Abstract
MicroRNAs are 20-24 nt long, single-stranded RNAs that repress gene expression. Dysregulation of miRNA expression is associated with many human diseases. Modulating the level of endogenous miRNA alters gene profiling and can achieve therapeutic benefits. Here, we reviewed currently used methods of altering miRNA activity in vivo. We focus on the delivery of miRNAs and miRNA inhibitors using recombinant adeno-associated virus (rAAV). In general, rAAV-mediated miRNA inhibition or overexpression provides a simple, efficient and informative way to study miRNA function in mammals. This method also provides the opportunity to explore potential miRNA therapeutics for many diseases.
Keywords: AAV, in vivo gene delivery, miRNA inhibition, miRNA overexpression
MicroRNAs (miRNAs) are small 20–24 nt RNAs that repress the expression of mRNAs by binding to the 3’UTR of the targeted mRNA. As a whole, miRNAs are predicted to regulate more than half of all mammalian protein-coding genes1. Based on their location in the genome, the genes that code for miRNA can be categorized into three groups: exonic miRNAs, intronic miRNAs and miRNAs embedded into protein-coding transcripts2.Most miRNAs are transcribed as primary miRNA (pri-miRNAs) by RNA polymerase II3, though some are transcribed by RNA polyIII4. A pri-miRNA contains a 7-methylguanosine cap at its 5′ end and a poly (A) tail at its 3′ end. It is cleaved by an intranuclear ribonuclease III (RNase III) enzyme, referred to as Drosha, to generate a precursor miRNA (pre-miRNA), which is a stem-loop molecule approximately 70 nt in length. Subsequently, Exportin-5 binds to the pre-miRNA and transports it into the cytoplasm. It is here that another RNase III, Dicer, processes the pre-miRNA into a mature miRNA. This miRNA is loaded into an RNA-induced silencing complex (RISC). Upon amalgamation this fully active protein-RNA aggregate is capable of repressing gene expression through the cleavage and/or degradation of mRNAs.
miRNA dysregulation in human diseases and miRNA therapeutics
In 1993, Ambros and his colleagues discovered the first miRNA, Lin-4, in Caenorhabditis elegans. Since this event, thousands of miRNA have been found and submitted to the miRNA database (http://www.mirbase.org). These miRNAs have been isolated from mammals and non-mammals; more than 2500 of which have been isolated from human5. The correlation between miRNA dysregulation and human disease was first reported by Calin et al. This pioneering study revealed that the loci for miR-15 and miR-16 were deleted in the majority of patients suffering from B cell chronic lymphocytic leukemia6. Accumulated data have demonstrated that miRNAs play important roles in almost all classes of human disease; including cancer, cardiovascular disease, diabetes, etc. For example, more than 50% of human miRNA-encoding genes are located in chromosomal locations associated with cancer or fragile sites on a genome-wide base 7. Let-7 is the first miRNA that was found to regulate the oncogene RAS expression by directly targeting its 3’UTR8. Further studies have shown that in non-small-cell lung cancer (NSCLC) mouse models, intratumoral injection of synthetically produced let-7 molecular mimics significantly reduces tumor burden9. In a cohort of 241 patients with hepatocellular carcinoma (HCC), it was shown that tumor tissues have reduced expression of miR-26 compared with noncancerous liver tissue from the same patient. Furthermore, in patients whose tumors have decreased miR-26 expression, lower levels of miR-26 correlate with shorter overall survival10. Subsequently, systemic delivery of miR-26a via adeno-associated virus vector 8 (AAV8)11, a vector known for its high liver tropism, dramatically suppresses the tumor progression in a murine liver cancer model12. In addition to the miRNA studies in cancer, Olson and his colleagues reported that they had found a signature pattern of miRNAs in cardiac hypertrophy and heart failure which initiated a wave of research focused on miRNA function in heart disease13. In a failing heart, miR-21 level is specifically increased in fibroblasts through the suppression of ERK-MAP kinase signaling pathway which triggers fibroblast motility and initiates the process of cardiac scarring. Scarring, or fibrosis, of the heart is an inappropriate physiological response that oftentimes is severely deleterious to the individual. In vivo silencing of miR-21 by antisense oligonucleotide inhibits interstitial fibrosis and corrects cardiac dysfunction in a TAC (Transverse aortic constriction) mouse model14. Genetic knockout (KO) of the cardiac-specific miRNA, miR-208a, can prevent pathological cardiac remodeling. Similarly, the anti-miR-208a oligonucleotide improved cardiac function and survival in a rat hypertension-induced heart failure model15,16. Another study found that mice who received anti-miR-208a oligonucleotide therapy confer resistance to diet-induced obesity and improved insulin responsiveness17. MiRNAs are also associated with metabolic diseases. MiR-375 is highly expressed in pancreatic islets and miR-375 KO mice are hyperglycemic18. MiR-33, an intronic miRNA located in the intron of SREBF-2 gene, cooperates with its SREBF-2 host gene to control cholesterol homeostasis19. Moreover, administration of anti-miR-33 oligonucleotide raises the plasma HDL level and represses the atherosclerosis in a hypercholesterolemia mouse model20. Using a similar approach, inhibition of the miR-33 family in non-human primates also raised plasma HDL and lower VLDL triglyceride levels21. MiR-122 antagomiR could be the first miRNA-target drug to treat human disease. MiR-122 is liver specific and highly expressed, constituting 70% of the total liver miRNA population22. The binding between miR-122 and the conserved 5’ untranslated region of the hepatitis C virus (HCV) genome protects the HCV from nucleolytic degradation and host innate immune response23,24. HCV load was dramatically reduced with the therapeutic use of miR-122 antagomiR to competitively bind endogenous miR-122. The clinical trial using miR-122 antagomiR to treat HCV patients has completed the Phase 2a stage, showing prolonged dose-dependent reductions in HCV RNA levels without evidence of viral resistance 25. All of these researches prove that modulation of miRNA is providing a new route of treatment against human diseases.
Strategies to investigate miRNA function in vivo
Though thousands of miRNAs have been discovered, few miRNA-target interactions have been experimentally validated, especially in vivo. To understand the complete spectrum of miRNA function, we need approaches to modulate miRNA activities in order to perform gain-and-loss of function studies. Genetic disruption of a miRNA gene provides a powerful strategy to study miRNA function, but many miRNAs share the same seed sequence, the 6–8 nt miRNA region that defines the target repertoire of a miRNA. Consequently, one member of a miRNA family may compensate for loss of another. Creating an animal model in which all members of a miRNA family are deleted is time consuming and expensive. In some cases, miRNA genetic ablation in mice are embryologically lethal, such as miR-17-19 cluster KO leads to 100% postnatal death with cardiac and lung defects26 and miR-126 KO results in around 50% embryonic death due to vascular ruptures and subsequent hemorrhage27.
Chemically modified miRNA mimics or anti-miRNA oligonucleotides (AMOs) complementary to mature miRNAs can be used to increase or decrease the activities of miRNAs respectively, in vitro and in vivo. Though effective, these oligonucleotides therapies are typically expensive and/or require proprietary modifications such as 2′-O-methyl, 2′-O-methoxyethyl, or 2′, 4′-methylene (LNA)28-30. Unfortunately, due to their transient lifespan of miRNA mimics and AMOs, treatment requires repeated administration to effectively express or suppress the cognate miRNAs. Current chemistries and formulations are limited in their success by the delivery of oligonucleotides to many tissues or organs, such as pancreas, muscle and brain.
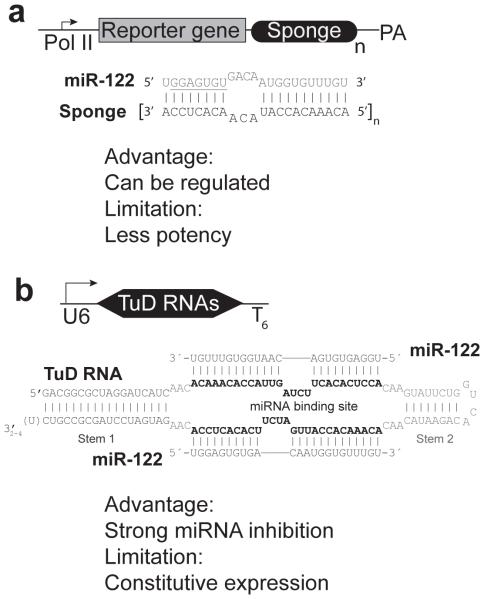
An additional strategy to lower miRNA activities is to use transcribed miRNA inhibitors referred to as miRNA “sponges”31 and “tough decoy RNAs (TuD RNAs)”32. The first alternative to AMOs are “sponges,” which are miRNA inhibitors that can be expressed in cells (Fig. 1a). They oftentimes contain multiple binding sites for a specific miRNA of interest and soak up endogenous miRNA like “sponges”31. To further improve the miRNA inhibition efficacy, Hideo IBA and his colleagues invented a more potent miRNA inhibitor known as TuD RNA (Fig. 1b). They did this by replacing the Pol II promoter in the “sponge” with the more robust Pol III promoter. Additionally, they optimized the secondary structures and the sequence of the miRNA-binding site32. Depletion of miR-223 using a sponge-expressing lentiviral vector to stably modify hematopoietic stem cells ex vivo, followed by bone marrow reconstitution in mice, produced a phenotype similar to that observed in a genetic miRNA knockout mouse33. This work successfully demonstrated that viral-vector delivered miRNA inhibitors can functionally knock down miRNAs in mice. However, the use of lentiviral vector-based miRNA inhibition for functional genomic studies and human therapy is limited due to the risk of insertional mutagenesis and the requirement for ex vivo manipulation. To create an efficient and simple way to study miRNA function in mice, Gao and his colleagues combined rAAV9 vector, which exhibits high tropism for mouse liver and heart11 with miRNA TuDs to inhibit specific miRNAs. One single dose of rAAV9 expressing anti-miR-122 or anti-let-7 TuD depleted the corresponding miRNA and thereby increased the expression of its mRNA targets in the liver and heart of adult mice. This miRNA inhibition lasted through the 25 weeks of monitoring without adverse side effects or any drop in efficacy overtime. High throughput sequencing of liver miRNAs from the treated mice demonstrated that the targeted miRNAs, but no other miRNAs, were depleted 34, indicating the specificity of target miRNA inhibition. Thus rAAV-mediated miRNA modulation is holding great promise on miRNA function study in vivo and potential therapeutics.
Figure 1.
(A) Sponge is tandem repeats of miRNA binding site after reporter gene driven by Pol II promoters. The imperfect paring between microRNA and sponge is diagrammed for miR-122. (B) Tough decoy RNAs (TuDs) contain two single-stranded miRNA binding sites, flanked by double-stranded stems that enhance stability and promote nuclear export. U6 promoter is used for high level expression of the miRNA inhibitors.
RAAV vector: a powerful in vivo gene delivery platform
The first human AAV (i.e. AAV serotype 2) was found as a contaminant of adenovirus preparation. Although 80-90% of the human population is AAV seropositive, infection has not been associated with any human disease. The wild-type AAV (wtAAV) contains a small non-enveloped capsid with a diameter of 26 nm and a 4.7 kb single-stranded (ss) genome. This genome contains two 145-nt inverted terminal repeats (ITRs) at each end and two open reading frames (ORF; Rep and Cap). The ITRs form two T-shaped structures at each end of the genome and contribute to AAV genome replication, packaging, integration and rescue from host genome 35,36. The Rep gene is involved in viral genome replication and encapsidation during AAV replication 37,38in the presence of a helper virus, such as adenovirus39 and herpes simplex virus40. The cap ORF provides structural proteins for the viral capsid which determine the tissue tropism and immune biology in viral infection41.
AAV can transduce dividing and non-dividing cells without causing known pathogenic consequence, which has made it a popular in vivo gene delivery tool. Long-term transgene expression is the major advantage of AAV vectors42. So far there are 12 AAV serotypes and more than 100 variants available to transfer foreign genes into the liver, pancreas, heart, lung, skeletal muscle and CNS efficiently43. The major barrier of translating AAV into human clinical trials is the host immune response which is relatively low however persistent though out preclinical models and clinical trials44, even though the immune responses to AAV vectors in mice is minimal.
A modified AAV, referred to as recombinant AAV (rAAV), which is made by taking the most useful aspects that wtAAV has to offer and removing the rest, has become the standard AAV vector. RAAV is designed using wtAAV as a template, but it is crafted brilliantly so that it is castrated, less risky and more predictable to work with. Though the two ITRs from the wild-type AAV remain in place, the original Rep and Cap genes of wtAAV are completely removed and are replaced with genes of interest45. WtAAV can integrate into the chromosome 19q13 in cultured human cell lines 46,47, but unlike wtAAV, rAAV genomes, due to their incapability to produce rep, do not integrate into this specific chromosomal site in cultured cells or infected mammals. Most rAAV genomes persist as episomes in cells48. Less than 0.1 to 0.5% of the genome randomly integrates into the host genome of mice and humans49-51. Currently rAAVs are used in many human gene therapy clinical trials, such as cystic fibrosis, muscular dystrophy, Parkinson’s, hemophilia B, and Leber’s congenital amaurosis44. The first rAAV-based commercial drug, Glybera, has been approved to treat lipoprotein lipase deficiency in Europe 52. RAAV vectors are regarded as the most efficient, long-lasting and safe somatic gene transfer vehicle in vivo.
RAAV-based in vivo toolkits for miRNA functional genomics and miRNA therapeutics
Though rAAV vectors have many advantages in in vivo gene delivery comparing with other viral vectors, the small size of AAV genome (< 4.7 kb) limits its applications on large gene replacement. Furthermore, to improve the in vivo efficacy of rAAVs transduction, the capability of rAAV genome is shortened even more to create what is known as self-complimentary rAAV vectors (scAAV). These vectors are essentially constructed by mutating one ITR. The mutated ITR is lack of the terminal resolution site for Rep endonuclease nicking during virus genome replication and enable continuing the synthesis of second strand. The failure of Rep nicking the mutated ITR generated two complimentary single stranded AAV genome and folds upon itself upon uncoating, creating a double stranded genome which is transcriptionally active in cells. When the virus genome released from scAAV vector in host cells, it bypasses the conversion of single-stranded genome to double-stranded genome to make the transgene expression more rapidly and efficiently. This optimization greatly enhanced the transduction efficacy of AAV in vivo 53,54. This mutation further reduces the AAV package capability to 2.5 kb. However, the small size of transcribed miRNA inhibitors (sponges < 0.2 kb; TuD RNAs including Pol III promoter< 0.5 kb) is well suited to scAAV delivery.
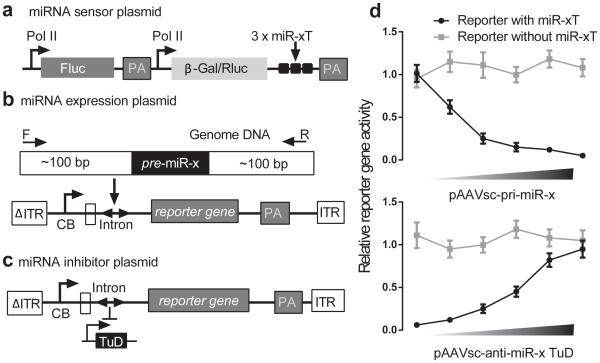
Based on the scAAV vectors, we can generate a library of rAAV vectors expressing miRNA inhibitors as well as companion vectors, over-expressing the corresponding miRNA. This toolkit will enable researchers to test the phenotypic effects of gain-or-loss of function in vivo. Before murine model study, these constructs can be validated in HEK 293 cells. Each miRNA toolkit contains 3 components: miRNA sensor plasmid, miRNA over-expression plasmid and miRNA inhibitor plasmid. The miRNA sensor plasmid is used to monitor the miRNA activity in cells. It contains two reporter genes. One reporter gene carries multiple sites complementary to miRNA, which allows the binding of cognate miRNA to abolish the reporter gene expression. The more miRNA in the target cells, the lower the expression of the reporter gene can be detected. The other reporter gene is served as a transfection control (Fig. 2a).
Figure 2.
Validation of miRNA toolkit in HEK293 cells. (A) miRNA sensor plasmid to monitor the miRNA activity. The strategy for construction of AAV plasmids expressing functional pri-miRNA fragments (B) and miRNA inhibitors, TuD RNAs (C). Cross validation of miRNA expression and inhibition plasmids (D). Top at (D) shows increasing amounts of a pri-miRNA producing vector inhibits expression of a miRNA sensor plasmid in HEK293 cells. The bottom shows the de-repression from anti-miR TuDs in a dose response manner when we fix the amount of pri-miR plasmids. ITR, inverted terminal repeat; ΔITR, mutated ITR; CB, chicken β-actin promoter with CMV enhancer; U6, U6 promoter; PA, poly (A); pre-miR, precursor miRNA; Fluc, Firefly luciferase; Rluc, Renilla luciferase; β-Gal, β-galactosidase; 3 × miR-xT, 3 miRNA perfect target sites.
In the standard practice of production of mature miRNAs, researchers integrate the pri-miRNA fragments into the 3’UTR of transgene or into the intron before the transgene (Fig. 2b).Basically, pri-miRNA fragments are isolated by polymerase chain reaction (PCR) from genomic DNA. The amplicons include the pre-miRNA and ~ 100 bp flanking sequence at both ends (Fig. 2b). We do not recommend embedding pri-miRNA fragments into the 3’UTR because the co-transcribed pri-miRNA will be processed by Drosha, resulting in the loss of reporter gene poly (A) and the consequent loss of its expression (Xie, et al., unpublished data).
In addition, the pri-miRNA fragments with the same design can be harbored by lentiviral vectors to suppress the development of non-small cell lung tumor by expressing the let-7 family 55or used to reduce hyperlipidemia and atherosclerosis by expressing miR-30c56.
We prefer the TuD RNAs in the miRNA inhibitor constructs because of their supremacy in potency among currently available plasmid-based miRNA inhibitors (Fig. 2c)34,57. In brief, miRNA over-expression constructs will be co-transfected with the corresponding miRNA sensor plasmid into HEK293 cells. The reduction of reporter gene will reflect the level of functional miRNA produced by both 293 cells endogenously and the miRNA over-expression construct. In the presence of miRNA inhibitor in the transfected HEK293 cells, the reemergence of reporter gene activity indicates the repression of miRNA activity. This is a result illustrating the repression of reporter gene from construct expressing miRNA (Fig. 2d top) and de-repression of reporter gene from miRNA inhibitor in the presence of ectopic miRNA (Fig. 2d bottom). Theoretically this strategy is able to cross-validate all the miRNA expression and inhibition constructs in HEK293 cells. For in vivo studies, in vitro validated miRNA expression and inhibition constructs can be packaged into different serotypes of AAV vectors by the conventional “ triple-transfection” method58. In brief, validated rAAV plasmid (Fig, 2 b and c), packaging plasmid which contains Rep and Cap genes, and adenovirus helper plasmid were co-transfected into HEK293 cells. After 2-3 days, cells were harvested for vector purification by ultracentrifugation on CsCl or iodixanol gradient and column chromatography59,60. In the packaging process, the vector genome flanked with 2 ITRs will be excised from the rAAV vector plasmid, replicated and packaged into AAV virions. Rep and Cap proteins expressed from the packaging plasmid and the adenovirus helper plasmid provides helper functions essential for rAAV rescue, replication and packaging. The serotype of AAV capsid determines tissue tropism, efficiency of transduction and immune biology of a rAAV vector.
In vivo delivery of ectopic miRNAs or their inhibitors is essential for the study of miRNA functional genomics and therapeutics. As we reviewed above, the advantages of tissue specific tropism, long-term expression and high transduction efficacy make AAV vectors ideal in vivo gene delivery tools. Even the TuD RNA expression driven by Pol III promoter cannot be regulated. By choosing different AAV serotype and by modifying the route of vector administration, manipulation of miRNA levels in target tissues can be achieved (Table 1).
Table 1.
Summary of delivering miRNA or inhibitors to targeted tissues by the combinations of AAV serotypes and route of vector administration
| Route of Vector Administration | |||
|---|---|---|---|
| Target Tissue | AAV Serotype | Neonates | Adults |
| Liver | AAV8 | Not suitable because of hepatocyte division |
Intravenous |
| Pancreas | AAV7, AAV8, or AAV9 |
Not efficient | Intravenous or retrograde bile duct injection |
| Heart | AAV9 | Intravenous or intrapericardial |
Intravenous |
| Lung | AAV5, AAV9, or rh.10 |
Intravenous | Intratracheal or intranasal |
| Skeletal Muscle | AAV1, AAV7, or AAV9 |
Intravenous of AAV9 only | Intramuscular or isolated limb perfusion (AAV9) |
| Brain | AAV9, rh.8, or rh.10 |
Intravenous or intracerebral ventricular |
Intravenous or intracranial |
Overall the combination of rAAV, a highly efficient in vivo gene delivery vehicle with optimized miRNA expression cassettes or inhibitors, will be valuable tools for miRNA functional genomic study and potential therapeutics.
RAAV-mediated miRNA Delivery and miRNA Therapeutics: case studies
The first remarkable breakthrough in miRNA-based therapy is using AAV vector delivered miR-26a to suppress liver cancer in an inducible-cMYC mouse model12. In the study, Kota et al found that miR-26a was the most down-regulated miRNA in the liver tumor resulting from the specific activation of cMYC oncogene in hepatocytes. They then built a scAAV construct carrying the pri-miR-26a fragment and packaged it into AAV8 vector, an AAV serotype so efficient in hepatic transfection that it can transduce nearly every hepatocyte in an individual mouse. After a single tail vein injection, there was a dramatic decrease of tumor burden, induced by a massive tumor-cell specific apoptosis. By putting back one miRNA into tumor cells, Kota et al achieved noticeable therapeutic benefit, leading the way in a novel and exciting therapeutic strategy for the treatment of liver cancer and the other diseases.
Another successful example of rAAV delivered miRNA therapy was accomplished through the use of rAAV9, which can cross blood brain barrier 61-63 . RAAV9 was used to deliver miR-196a to treat spinal-bulbar muscular atrophy (SBMA) 64. In this study, the authors compared the miRNA expression profile in the spinal cords of the diseased mice with those of wild type mice and found that miR-196a, along with 4 other miRNAs, were up-regulated more than two-fold during the advanced stage of this mouse model disease. The researchers found evidence that miR-196a was up-regulated in the diseased mice as a protective mechanism against the progression of the disease and decided that they would attempt to treat the disease by aiding the natural up-regulation of miR-196a by bolstering the amount of this miRNA with a delivery of exogenous miR-196a. After miR-196a was successfully delivered via rAAV9, they found the disease related gene expression was down-regulated, resulting in the improvement of not only mouse behavior but of body weight and mouse survival. The benefit from miR-196a over-expression also indicates endogenous miRNAs can be protective factors in the disease progress.
To understand the function of nearly 300 conserved miRNAs between humans and mice, we can continue to develop the field by producing hundreds of miRNA-KO mouse strains for future research or we can produce mouse miRNA toolkits as described above, applicable and adaptive for use in many animal and cell models. To accelerate analysis of miRNA function in mammals, Xie, et al., achieved similar phenotypes as miR-122 KO mice 65,66 by combining the advantages of rAAV vectors and TuD RNAs 34. Using the same approach, AAV delivered anti-miR-26a TuD delays the differentiation from myoblasts to myotubes67. It is likely that the desired results from the development of miRNA somatic KO mice can be obtained more quickly and easily by means of scAAV-delivered TuDs. There is also much to be learned from the up-regulation of miRNA; a goal that is difficult to be addressed by the creation of transgenic overexpression mouse libraries, but can be quite robustly accomplished with the construction of scAAV-delivered pri-miRNAs with a single bonus of rAAV injection.
Prospects and challenges
Over the past few years, miRNA-based therapeutics have achieved great success in many preclinical animal models, such as HCV29, HCC12, metabolic disorders21,56, and cardiovascular disease68. The first miRNA-targeted drug, miravirsen (miR-122 antisense oligonucleotide), completed its Phase 2a study recently25. The clinical data showed the patients were well tolerated, 2 to 3 logs reduction of HCV RNA and no signs of viral resistance. Miravirsen provides an additional option for the patients who are not responding to interferon therapy and avoids virus mutation because it targets the host gene miR-122 on which the virus relies, not on the virus itself. The clinical results are very attractive, but there are still concerns about the long term safety of miR-122 inhibition in patients. Every single miRNA regulates hundreds of target genes involved in multiple pathways. Modulation of miRNA may easily lead to unwanted outcomes. Indeed, miR-122 inhibition by AMOs or scAAV-delivered TuDs lowered high-density lipoprotein (HDL) and low-density lipoprotein (LDL) simultaneously28-30,34. However HDL is regarded as good cholesterol. The most severe concern is the correlation between low miR-122 level and HCC development in patients, although no direct causal link has been established 69-71. Furthermore, aged miR-122 KO mice developed HCC 65,66. This germ line depletion of miR-122 may not accurately reflect the real risk of liver cancer in adult HCV patients who only lose miR-122 during the miR-122 antisense oligonucleotide treatment, but it does warn of potential threats. AAV vector expression is stable for years in mice and humans. AAV delivered anti-miR-122 TuD may be used to evaluate the HCC risk of long-term miR-122 inhibition and other side effects.
MiR-26a based liver cancer therapy also confronts the same concern. AAV8 delivered miR-26a can result in tumor suppression in the mouse liver cancer model driven by cMYC gene12, but miR-26a also promotes cholangiocarcinoma growth by activating β-catenin72. The activation of Wnt/β-catenin is one of the major pathways involved in HCC73. In glioma, miR-26 was also reported as an oncomiR (miRNA associated with cancer) by directly repressing a well-known tumor suppressor74, PTEN. It is a legitimate concern that bolstering levels of miR-26a may worsen the pathogenesis in certain populations of HCC patients.
High unregulated expression of shRNAs delivered by rAAV has been reported to saturate endogenous cellular miRNA machinery and cause fatal effects in mice75,76. A surplus of rAAV-delivered shRNAs diminishes two crucial RNAi machineries, Exportin-5 and Argonaut-2 protein 76,77. To improve the safety, shRNAs were engineered into the position where the mature miRNA duplex is and flanked with native sequence to direct correct processing. By producing less unprocessed precursors and by increasing the effectiveness of processing, this procedure optimization reduced shRNA-mediated toxicity delivered by AAV in the CNS 78,79. However, this strategy also raised another risk from possibly disturbing another important RNAi machinery factor, Drosha, which is required for the miRNA shuttle strategy. In rAAV mediated miRNA replacement or enforcement, the impact of ectopic miRNAs on RNAi machinery and homeostasis of endogenous miRNAs as well as the off-target effects in targeted tissue and unwanted tissues have not been addressed. On the other hand, AAV delivered TuD RNA has demonstrated efficacy, specificity and safety of miRNA inhibition 34, but extensive studies particularly in large animal models are still required.
Overall miRNA therapeutics is an emerging field filled with lots of hope. Because of their roles as master regulators in many diseases, miRNAs can achieve previously unreachable medical benefit when compared with conventional mono-target therapeutics. Targeting of multiple genes is the strength of using miRNAs and miRNA inhibitors as therapeutics reagents, but the unique property is also its weakness. Without thoroughly understanding the miRNA functions, miRNA-based therapeutics will be double-edged swords in many cases.
Supplementary Material
Acknowledgment
This work was supported by Public Health Service grants P01 HL59407-11, P01AI100263-01 and 1R01NS076991from the National Institutes of Health to GG.
Abbreviations
- AAV
adeno-associated virus
- miRNA
microRNA
- pri-miRNA
primary miRNA
- pre-miRNA
precursor miRNA
- NSCLC
non-small-cell lung cancer
- HCC
hepatocellular carcinoma
- TAC
Transverse aortic constriction
- HCV
hepatitis C virus
- AMOs
anti-miRNA oligonucleotides
- TuD RNAs
tough decoy RNAs
- ITR
inverted terminal repeat
- scAAV
self-complimentary AAV
- HDL
high-density lipoprotein
- LDL
low-density lipoprotein
- SBMA
spinal and bulbar muscular atrophy
- RISC
RNA-induced silencing complexes
Footnotes
Disclosure
G.Gao is a founder of Voyager Therapeutics and holds equity in the company. G.Gao is an inventor on patents with potential royalties licensed to Voyager Therapeutics and other biopharmaceutical companies. J.X. and D.R.B have nothing to disclose.
References
- 1.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 2.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nature reviews. Molecular cell biology. 2005;6(5):376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 3.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. Rna. 2004;10(12):1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nature structural & molecular biology. 2006;13(12):1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 9.Trang P, Medina PP, Wiggins JF, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29(11):1580–1587. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji J, Shi J, Budhu A, et al. MicroRNA expression, survival, and response to interferon in liver cancer. The New England journal of medicine. 2009;361(15):1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao GP, Alvira MR, Wang L, Calcedo R, Johnston J. Wilson JM. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(18):11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kota J, Chivukula RR, O'Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137(6):1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Rooij E, Sutherland LB, Liu N, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(48):18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thum T, Gross C, Fiedler J, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456(7224):980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 15.Montgomery RL, Hullinger TG, Semus HM, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124(14):1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Rooij E, Sutherland LB, Qi XX, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316(5824):575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 17.Grueter CE, van Rooij E, Johnson BA, et al. A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell. 2012;149(3):671–683. doi: 10.1016/j.cell.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poy MN, Hausser J, Trajkovski M, et al. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(14):5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rayner KJ, Suarez Y, Davalos A, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328(5985):1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rayner KJ, Sheedy FJ, Esau CC, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. The Journal of clinical investigation. 2011;121(7):2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rayner KJ, Esau CC, Hussain FN, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478(7369):404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang J, Nicolas E, Marks D, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA biology. 2004;1(2):106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 23.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309(5740):1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Masaki T, Yamane D, McGivern DR, Lemon SM. Competing and noncompeting activities of miR-122 and the 5' exonuclease Xrn1 in regulation of hepatitis C virus replication. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(5):1881–1886. doi: 10.1073/pnas.1213515110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janssen HL, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting microRNA. The New England journal of medicine. 2013;368(18):1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 26.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhnert F, Mancuso MR, Hampton J, et al. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development. 2008;135(24):3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- 28.Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell metabolism. 2006;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Lanford RE, Hildebrandt-Eriksen ES, Petri A, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elmen J, Lindow M, Schutz S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 31.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nature methods. 2007;4(9):721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haraguchi T, Ozaki Y, Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic acids research. 2009;37(6):e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gentner B, Schira G, Giustacchini A, et al. Stable knockdown of microRNA in vivo by lentiviral vectors. Nature methods. 2009;6(1):63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- 34.Xie J, Ameres SL, Friedline R, et al. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nature methods. 2012;9(4):403–409. doi: 10.1038/nmeth.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarty DM, Ryan JH, Zolotukhin S, Zhou X, Muzyczka N. Interaction of the adeno-associated virus Rep protein with a sequence within the A palindrome of the viral terminal repeat. Journal of virology. 1994;68(8):4998–5006. doi: 10.1128/jvi.68.8.4998-5006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snyder RO, Im DS, Ni T, Xiao X, Samulski RJ, Muzyczka N. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. Journal of virology. 1993;67(10):6096–6104. doi: 10.1128/jvi.67.10.6096-6104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dubielzig R, King JA, Weger S, Kern A, Kleinschmidt JA. Adeno-associated virus type 2 protein interactions: formation of pre-encapsidation complexes. Journal of virology. 1999;73(11):8989–8998. doi: 10.1128/jvi.73.11.8989-8998.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kyostio SR, Owens RA, Weitzman MD, Antoni BA, Chejanovsky N, Carter BJ. Analysis of adeno-associated virus (AAV) wild-type and mutant Rep proteins for their abilities to negatively regulate AAV p5 and p19 mRNA levels. Journal of virology. 1994;68(5):2947–2957. doi: 10.1128/jvi.68.5.2947-2957.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoggan MD, Blacklow NR, Rowe WP. Studies of small DNA viruses found in various adenovirus preparations: physical, biological, and immunological characteristics. Proceedings of the National Academy of Sciences of the United States of America. 1966;55(6):1467–1474. doi: 10.1073/pnas.55.6.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buller RM, Janik JE, Sebring ED, Rose JA. Herpes simplex virus types 1 and 2 completely help adenovirus-associated virus replication. Journal of virology. 1981;40(1):241–247. doi: 10.1128/jvi.40.1.241-247.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rabinowitz JE, Samulski RJ. Building a better vector: the manipulation of AAV virions. Virology. 2000;278(2):301–308. doi: 10.1006/viro.2000.0707. [DOI] [PubMed] [Google Scholar]
- 42.Grieger JC, Samulski RJ. Adeno-associated virus vectorology, manufacturing, and clinical applications. Methods in enzymology. 2012;507:229–254. doi: 10.1016/B978-0-12-386509-0.00012-0. [DOI] [PubMed] [Google Scholar]
- 43.Asokan A, Schaffer DV, Samulski RJ. The AAV vector toolkit: poised at the clinical crossroads. Molecular therapy : the journal of the American Society of Gene Therapy. 2012;20(4):699–708. doi: 10.1038/mt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mingozzi F, High KA. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nature reviews. Genetics. 2011;12(5):341–355. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- 45.Buning H, Perabo L, Coutelle O, Quadt-Humme S, Hallek M. Recent developments in adeno-associated virus vector technology. The journal of gene medicine. 2008;10(7):717–733. doi: 10.1002/jgm.1205. [DOI] [PubMed] [Google Scholar]
- 46.Kotin RM, Siniscalco M, Samulski RJ, et al. Site-specific integration by adeno-associated virus. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(6):2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samulski RJ, Zhu X, Xiao X, et al. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. The EMBO journal. 1991;10(12):3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Penaud-Budloo M, Le Guiner C, Nowrouzi A, et al. Adeno-associated virus vector genomes persist as episomal chromatin in primate muscle. Journal of virology. 2008;82(16):7875–7885. doi: 10.1128/JVI.00649-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Malani N, Hamilton SR, et al. Assessing the potential for AAV vector genotoxicity in a murine model. Blood. 2011;117(12):3311–3319. doi: 10.1182/blood-2010-08-302729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaeppel C, Beattie SG, Fronza R, et al. A largely random AAV integration profile after LPLD gene therapy. Nature medicine. 2013;19(7):889–891. doi: 10.1038/nm.3230. [DOI] [PubMed] [Google Scholar]
- 51.Zhong L, Malani N, Li M, et al. Recombinant adeno-associated virus integration sites in murine liver after ornithine transcarbamylase gene correction. Human gene therapy. 2013;24(5):520–525. doi: 10.1089/hum.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bryant LM, Christopher DM, Giles AR, et al. Lessons learned from the clinical development and market authorization of Glybera. Human gene therapy. Clinical development. 2013;24(2):55–64. doi: 10.1089/humc.2013.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z, Ma HI, Li J, Sun L, Zhang J, Xiao X. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene therapy. 2003;10(26):2105–2111. doi: 10.1038/sj.gt.3302133. [DOI] [PubMed] [Google Scholar]
- 54.McCarty DM, Fu H, Monahan PE, Toulson CE, Naik P, Samulski RJ. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene therapy. 2003;10(26):2112–2118. doi: 10.1038/sj.gt.3302134. [DOI] [PubMed] [Google Scholar]
- 55.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(10):3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soh J, Iqbal J, Queiroz J, Fernandez-Hernando C, Hussain MM. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nature medicine. 2013;19(7):892–900. doi: 10.1038/nm.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bak RO, Hollensen AK, Primo MN, Sorensen CD, Mikkelsen JG. Potent microRNA suppression by RNA Pol II-transcribed 'Tough Decoy' inhibitors. Rna. 2013;19(2):280–293. doi: 10.1261/rna.034850.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao X, Li J, Samulski RJ. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. Journal of virology. 1998;72(3):2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grieger JC, Choi VW, Samulski RJ. Production and characterization of adeno-associated viral vectors. Nature protocols. 2006;1(3):1412–1428. doi: 10.1038/nprot.2006.207. [DOI] [PubMed] [Google Scholar]
- 60.Gao G, Sena-Esteves M. Molecular Cloning: A Laboratory Manual Introducing Genes into Mammalian Cells: Viral Vectors. Vol. 2. Cold Spring Harbor Laboratory Press; New York: 2012. pp. 1209–1313. [DOI] [PubMed] [Google Scholar]
- 61.Zhang H, Yang B, Mu X, et al. Several rAAV vectors efficiently cross the blood-brain barrier and transduce neurons and astrocytes in the neonatal mouse central nervous system. Molecular therapy : the journal of the American Society of Gene Therapy. 2011;19(8):1440–1448. doi: 10.1038/mt.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang B, Li S, Wang H, et al. Global CNS Transduction of Adult Mice by Intravenously Delivered rAAVrh.8 and rAAVrh.10 and Nonhuman Primates by rAAVrh.10. Molecular therapy : the journal of the American Society of Gene Therapy. 2014 doi: 10.1038/mt.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Foust KD, Nurre E, Montgomery CL, Hernandez A, Chan CM, Kaspar BK. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nature biotechnology. 2009;27(1):59–65. doi: 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miyazaki Y, Adachi H, Katsuno M, et al. Viral delivery of miR-196a ameliorates the SBMA phenotype via the silencing of CELF2. Nature medicine. 2012;18(7):1136–1141. doi: 10.1038/nm.2791. [DOI] [PubMed] [Google Scholar]
- 65.Hsu SH, Wang B, Kota J, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. The Journal of clinical investigation. 2012;122(8):2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsai WC, Hsu SD, Hsu CS, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. The Journal of clinical investigation. 2012;122(8):2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dey BK, Gagan J, Yan Z, Dutta A. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes & development. 2012;26(19):2180–2191. doi: 10.1101/gad.198085.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Rooij E, Olson EN. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nature reviews. Drug discovery. 2012;11(11):860–872. doi: 10.1038/nrd3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kutay H, Bai S, Datta J, et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. Journal of cellular biochemistry. 2006;99(3):671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28(40):3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsai WC, Hsu PW, Lai TC, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49(5):1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 72.Zhang J, Han C, Wu T. MicroRNA-26a promotes cholangiocarcinoma growth by activating beta-catenin. Gastroenterology. 2012;143(1):246–256. doi: 10.1053/j.gastro.2012.03.045. e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dahmani R, Just PA, Perret C. The Wnt/beta-catenin pathway as a therapeutic target in human hepatocellular carcinoma. Clinics and research in hepatology and gastroenterology. 2011;35(11):709–713. doi: 10.1016/j.clinre.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 74.Huse JT, Brennan C, Hambardzumyan D, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes & development. 2009;23(11):1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grimm D. The dose can make the poison: lessons learned from adverse in vivo toxicities caused by RNAi overexpression. Silence. 2011;2:8. doi: 10.1186/1758-907X-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grimm D, Streetz KL, Jopling CL, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441(7092):537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 77.Grimm D, Wang L, Lee JS, et al. Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver. The Journal of clinical investigation. 2010;120(9):3106–3119. doi: 10.1172/JCI43565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boudreau RL, Martins I, Davidson BL. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Molecular therapy : the journal of the American Society of Gene Therapy. 2009;17(1):169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McBride JL, Boudreau RL, Harper SQ, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(15):5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.