Abstract
Background
The Pediatric Eosinophilic Esophagitis Symptom Score (PEESS® v2.0) measures patient-relevant outcomes. However, whether patient-identified domains (dysphagia, gastrointestinal reflux disease (GERD), nausea/vomiting, and pain) align with clinical symptomology and histopathologic and molecular features of eosinophilic esophagitis (EoE) is unclear.
Objective
The purpose of this study was to determine if clinical features of EoE, measured through the PEESS® v2.0, associate with histopathologic and molecular features of EoE. This represents a novel approach for analysis of allergic diseases, given the availability of allergic tissue biopsy specimens.
Methods
We systematically recruited treated and untreated, pediatric patients with EoE (aged 2–18 years) and examined parent proxy–reported symptoms using the PEESS® v2.0. Clinical symptomology was collected by questionnaire. Esophageal biopsy samples were quantified for levels of eosinophils, eosinophil peroxidase (EPX) immunohistochemical staining, and mast cells. Molecular features were assessed by the EoE Diagnostic Panel (94 EoE-related gene transcripts). Associations between domain scores and clinical symptoms and biologic features were analyzed using Wilcoxon Rank Sum and Spearman correlation.
Results
The PEESS® v2.0 domains correlated to specific parent-reported symptoms: dysphagia (p = 0.0012), GERD (p = 0.0001), and nausea/vomiting (p < 0.0001). Pain correlated with multiple symptoms (p < 0.0005). Dysphagia correlated most strongly with overall histopathology, particularly in the proximal esophagus (p ≤ 0.0049). Markers of esophageal activity (EPX) were significantly associated with dysphagia (strongest r = .37; p = 0.02). Eosinophil levels were more associated with pain (r = 0.27; p=0.06) than for dysphagia (r = 0.24; p = 0.13). The dysphagia domain correlated the most with esophageal gene transcript levels, predominantly with mast cell–specific genes.
Conclusion
We have 1) established a validated, parent proxy–report measure for pediatric EoE — the PEESS® v2.0; 2) verified that parent-proxy effectively captures symptoms; 3) determined that the dysphagia domain most closely aligns with symptoms and tissue-based molecular biomarkers; 4) established that symptoms correlate EPX staining; and 5) observed association between mast cells and dysphagia.
Keywords: allergy, reflux, quality of life, surveys, mast cells, molecular genetics, pediatrics, microarray, patient-reported outcomes
Introduction
Eosinophilic esophagitis (EoE) is a chronic allergic inflammatory disease driven by food antigen exposure. Although a variety of histological features including the accumulation of eosinophils, mast cells and the deposition of their granule contents in the tissue, as well as epithelial hyperplastic and remodeling changes have been noted, the importance of each of these histological findings for discrete symptoms has been unclear. Clinical outcome endpoints such as disease-specific, patient-reported outcomes (PROs) are increasingly being recognized as essential for linking disease processes with key effector mechanisms and developing treatments that effectively improve clinically relevant features..(1, 2) We recently developed the Pediatric EoE Symptom Score (PEESS® v2.0) in an effort to identify and uniquely measure relevant outcomes that patients with EoE and their families identified as important.(3) Input from patients and their families established that the 20 questions from the PEESS® v2.0 could be consolidated into four major domains: dysphagia, gastrointestinal reflux disease (GERD), nausea/vomiting, and pain.(3) To further validate these domains, it is important to demonstrate that the domains align with clinical symptomology and histopathologic features.
EoE is typified by eosinophil-predominant infiltration of the esophagus (≥ 15 eosinophils per high-powered field [hpf] in at least one hpf in esophageal biopsies) that is not responsive to prolonged, high-dose acid suppression with proton pump inhibitors.(4) However, esophageal changes are not limited to the number of eosinophils and may also depend upon the extracellular content of eosinophil granule proteins such as eosinophil peroxidase (EPX), which may have a functional role in EoE.(5) In addition, a unique esophageal gene expression profile exists in EoE, and the magnitude of its expression is proportional to the quantity of inflammatory cells (e.g. eosinophils and mast cells).(6, 7) This early work implicates Th2 inflammatory responses (e.g. IL-13 and eotaxin-3), as well as the expression of mast cell–specific genes, in EoE pathogenesis.(7–10) Importantly, the gene expression profiles of a specific set of 94 genes can discriminate EoE from non-EoE.(11) However, whether the underlying clinical symptomology, histology, and molecular profiles relate to specific clinical manifestations has not been established.
The purpose of this study was 1) to validate patient-defined domains of the parent proxy–reported PEESS® v2.0 questionnaire; 2) to determine which histological features correlate most strongly and specifically with distinct clinical symptoms; and 3) to gain insight into disease pathophysiology by deeply probing molecular transcript expression as a function of distinct clinical symptoms.
Methods
Study Subjects
Pediatric patients had a confirmed diagnosis of EoE, which was defined as the presence of upper gastrointestinal tract symptoms and an endoscopy with ≥15 eosinophils/hpf in the proximal or distal esophageal tissue biopsies as per consensus recommendations.(12) Additional data collected included a parent-reported clinical symptom questionnaire and an endoscopic sample collection (see Supplementary Methods). Consent was performed by a study staff member and was completed either in same-day surgery or in the outpatient clinic. Assent was obtained for participants aged 11 to 17 years. This study was approved by the CCHMC Institutional Review Board.
Histological and Laboratory Methods
Biopsies obtained were evaluated for histologic features including eosinophil number, tryptase- and chymase-positive cells, and EPX-based immunohistochemistry (see Supplementary Methods). We performed PCR amplification of representative genes from total RNA extracted from distal esophageal biopsies (see Supplementary Methods).(11)
The PEESS® v2.0 and Domains
The PEESS® v2.0 is a content-validated metric that seeks to capture EoE-specific symptoms directly from children with EoE (8–18 years of age) and from their parents (2–18 years of age).12 From parent/participant interviews, four domains were established: dysphagia, gastroesophageal reflux disease (GERD), nausea/vomiting and pain (Table S1). The range for these PEESS® v2.0 scores was 0 to 100, with a higher score being indicative of more frequent and/or severe symptoms for total score, and the dysphagia, GERD, nausea/vomiting and pain domains. For further details, see the Supplementary Methods.
Statistical Analysis
To provide construct validation for the parent proxy–reported PEESS® v2.0 domain scores, we sought to demonstrate that the domain scores associated in an anticipated manner with the presence of specific clinical features obtained by interview and to determine whether any of the domains were associated with expected EoE biological features. A correlation corrected Bonferroni adjustment was made to establish the p value required for statistical significance (comparison between domains p ≤ 0.005; PEESS® v2.0 and clinical symptoms p ≤ 0.003, eosinophil measures p ≤ 0.03, and mast cells p ≤ 0.02). All analyses were performed in JMP v9.0 (SAS Institute, Cary, NC). Detailed statistical analyses are described in the Supplementary Methods.
Results
Patient Population
Overall, this pediatric cohort (n = 46) had a median age of 6.9 years and a median duration since initial diagnosis of 2.4 years (Table 1). Only three participants were untreated at the time of sample collection. Therapies were swallowed steroids alone (either fluticasone propionate or budesonide), diet modification alone (elemental diet, elemental diet and food, elimination diet; or food trial), or both swallowed steroids and diet modification (Table 1).
Table 1.
Characteristics of the pediatric, eosinophilic esophagitis study cohort
| Characteristics | Statistics |
|---|---|
| N | 46 |
| Age (year ± SD) | 8.2 ± 4.2 (median 6.9, range 2.4–17) |
| Age at diagnosis | 6.1 ± 4.4 (median 4.4, range 0.8–17) |
| Sex (% male)† | 100 |
| Race (% white)† | 97.8 |
| Duration since histological diagnosis (years ± SD) | 2.4 ± 1.6 (Range 0.2–7.3) |
| Treatment (%) | 93.5 |
| Diet only (%) | 30.4 |
| Swallowed steroids only (%) | 28.3 |
| Diet and swallowed steroids (%) | 34.8 |
| Peak eosinophil count (median [IQR, range]) | 35.5 (7.8–88.8, 0–295) |
| Disease activity* (% of cohort) | |
| Active disease count (eos ≥ 15 hpf) | 71.7 |
| Intermediate count (15 > eos ≥ 6 hpf) | 6.5 |
| Low count (6 > eos > 0) | 15.2 |
| None (eos = 0)‡ | 6.5 |
| Distal eosinophil count (median [IQR, range]) | 27.5 (7.8–77.3, 0–295) |
| Proximal eosinophil count (median [IQR, range]) | 4 (0–35, 0–295) |
Activity was based on the peak eosinophil count (e.g. maximum of the proximal and distal counts).
Eos, eosinophils; hpf, high-power field; IQR, interquartile; SD, standard deviation
The inclusion of only males and primarily whites was an inclusion criteria for the analysis set to minimize heterogeneity.
All indivduals with no eosinophils were treated.
Patient Characteristics and PEESS® v2.0 Scores
The parent reports correlated well with child reports with correlations ranging from 0.58 – 0.78 (Table 2). Overall, the PEESS® v2.0 total and domain scores showed modest elevations (median range 12.5–25.0) compared to the expectation of a score close to zero for those not reporting symptoms and a maximum score of 100 (Table 2). Of note, the pain domain score was significantly higher than the GERD domain, nausea/vomiting domain, and total scores (p < 0.0025 vs. pain domain for each).
Table 2.
Patient- and parent proxy–reported outcomes as a function of PEESS® v2.0 and its domains
| Parent (N = 46) |
Pediatric Patient* (n = 28) |
Spearman ρ* (p) |
|||
|---|---|---|---|---|---|
| PEESS® v2.0 | Median (IQR) | Range | Median (IQR) | Range | |
| Total score | 22.5 (6.7–31.4) | 0–57.5 | 28.8 (13.8–41.3) | 5–55.0 | 0.74 (0.0001) |
| Domain scores | |||||
| Dysphagia | 21.9 (9.4–33.6) | 0–65.6 | 34.4 (10.9–46.9) | 0–68.8 | 0.67 (0.0008) |
| GERD | 18.8 (0–26.6) | 0–81.3 | 25.0 (3.1–28.1) | 0–50.0 | 0.63 (0.0019) |
| Nausea/Vomiting | 12.5 (0–37.5) | 0–75.0 | 25.0 (3.1–43.8) | 0–81.3 | 0.58 (0.005) |
| Pain | 25.0 (12.5–43.8) | 0–100.0 | 37.5 (21.9–50.0) | 0–56.3 | 0.78 (< 0.0001) |
Only reflects children aged 8 or higher.
GERD, gastroesophageal reflux disease; IQR, interquartile; PEESS® v2.0, pediatric eosinophilic esophagitis symptom score version 2.0
PEESS® v2.0 Domains Associate with Clinical Symptomology
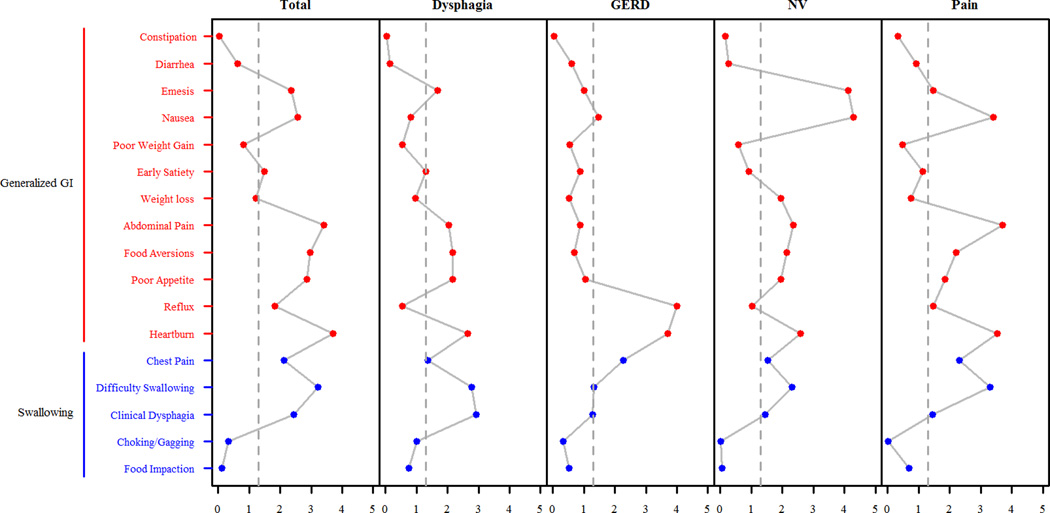
We sought to demonstrate the unique sensitivity of the domains in measuring disease-specific characteristics relative to patient symptomology. Participants had a diverse set of symptoms including a high proportion with upper gastrointestinal and allergic symptoms (Table 3, Table S2). To determine whether PEESS® v2.0 domains were related to specific gastrointestinal symptoms often associated with EoE, Wilcoxon rank sums were used to compare domain scores in individuals with and without specific symptoms (Table 3, Figure 1). The total score was associated with symptoms of difficulty swallowing, heartburn, poor appetite, food aversions, abdominal pain, and nausea (p ≤ 0.0027). However, as the total score is a composite measure of the four domains, it was important to examine associations with the individual domains. Of note, food impaction was not significantly associated with the dysphagia domain; however, this study was underpowered to detect an effect given the low frequency of food impaction in the study cohort (8.7%). In contrast, several symptoms were associated with multiple domains: difficulty swallowing was associated with the dysphagia and pain domains (p ≤ 0.0017, Table 3), heartburn was significantly associated with each of the four domains (p ≤ 0.0026), and nausea was associated with the nausea/vomiting and pain domains (p ≤ 0.0004). Symptoms associated with a single domain included clinical dysphagia (dysphagia domain, p = 0.0012), reflux (GERD domain, p = 0.0001), and emesis (nausea/vomiting domain, p < 0.0001). The PEESS® v2.0 domains were not associated with age, age at diagnosis, time since diagnosis, allergic status, asthma status, or eczema (p > 0.2).
Table 3.
Parent-reported clinical symptoms* in patients with eosinophilic esophagitis and associations with PEESS® v2.0 Score
| Clinical Symptom | N | Frequency (%) |
PEESS® v2.0 Score | |||||
|---|---|---|---|---|---|---|---|---|
| Total | Dysphagia domain |
GERD domain |
Nausea/Vomiting domain |
Pain domain |
||||
| Swallowing | Food Impaction | 46 | 8.7 | -- | -- | -- | -- | -- |
| Choking/gagging | 46 | 21.7 | -- | -- | -- | -- | -- | |
| Clinical Dysphagia | 46 | 28.3 | -- | 25.0 (0.0012) | -- | -- | -- | |
| Difficulty Swallowing | 46 | 21.7 | 22.9 (0.0006) | 29.4 (0.0017) | -- | -- | 25.0 (0.0005) | |
| Chest Pain | 46 | 19.6 | -- | -- | -- | -- | -- | |
| Generalized Gastrointestinal | Heartburn | 46 | 28.3 | 21.2 (0.0002) | 27.6 (0.0023) | 12.5 (0.0002) | 37.5 (0.0026) | 25.0 (0.0003) |
| Reflux | 45 | 33.3 | -- | -- | 14.6 (0.0001) | -- | -- | |
| Poor Appetite | 46 | 43.5 | 12.5 (0.0014) | -- | -- | -- | -- | |
| Food Aversions | 46 | 26.1 | 17 (0.0011) | -- | -- | -- | -- | |
| Weight Loss | 46 | 15.2 | -- | -- | -- | -- | -- | |
| Abdominal Pain | 46 | 47.8 | 15.6 (0.0004) | -- | -- | -- | 21.8 (0.0002) | |
| Early Satiety | 46 | 50.0 | -- | -- | -- | -- | -- | |
| Poor Weight Gain | 45 | 20.0 | -- | -- | -- | -- | -- | |
| Nausea | 46 | 43.5 | 13.1 (0.0027) | -- | -- | 34.4 (<0.0001) | 21.8 (0.0004) | |
| Emesis | 46 | 23.9 | -- | -- | -- | 43.8 (< 0.0001) | -- | |
| Diarrhea | 46 | 37.0 | -- | -- | -- | -- | -- | |
| Constipation | 46 | 17.4 | -- | -- | -- | -- | -- | |
| Bloody Stools** | 46 | 4.3 | -- | -- | -- | -- | -- | |
Symptoms are separated by swallowing and generalized gastrointestinal.
Those with significant differences (p < 0.003 are reported as median difference between those with and without the condition (p value).
The condition was too infrequent in the study cohort for comparative analyses.
-- non-significant
GERD, gastroesophageal reflux disease
Figure 1. The relationships between gastrointestinal symptoms and PEESS® v2.0 scores.
The x-axis represents the negative log10 p-value of the Wilcoxon Rank Sum test comparing individuals reporting the clinical symptom to those who do not report the clinical symptom. The x-axis columns in the figure are PEESS® v2.0 scores while the y-axis indicates clinical symptoms reported as yes/no. GERD, gastroesophageal reflux disease; GI, gastrointestinal; NV, nausea/vomiting domain.
PEESS® v2.0 Domains Associate with EoE Biological Features
Eosinophil Measures Most Strongly Correlate with the Dysphagia Domain
Most individuals (n = 33) had active EoE, and very few (n = 3) had biopsies without eosinophils. Peak eosinophil counts ranged from 0 to 205 eosinophils/hpf with a median count of 35.5 eosinophils/hpf (Table 1). Counts from the distal esophagus tended to be higher than from proximal counts (Table 1). The extracellular EPX staining demonstrated that most patients had some level of eosinophil activation and that this eosinophil activation was much more prominent in the distal than the proximal esophagus (Table S3).
Esophageal mucosal eosinophil counts did not exhibit statistically significant (p > 0.05) associations with the PEESS® v2.0 domain scores, but suggestive trends were identified (Figure 2). The strongest association with distal eosinophil count was with the pain domain (r = 0.28, p = 0.06) while the strongest association with proximal eosinophil count was with the dysphagia domain (r = 0.24, p = 0.13). While individuals histologically active disease (≥ 15 eosinophils/hpf) had higher domain scores than those with normal histology (0 eosinophils/hpf) with respect to the PEESS® v2.0 domain scores or total score (Table S4), these differences were not statistically significant, likely due to the small sample size in the normal histology group (n=3). Further, when we examined all possible comparisons between eosinophil classifications, no differences reached significance (p > 0.05). However, individuals in remission had the lowest PEESS® v2.0 scores (Table S4).
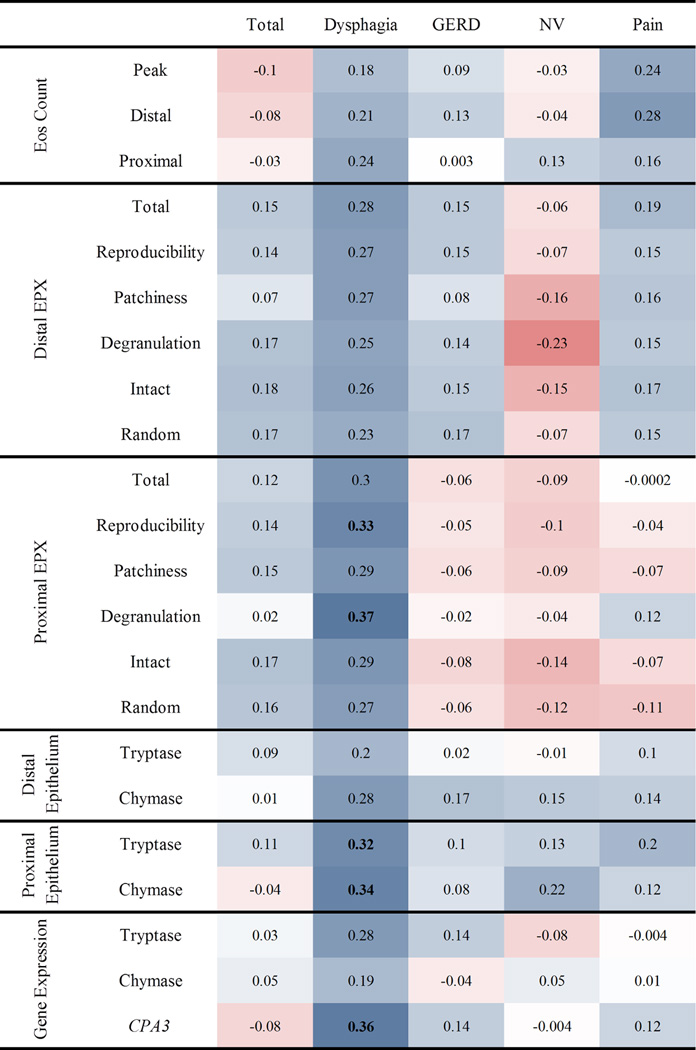
Figure 2. Spearman correlations between PEESS® v2.0 scores and disease parameters.
Strength of the association with a diagnostic subset of the eosinophilic esophagitis transcriptome is measured using Spearman’s ρ (text within cell). Darker red shades indicate stronger negative correlations, whereas darker blue shades indicate stronger positive correlations. Bolded values indicate correlations that are significant at p ≤ 0.05. CPA3, carboxypeptidase A3; Eos, eosinophil; EPX, eosinophil peroxidase; GERD, gastroesophageal reflux disease; NV, nausea/vomiting domain
Among all of the PEESS® v2.0 scores, the dysphagia domain score consistently exhibited the strongest correlation with EPX values for both distal and proximal biopsies (ρ = 0.23 – 0.37, p value = 0.019–0.17; Figure 2). The only correlations that reached nominal (p ≤ 0.05) significance were between EPX parameters of the proximal esophagus (degranulation and reproducibility) and the dysphagia domain. To determine whether the dysphagia domain correlates more strongly with the eosinophil markers than the other domains, we compared the median correlation coefficient of eosinophil counts and EPX of the dysphagia domain to the other domains. The dysphagia domain had higher median correlation than the total and domain scores for both distal (p ≤ 0.022) and proximal (p ≤ 0.0022) measures (Figure S1).
Mast Cell Markers and CPA3 Gene Expression Associate with the Dysphagia Domain
Tryptase+ and chymase+ mast cells were identified in proximal and distal esophageal biopsies (Table S3). In the proximal esophagus, both tryptase+ and chymase+ intraepithelial mast cells were significantly associated with the PEESS® v2.0 dysphagia domain (ρ = 0.34, p = 0.036 and ρ = 0.32, p = 0.041, respectively; Figure 1). The distal measures did not reach statistical significance. Notably, we found a strong correlation between dysphagia and CPA3 gene expression (ρ = 0.36, p = 0.02). The dysphagia domain also exhibited a correlative tendency with tryptase gene expression (ρ = 0.28) that did not reach statistical significance (p = 0.076), perhaps due to the small cohort size.
Diagnostic Subset of the EoE Transcriptome (EDP) Most Strongly Associates with the Dysphagia Domain
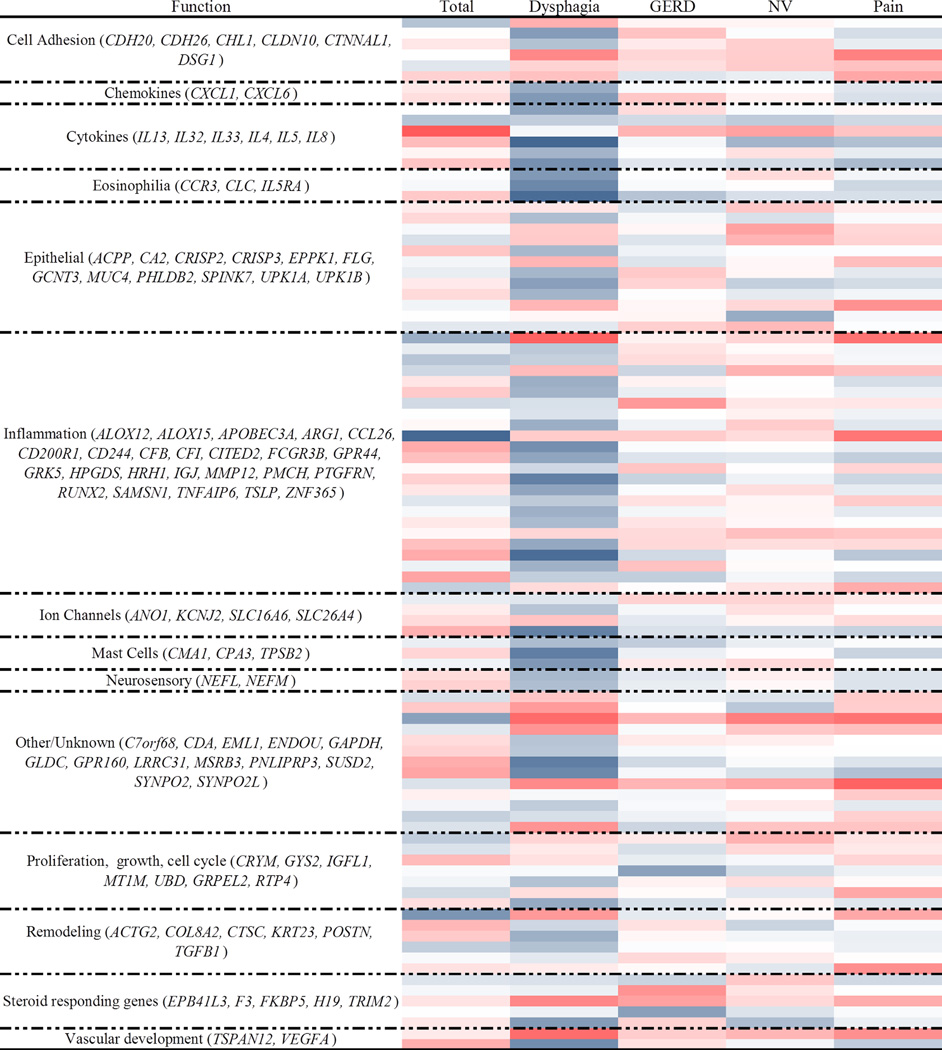
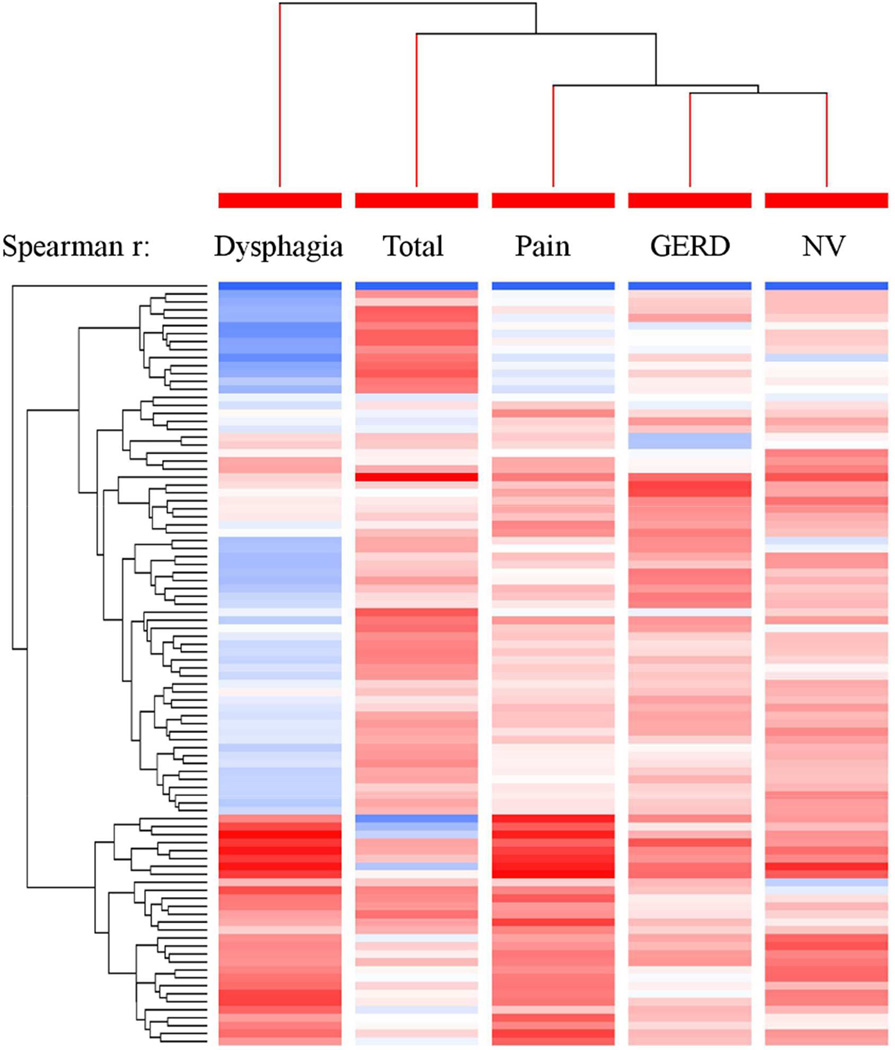
Overall, there was weak correlation between the domains and the genes (absolute median value ρ = 0.08, IQR 0.04–0.14, range 0–0.42) (Figure 3; Table S5). The dysphagia domain exhibited a significantly (p < 0.0001) higher magnitude of correlation with the EDP than the other domains and the total score (absolute median value ρ = 0.18, IQR 0.10–0.25 for the dysphagia domain; ρ = 0.07, IQR 0.04–0.12 for the total score; ρ = 0.06, IQR 0.02–0.10 for the GERD domain; ρ = 0.06, IQR 0.02–0.10 for the nausea/vomiting domain; and ρ = 0.08, IQR 0.04–0.12 for the pain domain). Focusing on the dysphagia domain and categories of genes, we observed that genes related to eosinophilia (IQR 0.28–0.41), chemokines (IQR 0.23–0.28), mast cells (0.10–0.36), neurosensory (0.18–0.20), cytokines (IQR 0.11–0.33) and inflammation (IQR 0.08–0.24) had positive Spearman correlation interquartile ranges that did not overlap zero, suggesting a positive relationship with the dysphagia domain. To determine how the domains relate to each other with respect to the gene expression patterns, we created a hierarchical tree using the Spearman correlation values (Figure 4), as indicated by tree-branch hierarchy. The PEESS® v2.0 dysphagia domain differed from the other domains but was most similar to the total score. These data suggest that the dysphagia domain is more effective at capturing biological processes underlying the EDP than the other PEESS® v2.0 domains or total score.
Figure 3. Spearman correlations between PEESS® v2.0 scores and a diagnostic subset of the eosinophilic esophagitis transcriptome.
Darker red shades indicate stronger negative correlations, whereas darker blue shades indicate stronger positive correlations. Correlations by functional groupings of genes.
Figure 4. The hierarchical relationships between domains based on gene expression profile correlations.
Using the Spearman r for the correlation between a diagnostic subset of the eosinophilic esophagitis (EoE) transcriptome (EoE diagnostic panel) gene expression and PEESS® v2.0 domain scores, we created a clustering tree representing the hierarchical order of the domain’s representativeness and plotted it with the Spearman r–based heat-diagram for the correlation at gene level. Darker red shades indicate stronger negative correlations, whereas darker blue shades indicate stronger positive correlations. The shorter the distance (tree-branch length), the more similar the expression correlation for each domain is.
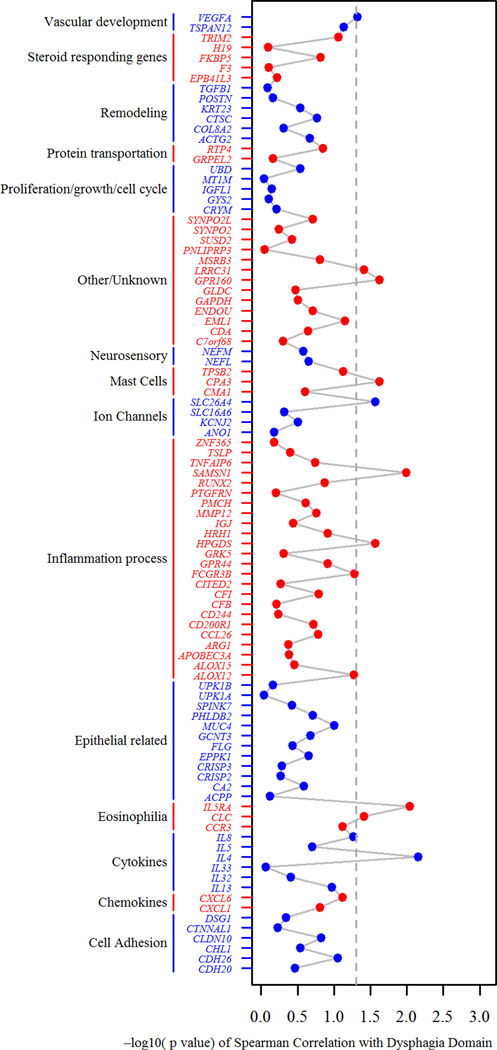
Genes that correlated with the dysphagia domain (Figure 5) include the vascular development gene VEGFA (ρ = 0.32, p = 0.048), the other/unknown category genes GPR160 (ρ = 0.36, p = 0.024) and LRRC31 (ρ = 0.33, p = 0.039), the mast cell gene CPA3 (ρ = 0.36, p = 0.024), the ion channel gene SLC26A4 (ρ = 0.35, p = 0.027), the inflammation genes SAMSN1 (ρ = 0.40, p = 0.01) and HPGDS (ρ = 0.35, p = 0.027), the eosinophil genes IL5RA (ρ = 0.42, p = 0.0092) and CLC (ρ = 0.33, p = 0.039), and the cytokine gene IL4 (ρ = 0.42, p = 0.0070).
Figure 5. Associations between the PEESS® v2.0 dysphagia domain and a diagnostic subset of the eosinophilic esophagitis transcriptome.
The x-axis represents the negative log10 p-value of the Spearman correlation between the dysphagia domain and a diagnostic subset of genes from the eosinophilic esophagitis (EoE) transcriptome (EoE diagnostic panel, EDP). The y-axis is organized by genes within functional groupings, color shading is for ease of interpretation only. Dashed line indicates p = 0.05.
Discussion
EoE has been well characterized biologically, but the severity and prevalence of signs and symptoms of this disease are less well documented. The development of therapeutics that improve the biological and clinical underpinnings of EoE in children are dependent upon availability of validated metrics for measuring clinical symptoms in pediatric patients with EoE.(1, 13, 14) This study demonstrated that parent proxy–reported PEESS® v2.0 domains are consistent with both clinical symptomology obtained by parent interview and EoE biological features, thus providing construct validity for these domains. Specifically, using a well-characterized, prospective cohort of treated and untreated pediatric patients with EoE, we were able to examine associations between parent proxy–reported PEESS® v2.0 domains and parent-reported clinical symptoms, EoE-specific gene expression and histology, thereby testing the validity of the domains. We found that parent proxy–reported PEESS® v2.0 domains correlated with parent-reported patient symptomology. Additionally, an EoE-specific gene expression panel (the EDP) most strongly correlated with the PEESS® v2.0 dysphagia domain. Furthermore, the PEESS® v2.0 dysphagia domain significantly correlated with esophageal histology (both EPX staining and mast cell levels) in the proximal esophagus. Notably, the EPX staining but not the eosinophil levels significantly correlated in the proximal esophagus, suggesting that it is the activity of the eosinophils rather than their absolute levels that correspond most as a marker of dysphagia. Collectively, these results support the value of the parent proxy–reported PEESS® v2.0 and it domains, especially dysphagia, and suggest a key role of eosinophil activity and mast cells in eliciting symptoms.
Most (93.5%) of our patient population was undergoing treatment, but they still exhibited signs of disease, both based on clinical symptoms and histology. The most common clinical symptoms included early satiety, abdominal pain and nausea. On the basis of these clinical symptoms, our patient population seems representative of EoE pediatric populations, as abdominal pain is one of the most commonly reported symptoms.(15–20) The lower frequency of dysphagia, food impaction, and emesis compared to other studies may be due to the high frequency of treated subjects.(21, 22) We also found that most of our patients had detectable eosinophils in their esophageal biopsies; only 6.5% were in remission (no eosinophils), and 71% had active disease (eosinophils/hpf ≥ 15/hpf). Though both dietary and swallowed steroid therapies have been noted to have high success rates,(23, 24) patients whose EoE has periodic flares and relapses may require multiple courses of treatment.(25, 26) Of note, given the study design, we were unable to explore the effect of different treatments on patient symptomology. Longitudinal studies will be required to capture the full symptomology of disease and response to treatment.
The parent proxy–reported PEESS® v2.0 demonstrated substantial disease burden across various domains. The PEESS® v2.0(27) addresses the concerns most pressing to patients with EoE and their families. Patients had previously identified four domains (dysphagia, GERD, nausea/vomiting and pain). Importantly, our patients exhibited moderate impairment across all domains, which is consistent with reported symptoms of EoE.(16, 17, 19, 28, 29) Reported clinical symptoms aligned well with related domains designed to capture the different aspects of disease, thus validating the domains. This is important because the impact of EoE clearly extends beyond dysphagia(30) but may not be captured by generic PROs.(27) These results provide tools that may be used to evaluate different aspects of EoE and determine the efficacy of different treatments on the quality of life of pediatric patients. This work will complement the recently developed symptom-based activity index for adults by Schoepfer and colleagues,(31) which aligns well with endoscopic changes seen less frequently in children (rings, furrows and strictures), as well as the recently developed PedsQL™ EoE Module.(13, 14)
Beyond clinical symptoms, we also found that one specific domain of the PEESS® v2.0, the dysphagia domain, correlated with a newly developed molecular diagnostic test for EoE, the EDP,(11) to a stronger degree than any of the other domains or the total score. Previous studies of EoE have identified sets of genes (~500) whose esophageal expression can discriminate EoE from non-EoE.(6, 10, 32, 33) This work led to the development of a smaller set of genes that provides strong discriminatory value (the EDP) for individuals who have ≥ 15 eosinophils/hpf in the esophagus compared to those with ≤2 eosinophils/hpf in the esophagus.(11) Thus, the PEESS® v2.0 dysphagia domain captures information associated with alterations in esophageal gene expression. Specifically, genes exhibiting association with dysphagia include IL4, IL5RA, CLC, SAMSN1, HPGDS, CPA3, GPR160, LRRC31, SLC26A4 and VEGFA. Although little is understood about the biology of LRRC31, SLC26A4 and GPR160, the other genes have strong ties to eosinophils (CLC, IL4, IL5RA and HPGDS) and mast cells (VEGFA, HPGDS, CPA3, SAMSN1, IL4 and IL5RA).(7, 34–38) These genes have deep and diverse implications, as CLC is a marker that is now being used in a new minimally invasive test, the EoE string test,(39) CPA3 is a specific marker of mast cells,(9, 40, 41) IL4 and IL5 are targets of relevant therapeutic humanized antibody therapies being developed (e.g. anti–IL-5/anti–IL-5 receptor, anti–IL-4 receptor),(42–44) HPGDS can be blocked by a variety of drugs (e.g. cyclooxygenase inhibitors)(42) and VEGFR highlights consideration of anti-VEGF and related angiogenesis-based therapeutics, particularly for treatment of the cardinal EoE symptom dysphagia. Furthermore, one gene (CPA3) has been previously associated with chronic pelvic pain,(45) thereby supporting a role in patient symptomology. Notably, only the PEESS® v2.0 dysphagia domain was associated with these tissue-based molecular biomarkers. It is interesting to speculate that dysphagia may be more reflective of pathophysiological mucosal responses, whereas pain and nausea/vomiting may reflect processes primarily outside of the mucosa. The failure to associate EoE gene transcripts with the other PEESS® v2.0 domains (pain, nausea/vomiting and GERD) may also explain why current treatments that alter the transcriptome do not resolve all symptoms.(46)
Importantly, we found that histologic markers of EoE, related to both eosinophil activity and mast cells, exhibited the strongest associations with the dysphagia domain. Though elevated esophageal eosinophil level is a hallmark of EoE, recent work also supports a role for mast cells.(7, 47–50) The association between dysphagia and mast cells is consistent with recent findings from adult studies.(51) In addition, both the eosinophil activity (EPX staining) and mast cell level from the proximal esophagus showed stronger associations than the distal esophagus with the PEESS® v2.0 dysphagia domain. The reason for these relationships is not known, but perhaps could be related to acid exposure in the esophagus which also contributes to distal esophageal eosinophilia. These data support the value of collecting proximal esophageal biopsies, as these may be more reflective of patient symptomology.
Lastly, unlike EPX staining, the absolute measures of eosinophils in the proximal esophagus were not significantly correlated with patient symptomology, suggesting that the presence of eosinophils alone is not the primary contributor for patient symptomology. The lack of association between dysphagia and eosinophil number is notable as the eosinophil level in the esophageal epithelium is considered the gold standard for the diagnosis of EoE. It is important to highlight that there was a trend between eosinophil levels and symptom scores, indicating that eosinophil levels alone may have a clinical correlation, but it is likely less contributory compared with the levels of their activation and levels of mast cells. It should also be noted that within the dysphagia domain, individuals with low eosinophil counts had a wide range of dysphagia scores, suggesting that an esophageal count of <15 eosinophils/hpf may still be clinically significant. These results are consistent with previous reports that demonstrated that individuals with low eosinophil counts (e.g. 5 eosinophils/hpf) were at risk for multiple endoscopies and chronic symptoms.(52, 53) Thus, other factors such as eosinophil activity and mast cell activity as well as other histologic alterations may also contribute to disease. These data suggest that additional evaluation of esophageal histologic changes may be valuable in understanding clinical symptoms.
In summary, patient-reported outcomes are increasingly recognized as important for the management of chronic conditions, but validated questionnaires for pediatric EoE are just now emerging. Previous work had validated 20 questions comprising the PEESS® v2.0, and pediatric patients with EoE and their parents indicated that these questions reflected four major domains (dysphagia, GERD, nausea/vomiting and pain). Herein, we have 1) established a validated report measure for pediatric EoE — the PEESS® v2.0. While this metric could be considered limited by parent report by proxy, we view this as a potential strength as it allows capture of symptoms in this key age range, particularly focused on an age window where approved drugs are needed. 2) Despite its potential limitation, we show that parent-proxy report is effective in capturing symptoms using this instrument; 3) demonstrated value of the four domains — dysphagia, GERD, pain and nausea/vomiting; 4) determined that the dysphagia domain most closely aligns with parent-reported symptoms and tissue-based molecular biomarkers; 5) established that symptoms correlate with the eosinophil activity marker of EPX staining; and 6) highlighted a relatively strong association between mast cells (and their markers) and dysphagia. Collectively, these results demonstrate that the parent proxy–reported PEESS® v2.0 is an objective measure of patient symptomology and may be used to help evaluate treatment response and better understand how biological changes associate with patient and parent perceptions of wellness.
Supplementary Material
Key message.
Parent proxy–reported PEESS® v2.0 domains correlate with symptomology.
The dysphagia domain correlates with EoE specific gene expression and esophageal histology, most notably identifying a link between markers of mast cells and eosinophil activity with symptomology.
Eosinophil activity, rather than absolute level, associate with dysphagia.
Acknowledgments
Support: This work has been generously supported by NIH R01 DK076893-03S1, The Campaign Urging Research for Eosinophilic Disease (CURED), the Buckeye Foundation and the Food Allergy Research and Education (FARE).
MER is a consultant for Immune Pharmaceuticals, Celsus Therapeutics and Receptos and has an equity interest in each. He is an inventor of eosinophilic esophagitis–related patents owned by Cincinnati Children’s Hospital Medical Center. He has a royalty interest in reslizumab, a drug being developed by Teva Pharmaceuticals.
MHC is a consultant with Meritage Pharma, Biogen Idec, Receptos, Regeneron, Novartis; a member of Consortium for Eosinophilic Gastrointestinal Disorders (CEGIR); a member of the Medical Advisory Panel of APFED; a member of the Executive Committee of TIGERS; a member of the Advisory Committee of REGID.
JWV holds the copyright and the trademark for the PedsQL™ and receives financial compensation from the Mapi Research Trust, which is a nonprofit research institute that charges distribution fees to for-profit companies that use the Pediatric Quality of Life Inventory™.
TW is a co-inventor for a pending patent owned by CCHMC based on the EDP gene expression panel described herein.
JJL acknowledges that the EPX antibody was made, in part, with a Sponsored Research grant from Schering Plough (which is now part of Merck).
PEP is on the speaker’s bureau for Abbott Nutrition and Nutricia.
Abbreviations
- EoE
Eosinophilic esophagitis
- PROs
patient reported outcomes
- PEESS® v2.0
Pediatric EoE Symptom Score
- GERD
gastrointestinal reflux disease
- hpf
high-powered field
- EPX
eosinophil peroxidase
- PCR
polymerase chain reaction
- RNA
ribonucleic acid
- EDP
EoE diagnostic panel
- CPA3
carboxypeptidase A3
- VEGFA
Vascular endothelial growth factor A
- GPR160
G protein-coupled receptor 160
- LRRC31
leucine rich repeat containing 31
- SLC26A4
solute carrier family 26 (anion exchanger), member 4
- SAMSN1
SAM domain, SH3 domain and nuclear localization signals 1
- HPGDS
hematopoietic prostaglandin D synthase
- IL5RA
interleukin 5 receptor, alpha
- CLC
Charcot-Leyden crystal galectin
- IL4
interleukin 4
- PedsQL
Pediatric Quality of Life InventoryTM
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions:
Obtain study funding (MER, JPF), study design (JPF, JPA, MER), study supervision (JTG), acquisition of data (JPF, JTG, ME, KM, PEP, JMG, AK), data analysis (LJM, JJL, TW), technical support (JTG, ME, KM), interpretation of data (LJM, MHC, JPA, JWV, TW, HH), drafting of the manuscript (LJM, MHC, JPA, HH), critical revision of manuscript (JPF, JJL, KAH, JWV, PEP, JMG, AK, TW, MER)
Conflicts of interest
LJM, JMG, AK, KAH, ME, HH, KM, JPA, JTG: None
References
- 1.Rothenberg ME, Aceves S, Bonis PA, Collins MH, Gonsalves N, Gupta SK, et al. Working with the US Food and Drug Administration: progress and timelines in understanding and treating patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2012;130(3):617–619. doi: 10.1016/j.jaci.2012.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fiorentino R, Liu G, Pariser AR, Mulberg AE. Cross-sector sponsorship of research in eosinophilic esophagitis: a collaborative model for rational drug development in rare diseases. J Allergy Clin Immunol. 2012;130(3):613–616. doi: 10.1016/j.jaci.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 3.Franciosi JP, Hommel KA, DeBrosse CW, Greenberg AB, Greenler AJ, Abonia JP, et al. Development of a validated patient-reported symptom metric for pediatric eosinophilic esophagitis: qualitative methods. BMC gastroenterology. 2011;11:126. doi: 10.1186/1471-230X-11-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins MH. Histopathology of eosinophilic esophagitis. Dig Dis. 2014;32(1–2):68–73. doi: 10.1159/000357012. [DOI] [PubMed] [Google Scholar]
- 5.Protheroe C, Woodruff SA, de Petris G, Mukkada V, Ochkur SI, Janarthanan S, et al. A novel histologic scoring system to evaluate mucosal biopsies from patients with eosinophilic esophagitis. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2009;7(7):749–755. e11. doi: 10.1016/j.cgh.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116(2):536–547. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abonia JP, Blanchard C, Butz BB, Rainey HF, Collins MH, Stringer K, et al. Involvement of mast cells in eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126(1):140–149. doi: 10.1016/j.jaci.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moawad FJ, Veerappan GR, Dias JA, Baker TP, Maydonovitch CL, Wong RK. Randomized controlled trial comparing aerosolized swallowed fluticasone to esomeprazole for esophageal eosinophilia. Am J Gastroenterol. 2013;108(3):366–372. doi: 10.1038/ajg.2012.443. [DOI] [PubMed] [Google Scholar]
- 9.Hsu Blatman KS, Gonsalves N, Hirano I, Bryce PJ. Expression of mast cell-associated genes is upregulated in adult eosinophilic esophagitis and responds to steroid or dietary therapy. J Allergy Clin Immunol. 2011;127(5):1307–1308. e3. doi: 10.1016/j.jaci.2010.12.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattacharya B, Carlsten J, Sabo E, Kethu S, Meitner P, Tavares R, et al. Increased expression of eotaxin-3 distinguishes between eosinophilic esophagitis and gastroesophageal reflux disease. Hum Pathol. 2007;38(12):1744–1753. doi: 10.1016/j.humpath.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Wen T, Stucke EM, Grotjan TM, Kemme KA, Abonia JP, Putnam PE, et al. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology. 2013;145(6):1289–1299. doi: 10.1053/j.gastro.2013.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liacouras CA, Furuta GT, Hirano I, Atkins D, Attwood SE, Bonis PA, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128(1):3–20. doi: 10.1016/j.jaci.2011.02.040. e6; quiz 1–2. [DOI] [PubMed] [Google Scholar]
- 13.Franciosi JP, Hommel KA, Bendo CB, King EC, Collins MH, Eby MD, et al. PedsQL eosinophilic esophagitis module: feasibility, reliability, and validity. J Pediatr Gastroenterol Nutr. 2013;57(1):57–66. doi: 10.1097/MPG.0b013e31828f1fd2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franciosi JP, Hommel KA, Greenberg AB, DeBrosse CW, Greenler AJ, Abonia JP, et al. Development of the Pediatric Quality of Life Inventory Eosinophilic Esophagitis module items: qualitative methods. BMC gastroenterology. 2012;12:135. doi: 10.1186/1471-230X-12-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orenstein SR, Shalaby TM, Di Lorenzo C, Putnam PE, Sigurdsson L, Mousa H, et al. The spectrum of pediatric eosinophilic esophagitis beyond infancy: a clinical series of 30 children. Am J Gastroenterol. 2000;95(6):1422–1430. doi: 10.1111/j.1572-0241.2000.02073.x. [DOI] [PubMed] [Google Scholar]
- 16.Baxi S, Gupta SK, Swigonski N, Fitzgerald JF. Clinical presentation of patients with eosinophilic inflammation of the esophagus. Gastrointest Endosc. 2006;64(4):473–478. doi: 10.1016/j.gie.2006.03.931. [DOI] [PubMed] [Google Scholar]
- 17.Hasosah MY, Sukkar GA, Alsahafi AF, Thabit AO, Fakeeh ME, Al-Zahrani DM, et al. Eosinophilic esophagitis in Saudi children: symptoms, histology and endoscopy results. Saudi journal of gastroenterology : official journal of the Saudi Gastroenterology Association. 2011;17(2):119–123. doi: 10.4103/1319-3767.77242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rezende ER, Barros CP, Ynoue LH, Santos AT, Pinto RM, Segundo GR. Clinical characteristics and sensitivity to food and inhalants among children with eosinophilic esophagitis. BMC research notes. 2014;7:47. doi: 10.1186/1756-0500-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sant'Anna AM, Rolland S, Fournet JC, Yazbeck S, Drouin E. Eosinophilic esophagitis in children: symptoms, histology and pH probe results. J Pediatr Gastroenterol Nutr. 2004;39(4):373–377. doi: 10.1097/00005176-200410000-00013. [DOI] [PubMed] [Google Scholar]
- 20.Miehlke S. Clinical features of eosinophilic esophagitis. Dig Dis. 2014;32(1–2):61–67. doi: 10.1159/000357011. [DOI] [PubMed] [Google Scholar]
- 21.Kagalwalla AF, Sentongo TA, Ritz S, Hess T, Nelson SP, Emerick KM, et al. Effect of six-food elimination diet on clinical and histologic outcomes in eosinophilic esophagitis. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2006;4(9):1097–1102. doi: 10.1016/j.cgh.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 22.Liacouras CA, Spergel JM, Ruchelli E, Verma R, Mascarenhas M, Semeao E, et al. Eosinophilic esophagitis: a 10-year experience in 381 children. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2005;3(12):1198–1206. doi: 10.1016/s1542-3565(05)00885-2. [DOI] [PubMed] [Google Scholar]
- 23.Arias A, Gonzalez-Cervera J, Tenias JM, Lucendo AJ. Efficacy of dietary interventions for inducing histologic remission in patients with eosinophilic esophagitis: a systematic review and meta-analysis. Gastroenterology. 2014;146(7):1639–1648. doi: 10.1053/j.gastro.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Boldorini R, Mercalli F, Oderda G. Eosinophilic oesophagitis in children: responders and non-responders to swallowed fluticasone. J Clin Pathol. 2013;66(5):399–402. doi: 10.1136/jclinpath-2012-201253. [DOI] [PubMed] [Google Scholar]
- 25.Papadopoulou A, Koletzko S, Heuschkel R, Dias JA, Allen KJ, Murch SH, et al. Management guidelines of eosinophilic esophagitis in childhood. J Pediatr Gastroenterol Nutr. 2014;58(1):107–118. doi: 10.1097/MPG.0b013e3182a80be1. [DOI] [PubMed] [Google Scholar]
- 26.Rodrigues M, D'Amico MF, Patino FR, Barbieri D, Damiao AO, Sipahy AM. Clinical manifestations, treatment, and outcomes of children and adolescents with eosinophilic esophagitis. J Pediatr (Rio J) 2013;89(2):197–203. doi: 10.1016/j.jped.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Franciosi JP, Hommel KA, DeBrosse CW, Greenberg AB, Greenler AJ, Abonia JP, et al. Quality of life in paediatric eosinophilic oesophagitis: what is important to patients? Child Care Health Dev. 2012;38(4):477–483. doi: 10.1111/j.1365-2214.2011.01265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noel RJ, Putnam PE, Collins MH, Assa'ad AH, Guajardo JR, Jameson SC, et al. Clinical and immunopathologic effects of swallowed fluticasone for eosinophilic esophagitis. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2004;2(7):568–575. doi: 10.1016/s1542-3565(04)00240-x. [DOI] [PubMed] [Google Scholar]
- 29.Eroglu Y, Lu H, Terry A, Tendler J, Knopes B, Corless C, et al. Pediatric eosinophilic esophagitis: single-center experience in northwestern USA. Pediatr Int. 2009;51(5):612–616. doi: 10.1111/j.1442-200X.2008.02796.x. [DOI] [PubMed] [Google Scholar]
- 30.Taft TH, Kern E, Keefer L, Burstein D, Hirano I. Qualitative assessment of patient-reported outcomes in adults with eosinophilic esophagitis. J Clin Gastroenterol. 2011;45(9):769–774. doi: 10.1097/MCG.0b013e3182166a5a. [DOI] [PubMed] [Google Scholar]
- 31.Schoepfer AM, Straumann A, Panczak R, Coslovsky M, Kuehni CE, Maurer E, et al. Development and Validation of a Symptom-Based Activity Index for Adults with Eosinophilic Esophagitis. Gastroenterology. 2014 doi: 10.1053/j.gastro.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu TX, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120(6):1292–1300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 33.Matoso A, Mukkada VA, Lu S, Monahan R, Cleveland K, Noble L, et al. Expression microarray analysis identifies novel epithelial-derived protein markers in eosinophilic esophagitis. Mod Pathol. 2013;26(5):665–676. doi: 10.1038/modpathol.2013.41. [DOI] [PubMed] [Google Scholar]
- 34.Shaik-Dasthagirisaheb YB, Varvara G, Murmura G, Saggini A, Potalivo G, Caraffa A, et al. Vascular endothelial growth factor (VEGF), mast cells and inflammation. International journal of immunopathology and pharmacology. 2013;26(2):327–335. doi: 10.1177/039463201302600206. [DOI] [PubMed] [Google Scholar]
- 35.Spik I, Brenuchon C, Angeli V, Staumont D, Fleury S, Capron M, et al. Activation of the prostaglandin D2 receptor DP2/CRTH2 increases allergic inflammation in mouse. J Immunol. 2005;174(6):3703–3708. doi: 10.4049/jimmunol.174.6.3703. [DOI] [PubMed] [Google Scholar]
- 36.Weller PF, Goetzl EJ, Austen KF. Identification of human eosinophil lysophospholipase as the constituent of Charcot-Leyden crystals. Proc Natl Acad Sci U S A. 1980;77(12):7440–7443. doi: 10.1073/pnas.77.12.7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uchida T, Nakao A, Nakano N, Kuramasu A, Saito H, Okumura K, et al. Identification of Nash1, a novel protein containing a nuclear localization signal, a sterile alpha motif, and an SH3 domain preferentially expressed in mast cells. Biochem Biophys Res Commun. 2001;288(1):137–141. doi: 10.1006/bbrc.2001.5722. [DOI] [PubMed] [Google Scholar]
- 38.Bjerke T, Gaustadnes M, Nielsen S, Nielsen LP, Schiotz PO, Rudiger N, et al. Human blood eosinophils produce and secrete interleukin 4. Respir Med. 1996;90(5):271–277. doi: 10.1016/s0954-6111(96)90098-0. [DOI] [PubMed] [Google Scholar]
- 39.Furuta GT, Kagalwalla AF, Lee JJ, Alumkal P, Maybruck BT, Fillon S, et al. The oesophageal string test: a novel, minimally invasive method measures mucosal inflammation in eosinophilic oesophagitis. Gut. 2013;62(10):1395–1405. doi: 10.1136/gutjnl-2012-303171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butterfield JH, Ackerman SJ, Weiler D, Eisenbrey AB, Gleich GJ. Effects of glucocorticoids on eosinophil colony growth. J Allergy Clin Immunol. 1986;78(3 Pt 1):450–457. doi: 10.1016/0091-6749(86)90032-1. [DOI] [PubMed] [Google Scholar]
- 41.Baines KJ, Simpson JL, Wood LG, Scott RJ, Fibbens NL, Powell H, et al. Sputum gene expression signature of 6 biomarkers discriminates asthma inflammatory phenotypes. J Allergy Clin Immunol. 2014;133(4):997–1007. doi: 10.1016/j.jaci.2013.12.1091. [DOI] [PubMed] [Google Scholar]
- 42.Kern E, Hirano I. Emerging drugs for eosinophilic esophagitis. Expert opinion on emerging drugs. 2013;18(3):353–364. doi: 10.1517/14728214.2013.829039. [DOI] [PubMed] [Google Scholar]
- 43.Corren J. Inhibition of interleukin-5 for the treatment of eosinophilic diseases. Discovery medicine. 2012;13(71):305–312. [PubMed] [Google Scholar]
- 44.Abonia JP, Putnam PE. Mepolizumab in eosinophilic disorders. Expert review of clinical immunology. 2011;7(4):411–417. doi: 10.1586/eci.11.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roman K, Done JD, Schaeffer AJ, Murphy SF, Thumbikat P. Tryptase-PAR2 axis in experimental autoimmune prostatitis, a model for chronic pelvic pain syndrome. Pain. 2014;155(7):1328–1338. doi: 10.1016/j.pain.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rothenberg ME, Wen T, Greenberg A, Alpan O, Enav B, Hirano I, et al. Intravenous anti–IL-13 mAb QAX576 for the treatment of eosinophilic esophagitis. J Allergy Clin Immunol. doi: 10.1016/j.jaci.2014.07.049. (0). [DOI] [PubMed] [Google Scholar]
- 47.Aceves SS, Chen D, Newbury RO, Dohil R, Bastian JF, Broide DH. Mast cells infiltrate the esophageal smooth muscle in patients with eosinophilic esophagitis, express TGF-beta1, and increase esophageal smooth muscle contraction. The Journal of allergy and clinical immunology. 2010;126(6):1198–1204. doi: 10.1016/j.jaci.2010.08.050. [DOI] [PubMed] [Google Scholar]
- 48.Schlag C, Pfefferkorn S, Brockow K, Haller B, Slotta-Huspenia J, Schulz S, et al. Serum Eosinophil Cationic Protein is Superior to Mast Cell Tryptase as Marker for Response to Topical Corticosteroid Therapy in Eosinophilic Esophagitis. Journal of clinical gastroenterology. 2014;48(7):600–606. doi: 10.1097/01.mcg.0000436439.67768.8d. [DOI] [PubMed] [Google Scholar]
- 49.Zhang S, Wu X, Yu S. Prostaglandin D2 receptor D-type prostanoid receptor 2 mediates eosinophil trafficking into the esophagus. Dis Esophagus. 2014;27(6):601–606. doi: 10.1111/dote.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niranjan R, Mavi P, Rayapudi M, Dynda S, Mishra A. Pathogenic role of mast cells in experimental eosinophilic esophagitis. American journal of physiology Gastrointestinal and liver physiology. 2013;304(12):G1087–G1094. doi: 10.1152/ajpgi.00070.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arias A, Lucendo AJ, Martinez-Fernandez P, Gonzalez-Castro AM, Fortea M, Gonzalez-Cervera J, et al. Dietary Treatment Modulates Mast Cell Phenotype, Density, and Activity in Adult Eosinophilic Esophagitis. Clin Exp Allergy. 2015 doi: 10.1111/cea.12504. [DOI] [PubMed] [Google Scholar]
- 52.DeBrosse CW, Collins MH, Buckmeier Butz BK, Allen CL, King EC, Assa'ad AH, et al. Identification, epidemiology, and chronicity of pediatric esophageal eosinophilia, 1982–1999. J Allergy Clin Immunol. 2010;126(1):112–119. doi: 10.1016/j.jaci.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeBrosse CW, Franciosi JP, King EC, Butz BK, Greenberg AB, Collins MH, et al. Long-term outcomes in pediatric-onset esophageal eosinophilia. J Allergy Clin Immunol. 2011;128(1):132–138. doi: 10.1016/j.jaci.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.