI-resistant EGFR mutant (L858R/T790M) had a higher degree of sialylation and phosphorylation at Y1068, Y1086, and Y1173 than the TKI-sensitive EGFR. In addition, although sialylation in the TKI-resistant mutants suppresses EGFR tyrosine phosphorylation, with the most significant effect on the Y1173 site, the sialylation effect is not strong enough to stop cancer progression by inhibiting the phosphorylation of these three sites. These findings were supported further by the observation that the L858R/T790M EGFR mutant, when treated with sialidase or sialyltransferase inhibitor, showed an increase in tyrosine phosphorylation, and the sensitivity of the corresponding resistant lung cancer cells to gefitinib was reduced by desialylation and was enhanced by sialylation.
Epidermal growth factor receptor (EGFR), one of the most studied receptor tyrosine kinases, is a drug target for cancer therapy, because its kinase activity correlates with tumorigenicity (1). Under normal conditions, EGFR forms dimers upon ligand binding and induces kinase activation (2–6). The conformational change of EGFR from tethered to extended form induced by ligand binding involves the exposure of the interface, followed by dimerization, activation, and autophosphorylation (7). The phosphorylation code of EGFR determines the propensity of the downstream signaling network to regulate cell proliferation, survival, migration, and angiogenesis (8, 9).
In a significant fraction of patients with nonsmall cell lung cancer (NSCLC), especially patients in Asia and those with the adenocarcinoma subtype, mutations in the kinase domain of EGFR cause constitutive activation and have been identified as an important factor in EGFR dysregulation (10, 11). Particularly, mutation from leucine to arginine at position 858 (L858R) and, less significantly, deletion of exon 19 that eliminates four amino acids (LREA) account for ∼90% of the mutations involved in the constitutive activation of EGFR. These mutations are commonly found in patients with increased sensitivity to EGFR tyrosine kinase inhibitors (TKIs) such as gefitinib and erlotinib (12–14). However, most patients with such mutations show resistance within months after TKI therapy, and >50% of them develop a second EGFR mutation, T790M, which confers TKI resistance by increasing the affinity for ATP and decreasing the affinity for TKIs (15–17).
Studies have demonstrated that the glycans on EGFR participate in the regulation of EGFR function. The number of N-glycans and the degree of branching can regulate the cell-surface expression of EGFR in response to N-acetyl-d-glucosamine (GlcNAc) supplementation (18). In addition, studies with site-directed mutagenesis indicate that the glycans on Asn420 and 579 prevent EGFR from ligand-independent dimerization (19–21), and knocking down/out fucosyltransferase 8, the enzyme responsible for the core fucosylation, attenuates EGFR phosphorylation and EGF binding (22, 23). Moreover, our previous study revealed that sialylation and fucosylation suppress EGFR dimerization, autophosphorylation, and EGF-induced lung cancer cell invasion (24).
Here, we investigated the effect of sialylation on EGFR dimerization to understand how extracellular sialylation influences intracellular phosphorylation in both wild-type and mutant EGFR. Our biochemical data demonstrated that sialylation could suppress EGFR dimerization by attenuating its association with EGF, and sialylation could significantly and selectively suppress tyrosine phosphorylation and affect the levels of phosphoserine and phosphothreonine on EGFR. In EGFR mutants, especially L858R/T790M, sialylation was observed to have a selective effect on EGFR phosphorylation, and inhibition of sialylation resulted in increased phosphorylation and resistance to gefitinib in this TKI-resistant lung cancer cell line. Further study of these findings should provide a better understanding of EGFR-mediated phosphorylation and disease progression affected by glycosylation and lead to the development of a new therapeutic strategy.
Results
Preparation of Soluble EGFR and Its Desialylated Form from 293F Cells for Dimerization and EGF-Binding Studies.
To study the effect of sialylation on EGFR activation, the extracellular domain of EGFR was overexpressed in 293F cells and was affinity purified for biochemical assays. The desialylated soluble EGFR (sEGFR) was prepared by sialidase treatment to remove the α2,3-, α2,6-, and α2,8-linked sialic acid residues before affinity purification. The removal of sialic acids on each glycosylation site was monitored by mass spectrometry through matching with the calculated masses of both tryptic peptide fragments and glycans and by the appearance of fragmented glycans in MS/MS spectra (24). The results showed that all the sialic acid residues of sEGFR were removed except (perhaps because steric hindrance) those at Asn328 (Fig. S1).
To investigate the effect of sialylation on EGFR dimerization, multiangle laser light scattering (MALLS) was used to determine the kinetics of EGFR dimerization. Various concentrations of sEGFR with or without sialidase treatment were preincubated with EGF in a 1:1.1 molar ratio and then were analyzed by MALLS. The average molecular mass (MM) at each concentration was calculated and converted into the dimerization ratio.
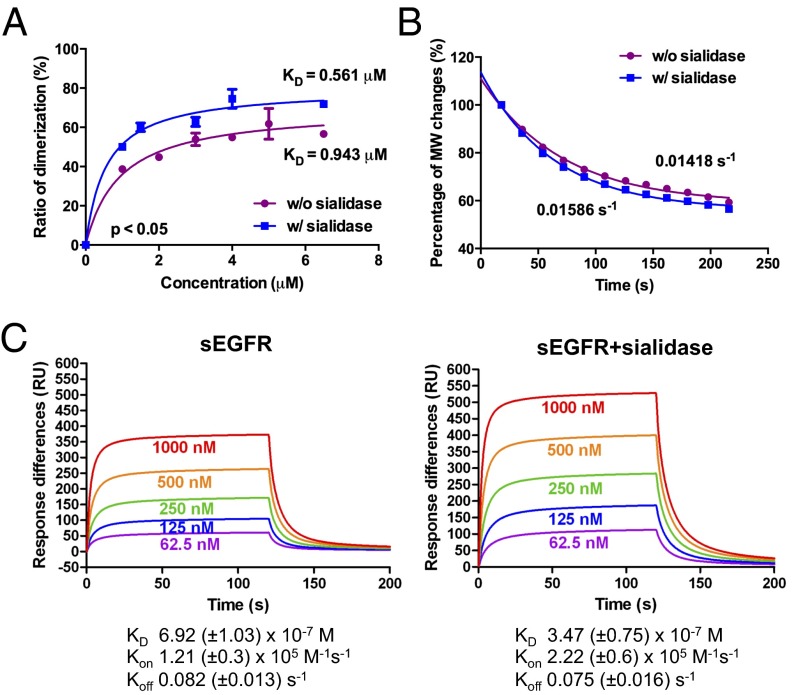
Dimerization of sEGFR was induced in an EGF dose-dependent manner (Fig. 1A). Compared with sEGFR without sialidase treatment, the desialylated sEGFR showed a higher degree of dimerization, especially in the slope phase of the fitting curve. Statistical analysis with one-site specific binding showed that the dissociation constant (Kd) for EGF-induced dimerization of sEGFR is 0.943 μM, which is consistent with previous studies (the Kd of EGFR dimerization ranges from 0.6–3.8 μM with different approaches) (2, 25, 26). Moreover, the Kd value for EGF-induced dimerization of desialylated sEGFR was 0.561 μM, twofold higher than that for sEGFR. These data confirmed the suppression effect of EGFR sialylation on its dimerization. To dissect the impact of sialylation on dimerization further, the dissociation rate of EGFR dimers was measured. Samples were prepared in a saturated dimerization concentration (11 μM) and then were injected into MALLS to monitor the changes in average MM upon diffusion in the buffer system. The change of monomer–dimer stoichiometry was analyzed. As shown in Fig. 1B, the best-fitting curves showed that the dissociation rate of the desialylated sEGFR dimer (0.01586/s) is similar to the sialylated sEGFR dimer (0.01418/s), suggesting that sialylation on EGFR mainly regulates the rate of association, not its dissociation. Because previous studies indicated that the lack of glycans on specific glycosylation sites could induce ligand-independent EGFR dimerization (20, 21), we next examined whether desialylation could induce ligand-independent dimerization using MALLS analysis. The MM of sEGFR with or without desialylation in different concentrations was measured; no significant change was observed (Fig. S2), indicating that desialylation of EGFR does not induce spontaneous dimerization.
Fig. 1.
Sialylation suppresses EGFR dimerization and the EGF-binding ability of sEGFR. (A) EGF-induced dimerization of EGFR. The average MM of sEGFR at various concentrations was measured in the presence of EGF and transformed into percentage of dimerization (n = 3). Data were analyzed by nonlinear curve-fitting using GraphPad Prism software; the Kd for each sample is listed. (B) Dissociation of dimerized sEGFR. Dimerized sEGFR was prepared by incubating EGF and sEGFR at a saturated concentration. The decrement in MM was measured in gradual dilution condition and analyzed with nonlinear curve fitting. The purple trace represents sEGFR; the blue trace represents sEGFR with sialidase treatment. (C) SPR study of sEGFR binding to EGF. The binding constants of sEGFR to immobilized EGF were measured by SPR with various concentrations of sEGFR (n = 4). The calculated kinetics parameters (Kd, Kon, and Koff) of both sEGFR and desialylated sEGFR are shown.
We next investigated whether sialylation could regulate EGFR–EGF interaction, because other studies have suggested that glycosylation could affect such interaction (23, 27). Surface plasmon resonance (SPR) was used to measure the binding of sEGFR (with or without desialylation) to EGF, which was immobilized onto CM5 BIAcore biosensor chips. The kinetic parameters, including the Kd and the association and dissociation rate constants (Kon and Koff) of EGFR to EGF, were determined (Fig. 1C). The results showed that the Kd of sEGFR was 692 ± 103 nM, whereas the Kd of desialylated sEGFR was 347 ± 75 nM, a nearly twofold increase in affinity. The Kon of desialylated sEGFR was higher than that of sialylated sEGFR, but there was no significant difference in the Koff between these two types of EGFR. Therefore, sialylation on EGFR reduced its association with EGF. Because high levels of sialylation on the three glycosylation sites (N32, N151, and N389) near the EGF-binding surface were observed (Fig. S1), the negative charges of sialic acid residues might have a negative impact on the electrostatic interaction between EGF and EGFR (28). In addition, the Kd for EGF binding to sEGFR measured here is different from that previously reported (175 ± 5.8 nM) (2), perhaps because of the differences in glycosylation (complex type from human cells vs. high-mannose type from insect cells).
Effect of Sialylation on the Autophosphorylation of EGFR Expressed in 293T Cells and EGFR Wild-Type CL1-0 and CL1-5 Cancer Cells.
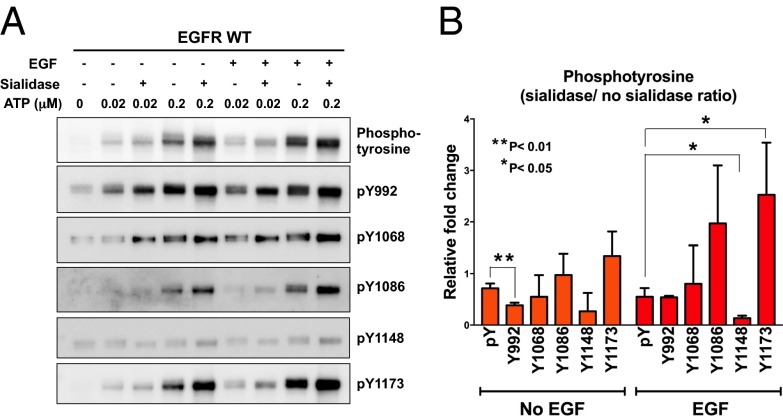
The results from kinetics studies revealed that sialylation on EGFR could negatively regulate ligand-induced EGFR dimerization, which is critical for EGFR activation and autophosphorylation. To investigate the sialylation effect further, recombinant full-length EGFR (flEGFR) was transiently expressed in 293T cells and purified for the phosphorylation assay in vitro, and the phosphorylation profiles of flEGFR and desialylated flEGFR were examined by the antibodies that recognize site-specific tyrosine phosphorylation. As shown in Fig. 2, the desialylated flEGFR exhibited higher levels of phosphorylation on Y992, Y1068, Y1086, and Y1173 and a slight increase (<0.5-fold) in Y1148 phosphorylation. This result suggested that sialylation or desialylation of EGFR might have a selective effect on the phosphorylation of specific tyrosine residues, and, as shown previously, sialylation of EGFR suppresses its autophosphorylation through inhibition of EGFR dimerization. Interestingly, without EGF stimulation, sialylation still suppressed EGFR tyrosine phosphorylation, especially on Y1086 and Y1173.
Fig. 2.
Phosphorylation profiling of EGFR. (A) Purified flEGFR and desialylated flEGFR were treated with or without EGF at two concentrations of ATP (0.02 and 0.2 μM). The level of phosphorylation was analyzed by site-specific anti-EGFR phosphotyrosine antibodies (n = 3). (B) Semiquantitative results for the phosphorylation level of flEGFR incubated with 0.2 μM ATP. Relative fold change of phosphotyrosines between flEGFR and desialylated flEGFR was calculated. Error bars represent SD values. P values were calculated by paired t test. *P < 0.05; **P < 0.01.
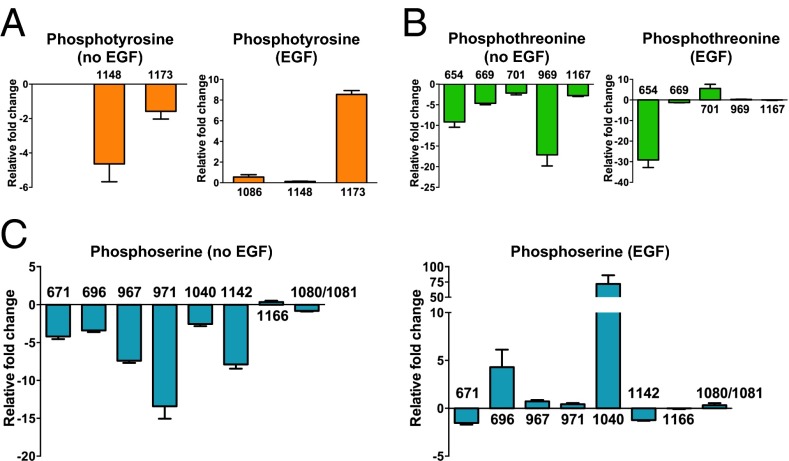
In addition to tyrosine phosphorylation, the phosphorylation of serine or threonine residues on EGFR is known to modulate EGFR signaling. To understand further how sialylation regulates EGFR phosphorylation, we performed mass spectrometry analysis to investigate the phosphorylation pattern of EGFR comprehensively. We have developed a label-free quantification strategy that combines highly efficient protein enrichment, immobilized metal affinity chromatography (IMAC), and high-resolution mass spectrometry to characterize EGFR phosphopeptides. The EGFR proteins from two cancer cell lines, CL1-0 (mild), CL1-5 and (aggressive), and sialidase-treated CL1-5, in starved or EGF-stimulated condition, were immunoprecipitated by covalently immobilized anti-EGFR mAb. The EGFR derived from these two cell lines was eluted in an acidic condition and subjected to phosphopeptide enrichment by IMAC following trypsin digestion. The phosphopeptides then were identified and quantified by mass spectrometry (Table S1). The quantification of phosphopeptides was verified further by sequential window acquisition of theoretical mass spectra (SWATH) (Fig. S3). Sixteen phosphosites were identified: three phosphotyrosines, eight phosphoserines, and five phosphothreonines. Some phosphosites showed different EGF responsiveness in CL1-0 and CL1-5 cells (Fig. S4). For example, pY1148 and pY1173 were induced by EGF only in CL1-0 but not in CL1-5 cells; the phosphorylation of two threonine residues (pT701 and pT969) and four serine residues (pS696, pS967, pS971, and pS1142) was suppressed dramatically by EGF treatment in CL1-5 in comparison with CL1-0 cells. Removal of sialic acid residues by sialidase (Fig. S5B) altered the responsiveness of EGF-induced phosphorylation to similar degrees in CL1-5 and CL1-0 cells, indicating that cell-surface sialylation is specifically involved in regulating EGFR phosphorylation. To link phosphorylation and sialylation, the relative change in identified phosphosites after the removal of cellular sialic acid was calculated, with positive and negative changes representing the suppression and enhancement effects of sialylation on phosphorylation, respectively. As shown in Fig. 3A, under EGF stimulation, sialylation of EGFR suppressed the phosphorylation on Y1173 but had no significant effect on pY1086 and pY1148. Surprisingly, sialylation also site-specifically regulated EGFR serine/threonine phosphorylation (Fig. 3 B and C). Four phosphoserine sites (pS696, pS967, pS971, and pS1040) and one phosphothreonine site (pT701) were suppressed by sialylation in an EGF-dependent manner; in particular, phosphorylation on pS1040 was increased by around 75-fold when desialylated. On the contrary, phosphorylation on pS671 and pS1142 was enhanced by sialylation, and phosphorylation on T654 was reduced dramatically (∼30-fold) in the desialylated condition.
Fig. 3.
Identification of EGFR phosphorylation in the lung cancer cell line CL1-5. The intensities of identified phosphopeptides containing phosphotyrosines (A), phosphothreonines (B), and phosphoserines (C) are shown. The EGFR phosphopeptides derived from EGF-treated or untreated cells were identified by mass spectrometry, and the intensity of phosphopeptides was quantified based on a label-free strategy and normalized with the sum of intensity of the three most abundant EGFR peptides. The relative fold change of each sample was calculated by dividing the intensity of normalized EGFR phosphopeptides from sialidase-treated cells by the intensity of normalized EGFR phosphopeptides of untreated cells. The positive (fold change >0) or negative (fold change <0) effect of desialylation on EGFR phosphorylation is indicated (n = 4). Error bars represent SD values.
Sialylation also had a regulatory effect on EGFR phosphorylation without EGF stimulation, and desialylation reduced the phosphorylation of Y1148 and Y1173 (Fig. 3A). Desialylation had a negative impact on the phosphorylation of serines and threonines when there was no EGF stimulation (Fig. 3 B and C). Given these observations, we conclude that cellular sialylation may regulate EGFR phosphorylation by modulating the activity of other kinases responsible for EGFR phosphorylation in addition to suppressing EGFR autophosphorylation directly.
Effect of EGFR Sialylation on the Autophosphorylation of EGFR from TKI-Sensitive and -Resistant Mutants.
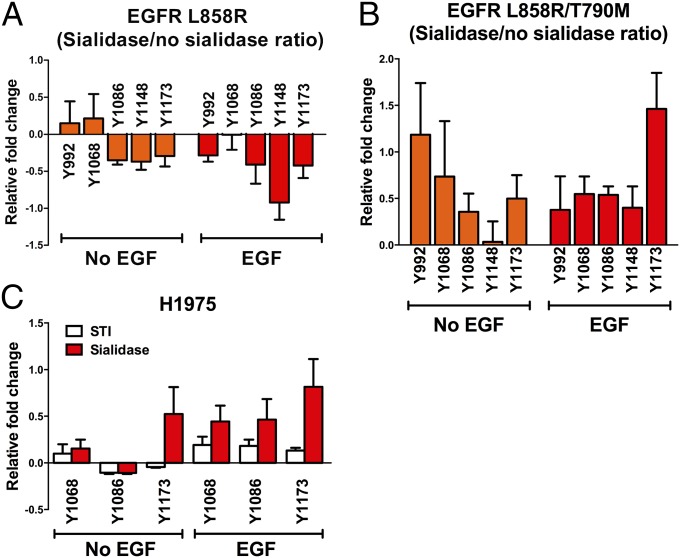
Previous in vitro studies have shown that dimerization of the kinase domain is essential to maintain the activity of the oncogenic mutants of EGFR such as the TKI-sensitive mutant L858R (29–31). Moreover, EGF is capable of promoting the phosphorylation of EGFR mutants in many cell-based experiments. These observations collectively indicate that dimerization of EGFR is involved in the constitutive activation of EGFR mutants. Because it has been demonstrated that sialylation suppressed EGFR dimerization, we next investigated the impact of EGFR sialylation on the phosphorylation of EGFR mutants. First, an in vitro phosphorylation assay was performed to analyze the change in tyrosine phosphorylation on the flEGFR L858R and flEGFR L858R/T790M (TKI-resistant) mutants when treated with sialidase (Fig. 4 and Figs. S5A and S6). As shown in Fig. 4A, sialylation was less effective in regulating the phosphorylation of EGFR L858R, but its effect of sialylation on the TKI-resistant mutant L858R/T790M was significant. All phosphotyrosines with or without EGF stimulation were suppressed by sialylation, with most significant effect on Y1173 under EGF treatment (Fig. 4B).
Fig. 4.
Effect of sialylation on tyrosine phosphorylation in EGFR mutants. (A and B) The EGFR mutant proteins EGFR L858R (A) and EGFR L858R/T790M (B) were purified for the in vitro phosphorylation assay. The relative fold change of tyrosine phosphorylation in each phosphopeptide was calculated by dividing the intensity of phosphorylation in sialidase-treated EGFR by the intensity of phosphorylation in untreated EGFR. The positive (fold change >0) or negative (fold change <0) effect of desialylation on EGFR phosphorylation is indicated (n = 3). Error bars represent SD values. Representative Western blots are shown in Fig. S6. (C) Tyrosine phosphorylation (pY1068, pY1086, and pY1173) of H1975 cells treated with STI or sialidase is shown. The relative intensities of phosphosites were normalized to their individual amounts of EGFR.
To examine the effect of sialylation on the phosphorylation of EGFR mutants at the cellular level, the TKI-resistant cell line H1975 with L858R/ T790M mutations on the EGFR was treated with a sialyltransferase inhibitor (STI) (32) or sialidase to reduce surface sialylation, and the level of three phosphotyrosines, pY1068, pY1086, and pY1173, which showed a high degree of suppression by sialylation (>0.5-fold), was examined (Fig. 4C and Figs. S5C and S7A). Generally, as consistent with the results in the in vitro phosphorylation assay, in the presence or absence of EGF stimulation, the level of phosphorylation on these tyrosine residues, except for Y1086 in the absence of EGF, was elevated upon attenuation of cellular sialylation, with a more significant effect on Y1173 phosphorylation.
Effect of Sialylation on Gefitinib Sensitivity in Gefitinib-Resistant Cancer Cells.
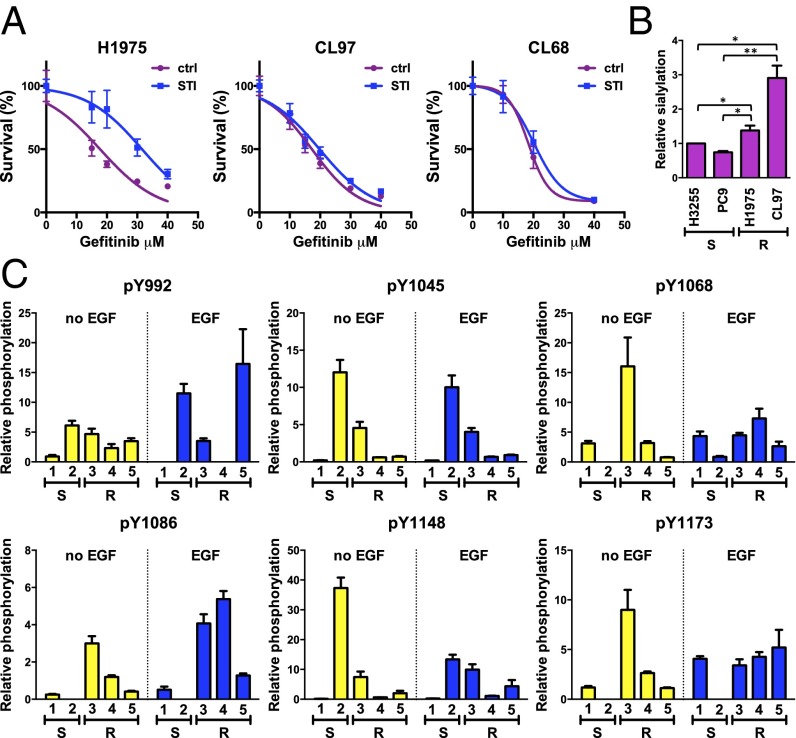
Based on the inhibitory effect of sialylation on the phosphorylation of EGFR mutant L858R/T790M, we hypothesized that sialylation also might influence the responsiveness of cells toward TKI by attenuating the overall signaling output of EGFR. To address this possibility, the sensitivity toward gefitinib in the presence of STI was measured in three TKI-resistant cell lines, H1975 (EGFR L858R/T790M), CL68 (EGFR Del19/T790M), and CL97 (EGFR G719A/T790M). H1975 cells treated with STI showed a significantly higher resistance to gefitinib under concentrations ranging from 15–30 μM (Fig. 5A), but the effect on gefitinib-mediated inhibition of cell growth in CL97 and CL68 cells was not significant.
Fig. 5.
Effect of sialylation on gefitinib sensitivity and EGFR phosphorylation in lung cancer cell lines with EGFR mutations. (A) Proliferation of TKI-resistant lung cancer cell lines with or without STI treatment in the presence of gefitinib. The proliferation assay was performed as described in SI Materials and Methods. (B) Levels of sialylation on EGFR in lung cancer cell lines. Sialylation was analyzed by a lectin pull-down experiment with SNA as described in SI Materials and Methods. Error bars represent SD values. S, TKI sensitive; R, TKI resistant. P values were calculated by paired t test. *P < 0.05; **P < 0.01. (C) Profiling of EGFR phosphorylation in lung cancer cell lines. The levels of site-specific phosphorylation of EGFR were detected by immunoblotting with antibodies recognizing specific phosphosites, and the relative phosphorylation was calculated by normalization to the intensity of A549 cells with EGF treatment. Error bars represent SD values. Representative Western blots are shown in Fig. S7C. Cell lines examined were 1, H3255; 2, PC9; 3, H1975; 4, CL97; 5, CL68. S, TKI sensitive; R, TKI resistant.
To correlate further the EGFR sialylation with its phosphorylation status in lung cancer cells harboring different genotypes of EGFR, two TKI-sensitive cell lines (PC9 and H3255) and three TKI-resistant cell lines with the EGFR T790M mutation (H1975, CL68, and CL97) were examined for the level of EGFR sialylation and tyrosine phosphorylation on EGFR. The level of sialylation on EGFR in different cell lines was examined by Sambucus nigra lectin (SNA) pull-down assay, and the results showed that the TKI-resistant cell lines had higher levels of sialylation on EGFR than did the TKI-sensitive cell lines (Fig. 5B). To quantify the level of tyrosine phosphorylation on EGFR in each cell line properly, the amount of EGFR input in each cell line was carefully adjusted to a similar level (Fig. S7B) and then was probed for tyrosine phosphorylation. The results showed that without EGF stimulation the levels of phosphorylation at Y1068, Y1086, and Y1173 were up-regulated in the TKI-resistant cells harboring the T790M mutation, but in the presence of EGF only the phosphorylation at Y1086 remained significantly higher than that of the TKI-sensitive cells (Fig. 5C and Fig. S7C). However, we could not observe a good correlation between EGFR sialylation and gefitinib sensitivity in all of the cell lines examined, indicating that the suppression effect of sialylation on EGFR phosphorylation is insufficient to combat tumorigenesis.
Discussion
The activation of EGFR depends on intermolecular dimerization between two kinase domains and is triggered by dimerization of the two extracellular domains. Because sialylation attenuates the dimerization of EGFR extracellular domain, it is not surprising that all the EGFR autophosphorylation sites are down-regulated when EGFR is highly sialylated. A study suggested that the elevated kinase activity of the EGFR L858R mutant is caused primarily by the suppression of the intrinsic disorder of the kinase domain that thus facilitates the kinase domain dimerization (31). A more recent study based on the crystal structures showed that neither the L858R nor the L858R/T790M mutant was in the constitutively active conformation, but the dynamic nature of these mutants led to a greater activity even in their monomeric forms (33). Therefore the effect of sialylation on autophosphorylation would not be expected to be as prominent in the L858R or L858R/T790M EGFR mutant as in the wild-type EGFR. However, in our in vitro and in vivo studies we observed site-specific suppression of pY1173 by sialylation, especially under EGF stimulation, in the L858R/T790M mutant. It has been reported that the rates of autophosphorylation in the wild-type EGFR and EGFR L858R mutant are different, suggesting that different EGFR kinases (wild-type or mutants) have different preferences for phosphorylation sites (34). Although the mechanism remains unknown, we speculate that sialylation changes the phosphorylation propensity toward Y1173 in EGFR L858R/T790M. This notion is supported by the observation that the phosphorylation of Y1173 is more dependent on EGF-induced dimerization than are the other phosphosites (Fig. S3); therefore, sialylation suppressed the phosphorylation of Y1173 more significantly. In addition, sialylation also was reported to induce a conformational alteration of other glycoproteins, including MUC1 (35).
EGFR signaling is a complicated network regulated by its phosphorylation. According to the PhosphoSitePlus database (36), more than 50 EGFR phosphosites have been determined by mass spectrometry and other methods. Phosphorylation on each site has a distinct function in regulating the downstream signaling, the kinase activity, and receptor internalization. In addition to tyrosine phosphorylation, many serine and threonine residues are known to be phosphorylated in EGFR, indicating the complex nature of the EGFR signaling network. In this study, we found that sialylation of EGFR regulates the phosphorylation of EGFR, including tyrosine and serine/threonine phosphorylation, in lung cancer cells. Although the precise effect of sialylation on phosphorylation is not well understood, it also may affect other intermolecular interactions, as reported in other related studies. For example, GM3, the ganglioside containing the sialyllactose epitope, has been reported to interact with EGFR and inhibit its kinase activity in a model supplemented with the GM3 glycolipid, and treatment with neuraminidase can rescue the autophosphorylation of EGFR (37). In addition, galectin-3 also can regulate the cellular trafficking and the level of surface EGFR through binding to the glycans on EGFR, and the binding can be blocked by sialylation on EGFR (38, 39).
Studies have shown that distinct EGFR downstream signaling can be initiated by different patterns of EGFR phosphorylation. Therefore investigating the phosphorylation profiles of EGFR is important for understanding the regulation of cellular functions. Because the up-regulation of phosphotyrosines was observed in lung cancer cells with EGFR T790M mutation, the relationship between EGFR genotype and its phosphorylation patterns and the contribution of EGFR phosphorylation in TKI resistance should be studied further. It has been shown that specific EGFR downstream signaling pathways can be elicited by phosphorylation on specific sites. For example, the phosphorylation of Y1068 can recruit GAB-1 or growth factor receptor-bound protein 2 (Grb-2) to activate survival signals (40), whereas the phosphorylation of Y1173 is responsible for eliciting the activation of ERK via inhibition of the SH2 domain-containing transforming protein (SHC) and Grb2 (41, 42). Furthermore, the up-regulation of pY1173 also has been reported in patients who have NSCLC with EGFR mutation, and Akt, MAPK, and Stat3 signaling is higher in pY1173-positive patients (43). It also has been shown that patients with stage IIIb and IV NSCLC with positive pY1173 staining have a shorter superior progression-free survival rate than patients with negative pY1173 staining (44). These data suggest that site-specific phosphorylation of EGFR plays an important role in the maintenance of TKI resistance and that targeting these selective EGFR phosphorylations could be a future direction for drug discovery. Because sialylation modulates the phosphorylation of EGFR, it is possible that sialylation can regulate TKI sensitivity in cells (Fig. 6). Similarly, glycosylation with the bisecting GlcNAc on N-glycans inhibits mammary tumor progression (45). Our preliminary data also revealed that a sialic acid-containing glycolipid, SSEA4, is up-regulated in the TKI-resistant mutants of lung cancer cell lines, compared with the cells with wild-type EGFR (Fig. S8). All these observations suggest a new strategy for lung cancer therapy, possibly using a combination approach (46).
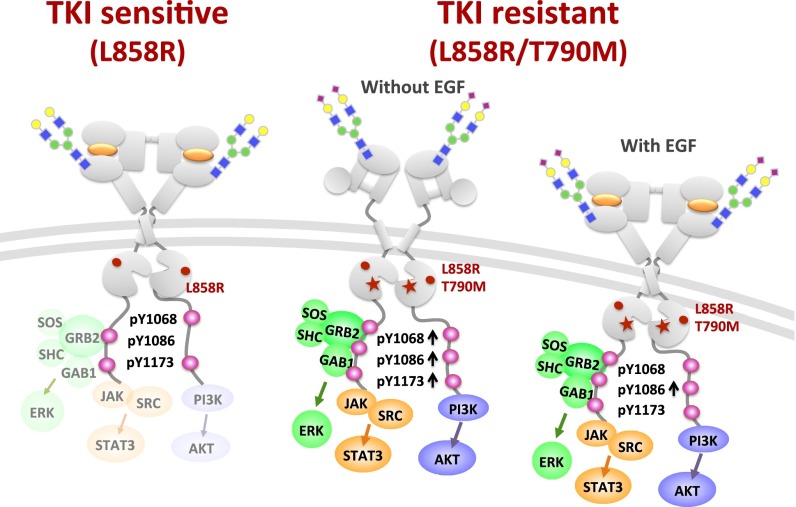
Fig. 6.
TKI-sensitive and -resistant EGFRs and their sialylation and phosphorylation on Y1068, Y1086, and Y1173. Compared with the TKI-sensitive L858R mutant, the TKI-resistant L858R/T790M mutant showed a higher level of phosphorylation at Y1068, Y1086, and Y1173 in the absence of EGF; with EGF, Y1086 showed a higher level of phosphorylation. Note that in the absence of EGF the kinase domain of EGFR L858R and L858R/T790M mutants can dimerize to activate the downstream signaling.
In summary, this study shows the complexity of EGFR sialylation and phosphorylation. Compared with the TKI-sensitive lung cancer-cell mutant L858R, the TKI-resistant lung cancer-cell mutant L858R/T790M has a higher degree of phosphorylation at Y1086 with EGF stimulation and also has higher phosphorylation at Y1068, Y1086, and Y1173 without EGF stimulation. Although sialylation is induced to suppress the phosphorylation of EGFR, the effect of suppression is not strong enough to inhibit the downstream signaling of cancer progression. Development of new-generation TKIs to inhibit the phosphorylation of these sites could overcome the problem of drug resistance.
Materials and Methods
Cell Lines.
The A549 (wild-type), H3255 (L858R), and H1975 (L858R/T790M) cell lines were obtained from ATCC; the PC9 (exon 19 deletion, Del19) cell line was obtained from RIKEN BioResource Center. CL1-0 and CL1-5 (both wild-type) cell lines were as described previously (47), and CL68 (Del19/T790M), CL83 (wild-type), and CL97 (G719A/T790M) cell lines were established from patients who provided informed consent and with the approval of the institutional review board (National Taiwan University Hospital Research Ethics Committee). Among these cell lines, H3255 and PC9 are gefitinib sensitive, and H1975, CL68 and CL97 are gefitinib resistant.
Determination of MM by MALLS Measurement.
MALLS measurements were made with a system composed of a multiangle laser light-scattering photometer (DAWN HELEOS II; Wyatt Technology), a differential refractive index detector (Optilab T-Rex; Wyatt Technology), and a generic UV-absorbance detector equilibrated with 50 mM sodium phosphate buffer at a flow rate of 0.07 mL/min. Samples (0.2 mL) were manually applied to the sample injector conjugated with 0.1 μm Anotop filter (Whatman), and data collection and processing were performed by ASTRA software (Wyatt Technology). The differential refractive index increment (dn/dc) of sEGFR was estimated by the saccharide–protein conjugation method (protein: 0.185; saccharide: 0.147). MM was calculated according to the scattered light intensity, and protein concentration was measured by UV absorbance within a 0.2-min interval of the signal peak. To measure EGF-induced EGFR dimerization, sEGFR was preincubated with EGF in a molar ratio of 1:1.1 for 30 min at 37 °C. The percentage of dimerization was calculated by the following formula: dimerization ratio = (observed MM − monomer MM)/(dimer MM − monomer MM).
In Vitro EGFR Phosphorylation Assay.
The EGFR protein was purified from 293T cells transiently overexpressing FLAG-tagged wild-type or mutant EGFR. Plasmid DNA-transfected 293T cells were lysed with lysis buffer [20 mM Tris (pH 7.4), 400 mM NaCl, 10% (vol/vol) glycerol, 1 mM EDTA, 0.5 mM DTT, 0.2% Triton X-100] and were pretreated with phosphatase (Promega) at 37 °C for 30 min to remove the phosphorylation in cellular proteins. The phosphatase-treated cell lysates were incubated further with sialidase (α2,3/6/8-sialidase; Roche) at 4 °C overnight. The FLAG-tagged wild-type or mutant EGFR then were purified with anti-FLAG M2 agarose (Sigma-Aldrich) and were eluted with 3× FLAG peptide (Sigma-Aldrich) in elution buffer [20 mM Tris (pH 7.4), 400 mM NaCl, 10% (vol/vol) glycerol, 1 mM EDTA, 0.5 mM DTT, 0.1% Triton X-100, 0.1 mg/mL 3× FLAG peptide]. FLAG-tagged wild-type or mutant EGFR protein (0.5 μg) was premixed with 0.1 mg/mL EGF in tyrosine kinase reaction buffer [25 mM Hepes (pH 7.0), 150 mM NaCl, 2 mM MnCl2, 1 mM Tris(2-carboxyethyl)phosphine, 0.1 mg/mL BSA] for 5 min at room temperature, followed by the addition of ATP and further incubation at room temperature for 10 min. Reactions were stopped by the addition of 4× protein sample buffer (Life Technologies) containing 5% (vol/vol) of 2-mercaptoethanol. Samples were separated by SDS/PAGE and subjected to immunoblotting with antibodies specific for EGFR phosphosites (Cell Signaling).
Supplementary Material
Acknowledgments
We thank Professor Jin-Yuan Shih (National Taiwan University Hospital) for the EGFR constructs and the Mass Spectrometry Core Facility at the Genomics Research Center, Academia Sinica, Taiwan for glycan mapping of the sEGFR. This research was supported by the Ministry of Science and Technology and the Genomics Research Center, Academia Sinica, Taiwan.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507329112/-/DCSupplemental.
References
- 1.De Luca A, et al. The role of the EGFR signaling in tumor microenvironment. J Cell Physiol. 2008;214(3):559–567. doi: 10.1002/jcp.21260. [DOI] [PubMed] [Google Scholar]
- 2.Dawson JP, et al. Epidermal growth factor receptor dimerization and activation require ligand-induced conformational changes in the dimer interface. Mol Cell Biol. 2005;25(17):7734–7742. doi: 10.1128/MCB.25.17.7734-7742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferguson KM, et al. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol Cell. 2003;11(2):507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 4.Garrett TP, et al. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell. 2002;110(6):763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- 5.Lemmon MA. Ligand-induced ErbB receptor dimerization. Exp Cell Res. 2009;315(4):638–648. doi: 10.1016/j.yexcr.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogiso H, et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110(6):775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson KM. Structure-based view of epidermal growth factor receptor regulation. Annu Rev Biophys. 2008;37:353–373. doi: 10.1146/annurev.biophys.37.032807.125829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Citri A, Yarden Y. EGF-ERBB signalling: Towards the systems level. Nat Rev Mol Cell Biol. 2006;7(7):505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 9.Morandell S, Stasyk T, Skvortsov S, Ascher S, Huber LA. Quantitative proteomics and phosphoproteomics reveal novel insights into complexity and dynamics of the EGFR signaling network. Proteomics. 2008;8(21):4383–4401. doi: 10.1002/pmic.200800204. [DOI] [PubMed] [Google Scholar]
- 10.Mitsudomi T, Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J. 2010;277(2):301–308. doi: 10.1111/j.1742-4658.2009.07448.x. [DOI] [PubMed] [Google Scholar]
- 11.Sharma SV, Settleman J. ErbBs in lung cancer. Exp Cell Res. 2009;315(4):557–571. doi: 10.1016/j.yexcr.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 12.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 13.Paez JG, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 14.Pao W, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101(36):13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yun CH, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci USA. 2008;105(6):2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 17.Pao W, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau KS, et al. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129(1):123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 19.Yokoe S, et al. The Asn418-linked N-glycan of ErbB3 plays a crucial role in preventing spontaneous heterodimerization and tumor promotion. Cancer Res. 2007;67(5):1935–1942. doi: 10.1158/0008-5472.CAN-06-3023. [DOI] [PubMed] [Google Scholar]
- 20.Whitson KB, et al. Functional effects of glycosylation at Asn-579 of the epidermal growth factor receptor. Biochemistry. 2005;44(45):14920–14931. doi: 10.1021/bi050751j. [DOI] [PubMed] [Google Scholar]
- 21.Tsuda T, Ikeda Y, Taniguchi N. The Asn-420-linked sugar chain in human epidermal growth factor receptor suppresses ligand-independent spontaneous oligomerization. Possible role of a specific sugar chain in controllable receptor activation. J Biol Chem. 2000;275(29):21988–21994. doi: 10.1074/jbc.M003400200. [DOI] [PubMed] [Google Scholar]
- 22.Li W, et al. Down-regulation of trypsinogen expression is associated with growth retardation in alpha1,6-fucosyltransferase-deficient mice: Attenuation of proteinase-activated receptor 2 activity. Glycobiology. 2006;16(10):1007–1019. doi: 10.1093/glycob/cwl023. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, et al. Core fucosylation regulates epidermal growth factor receptor-mediated intracellular signaling. J Biol Chem. 2006;281(5):2572–2577. doi: 10.1074/jbc.M510893200. [DOI] [PubMed] [Google Scholar]
- 24.Liu YC, et al. Sialylation and fucosylation of epidermal growth factor receptor suppress its dimerization and activation in lung cancer cells. Proc Natl Acad Sci USA. 2011;108(28):11332–11337. doi: 10.1073/pnas.1107385108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odaka M, Kohda D, Lax I, Schlessinger J, Inagaki F. Ligand-binding enhances the affinity of dimerization of the extracellular domain of the epidermal growth factor receptor. J Biochem. 1997;122(1):116–121. doi: 10.1093/oxfordjournals.jbchem.a021718. [DOI] [PubMed] [Google Scholar]
- 26.Ferguson KM, Darling PJ, Mohan MJ, Macatee TL, Lemmon MA. Extracellular domains drive homo- but not hetero-dimerization of erbB receptors. EMBO J. 2000;19(17):4632–4643. doi: 10.1093/emboj/19.17.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soderquist AM, Carpenter G. Glycosylation of the epidermal growth factor receptor in A-431 cells. The contribution of carbohydrate to receptor function. J Biol Chem. 1984;259(20):12586–12594. [PubMed] [Google Scholar]
- 28.Sanders JM, Wampole ME, Thakur ML, Wickstrom E. Molecular determinants of epidermal growth factor binding: A molecular dynamics study. PLoS ONE. 2013;8(1):e54136. doi: 10.1371/journal.pone.0054136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yun CH, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11(3):217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, et al. Mechanistic insights into the activation of oncogenic forms of EGF receptor. Nat Struct Mol Biol. 2011;18(12):1388–1393. doi: 10.1038/nsmb.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shan Y, et al. Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell. 2012;149(4):860–870. doi: 10.1016/j.cell.2012.02.063. [DOI] [PubMed] [Google Scholar]
- 32.Rillahan CD, et al. Global metabolic inhibitors of sialyl- and fucosyltransferases remodel the glycome. Nat Chem Biol. 2012;8(7):661–668. doi: 10.1038/nchembio.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gajiwala KS, et al. Insights into the aberrant activity of mutant EGFR kinase domain and drug recognition. Structure. 2013;21(2):209–219. doi: 10.1016/j.str.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 34.Kim Y, et al. Temporal resolution of autophosphorylation for normal and oncogenic forms of EGFR and differential effects of gefitinib. Biochemistry. 2012;51(25):5212–5222. doi: 10.1021/bi300476v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsushita T, et al. Site-specific conformational alteration induced by sialylation of MUC1 tandem repeating glycopeptides at an epitope region for the anti-KL-6 monoclonal antibody. Biochemistry. 2013;52(2):402–414. doi: 10.1021/bi3013142. [DOI] [PubMed] [Google Scholar]
- 36.Hornbeck PV, et al. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43(Database issue):D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coskun Ü, Grzybek M, Drechsel D, Simons K. Regulation of human EGF receptor by lipids. Proc Natl Acad Sci USA. 2011;108(22):9044–9048. doi: 10.1073/pnas.1105666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merlin J, et al. Galectin-3 regulates MUC1 and EGFR cellular distribution and EGFR downstream pathways in pancreatic cancer cells. Oncogene. 2011;30(22):2514–2525. doi: 10.1038/onc.2010.631. [DOI] [PubMed] [Google Scholar]
- 39.Zhuo Y, Bellis SL. Emerging role of alpha2,6-sialic acid as a negative regulator of galectin binding and function. J Biol Chem. 2011;286(8):5935–5941. doi: 10.1074/jbc.R110.191429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito T, et al. Differential activation of epidermal growth factor (EGF) receptor downstream signaling pathways by betacellulin and EGF. Endocrinology. 2004;145(9):4232–4243. doi: 10.1210/en.2004-0401. [DOI] [PubMed] [Google Scholar]
- 41.Batzer AG, Rotin D, Ureña JM, Skolnik EY, Schlessinger J. Hierarchy of binding sites for Grb2 and Shc on the epidermal growth factor receptor. Mol Cell Biol. 1994;14(8):5192–5201. doi: 10.1128/mcb.14.8.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rozakis-Adcock M, et al. Association of the Shc and Grb2/Sem5 SH2-containing proteins is implicated in activation of the Ras pathway by tyrosine kinases. Nature. 1992;360(6405):689–692. doi: 10.1038/360689a0. [DOI] [PubMed] [Google Scholar]
- 43.Zimmer S, et al. Epidermal growth factor receptor mutations in non-small cell lung cancer influence downstream Akt, MAPK and Stat3 signaling. J Cancer Res Clin Oncol. 2009;135(5):723–730. doi: 10.1007/s00432-008-0509-9. [DOI] [PubMed] [Google Scholar]
- 44.Wang F, et al. Phosphorylated EGFR expression may predict outcome of EGFR-TKIs therapy for the advanced NSCLC patients with wild-type EGFR. J Exp Clin Cancer Res. 2012;31:65–74. doi: 10.1186/1756-9966-31-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song Y, Aglipay JA, Bernstein JD, Goswami S, Stanley P. The bisecting GlcNAc on N-glycans inhibits growth factor signaling and retards mammary tumor progression. Cancer Res. 2010;70(8):3361–3371. doi: 10.1158/0008-5472.CAN-09-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brugger W, Thomas M. EGFR-TKI resistant non-small cell lung cancer (NSCLC): New developments and implications for future treatment. Lung Cancer. 2012;77(1):2–8. doi: 10.1016/j.lungcan.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 47.Chu YW, et al. Selection of invasive and metastatic subpopulations from a human lung adenocarcinoma cell line. Am J Respir Cell Mol Biol. 1997;17(3):353–360. doi: 10.1165/ajrcmb.17.3.2837. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.