Abstract
Background
The metabolic effect of intratumor cholesteryl ester (CE) in breast cancer remains poorly understood. The objective was to analyze the relationship between intratumor CE content and clinicopathological variables in human breast carcinomas.
Methods
We classified 30 breast carcinoma samples into three subgroups: 10 luminal-A tumors (ER+/PR+/Her2-), 10 Her-2 tumors (ER-/PR-/Her2+), and 10 triple negative (TN) tumors (ER-/PR-/Her2-). We analyzed intratumor neutral CE, free cholesterol (FC) and triglyceride (TG) content by thin layer chromatography after lipid extraction. RNA and protein levels of lipid metabolism and invasion mediators were analyzed by real time PCR and Western blot analysis.
Results
Group-wise comparisons, linear regression and logistic regression models showed a close association between CE-rich tumors and higher histologic grade, Ki-67 and tumor necrosis. CE-rich tumors displayed higher mRNA and protein levels of low-density lipoprotein receptor (LDLR) and scavenger receptor class B member 1 (SCARB1). An increased expression of acetyl-Coenzyme A acetyltransferase 1 (ACAT1) in CE-rich tumors was also reported.
Conclusions
Intratumor CE accumulation is intimately linked to proliferation and aggressive potential of breast cancer tumors. Our data support the link between intratumor CE content and poor clinical outcome and open the door to new antitumor interventions.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-015-1469-5) contains supplementary material, which is available to authorized users.
Keywords: ACAT1, Breast cancer, CD36, Cholesteryl ester, LDL receptor, SCARB1
Background
The homeostasis of lipids is deregulated in several tumors [1, 2] including breast cancer [3, 4]. The risk of breast cancer increases in association with high fat diets, overweight, obesity, and type 2 diabetes, conditions characterized by an anomalous lipid profile [5–7]. Plasma levels of total cholesterol, low-density lipoprotein (LDL) and triglycerides (TG) are frequently higher, and high density lipoproteins (HDL) lower, in patients with breast cancer [8, 9]. The expression of lipid metabolism genes is also changed in breast tumors compared to the normal adjacent tissue [10]. Dyslipidemia favors mammary tumor growth and metastasis in in vivo models [11], and LDL and HDL stimulate proliferation and migration in in vitro breast tumor cell models [12–15]. Taken together, these results show a close relationship between the deregulation of lipid homeostasis and breast cancer.
Plasma lipoproteins are a source of fatty acids (FA) and cholesteryl esters (CE) for tumor cells. FA oxidation is the main source of energy for prostate and pancreatic tumors [16, 17]. Availability of intratumoral CE reduces de novo lipid synthesis, favors membrane biogenesis, induces lipid raft formation and alters tumor cell signaling, essential processes for tumor proliferation, invasiveness and survival [18–20]. In concordance with these data, the inhibition of CE synthesis has anticancer effects [21, 22].
Breast tumor subtypes represent different molecular entities that show great heterogeneity in their tumorigenesis, aggressiveness and malignancy and a significant disparity in clinical outcomes and management [23–27]. Furthermore, most investigations about deregulated lipid metabolism in breast neoplasms have been performed in vitro and animal models rather than in human samples. This makes it difficult to extrapolate results for clinical practice. Complementary knowledge based on translational approaches is thus required to improve diagnosis and treatment of breast cancer. We hypothesized that intratumor CE levels are associated with clinicopathological variables in human breast carcinomas. The objective of this study was thus to analyze the relationship between intratumor CE content, lipid metabolism mediators, invasion markers, and clinicopathological parameters.
Material and Methods
Patient and tumor samples
The clinical-pathological features of patients and tumor samples are summarized in Table 1. Thirty tumor samples were selected retrospectively (from May 2006 to November 2012) from the Tumor Tissue Bank in the Pathology Department at Hospital Santa Creu i Sant Pau, Barcelona, Spain. Tumor samples were classified in three subgroups: 10 cases were Luminal-A tumors (ER+/PR+/Her2-), 10 Her-2 tumors (ER-/PR-/Her2+) and 10 TN tumors (ER-/PR-/Her2-). For this comparative clinicopathological and molecular study, we selected a similar number of each subtype of breast carcinomas. Exclusion criteria were patients with mutations in BRCA1 and BRCA2 genes. Patients were staged according to the TNM staging system and tumors were processed and studied according to the standard protocols [28, 29]. The Ki-67 index was also evaluated by immunohistochemistry. Patients’ medical history and clinical evaluation were fully reviewed. The presence of dyslipidemia and menopause was based on explicit diagnosis in the medical history, assuming that, to diagnose the condition, clinicians have followed the criteria admitted to each time. Patients were treated according to our institution guidelines, essentially first treatment was surgery for patients with stages I and II, followed by adjuvant chemotherapy, endocrine therapy and radiotherapy when indicated.
Table 1.
Clinical and pathological characteristics of patients and tumor samples
| Variables | N = 30 |
|---|---|
| Age (years) | 62.5 (48.8-76.0) |
| Clinicopathological parameters | |
| Breast carcinoma type | |
| Luminal-A (%) | 10 (33) |
| Her-2 (%) | 10 (33) |
| TN (%) | 10 (33) |
| Histologic types | |
| Invasive ductal carcinoma (%) | 26 (87) |
| Medullary carcinoma (%) | 2 (7) |
| Papillary carcinoma (%) | 1 (3) |
| Mixed carcinoma (%) | 1 (3) |
| Nottingham combined histologic grade | |
| Grade I (%) | 6 (20) |
| Grade II (%) | 4 (13) |
| Grade III (%) | 20 (67) |
| TNM staging | |
| Stage IA (%) | 7 (23) |
| Stage IIA (%) | 14 (47) |
| Stage IIB (%) | 4 (13) |
| Stage IIIA (%) | 2 (7) |
| Stage IIIB (%) | 1 (3) |
| Stage IV (%) | 1 (3) |
| Unknown (%) | 1 (3) |
| Tumor size (cm) | 3.00 (2.00-4.00) |
| Lymph node affected (%) | 10 (33) |
| Vascular invasion (%) | 4 (13) |
| Tumor necrosis (%) | 12 (40) |
| Mean Ki-67 (%) | 20 (5–53) |
| Ki-67 over 20 (%) | 15 (50) |
| Other conditions | |
| Dyslipidemia (%) | 11 (38) |
| Menopause (%) | 15 (50) |
| Intratumor lipid parameters | |
| CE (μg/mg protein) | 3.14 (1.45-4.65) |
| TG (μg/mg protein) | 5.91 (2.87-13.95) |
| Free Chl (μg/mg protein) | 4.97 (4.47-5.50) |
Data are presented as medians (interquartile ranges) for continuous variables and as frequencies (percentages) for categorical variables
CE: Cholesteryl Esters; Free Chl: Free Cholesterol; Her-2: Human Epidermal Growth Factor Receptor 2; TG: Triglycerides; TN: Triple Negative
This study was conducted according to the Declaration of Helsinki principles, with approval from the Clinical Research Ethics Committee at Institut d’Investigacions Biomèdiques Sant Pau. Written informed consent was obtained from all patients
Tumor sample collection
Tumor tissue samples were obtained from surgical specimens, mastectomy or lumpectomy, rapidly embedded in OCT (Tissue-Tek, Sakura, Europe, Alphen den Rijn, The Netherlands) and frozen using a histobath (Thermo Shandon, Pittsburgh, PA, USA). A macrodisecction of the samples was performed to remove fatty tissue and exclusively collect tumoral tissue. Immunohistochemistry was carried out using the EnVision Flex™ detection system (Dako/Agilent Technologies; Carpinteria, CA), following the manufacturer´s instructions.
RNA extraction and cDNA synthesis
Total RNA was extracted from frozen tumoral tissue using TriPure isolation reagent (Roche Molecular Biochemicals) and the RNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Extracted RNA was eluted in 25 μL of nucleases-free water. RNA yield and quality were assessed by agarose electrophoresis and spectrophotometry, and then stored at −80 °C until use. RNA was digested with DNase I (Invitrogen). One μg of total RNA was used for cDNA synthesis according to the protocol provided with the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). Recombinant RNasin Ribonuclease Inhibitor (Applied Biosystems) was added to prevent RNase-mediated degradation. The cDNA was also stored at –20C.
Gene expression analyses by RT-PCR
The expression of different genes involved in lipid metabolism and tumor invasion was analyzed at mRNA level by quantitative real-time reverse transcriptase-polymerase chain reaction (q-RT-PCR). Specific primer and fluorescent TaqMan probes were selected within a list of predesigned assays (Table 2). 18srRNA (4319413E) was used as a housekeeping gene. We mixed 5 μl of single-stranded cDNA (equivalent to 100 ng of total RNA) with 1 μl of 20x TaqMan Gene Expression Assays for each Assay-on-Demand, 10 μl of TaqMan Universal PCR Master Mix, and 4 μl of nucleases-free water. After gentle mixing, the mixture was transferred into a real-time PCR microplate. The Real-time PCR microplate was sealed, centrifuged, and placed in the sample block of an Applied Biosystems 7300 Real Time PCR System (Applied Biosystems). The thermal cycling conditions were 2 min at 50 °C and 10 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. Expression levels were measured in triplicate. The threshold cycle (Ct) values were normalized to the housekeeping gene.
Table 2.
Mediators of tumor lipid homeostasis and invasion
| Marker | Abbreviation | Probe ID |
|---|---|---|
| Tumor Lipid Homeostasis | ||
| Lipid internalization | ||
| Low density lipoprotein receptor-related protein 1 | LRP1 | Hs00233899_m1 |
| Cluster of Differentiation 36 | CD36 | |
| Very low-density lipoprotein receptor | VLDLR | Hs01045922_m1 |
| Low-density lipoprotein receptor | LDLR | Hs00181192_m1 |
| Scavenger receptor class B member 1 | Gene:SCARB1 | |
| Protein: SR-BI | ||
| Lipid efflux | ||
| ATP-binding cassette transporter | ABCA1 | Hs01059118_m1 |
| Cholesterol synthesis | ||
| 3-hydroxy-3-methyl-glutaryl-CoA reductase | HMG-CoAR | Hs00168352_m1 |
| Fatty acid synthesis | ||
| Fatty acid synthase | FASN | Hs01005622_m1 |
| Cholesterol esterification | ||
| Acetyl-Coenzyme A acetyltransferase 1 | ACAT1 | Hs00608002_m1 |
| Cholesterol management | ||
| Caveolin-1 | CAV1 | Hs00971716_m1 |
| Liver X receptor α | LXR-α | Hs00172885_m1 |
| Sterol regulatory element-binding transcription factor 1 | SREBP1 | Hs00231674_m1 |
| Sterol regulatory element-binding transcription factor 2 | SREBP2 | Hs00190237_m1 |
| Tumor Invasion | ||
| Matrix metalloproteinase-2 | MMP-2 | Hs00234422_m1 |
| Matrix metalloproteinase-9 | MMP-9 | Hs00234579_m1 |
| Tissue inhibitor of metalloproteinase 1 | TIMP1 | Hs01092511_m1 |
| Cathepsin S | CTSS | Hs00175407_m1 |
Western blotting
Total protein was extracted from tumoral frozen tissue using TriPure isolation reagent (Roche Molecular Biochemicals). Proteins were analyzed by Western blot analysis under non-reducing (LRP1 and VLDLR) and reduced (LDLR, SR-BI, CTSS and MMP9) conditions on polyacrylamide gels for SDS-PAGE. The samples were electrotransferred to nitrocellulose, and the membranes were saturated at room temperature for 1 hour in TTBS (20 mM Tris–HCl pH 7.5, 500 mM NaCl, 0.01 % Tween 20, and 5 % non-fat milk). Western blot analysis was performed with specific monoclonal antibodies against LRP1 (10R-L107c, Fitzgerald), VLDLR (sc-18824, Santa Cruz Biotechnology), LDLR (ab52818, Abcam), SR-BI (sc-67098, Santa Cruz Biotechnology), CTSS (sc-271619, Santa Cruz Biotechnology), and MMP9 (AB805, Chemicon International). Equal protein loading in each lane was verified by staining filters with Pounceau. A reference sample was applied in all blots and the value of the reference sample was used to normalize the other samples of the same blot.
Lipid extraction and semi-quantitative analysis of cholesteryl ester, free cholesterol and triglyceride content of tumor samples
An aliquot of the frozen tumoral tissue was homogenized in NaOH 0.1 M, and CE, TG and free cholesterol (FC) were partitioned by thin layer chromatography after lipid extraction as previously described [30, 31]. The different concentrations of standards (a mixture of cholesterol, cholesterol palmitate and triglycerides) were applied to each plate. The chromatographic developing solution was heptane/diethylether/acetic acid (74:21:4, vol/vol/vol). The spots corresponding to CE, TG and FC were quantified by densitometry against the standard curve of cholesterol palmitate, triglycerides and cholesterol, respectively, using a computing densitometer.
Statistical analysis
Statistical analysis was performed using SPSS 15.0 statistical software (SPSS Inc, Chicago, IL). Descriptive statistics were used to characterize the study population and to describe the clinicopathological variables. Data are presented as frequencies (percentages) for categorical variables, and medians (interquartile ranges) for continuous variables. Categorical variables were compared between groups using Pearson´s Chi square test or Fisher’s exact test as required. Continuous variables were compared between groups with the Krusall-Wallis test or non-parametric Mann–Whitney U test for medians, as required. In comparisons where differences between groups were detected with the Krusall-Wallis test, the Mann–Whitney U test for medians was used with Bonferroni correction to test for differences in pairs (0.05/3 = 0.017). Correlations were performed with Spearman's rank test. Linear regression analysis was performed to explore the association between Ki-67 levels and intratumor CE content. Several models were developed: unadjusted (model 1) and adjusted by age (model 2). Logistic regressions were performed to examine the associations between categorical clinicopathological parameters and intratumor CE content. Model 1 was unadjusted. To establish whether the observed association could be influenced by possible confounders, model 2 was adjusted by age and neoadjuvant therapy. Differences were considered statistically significant when p < 0.05.
RESULTS
Clinicopathological findings according to receptor status
Table 3 summarizes clinicopathological characteristics according to receptor status. Her-2 and TN tumors were the most prevalent in grade III and Ki-67 over 20 %. Tumor necrosis was more prevalent in Her-2 and TN than in Luminal-A tumors but did not reach statistical significance (p = 0.054). There were no differences in TNM stage, tumor size, number of lymph nodes affected and vascular invasion markers between groups. Concerning other conditions, dyslipidemia was more prevalent in patients with TN carcinomas than in those with Luminal-A or Her-2 tumors.
Table 3.
Clinical and pathological characteristics according to breast carcinoma type
| Variables | Luminal-A | Her-2 | TN | |
|---|---|---|---|---|
| N = 10 | N = 10 | N = 10 | p-value | |
| Age (years) | 66.5 (46.8-85.3) | 54.0 (49.8-63.0) | 68.5 (50.8-76.0) | 0.477 |
| Clinicopathological parameters | ||||
| Nottingham combined histologic grade | <0.001* | |||
| Grade I (%) | 5 (50) | 0 (0) | 1 (10) | |
| Grade II (%) | 4 (40) | 0 (0) | 0 (0) | |
| Grade III (%) | 1 (10) | 10 (100) | 9 (90) | |
| TNM Stage | 0.779 | |||
| Stage I-II (%) | 9 (90) | 8 (80) | 8 (80) | |
| Stage III-IV (%) | 1 (10) | 1 (10) | 2 (20) | |
| Unknown (%) | 0 (10) | 1 (10) | 0 (10) | |
| Tumor size (cm) | 2.25 (1.88-3.62) | 2.75 (1.73-4.12) | 3.25 (2.57-4.25) | 0.349 |
| Lymph node affected (%) | 4 (40) | 3 (30) | 3 (30) | 0.861 |
| Vascular invasion (%) | 1 (10) | 2 (20) | 1 (10) | 0.749 |
| Tumor necrosis (%) | 1 (10) | 6 (60) | 5 (50) | 0.054 |
| Ki-67 (%) | 5.00 (4.50-7.75) | 30.00 (12.50-42.50)a | 65.00 (21.25-82.50) b | 0.001* |
| Ki-67 over 20 (%) | 0 (0) | 7 (70) | 8 (80) | 0.001* |
| Other conditions | ||||
| Dyslipidemia (%) | 2 (20) | 1 (10) | 8 (80) | 0.003* |
| Menopause (%) | 5 (50) | 6 (60) | 4 (40) | 0.537 |
Data are presented as medians (interquartile ranges) for continuous variables and as frequencies (percentages) for categorical variables
p-values by Krusall-Wallis test, Pearson´s Chi square test or Fisher’s exact test when corresponding
*Statistically significant
For continuous variables U Mann–Whitney test was performed when Krusall-Wallis test was significant: aLuminal-Avs.Her-2; bLuminal-Avs. TN; c Her-2vs. TN
CE: Cholesteryl Esters; Free Chl: Free Cholesterol; Her-2: Human Epidermal Growth Factor Receptor 2; TG: Triglycerides; TN: Triple Negative
Thin layer chromatography after lipid extraction showed that intratumoral CE accumulation was significantly higher in Her-2 and TN tumors than in Luminal-A tumors: Her-2: 3.55 (2.78-7.85) & TN: 5.11 (1.69-6.79 vs. Luminal-A: 1.38 (1.09-3.24) (Fig. 1a and b). In contrast, there were no differences in intratumor TG (Fig. 1a and c) or FC (Fig. 1a and d) content between groups. Immunhistochemical studies showed the presence of cytoplasmic vacuoles (asterisks) in Her-2 and TN breast tumors cells (Fig. 1e).
Fig. 1.
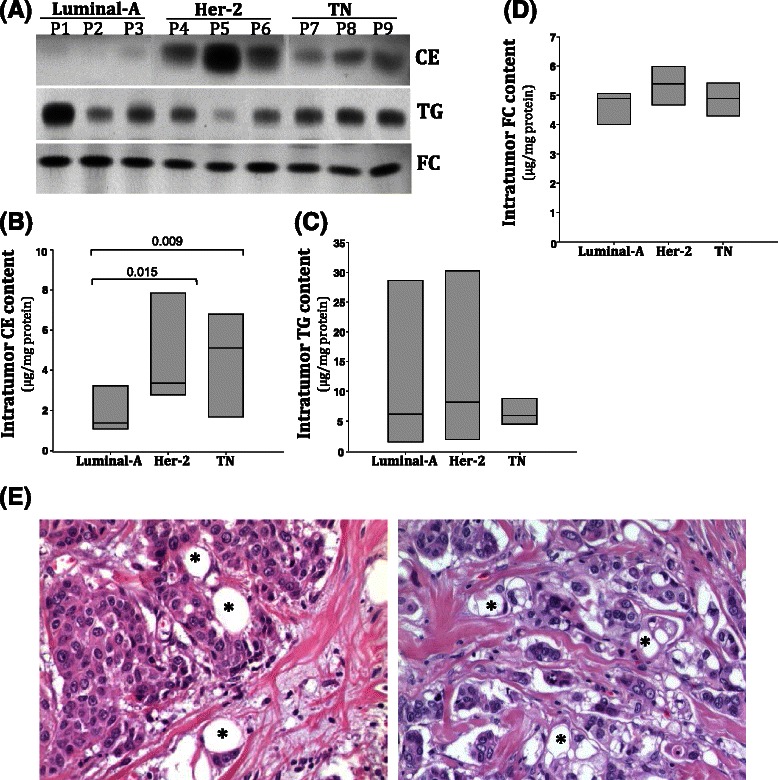
Intratumor cholesteryl ester (CE), triglyceride (TG) and free cholesterol content (FC) in Luminal-A, Her-2 and TN breast tumors. (a) Representative thin layer chromatography showing CE, TG and FC bands in three luminal-A, Her-2 and TN representative tumors. Bar graphs show medians (interquartile ranges) of intratumor CE (b), TG (c) and FC (d) content in luminal-A (n = 10), Her-2 (n = 10) and TN (n = 10) groups. (e), Representative images showing a high content of intratumor vacuoles (asterisk) in Her-2 and TN breast tumors
Molecular analysis according to receptor status
We analyzed the expression of: a) genes involved in lipid metabolism including lipid uptake, cholesterol synthesis, fatty acid synthesis, cholesterol esterification, cholesterol management and cholesterol efflux; and b) genes involved in tumor invasiveness (Table 2). Real time PCR results showed that LDLR and ACAT1 expression was significantly higher in Her-2 than in luminal-A tumors (Additional file 1: Table S1). No differences between tumor subtypes were observed in other mediators of lipid uptake, cholesterol synthesis, fatty acid synthesis, CE synthesis, cholesterol management, or cholesterol efflux. No differences were reported in markers of invasion.
Clinicopathological findings according to intratumor CE content
To explore the associations between intratumor CE levels and tumor malignancy, the population was divided into statistical tertiles of intratumor CE content: tertile 1: 0.16-2.59 (μg/mg protein), tertile 2: 2.60-5.16 (μg/mg protein), and tertile 3: 5.17-9.92 (μg/mg protein). Tumor samples in tertile 3 were considered the CE-rich group. The remaining tissue samples were included in the control CE group. Table 4 summarizes clinicopathological characteristics according to intratumor CE levels. Regarding receptor status, the highest CE tertile was exclusively composed of Her-2 (40 %) and TN (60 %) carcinomas. Luminal-A tumors were all included in the group with lower CE content. According to histological grade, all tumors in the highest tertile of CE content were grade III. Tumor necrosis and Ki-67 were higher in the CE-rich group than in the control group (80 % vs 20 % and 90 % vs 30 %). Ki-67 levels were higher in tumors of tertile 3 than in tumors of tertiles 1 and 2. There were no statistically significant differences between groups in TNM stage, tumor size, number of lymph nodes affected or vascular invasion markers. There were no differences according dyslipidemia or menopause status between groups. There were no differences in TG or free cholesterol levels between the CE-rich group and other groups.
Table 4.
Clinical and pathological characteristics according to intratumor cholesteryl esters content tertile
| Variables | Control N = 20 |
CE-rich N = 10 |
p-value |
|---|---|---|---|
| Age (years) | 65.5 (49.8-78.3) | 53 (45.5-73.0) | 0.267 |
| Clinicopathological parameters | |||
| Breast carcinoma type | 0.015* | ||
| Luminal-A (%) | 10 (50) | 0 (0) | |
| Her-2 (%) | 6 (30) | 4 (40) | |
| TN (%) | 4 (20) | 6 (60) | |
| Nottingham combined histologic grade | 0.024* | ||
| Grade I (%) | 6 (30) | 0 (0) | |
| Grade II (%) | 4 (20) | 0 (0) | |
| Grade III (%) | 10 (50) | 10 (100) | |
| TNM Stage | 0.636 | ||
| Stage I-II (%) | 17 (85) | 8 (80) | |
| Stage III-IV (%) | 3 (15) | 1 (10) | |
| Unknown (%) | 0 (10) | 1 (10) | |
| Tumor size (units) | 2.40 (1.85-3.87) | 3.25 (2.57-4.00) | 0.231 |
| Lymph node affected (%) | 7 (35) | 3 (30) | 0.560 |
| Vascular invasion (%) | 2 (10) | 2 (20) | 0.407 |
| Tumor necrosis (%) | 4 (20) | 8 (80) | 0.003* |
| Ki-67 (%) | 8.50 (5.00-28.75) | 65.00 (37.50-82.50) | <0.001* |
| Ki-67 over 20 (%) | 6 (30) | 9 (90) | 0.003* |
| Other conditions | |||
| Dyslipidemia (%) | 7 (35) | 4 (40) | 0.466 |
| Menopause(%) | 11 (55) | 4 (40) | 0.603 |
| Intratumor lipid parameters | |||
| CE (μg/mg protein) | 2.59 (1.29-3.30) | 7.30 (5.94-8.67) | <0.001* |
| TG (μg/mg protein) | 4.31 (2.07-18.70) | 7.36-14.38) | 0.502 |
| Free Chl (μg/mg protein) | 4.89 (4.24-5.25) | 5.18 (4.71-6.29) | 0.155 |
Data are presented as medians (interquartile ranges) for continuous variables and as frequencies (percentages) for categorical variables
p-values by Mann–Whitney U test or by Fisher’s exact test. *Statistically significant
CE: Cholesteryl Esters; Free Chl: Free Cholesterol; Her-2: Human Epidermal Growth Factor Receptor 2; TG: Triglycerides; TN: Triple Negative
To further explore the relationships between the intratumoral CE accumulation and clinicopathological parameters, we analyzed the association between intratumoral CE accumulation and the clinicopathological categorical variables: histologic grade (III vs. I-II), TNM stage (III-IV vs. I-II), tumor size > 3 cm, lymph node affected, vascular invasion, tumor necrosis and Ki-67 > 20 %. Logistic regression models showed a direct association between intratumoral CE accumulation and higher histologic grade, tumor necrosis and Ki-67 > 20 % (Tables 5 and 6). To analyze the influence of age on the associations between the intratumor CE content and clinicopathological parameters, logistic regressions were adjusted for these covariables and the association between CE levels and histologic grade, tumor necrosis and Ki-67 > 20 %, remained statistically significant (Table 5). No associations between intratumor CE content and TNM Stage, tumor size, lymph node affected or vascular invasion were reported.
Table 5.
Association between intratumor cholesteryl ester content and Ki-67
| B (95 % CI) | β | p value | |
|---|---|---|---|
| Model 1 | |||
| CE | 7.323 (3.981-10-666) | 0.662 | <0.001* |
| Model 2 | |||
| CE + Age | 7.353 (3.731-10.975) | 0.665 | <0.001* |
B: Beta; CI: Confidence Interval; β: Standardized betas
Model 1: Unadjusted; Model 2: Adjusted by age
*Statically significant
CE: Cholesteryl Esters
Table 6.
Association between intratumor cholesteryl esters content and clinicopathological parameters
| Model 1 | Model 2 | |||
|---|---|---|---|---|
| OR (95 % CI) | p-value | OR (95 % CI) | p-value | |
| Histologic grade (III vs. I-II) | 2.115 (1.134-3.945) | 0.018* | 2.084 (1.092-3.978) | 0.037* |
| TNM Stage (III-IV vs. I-II) | 0.693 (0.332-1.447) | 0.329 | 0.675 (0.282-1.617) | 0.378 |
| Tumor size >3 cm (%) | 1.056 (0.794-1.404) | 0.709 | 0.999 (0.716-1.394) | 0.997 |
| Lymph node affected | 0.811 (0.569-1.158) | 0.249 | 0.808 (0.561-1.164) | 0.808 |
| Vascular invasion | 1.112 (0.723-1.711) | 0.629 | 1.098 (0.715-1.685) | 0.670 |
| Tumor necrosis | 1.718 (1.123-2.630) | 0.013* | 1.906 (1.118-3.249) | 0.018* |
| Ki-67 over 20 % | 1.781 (1.120-2.831) | 0.015* | 1.746 (1.078-2.829) | 0.024* |
OR: Odds Ratio. CI: Confidence Interval
Model 1: Unadjusted; Model 2: Adjusted by age and neoadjuvant therapy
*Statically significant
Molecular analysis according to intratumor CE content
CE-rich tumors presented higher LDLR (Fig. 2a) and SCARB1 (Fig. 2b) mRNA expression than controls. In contrast, CD36 mRNA expression was downregulated in CE-rich tumors compared to controls (Fig. 2c). A statistically significant correlation between LDLR expression levels and intratumor CE content was observed (Fig. 2d). The correlation between intratumor CE content and LDLR (positive) (Fig. 2e) or CD36 (negative) (Fig. 2f) did not reach statistical significance. In line with real time PCR analysis, western blot analysis showed that protein levels or LDLR (Fig. 3a and b) and SCARB1 (Fig. 3a and c) were significantly higher in CE-rich breast tumors than in controls. Real time PCR experiments also showed higher ACAT1 mRNA expression levels in CE-rich tumors than in the control group (Fig. 4a). ACAT mRNA expression was positively correlated with LDLR (Fig. 4b) and SCARB1 (Fig. 4c) mRNA expression. There were no differences in the expression of lipid mediators implicated in cholesterol synthesis (HMG-CoAR), cholesterol management (CAV1, LXR-α, SREBP1 and SREBP2) and cholesterol efflux (ABCA1) between groups. No differences were observed in invasiveness markers (MMP2, MMP9, TIMP and CTSS) between groups (Additional file 1: Table S2).
Fig. 2.
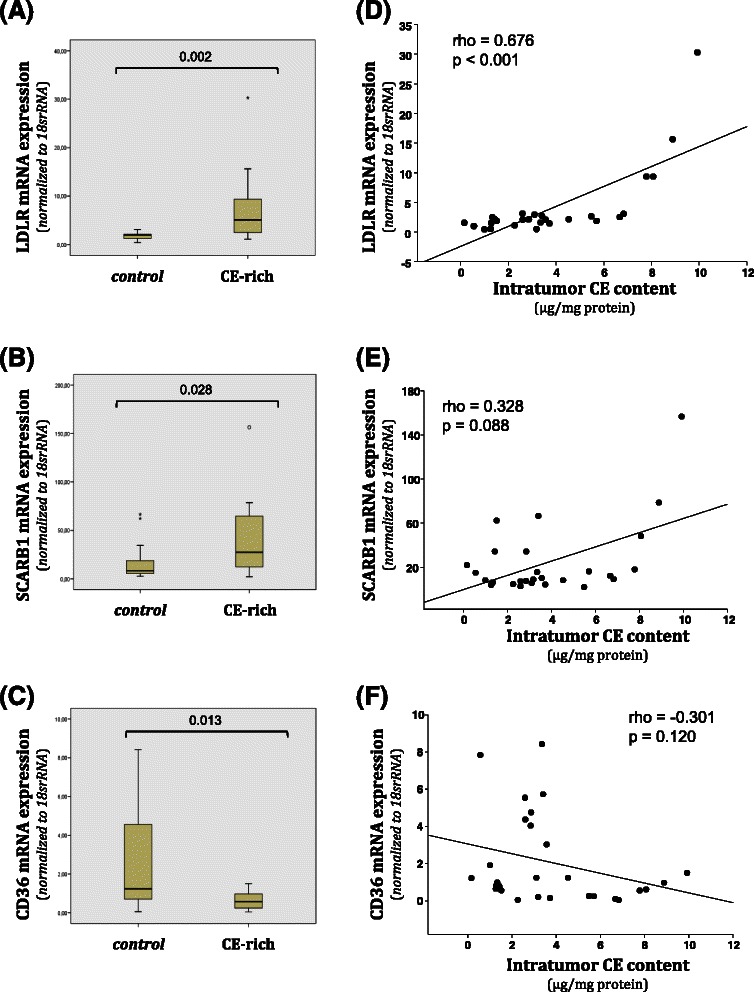
Analysis of LDLR, SCARB1 and CD36 mRNA expression in control and CE-rich groups and correlation with intratumor CE-content. Real-time PCR analysis of LDLR, SCARB1 and CD36 mRNA expression. Data were processed by especially designed software based on the Ct value of each sample and normalized to 18srRNA. Bar graphs showing medians (interquartile ranges) of normalized values of LDLR (a), SCARB1 (b) and CD36 (c) mRNA expression in control (N = 20) and CE-rich (N = 10) groups. Correlations between intratumor CE content and LDLR (d), SCARB1 (e) and CD36 (f) mRNA expression in breast tumors
Fig. 3.
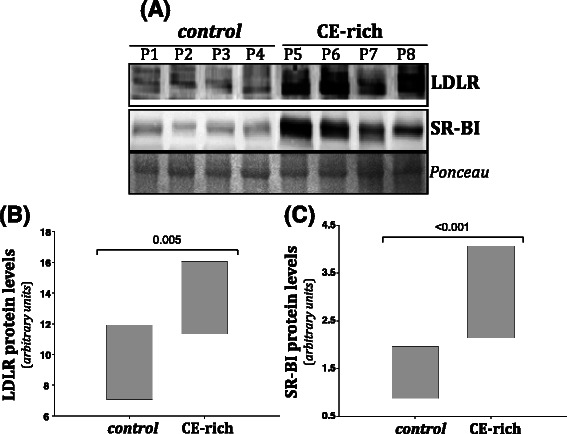
Analysis of LDLR and SR-BI protein levels in control and CE-rich groups. Representative Western blot analysis showing LDLR and SR-BI protein levels in breast tumors (a). Ponceau staining was used as loading control. Bar graphs showing medians (interquartile ranges) of normalized LDLR (b) and SCARB1 (c) protein levels in control (N = 20) and CE-rich (N = 10) groups
Fig. 4.
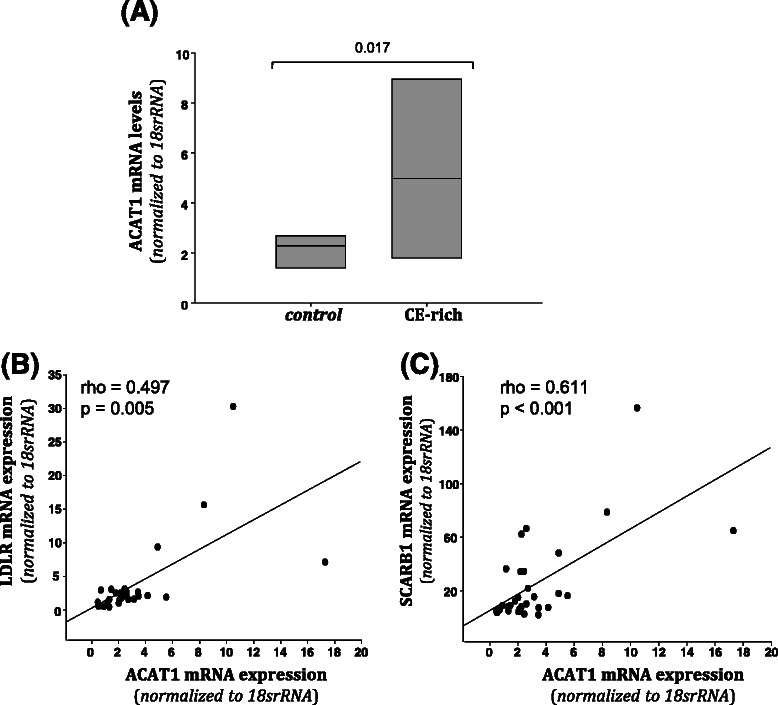
Analysis of ACAT mRNA expression in control and CE-rich groups and its correlation with intratumor CE-content. Real-time PCR analysis of ACAT mRNA expression (a). Data were processed using a specially designed software based on Ct value of each sample and normalized to 18srRNA. Bar graphs showing medians (interquartile ranges) of normalized values of ACAT mRNA expression in control (N = 20) and CE-rich (N = 10) groups. Correlations between ACAT and LDLR mRNA expression (b) and between ACAT and SCARB1 (c) mRNA expression in breast tumors
Discussion
This investigation demonstrated an important association of intratumor cholesteryl ester (CE) accumulation with tumor proliferation but not with invasiveness. We also found that mechanisms potentially involved in intratumoral CE accumulation may be lipid internalization through the overexpression of LDLR and SCARB1 receptors and cholesterol esterification through the upregulation of ACAT1 enzyme.
Group-wise comparisons, linear regression and logistic regression models showed that human breast tumors in the highest tertile of CE content, CE-rich tumors, were intimately associated with higher breast tumor aggressive potential. Our results also showed a close relationship between intratumoral CE accumulation and Ki-67, a well-known marker of proliferation, associated with shorter disease-free survival and overall survival and more frequent tumor recurrence [32]. In addition, we found that intratumoral CE accumulation was intimately associated with higher tumor necrosis, an independent prognostic variable of progression-free and cancer-specific survival [33]. Our data support a close link between breast intratumor CE accumulation and tumor proliferation, indicating that intratumor CE accumulation is associated with poor clinical outcome of patients with breast cancer. These results are in line with previous findings showing an association between intratumor neutral lipids and tumor malignancy [20, 34]. At present we could only speculate about the role of CE in tumor proliferation and aggressive potential; however, it has been proposed that stored CE may reduce the energetically costly lipid synthesis, increase membrane synthesis and rigidity, and induce a pro-tumorigenic signaling, all mechanisms linked to the malignancy of breast cancer [2, 18, 20].
Results for the present study showed a tight correlation between intratumor CE accumulation and LDLR expression and that both LDLR mRNA and protein levels were significantly elevated in CE-rich breast carcinomas. These results suggest that LDLR overexpression facilitates an exacerbated LDL-cholesterol uptake by breast tumor cells. A crucial question is why LDLR, a receptor that is usually downregulated by intracellular cholesterol [35], remains upregulated despite elevated intratumor CE levels. It has been proposed that increased ACAT activity, may contribute to cholesterol removal from endoplasmic reticulum by promoting the cholesterol esterification rate [36]. Cholesterol of the endoplasmic reticulum represses the activation of SREBPs, key transcription factors involved in the positive modulation of LDLR and other lipogenic genes such as HMG-CoA reductase. Our results show that the expression of SREBP-1, SREBP-2, LDLR or HMG-CoA reductase was not significantly reduced in breast CE-rich tumors in which ACAT was upregulated. These results indicate that intracellular cholesterol could not repress SREBP or SREBP-modulated genes because cholesterol is rapidly esterified through ACAT in tumor cells. Consistently, we found a close and direct correlation between ACAT and LDLR overexpression in breast tumors. Therefore, as previously suggested in breast cancer cell lines [36], overexpression of ACAT in human breast cancer maintains low endoplasmic reticulum cholesterol levels, thereby allowing LDL cholesterol to enter the tumor. Our results show that, both LDLR and ACAT overexpression contribute to tumor malignancy in breast cancer patients by promoting intratumoral CE accumulation. Our data are in agreement with previous in vitro studies highlighting the pivotal role of LDLR [36], and ACAT [19, 37] overexpression in massive cholesterol internalization, in particular in ER- breast cell lines. Here, we also show a significant upregulation of another lipoprotein receptor, SCARB1, in human CE-rich breast carcinomas. SCARB1 is a cell-surface glycoprotein that mediates LDL-CE [38], and HDL-CE [39], selective uptake. According to our results, SCARB1 levels, similar to LDLR levels, are significantly elevated in CE-rich breast carcinomas and significantly correlated with ACAT expression. Moreover, SCARB1 has been reportedly upregulated by hypercholesterolemia in a breast cancer mice model [40]. Taken together, these results suggest that SCARB1, besides LDLR, may actively contribute to human breast tumor CE accumulation and aggressive potential. In line with this, it has been reported that SR-BI knockdown inhibits cellular proliferation in in vitro [10], and in vivo breast cancer models [18]. These results suggest that LDLR and SCARB1 overexpression combined with ACAT overexpression are key mechanisms for intratumor CE accumulation in human breast carcinomas.
Differently to LDLR and SCARB1 upregulation, CD36 mRNA expression was significantly downregulated in more aggressive CE-rich breast cancer tumors. This is an interesting finding since the loss of CD36 has been proposed as one of the early events in breast carcinogenesis [41]. CD36 is lower in high density breast tissue than in normal density tissue [42]. Notably, normal adjacent tissue from TN tumors exhibited significantly lower CD36 expression levels than ER+ or Her-2 tumors, suggesting that low expression levels of CD36 may predispose to TN tumors and to poor disease outcomes [42]. Despite the capacity of CD36 to bind native and modified lipoproteins [43], CD36 does not seem to contribute to intratumor CE accumulation in breast cancer. Further studies are required to know why CD36 is downregulated in CE-rich breast cancer carcinomas.
Previous in vitro studies showed that modified lipoproteins induced breast tumor proliferation, migration and invasiveness [12, 13, 44]. Blockade of CE entry and biosynthesis led to decreased cell migration [18, 36]. In contrast, previous results from our group showed that intracellular CE accumulation decreased human vascular cell migration even in the presence of the hypoxic stimulus [45, 46]. In line with the negative effect of intracellular CE content on vascular cell migration, we could not detect a relation of intratumoral CE accumulation with migration and invasion markers (MMP9, MMP2, TIMP and CTSS) in breast tumors, nor with clinicopathological characteristics of invasion (TNM stage, number of lymph nodes affected, and vascular invasion). Further investigation is required to explain why intratumor CE do not show association with invasiveness despite CE-rich tumors have a high aggressive potential,
Lipid profiling classifications have the advantage over molecular classifications which allows better stratification of breast cancer malignancy [47]. Our study shows that intratumor CE accumulation is a good indicator of human breast cancer proliferation. We suggest thus that intratumor CE would be a marker of aggressive potential after mastectomy or lumpectomy and that cholesterol internalization and esterification mediators are potential targets to counteract intratumor CE accumulation (summarized in Fig. 5).
Fig. 5.
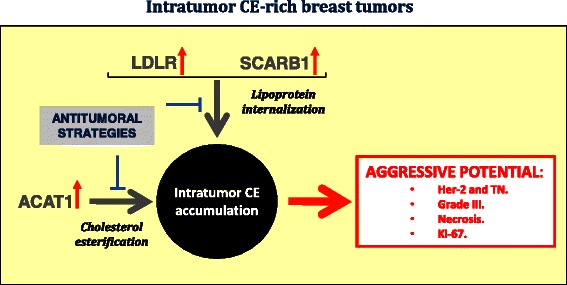
Representative scheme showing potential mechanisms involved in intratumor CE accumulation in breast cancer patients and their relation with breast cancer aggressive potential
A larger study sample would have been desirable and our findings should be reproduced in a significantly larger cohort. A detailed description of cause-effect mechanisms cannot be provided when working with human samples from breast cancer tumors.
Conclusion
In conclusion, our results highlight intratumor CE accumulation as a potential indicator of proliferation in human breast cancer. LDLR and SCARB1 overexpression besides ACAT1 activation may be crucial players causing intratumoral CE accumulation. These results thus open the door to new anticancer therapeutic strategies.
Acknowledgements
The authors declare that they have no conflict of interest. This work was supported by grants from Instituto Salud Carlos III, FIS PI11/00747, Fondo Europeo de Desarrollo Regional (F.E.D.E.R) to VLl-C, a grant from the Instituto de Salud Carlos III (ISCIII, PI10/0307) to DE, a Pfizer award to AB, Red temática de Cáncer (RD12/0036/0042) and Red Temática de Investigación Cooperativa de Cáncer (RD12/0036/0076). This work was supported by the Instituto de Salud Carlos III Grant Sara Borrell to DdG-C (CD14/00109). The authors thank Carolyn Newey (IIB Sant Pau) for editorial assistance.
Additional file
Expression level of genes involved in lipid metabolism.
Footnotes
David de Gonzalo-Calvo and Laura López-Vilaró equally contributed to this work
Competing interest
The authors declare no conflict of interest
Authors´ contributions
AB, EL and VLl-C conceived the study, and participated in its design and coordination. LL-V, LN, MP-O, TV and DE collected samples, conducted the research and analyzed data. DdG-C performed the statistical analysis. DdGC, LB, EL and VLl-C wrote the manuscript. LL-V helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
David de Gonzalo-Calvo, Email: david.degonzalo@gmail.com.
Laura López-Vilaró, Email: llopezv@santpau.cat.
Laura Nasarre, Email: lnasarre@csic-iccc.org.
Maitane Perez-Olabarria, Email: mperez@santpau.cat.
Tania Vázquez, Email: tvazquez@santpau.cat.
Daniel Escuin, Email: descuin@santpau.cat.
Lina Badimon, Email: lbadimon@csic-iccc.org.
Agusti Barnadas, Email: abarnadas@santpau.cat.
Enrique Lerma, Email: elerma@santpau.cat.
Vicenta Llorente-Cortés, Phone: +34 935565888, Email: cllorente@csic-iccc.org.
References
- 1.Santos CR, Schulze A. Lipid metabolism in cancer. Febs J. 2012;279:2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 2.Wu Q, Ishikawa T, Sirianni R, Tang H, McDonald JG, Yuhanna IS, Thompson B, Girard L, Mineo C, Brekken RA, et al. 27-Hydroxycholesterol promotes cell-autonomous, ER-positive breast cancer growth. Cell Rep. 2013;5:637–645. doi: 10.1016/j.celrep.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danilo C, Frank PG. Cholesterol and breast cancer development. Curr Opin Pharmacol. 2012;12:677–682. doi: 10.1016/j.coph.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 4.Nelson ER, Wardell SE, Jasper JS, Park S, Suchindran S, Howe MK, Carver WJ, Pillai RV, Sullivan PM, Sondhi V, et al. 27-Hydroxycholesterol links hypercholesterolemia and breast cancer pathophysiology. Science. 2013;342:1094–1098. doi: 10.1126/science.1241908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bianchini F, Kaaks R, Vainio H. Overweight, obesity, and cancer risk. Lancet Oncol. 2002;3:565–574. doi: 10.1016/S1470-2045(02)00849-5. [DOI] [PubMed] [Google Scholar]
- 6.Crujeiras AB, Diaz-Lagares A, Carreira MC, Amil M, Casanueva FF. Oxidative stress associated to dysfunctional adipose tissue: a potential link between obesity, type 2 diabetes mellitus and breast cancer. Free Radic Res. 2013;47:243–256. doi: 10.3109/10715762.2013.772604. [DOI] [PubMed] [Google Scholar]
- 7.Pelton K, Coticchia CM, Curatolo AS, Schaffner CP, Zurakowski D, Solomon KR, Moses MA. Hypercholesterolemia induces angiogenesis and accelerates growth of breast tumors in vivo. Am J Pathol. 2014;184:2099–2110. doi: 10.1016/j.ajpath.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franky Dhaval S, Shilin Nandubhai S, Pankaj Manubhai S, Patel HR, Prabhudas Shankerbhai P. Significance of alterations in plasma lipid profile levels in breast cancer. Integr Cancer Ther. 2008;7:33–41. doi: 10.1177/1534735407313883. [DOI] [PubMed] [Google Scholar]
- 9.Ray G, Husain SA. Role of lipids, lipoproteins and vitamins in women with breast cancer. Clin Biochem. 2001;34:71–76. doi: 10.1016/S0009-9120(00)00200-9. [DOI] [PubMed] [Google Scholar]
- 10.Cao WM, Murao K, Imachi HYX, Abe H, Yamauchi A, Niimi M, Miyauchi A, Wong NC, Ishida T. A mutant high-density lipoprotein receptor inhibits proliferation of human breast cancer cells. Cancer Res. 2004;64:1515–1521. doi: 10.1158/0008-5472.CAN-03-0675. [DOI] [PubMed] [Google Scholar]
- 11.Alikhani N, Ferguson RD, Novosyadlyy R, Gallagher EJ, Scheinman EJ, Yakar S, LeRoith D. Mammary tumor growth and pulmonary metastasis are enhanced in a hyperlipidemic mouse model. Oncogene. 2013;32:961–967. doi: 10.1038/onc.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan B, Ren H, He Y, Lv X, Ma Y, Li J, Huang L, Yu B, Kong J, Niu C, et al. HDL of patients with type 2 diabetes mellitus elevates the capability of promoting breast cancer metastasis. Clin Cancer Res. 2012;18:1246–1256. doi: 10.1158/1078-0432.CCR-11-0817. [DOI] [PubMed] [Google Scholar]
- 13.Pan B, Ren H, Ma Y, Liu D, Yu B, Ji L, Pan L, Li J, Yang L, Lv X, et al. High-density lipoprotein of patients with type 2 diabetes mellitus elevates the capability of promoting migration and invasion of breast cancer cells. Int J Cancer. 2012;131:70–82. doi: 10.1002/ijc.26341. [DOI] [PubMed] [Google Scholar]
- 14.Rodrigues Dos Santos C, Domingues G, Matias I, Matos J, Fonseca I, de Almeida JM, Dias S. LDL-cholesterol signaling induces breast cancer proliferation and invasion. Lipids Health Dis. 2014;13:16. doi: 10.1186/1476-511X-13-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rotheneder M, Kostner GM. Effects of low- and high-density lipoproteins on the proliferation of human breast cancer cells in vitro: differences between hormone-dependent and hormone-independent cell lines. Int J Cancer. 1989;43:875–879. doi: 10.1002/ijc.2910430523. [DOI] [PubMed] [Google Scholar]
- 16.Khasawneh J, Schulz MD, Walch A, Rozman J, Hrabe de Angelis M, Klingenspor M, Buck A, Schwaiger M, Saur D, Schmid RM, et al. Inflammation and mitochondrial fatty acid beta-oxidation link obesity to early tumor promotion. Proc Natl Acad Sci U S A. 2009;106:3354–3359. doi: 10.1073/pnas.0802864106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y. Fatty acid oxidation is a dominant bioenergetic pathway in prostate cancer. Prostate Cancer Prostatic Dis. 2006;9:230–234. doi: 10.1038/sj.pcan.4500879. [DOI] [PubMed] [Google Scholar]
- 18.Danilo C, Gutierrez-Pajares JL, Mainieri MA, Mercier I, Lisanti MP, Frank PG. Scavenger receptor class B type I regulates cellular cholesterol metabolism and cell signaling associated with breast cancer development. Breast Cancer Res. 2013;15:R87. doi: 10.1186/bcr3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paillasse MR, de Medina P, Amouroux G, Mhamdi L, Poirot M, Silvente-Poirot S. Signaling through cholesterol esterification: a new pathway for the cholecystokinin 2 receptor involved in cell growth and invasion. J Lipid Res. 2009;50:2203–2211. doi: 10.1194/jlr.M800668-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tosi MR, Tugnoli V. Cholesteryl esters in malignancy. Clin Chim Acta. 2005;359:27–45. doi: 10.1016/j.cccn.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 21.de Medina P, Boubekeur N, Balaguer P, Favre G, Silvente-Poirot S, Poirot M. The prototypical inhibitor of cholesterol esterification, Sah 58–035 [3-[decyldimethylsilyl]-n-[2-(4-methylphenyl)-1-phenylethyl]propanamide], is an agonist of estrogen receptors. J Pharmacol Exp Ther. 2006;319:139–149. doi: 10.1124/jpet.106.104349. [DOI] [PubMed] [Google Scholar]
- 22.de Medina P, Genovese S, Paillasse MR, Mazaheri M, Caze-Subra S, Bystricky K, Curini M, Silvente-Poirot S, Epifano F, Poirot M. Auraptene is an inhibitor of cholesterol esterification and a modulator of estrogen receptors. Mol Pharmacol. 2010;78:827–836. doi: 10.1124/mol.110.065250. [DOI] [PubMed] [Google Scholar]
- 23.Berrada N, Delaloge S, Andre F. Treatment of triple-negative metastatic breast cancer: toward individualized targeted treatments or chemosensitization? Ann Oncol. 2010;21:Suppl 7. doi: 10.1093/annonc/mdq279. [DOI] [PubMed] [Google Scholar]
- 24.Guedj M, Marisa L, de Reynies A, Orsetti B, Schiappa R, Bibeau F, MacGrogan G, Lerebours F, Firetti P, et al. A refined molecular taxonomy of breast cancer. Oncogene. 2012;31:1196–1206. doi: 10.1038/onc.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodenhiser DI, Andrews JD, Vandenberg TA, Chambers AF. Gene signatures of breast cancer progression and metastasis. Breast Cancer Res. 2011;13:201. doi: 10.1186/bcr2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sorlie T, Wang Y, Xiao C, Johnsen H, Naume B, Samaha RR, BOrresen-Dale AL. Distinct molecular mechanisms underlying clinically relevant subtypes of breast cancer: gene expression analyses across three different platforms. BMC Genomics. 2006;7:127. doi: 10.1186/1471-2164-7-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Catasus L, Gallardo A, Llorente-Cortes V, Escuin D, Munoz J, Tibau A, Peiro G, Barnadas A, Lerma E. Low-density lipoprotein receptor-related protein 1 is associated with proliferation and invasiveness in Her-2/neu and triple-negative breast carcinomas. Hum Pathol. 2011;42:1581–1588. doi: 10.1016/j.humpath.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 29.Tavassoéli F, Deville P. Pathology and Genetics Tumours of the Breast and Female Genital Organs. 2003. [Google Scholar]
- 30.Llorente-Cortés V, Martínez-González J, Badimon L. Esterified cholesterol accumulation induced by aggregated LDL uptake in human vascular smooth muscle cells is reduced by HMG-CoA reductase inhibitors. Arterioscler Thromb Vasc Biol. 1998;18:738–746. doi: 10.1161/01.ATV.18.5.738. [DOI] [PubMed] [Google Scholar]
- 31.Llorente-Cortés V, Martínez-González J, Badimon L. LDL receptor-related protein mediates uptake of aggregated LDL in human vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2000;20:1572–1579. doi: 10.1161/01.ATV.20.6.1572. [DOI] [PubMed] [Google Scholar]
- 32.de Azambuja E, Cardoso F, de Castro G, Jr CM, Mano MS, Durbecq V, Sotiriou C, Larsimont D, Piccart-Gebhart MJ, Paesmans M. Ki-67 as prognostic marker in early breast cancer: a meta-analysis of published studies involving 12,155 patients. Br J Cancer. 2007;96:1504–1513. doi: 10.1038/sj.bjc.6603756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollheimer MJ, Kornprat P, Lindtner RA, Harbaum L, Schlemmer A, Rehak P, Langner C. Tumor necrosis is a new promising prognostic factor in colorectal cancer. Hum Pathol. 2010;41:1749–1757. doi: 10.1016/j.humpath.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 34.Hilvo M, Denkert C, Lehtinen L, Müller B, Brockmöller S, Seppänen-Laakso T, Budczies J, Bucher E, Yetukuri L, Castillo S, et al. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res. 2011;71:3236–3245. doi: 10.1158/0008-5472.CAN-10-3894. [DOI] [PubMed] [Google Scholar]
- 35.Francke U, Brown MS, Goldstein JL. Assignment of the human gene for the low density lipoprotein receptor to chromosome 19: synteny of a receptor, a ligand, and a genetic disease. Proc Natl Acad Sci U S A. 1984;81:2826–2830. doi: 10.1073/pnas.81.9.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antalis CJ, Uchida A, Buhman KK, Siddiqui RA. Migration of MDA-MB-231 breast cancer cells depends on the availability of exogenous lipids and cholesterol esterification. Clin Exp Metastasis. 2011;28:733–741. doi: 10.1007/s10585-011-9405-9. [DOI] [PubMed] [Google Scholar]
- 37.Antalis CJ, Arnold T, Rasool T, Lee B, Buhman KK, Siddiqui RA. High ACAT1 expression in estrogen receptor negative basal-like breast cancer cells is associated with LDL-induced proliferation. Breast Cancer Res Treat. 2010;122:661–670. doi: 10.1007/s10549-009-0594-8. [DOI] [PubMed] [Google Scholar]
- 38.Swarnakar S, Temel RE, Connelly MA, Azhar S, Williams DL. Scavenger receptor class B, type I, mediates selective uptake of low density lipoprotein cholesteryl ester. J Biol Chem. 1999;274:29733–29739. doi: 10.1074/jbc.274.42.29733. [DOI] [PubMed] [Google Scholar]
- 39.Pussinen PJ, Karten B, Wintersperger A, Reicher H, McLean M, Malle E, Sattler W. The human breast carcinoma cell line HBL-100 acquires exogenous cholesterol from high-density lipoprotein via CLA-1 (CD-36 and LIMPII analogous 1)-mediated selective cholesteryl ester uptake. Biochem J. 2000;349:559–566. doi: 10.1042/bj3490559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Llaverias G, Danilo C, Mercier I, Daumer K, Capozza F, Williams TM, Sotgia F, Lisanti MP, Frank PG. Role of cholesterol in the development and progression of breast cancer. Am J Pathol. 2011;178:402–412. doi: 10.1016/j.ajpath.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seewaldt VL. Cancer: Destiny from density. Nature. 2012;490:490–491. doi: 10.1038/490490a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeFilippis RA, Chang H, Dumont N, Rabban JT, Chen YY, Fontenay GV, Berman HK, Gauthier ML, Zhao J, Hu D, et al. CD36 repression activates a multicellular stromal program shared by high mammographic density and tumor tissues. Cancer Discov. 2012;2:826–839. doi: 10.1158/2159-8290.CD-12-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silverstein RL, Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci Signal. 2009;2:re3. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soto-Guzman A, Navarro-Tito N, Castro-Sanchez L, Martinez-Orozco R, Salazar EP. Oleic acid promotes MMP-9 secretion and invasion in breast cancer cells. Clin Exp Metastasis. 2010;27:505–515. doi: 10.1007/s10585-010-9340-1. [DOI] [PubMed] [Google Scholar]
- 45.Otero-Viñas M, Llorente-Cortés V, Peña E, Padro T, Badimon L. Aggregated low density lipoproteins decrease metalloproteinase-9 expression and activity in human coronary smooth muscle cells. Atherosclerosis. 2007;194:326–333. doi: 10.1016/j.atherosclerosis.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 46.Revuelta-López E, Castellano J, Roura S, Galvez-Monton C, Nasarre L, Benitez S, Bayes-Genis A, Badimon L, Llorente-Cortes V. Hypoxia induces metalloproteinase-9 activation and human vascular smooth muscle cell migration through low-density lipoprotein receptor-related protein 1-mediated pyk2 phosphorylation. Arterioscler Thromb Vasc Biol. 2013;33:2877–2887. doi: 10.1161/ATVBAHA.113.302323. [DOI] [PubMed] [Google Scholar]
- 47.Nieva C, Marro M, Santana-Codina N, Rao S, Petrov D, Sierra A. The lipid phenotype of breast cancer cells characterized by Raman microspectroscopy: towards a stratification of malignancy. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046456. [DOI] [PMC free article] [PubMed] [Google Scholar]


