Abstract
The current study examined the changes in striatal gene network structure induced by short-term selective breeding from a heterogeneous stock for haloperidol response. Brain (striatum) gene expression data were obtained using the Illumina WG 8.2 array, and the datasets from responding and non-responding selected lines were independently interrogated using a weighted gene coexpression network analysis (WGCNA). We detected several gene modules (groups of coexpressed genes) in each dataset; the membership of the modules was found to be largely concordant, and a consensus network was constructed. Further validation of the network topology showed that using approximately 35 samples is sufficient to reliably infer the transcriptome network. An in-depth analysis showed significant changes in network structure and gene connectivity associated with the selected lines; these changes were validated using a bootstrapping procedure. The most dramatic changes were associated with a gene module richly annotated with neurobehavioral traits. The changes in network connectivity were concentrated in the links between this module and the rest of the network, in addition to changes within the module; this observation is consistent with recent results in protein and metabolic networks. These results suggest that a network-based strategy will help identify the genetic factors associated with haloperidol response.
Keywords: Catalepsy, gene coexpression network, gene connectivity, haloperidol, heterogeneous stock, short-term selective breeding, striatum
Previously, two independent selections for haloperidol response were conducted starting from heterogeneous stock (HS) mice (Hitzemann et al. 1991, 1994). Both selections were bi-directional, and the segregation of the lines was rapid such that by generation 5 (S5), they differed 10- to 20-fold in haloperidol and related antipsychotic drugs response (e.g. raclopride). The selections detected correlated responses to selection that included differences in density of midbrain dopamine receptors (Hitzemann et al. 1991; Qian et al. 1993), number of midbrain dopamine neurons (Hitzemann et al. 1994) and number of striatal cholinergic neurons (Dains et al. 1996).
This study takes a systems approach to understanding the genetic and transcription changes associated with haloperidol selection. First, we quantify the genetic differences emerging between the selected lines. Subsequently, we focus on the gene coexpression network differences and we apply to the data the weighted gene coexpression network analysis (WGCNA; Zhang & Horvath 2005). Short-term selective breeding (STSB; Belknap et al. 1997) was used to produce the selected lines. The founder HS4 population (Malmanger et al. 2006) was formed by crossing the C57BL/6J (B6), DBA/2J (D2), BALB/cJ (C) and LP/J (LP) strains. The D2 and C strains are haloperidol responsive (ED50 < 0.6 mg/kg), whereas the B6 and LP strains are haloperidol non-responsive (ED50 > 4 mg/kg); in fact the LP strain is frequently non-responsive to haloperidol doses of >10 mg/kg (Hitzemann et al. 1995; Kanes et al. 1996). Gene coexpression analyses have provided insights into the functional organization of the transcriptome in several species, including yeast (Carlson et al. 2006), mouse (Fuller et al. 2007) and primates (Oldham et al. 2006); interacting proteins often are encoded by coexpressed genes (Iancu et al. 2010; Oldham et al. 2006). Furthermore, there is evidence that differences between coexpression networks are more significant from a systems biology perspective than expression level changes of individual genes (Oti et al. 2008). The WGCNA paradigm has been reviewed and compared with alternative approaches such as Bayesian networks (Zhao et al. 2010). Importantly, the WCGNA assumes that the underlying metabolic networks are scale free and follow a power-law distribution (Barabasi & Albert 1999; Ravasz et al. 2002). Analysis of these metabolic networks showed that of particular importance are the relatively ‘few connected nodes that participate in a large number of metabolic reactions’ (Zhao et al. 2010); these nodes are denoted in literature as network hubs. A distinct category, nodes that link different parts of the network together, has been shown to be essential in protein interaction networks; these nodes are known as network connectors (Guimera & Nunes Amaral 2005). The goal of this study was to identify network changes that differentiate the selected lines. Haloperidol response is ideal for this application because the mechanism of haloperidol action, the blockade of D2 dopamine receptors, is well established (Hitzemann et al. 2003; Qian et al. 1993).
Methods
Animals
Details describing the formation of the HS4 colony are provided elsewhere (Malmanger et al. 2006). Animals from the G25 generation were used as the founders for STSB. Two males and females from each of the 48 families were phenotyped for haloperidol response as described in Rasmussen et al. (1999). Briefly, on day 1 all animals were administered 4 mg/kg i.p. of haloperidol; the catalepsy response (maintaining a fixed rearing posture for 30 seconds) was measured 15 min later. This test characterized the animals as haloperidol responders (R) and non-responders (NR). One week later, the R animals were administered 1 mg/kg of haloperidol, and the NR animals were administered 7 mg/kg of haloperidol. Response and non-response to these challenges yielded two additional categories: very responsive (RR) and very non-responsive (NN) animals. Sixteen families of RR and NN animals were bred to produce the first selection generation. This procedure was repeated through three generations of breeding to produce the high (responsive) and the low (non-responsive) selected lines. The general strategy for STSB is found in Belknap et al. (1997). Selected lines were genotyped (~48/group) using the panel of 768 single nucleotide polymorphisms (SNPs) that has been previously described (Malmanger et al. 2006). All animal care, breeding and testing procedures were approved by the Laboratory Animal Users Committees at the Veterans Affairs Medical Center, Portland, OR, USA, and at the Oregon Health and Science University, Portland, OR, USA.
Gene expression data processing
Our data preprocessing steps closely follow the procedure described previously (Iancu et al. 2010). Gene expression data for the high and low selected lines were obtained from the striatum using the Illumina WG 8.2 array exactly as described by the manufacturer. The dissection of the striatum and the details of sample preparation for hybridization are found in (Malmanger et al. 2006). Data were imported into the R application environment (http://www.r-project.org) and outlier samples were removed.
Data were culled for any probes without Entrez identifiers and also for any probe that overlapped with known SNPs in the founding strains, using the publicly available Wellcome Trust Sanger Institute database of known polymorphisms (http://www.sanger.ac.uk/resources/mouse/genomes/). Probes not expressed above background (P < 0.01) in at least a quarter of the samples were removed and the data were quantile normalized. All catalepsy samples were processed during the same experiment, and therefore no adjustment for batch effects was performed. Striatal gene expression data from the ancestral HS4 population at generation G19 was previously obtained using the Illumina WG 6.1 array; this dataset have been previously described (Iancu et al. 2010). eQTL detection was performed on this data using HAPPY (Mott et al. 2000), a QTL detection tool designed for association studies in outbred mouse populations. Probes detected as eQTLs in the G19 animals were subsequently evaluated for differential expression in the catalepsy datasets using a competitive group comparison approach (Goeman et al. 2004). This procedure evaluates predetermined groups of genes (in our case eQTL genes) for group-wise differential expression.
Quantification of genetic variability and genetic differences
For computing genetic distances between individuals, each genome was encoded as a long vector with entries in the range 0, 1, 2 based on the allelic content at a marker. Once all pairwise distances were computed, we evaluated whether within-group average distances were significantly smaller than between-group distances. Statistical significance was evaluated using a bootstrapping approach that randomized group labels. This analysis was performed at the whole genome level and at individual genomic intervals. Our approach follows the analysis of molecular variance (AMOVA) procedure (Excoffier et al. 1992) and uses the ‘vegan’ R package (http://cran.r-project.org/web/packages/vegan/index.html).
Construction of the gene coexpression networks
For the high and low selected line datasets, a series of steps were performed for constructing a gene coexpression network (Langfelder & Horvath 2008; Zhang & Horvath 2005). For each probe, we estimated coefficient of variation (CV) and the network connectivity, which is defined below. Because network construction is more robust if probes have high CV and high connectivity (Fuller et al. 2007), we retained only the probes in the top two quantiles for both connectivity and CV; this corresponded with a CV = 0.015 threshold. The resulting intersection resulted in 3924 probes, all of which mapped to distinct Entrez identifiers.
The first step in constructing the transcriptome network is computation of the absolute value of the Pearson correlation coefficient between probe pairs. The Pearson correlation matrix was subsequently transformed into an adjacency matrix (A) using a power function. The connection strength aij between probes xi and xj then becomes aij = |corr(xi, xj)|β; β = 10 was selected in accordance with the scale-free topology criterion (Zhang & Horvath 2005).
Gene modules were characterized as groups of genes with high ‘topological overlap’ (TO), a concept defined for weighted networks by Zhang and Horvath (2005). The TO between two genes i, j was computed as ωij (lij + aij)/(min{ki, kj} + 1 – aij) where represents the number of genes connected to both gene i and gene j, whereas u indexes all the genes in the network. Total node connectivity was computed as , where j iterates over all other nodes in the network. For in module connectivity, j iterates over all nodes in the same module, whereas for out module connectivity j iterates over all nodes in the other modules.
Validation of gene modules membership and network structure
For the construction of the consensus network, the topological overlap between two probes was set to the minimum of the topological overlaps of the high and low networks: TOconsensus = min(TOLow, TOHigh). Consensus modules were obtained by clustering the consensus similarity matrix. The members in the detected consensus modules were compared against same-size random groups of genes using a bootstrapping procedure; this analysis showed that gene modules had significantly higher TO than randomly selected groups of genes, thus validating network modularity. The consistency of module membership between the high and low selected lines was evaluated using a cross-tabulation-based measure (Langfelder et al. 2011). Modules were detected independently in the high and low networks, and the module overlap was evaluated using Fisher's exact test for each pair of high/low modules. The overall structure of the network was further evaluated for robustness in regard to data subsampling. Network TO matrices were constructed in each of the datasets using smaller subsets of the available samples. Between 6 and 40 samples were selected, and TO matrices were constructed from these samples. Matrix correlations with the full data TO matrix were computed, using the Mantel procedure (Mantel 1967).
Comparison of high and low networks
The statistical significance of changes in expression and connectivity was evaluated using a permutation procedure, following the general procedure outlined in Langfelder et al. (2011). The expression data were randomly divided into groups of similar size (n = 40 samples), without regard to phenotypic group assignments. These randomly assigned expression samples were used to construct coexpression networks, and the differences in expression and connectivity between these random networks were computed (N = 1000 permutations) and subsequently normalized with respect to mean and standard deviation. These standardized values were used to generate an empirical distribution of differences in expression and connectivity that arise purely by chance. Differences in expression and connectivity between the high and low networks were evaluated for statistical significance against this bootstrap-generated empirical distribution.
Results
Selection of the high and low haloperidol-responsive lines
Two males and females from each of the 48 HS4 families were phenotyped for haloperidol response using a two-step procedure (Rasmussen et al. 1999). The procedure assigned the mice to four response categories ranging from very responsive (RR) to responsive (R) to non-responsive (N) to very non-responsive (NN) (Fig. 1a). Sixteen breeding pairs each were taken from the RR and NN to produce the first selection generation (S1). The phenotyping and selection continued in a similar fashion for two additional generations. The segregation of the high (responsive) and low (non-responsive) lines over the three generations of selection is illustrated in Fig. 1b–d. The data illustrate that at S3 there were no R or RR mice in the low line and no N or NN mice in the high line. The S3 parents were bred for three rounds to produce sufficient progeny for all subsequent testing. As determined by the ‘up and down’ method (Dixon 1965), the haloperidol ED50s in the high and low lines were 0.35 and 11 mg/kg, respectively.
Figure 1. Dynamics of selective breeding.
(a) Distribution of haloperidol response in the founder HS4 population. Animals were tested as described in Rasmussen et al. (1999) to yield four response categories: very non-responsive (NN), non-responsive (N), responsive (R) and very responsive (RR). (b) Distribution of haloperidol response in the S1 generation. Red = low (non-responding) line; blue = high (responding) line. (c, d) Response distribution in the S2 and S3 generations.
Genetic differences emerging in the selected lines
The founders and the selected lines were genotyped using a SNP panel described elsewhere (Malmanger et al. 2006). Genetic variability on the basis of pairwise genetic distances was calculated using the AMOVA approach (Excoffier et al. 1992). As illustrated in Fig. 2, variability was higher in the founder animals compared with both selected lines, indicating loss of variability during selection. Furthermore, using the top 100 most variable probes, we computed transcriptional distance between individuals (Euclidean distance). The Mantel test of matrix correlation (Mantel 1967) showed a small but statistically significant correlation between genetic distances and transcription distances: coefficient of correlation 0.06, P < 0.035. This relationship illustrates the fact that genetic variability is one of the factors generating transcriptional variability.
Figure 2. Multidimensional scaling plot of pairwise genetic distances between mice.
Distances were constructed using polymorphisms at all available genomic markers. More genetic variability in the founder population (green) results in larger genomic distances between the individuals, signifying greater diversity than in the selected lines (red and blue). The high and low lines appear distinct but also more clustered because of smaller distances between individuals compared with the larger variability in the founder animals.
Using the AMOVA measure of genetic difference (see Methods), we quantified the level of genetic difference between the selected lines at each interval. Out of the 666 genotyped locations available for the catalepsy datasets, 180 showed significant divergence between the selected lines (P < 0.01); the genetic differences on chromosome 9 are illustrated in Fig. 3. Some of the significant genetic differences colocalized with previously detected catalepsy QTLs (Kanes et al. 1996; Rasmussen et al. 1999) on chromosome 9 (Fig. 3) as well as chromosomes 1 and 14 (see Supporting Information Figs. S5 and S6).
Figure 3. Genetic effects on gene expression, chromosome 9.
Black: genetic differences between the high and low lines, –log10(P) for AMOVA P values. Red: strength of evidence for persistent genetic effect on gene expression, –log10(P), generated from gene set analysis P values. Green: peak location of previously detected catalepsy QTL on chromosome 9.
A number of genomic locations diverging between the high and low groups were associated with expression level differences (eQTLs) in the G19 HS4 animals. Detection of eQTLs implies that differences in allelic content influence gene expression levels. Because the allelic content at these locations was also different between the high and low groups, we inquired whether the genetic effects on gene expression persisted over the generations of breeding and selection. In the catalepsy data, the eQTL probes were evaluated for differential expression. Although none of the strong eQTLs (FDR < 0.05) were among the most differentially expressed probes (FDR < 0.05), a more sensitive gene set analysis (Goeman et al. 2004) of the eQTL probes as a group showed evidence of differential expression. For each genomic interval, we collected the HS4 G19 eQTL probes (P < 0.01) and we evaluated whether their collection of differential expression P values (in the catalepsy datasets) were lower than that of identical sized, randomly selected probes. The strength of evidence for eQTLs displaying differential expression is illustrated in Fig. 3. In particular, the catalepsy QTL on chromosome 9 overlapped with a genomic location that was different between the high and low groups; the probes modulated by this eQTL also displayed differential expression.
Gene coexpression networks construction and validation
Gene coexpression networks were constructed as described elsewhere (Iancu et al. 2010; Langfelder & Horvath 2008). This process showed several distinct modules or groups of genes with similar expression profiles. Genes that were not assigned to modules were denoted with the gray color in each network. Using this methodology, we detected 16 modules in the high line and 18 modules in the low line. We do not regard the exact number of modules as essential because it is dependent on the parameters of the clustering procedure. The results of the clustering procedure are illustrated in Supporting Information Fig. S1. For each module, the average TO was significantly higher than what can be expected by chance (P < 10–4), whereas for the gray genes the TO was lower than for random groups of genes (P < 10–4).
The extent of overlap between the high and low networks was evaluated using two distinct but related network and module preservation measures: a cross-tabulation-based measure and a density preservation measure (Langfelder et al. 2011). The cross-tabulation measure evaluates whether independent clustering procedures give rise to similar groupings of genes in the high and low gene networks. As illustrated in Supporting Information Fig. S2, the majority of the gene modules in one network had a strong counterpart in the other. In some cases, modules in one network appeared to split in the other network; for example, the pink module in the low line network was divided between the brown and yellow modules in the high line network. The genes that were not assigned to any module (denoted by gray in both networks) were also largely preserved.
A density-based module preservation measure (Langfelder et al. 2008) was also used. First, a consensus network was constructed based on the consensus topological overlap matrix (TOM), as detailed in Methods. The consensus modules were obtained by clustering the consensus TO, which is the minimum TO value from the two networks. Using the minimum TO value ensures that genes connected in the consensus network are connected in both the high and the low networks; however, differences in the strength of the connection are of interest and will be quantified in subsequent sections. Clustering the consensus network resulted in 18 network modules; the density-based module preservation evaluation confirmed that the consensus modules showed evidence of coexpression in both the high and low networks. The consensus network was further evaluated in regard to stability to data subsampling. Our goal was to determine how many microarray samples are necessary for robust network construction. We selected smaller numbers of microarray samples (between 6 and 40) in each dataset, and we constructed TO network matrices based on these smaller data samples. If the network construction is stable and robust, then these smaller datasets should converge to the full-data TO matrix as the number of samples increases. We computed the similarity between the TO matrices using the Mantel correlation coefficient (Mantel 1967). The results of our resampling procedure are presented in Fig. 4; these data suggest that it is possible to infer the correct network structure using 35 samples because the correlation coefficient using 35 samples reaches values near 0.9.
Figure 4. Results of subsampling procedure used to determine the minimum number of samples necessary for robust network construction.
Network matrices were constructed using between 6 and 40 random samples from the high line (blue) or low line (red). The matrices were correlated with the network matrix constructed using all available samples. This procedure was repeated 40 times for each subsampling, and the average correlation is reported. As the number of samples increases above 25, the correlation rises above 0.8; for 35 samples the correlation is about 0.95.
GO annotation of the consensus modules
GO annotation (Alexa et al. 2006; Ashburner et al. 2000; Ghazalpour et al. 2006) was used to characterize the 18 consensus modules; the details for each of the modules are found in Supporting Information Table S1. For reasons outlined in the next section, the turquoise and blue modules were of special importance. Some of the most highly represented biological processes or molecular function GO categories associated with the turquoise module were G-protein coupled receptor protein signaling pathways and activity, locomotion, behavior, regulation of biological processes, peptide and neurotransmitter receptor activity, receptor binding, plasma membrane and extracellular region. For the blue module, the most highly represented GO categories were electron transport chain function, respiratory chain function and mitochondrial function.
Differential network analysis of the selected lines
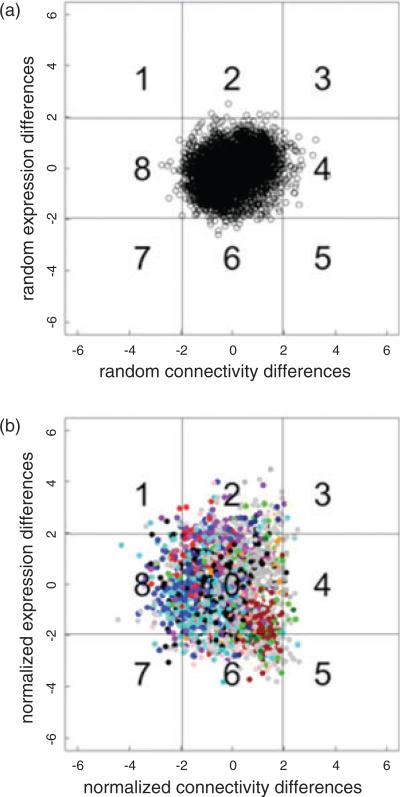
The overall goal of our differential network analysis was the detection of gene coexpression changes that can be attributed to selection; the amplitude of these changes was compared with coexpression changes that can occur by pure chance in networks that include different samples. For each gene within the consensus modules, the difference in expression and connectivity between the high and the low line networks was computed. Note that connectivity can be defined in three different contexts: in-module connectivity, out-module connectivity and total connectivity (Dong & Horvath 2007; Guimera & Nunes Amaral 2005). As outlined in Methods, the statistical significance of the observed changes was evaluated using a permutation procedure. The normalized expression and connectivity changes observed comparing random-assignment networks were used to populate a grid with boundaries of two standard deviations (Fig. 5a). A similar grid was populated with data obtained by comparing the high and low line networks. These data are illustrated in Fig. 5b; each gene is color coded as to module membership. The connectivity represented in Fig. 5a,b is out-module connectivity, i.e. the connectivity of genes in one module to genes in other modules. Fisher's exact test was used to compare the data in Fig. 5b; the comparison was at the level of module genes. These results are summarized in Fig. 6. Six modules showed significant differences; however, only the turquoise module showed differences in network out-module connectivity – higher connectivity in the low line. Repeating the analysis for in-module connectivity and total network connectivity, a significant change in connectivity was found for the blue module (Supporting Information Figs. S3 and S4); again connectivity was higher in the low line. Syn1, which encodes for synapsin 1 and is found in the turquoise module, illustrates a gene that shows both inand out-module changes. As illustrated in Fig. 7a, in the low line network, Syn1 is strongly connected to members of the red, black, purple and light cyan modules. By contrast, these out-module connections are markedly reduced in the high line network (Fig. 7b). Similarly, the in-module connections of Syn1 to other members of the turquoise module were reduced in the high line. Results on total connectivity were very similar to the results for in-module connectivity as in-module connectivity is often the major component of total gene connectivity; the connectivity results for all genes are available in Supporting Information Table S2.
Figure 5. Differences in out-module network connectivity (x-axis) and expression level (y-axis) between high and low selected line datasets.
Differences are normalized based on empirical distribution of differences between random networks. Sector lines correspond to differences greater than 1.97 standard deviations. Genes were color coded as to module membership. Sectors 1, 7 and 8 – higher out-module connectivity in the low line; sectors 3, 4 and 5 – higher connectivity in the high line; sectors 5, 6 and 7 – higher expression in the low line; sectors 1, 2 and 3 – higher expression in the high line. (a) Differences between random networks. (b) Differences between networks inferred using high versus low line datasets.
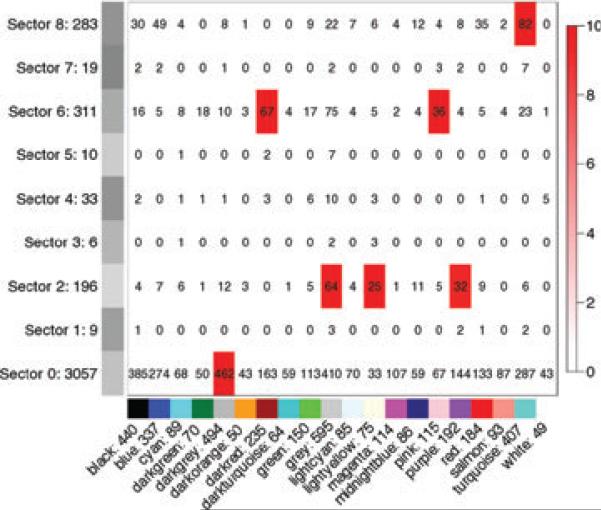
Figure 6.
Membership overlap between consensus modules (horizontal axis) and differentially connected or expressed genes sectors (vertical axis).
Figure 7. Differences in the connectivity profiles of Syn1, a member of the turquoise module.
Network genes are arranged on a circle, grouped by module. Pairwise correlation between genes has been thresholded for visual clarity; genes highly correlated with Syn1 are connected by an edge. (a) Low line; (b) high line. Note that in the high line the edges from Syn1 to genes in the non-turquoise modules (out-module connectivity) are markedly reduced as are the number of in-module edges.
Four modules showed significant changes in differential expression that were independent of the changes in connectivity. The dark red and pink modules were enriched in genes with higher expression in the low line, whereas the purple and light yellow modules were enriched in genes with higher expression in the high line. Details on these genes are found in Supporting Information Table S2. Here, we simply note that Drd2 is a member of the purple module and showed a modest difference in expression (21%). Details on the GO annotation for these modules are found in Supporting Information Table S1.
Discussion
Although haloperidol-induced catalepsy has been widely studied (~1200 publications), the genetics of this highly heritable phenotype (see, e.g. Kanes et al. 1996) remains unclear. Fink et al. (1982) and Severson et al. (1981) appear to have been the first to document the marked differences in catalepsy response among inbred mouse strains. Subsequently, more than 40 standard inbred and recombinant inbred strains have been tested for the catalepsy response (Barykina et al. 2002; Fowler et al. 2001; Hitzemann et al. 1995; Kanes et al. 1993, 1996). Differences in the measuring techniques led to some differences in the strain distribution patterns; however, the range of response differences among inbred strains often differs more than 50-fold (e.g. Kanes et al. 1993). Quantitative trait loci (QTL) analyses have detected catalepsy QTLs on mouse chromosomes 1, 9 and 14 (Hofstetter et al. 2008; Kanes et al. 1996; Patel & Hitzemann 1999; Rasmussen et al. 1999); a major locus for pinch-induced catalepsy has been recently detected on chromosome 13 (Kulikov et al. 2008). The haloperidol-related QTLs were also detected here, but not the pinch-induced catalepsy locus on chromosome 13. This suggests either the different genetic basis for the two mechanisms of catalepsy or, alternatively, the different genetic backgrounds (CBA/lacJ and AKR/J were used by Kulikov et al. 2008). Interval-specific congenic strains were used to reduce the chromosome 1 QTL to a 5.9 Mbp interval; on the basis of differential expression between the B6 and D2 strains, Parp1 emerged as a candidate quantitative trait gene (Hofstetter et al. 2008). However, here Parp1 was not differentially expressed; further and despite the considerable power to detect differential expression (N = >40/line), only eight transcripts were differentially expressed (FDR < 0.01, data not shown).
Given the high heritability and rapid segregation, there is an expectation that the lines will be differentiated by a few QTLs with large effect sizes. However, at least two factors complicate the interpretation of QTLs. First, the effects of genetic drift during the STSB lead to an inflation of false positive errors unless the significance threshold is drastically adjusted (Hitzemann et al. 2008). Second, outbred mouse populations have complex genetic structure, including uneven relatedness; it is even possible for distant genetic markers to have higher levels of linkage than adjacent markers (Ghazalpour et al. 2008). Under these conditions, the STSB, beginning with a relatively large founder population, is able to exert enormous selection pressure and segregate numerous loci. Many of these genes do not have direct effects but are simply in linkage with causative genes. Therefore, the traditional QTL analysis will not be fully informative. We originally assumed that causative polymorphisms affect expression in both the ancestral population and selected lines. However, only one catalepsy selection QTLs overlapped with ancestral eQTLs. This implies that selection transcriptome effects are more subtle and complex. Our approach to address these questions is the WGCNA, which emphasizes network structure and connectivity.
To our knowledge, this is the first application of differential network analysis to animals selectively bred for a behavioral trait. The catalepsy response suits this application given that the mechanism of haloperidol action, blockade of striatal D2 dopamine receptors, is well established. Although direct injection of haloperidol into the striatum produces catalepsy, it does not necessarily follow that the striatum is the sole locus of the differential response. Other components of the basal ganglia circuit, e.g. the subthalamic nucleus, could also be involved and are worthy of future study. Gene coexpression could be generated by coregulation, although spatial colocalization within the striatum could also cause coexpression (Iancu et al. 2010); the importance of spatial aspects of gene coexpression has recently been demonstrated in the context of pinch-induced catalepsy, where selection led to changes in the coexpression across several brain structures including the striatum (Kondaurova et al. 2011).
Our results suggest that selection induces significant changes in the coexpression network structure; importantly, these changes are distributed among a multitude of genes, but preferentially affecting only a few of the modules. We evaluate the importance of individual gene by measuring its network connectivity: genes with high connectivity or large changes in connectivity are of higher interest. Figure 6 summarizes the key findings. Turquoise out-module connectivity was significantly reduced in the high line. The turquoise module is large and is significantly enriched in GO categories including locomotion, behavior, peptide and neurotransmitter receptor activity, and receptor binding. The observation that selection affects this module is not surprising given the mechanism of haloperidol action. However, the modules were constructed in an unbiased fashion and that significance of the change in connectivity for the turquoise module far exceeds random chance. The decreased out-module connectivity in the high line suggests that enhanced haloperidol response is associated with a disruption of inter-module communication, which may in turn reduce the ability of the striatum to compensate for the effects of haloperidol. The change in out-module connectivity of several turquoise genes, e.g. Syn1, is particularly intriguing in light of studies suggesting that connectivity between modules is a highly relevant network feature (Guimera & Nunes Amaral 2005). It has also been suggested that targeting between module links might yield promising clinical drug targets (Guimera et al. 2007). It is of interest to note that a number of genes in the turquoise module are associated with striatal/dopamine function and haloperidol response; these include a cluster of genes associated with cholinergic function: Chat, Chrm3, Chrna4 and Slc18a3. Dains et al. (1996) observed in the BXD recombinant inbred series the expected positive relationship between the number of striatal cholinergic neurons and haloperidol response. Furthermore, Rasmussen et al. (1999) detected a strong QTL for haloperidol response on chromosome 14; the Chat locus falls within the QTL interval.
Our results show that selection led to a significant reduction within the high line of blue in-module connectivity (see Supporting Information Fig. S4). The blue module is enriched in genes associated with mitochondrial function (see Supporting Information Table S1). Burkhardt et al. (1993) found that haloperidol and two other typical antipsychotic drugs, chlorpromazine and thiothixene, inhibited complex 1 in rat brain mitochondria in vitro. The atypical antipsychotic, clozapine, also inhibited complex 1 but at a much higher concentration. Somewhat similar results were obtained by Modica-Napolitano et al. (2003) using isolated rat liver mitochondria. Several lines of evidence, including human studies, neurotoxin models and genetic models, suggest an association between mitochondrial dysfunction and Parkinson's disease (Zhu & Chu 2010). The finding that differential connectivity within the blue (mitochondrial) module was associated with enhanced haloperidol response is consistent with these observations.
Four modules exhibited significant differential expression. As with the changes in connectivity, these changes are important at the module level rather than at the level of individual genes and are significant only because they are considered collectively. Two modules, dark red and pink, were enriched with genes showing higher expression in the low line. GO terms associated with the dark red module focused on catabolic activity and ligase activity; terms associated with the pink module included the regulation of glucose metabolism and regulation of neurotransmitter levels. Two modules (purple and light yellow) were enriched in genes showing increased expression in the high line. GO annotation for both modules was poor. GO terms associated with the purple module were phosphoric diester hydrolase activity and transcription activator activity; the only GO term associated with the light yellow module was transferase activity. The purple module is of interest in that it contains Drd2, which falls within a previously detected QTL. The purple module includes a number of other genes known to be important to striatal signaling, e.g. Pde2a (Lin et al. 2010) which in turn aligns this module with the turquoise module; as noted in Fig. 6, the connectivity from Syn1 (turquoise module) to the purple module is markedly reduced in the high line. These observations suggest that selection effects go beyond changes in expression, and network connectivity provides a scaffold for propagation of genetic effects to differences in haloperidol response.
The current study illustrates that WGCNA effectively interrogates the transcriptome from STSB lines. The major advantage in using network approaches is that it allows a more comprehensive evaluation of changes in gene expression, under the assumption that genes and groups of genes with significant coexpression changes are more likely to also be associated with variations in the complex trait. It appears that this approach is suitable for other behavioral traits, especially in well-defined associated brain regions. The changes in connectivity were confined to modules highly likely to be involved in haloperidol response. The relationship of haloperidol response to differentially expressed modules is somewhat less clear, but the data provide new targets for investigation. The question arises as to how best to confirm the complex data generated by the WGCNA. We have considered this issue carefully. Selection data are best confirmed by an independent selection. However, if the network structure is relatively consistent across founding populations (see Iancu et al. 2010), it then follows that the confirming selection data should come from different founder populations. These studies are currently underway.
Supplementary Material
Acknowledgments
This study was supported in part by a grant from the United States Public Health Service, MH 51372 and from a Veterans Affairs Medical Service research grant. The authors also wish to thank Dr Kristin Demarest for her help with the manuscript.
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article:
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Barykina NN, Chuguy VF, Alekhina TA, Kolpakov VG, Maksiutova AV, Kulikov AV. Effects of thyroid hormone deficiency on behavior in rat strains with different predisposition to catalepsy. Physiol Behav. 2002;75:733–737. doi: 10.1016/s0031-9384(02)00662-5. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Richards SP, O'Toole LA, Helms ML, Phillips TJ. Short-term selective breeding as a tool for QTL mapping: ethanol preference drinking in mice. Behav Genet. 1997;27:55–66. doi: 10.1023/a:1025615409383. [DOI] [PubMed] [Google Scholar]
- Burkhardt C, Kelly JP, Lim YH, Filley CM, Parker WD., Jr Neuroleptic medications inhibit complex I of the electron transport chain. Ann Neurol. 1993;33:512–517. doi: 10.1002/ana.410330516. [DOI] [PubMed] [Google Scholar]
- Carlson MR, Zhang B, Fang Z, Mischel PS, Horvath S, Nelson SF. Gene connectivity, function, and sequence conservation: predictions from modular yeast co-expression networks. BMC Genomics. 2006;7:40. doi: 10.1186/1471-2164-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dains K, Hitzemann B, Hitzemann R. Genetics, neuroleptic response and the organization of cholinergic neurons in the mouse striatum. J Pharmacol Exp Ther. 1996;279:1430–1438. [PubMed] [Google Scholar]
- Dixon WJ. The up-and-down method for small samples. J Am Stat Assoc. 1965;60:967–978. [Google Scholar]
- Dong J, Horvath S. Understanding network concepts in modules. BMC Syst Biol. 2007;1:24. doi: 10.1186/1752-0509-1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink JS, Swerdloff A, Reis DJ. Genetic control of dopamine receptors in mouse caudate nucleus: relationship on cataleptic response to neuroleptic drugs. Neurosci Lett. 1982;32:301–306. doi: 10.1016/0304-3940(82)90311-1. [DOI] [PubMed] [Google Scholar]
- Fowler SC, Zarcone TJ, Vorontsova E. Haloperiodol-induced microcatalepsy differs in CD-1, BALB/c, and C57BL/6 mice. Exp Clin Psychopharmacol. 2001;9:277–284. doi: 10.1037//1064-1297.9.3.277. [DOI] [PubMed] [Google Scholar]
- Fuller TF, Ghazalpour A, Aten JE, Drake TA, Lusis AJ, Horvath S. Weighted gene coexpression network analysis strategies applied to mouse weight. Mamm Genome. 2007;18:463–472. doi: 10.1007/s00335-007-9043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour A, Doss S, Zhang B, Wang S, Plaisier C, Castellanos R, Brozell A, Schadt EE, Drake TA, Lusis AJ, Horvath S. Integrating genetic and network analysis to characterize genes related to mouse weight. PLoS Genet. 2006;2:e130. doi: 10.1371/journal.pgen.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour A, Doss S, Kang H, Farber C, Wen PZ, Brozell A, Castellanos R, Eskin E, Smith DJ, Drake TA, Lusis AJ. High-resolution mapping of gene expression using association in an outbred mouse stock. PLoS Genet. 2008;4:e1000149. doi: 10.1371/journal.pgen.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- Guimera R, Nunes Amaral LA. Functional cartography of complex metabolic networks. Nature. 2005;433:895–900. doi: 10.1038/nature03288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimera R, Sales-Pardo M, Amaral LA. A network-based method for target selection in metabolic networks. Bioinformatics. 2007;23:1616–1622. doi: 10.1093/bioinformatics/btm150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzemann R, Dains K, Bier-Langing CM, Zahniser NR. On the selection of mice for haloperidol response and non-response. Psychopharmacology (Berl) 1991;103:244–250. doi: 10.1007/BF02244211. [DOI] [PubMed] [Google Scholar]
- Hitzemann B, Dains K, Kanes S, Hitzemann R. Further studies on the relationship between dopamine cell density and haloperidol-induced catalepsy. J Pharmacol Exp Ther. 1994;271:969–976. [PubMed] [Google Scholar]
- Hitzemann R, Qian Y, Kanes S, Dains K, Hitzemann B. Genetics and the organization of the basal ganglia. Int Rev Neurobiol. 1995;38:43–94. doi: 10.1016/s0074-7742(08)60524-3. [DOI] [PubMed] [Google Scholar]
- Hitzemann R, Hitzemann B, Rivera S, Gatley J, Thanos P, Shou LL, Williams RW. Dopamine D2 receptor binding, Drd2 expression and the number of dopamine neurons in the BXD recombinant inbred series: genetic relationships to alcohol and other drug associated phenotypes. Alcohol Clin Exp Res. 2003;27:1–11. doi: 10.1097/01.ALC.0000047862.40562.27. [DOI] [PubMed] [Google Scholar]
- Hitzemann R, Malmanger B, Belknap J, Darakjian P, McWeeney S. Short-term selective breeding for high and low prepulse inhibition of the acoustic startle response; pharmacological characterization and QTL mapping in the selected lines. Pharmacol Biochem Behav. 2008;90:525–533. doi: 10.1016/j.pbb.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstetter JR, Hitzemann RJ, Belknap JK, Walter NA, McWeeney SK, Mayeda AR. Characterization of the quantitative trait locus for haloperidol-induced catalepsy on distal mouse chromosome 1. Genes Brain Behav. 2008;7:214–223. doi: 10.1111/j.1601-183X.2007.00340.x. [DOI] [PubMed] [Google Scholar]
- Iancu OD, Darakjian P, Walter NA, Malmanger B, Oberbeck D, Belknap J, McWeeney S, Hitzemann R. Genetic diversity and striatal gene networks: focus on the heterogeneous stock-collaborative cross (HS-CC) mouse. BMC Genomics. 2010;11:585. doi: 10.1186/1471-2164-11-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanes SJ, Hitzemann BA, Hitzemann RJ. On the relationship between D2 receptor density and neuroleptic-induced catalepsy among eight inbred strains of mice. J Pharmacol Exp Ther. 1993;267:538–547. [PubMed] [Google Scholar]
- Kanes S, Dains K, Cipp L, Gatley J, Hitzemann B, Rasmussen E, Sanderson S, Silverman M, Hitzemann R. Mapping the genes for haloperidol-induced catalepsy. J Pharmacol Exp Ther. 1996;277:1016–1025. [PubMed] [Google Scholar]
- Kondaurova EM, Naumenko VS, Sinyakova NA, Kulikov AV. Map3k1, Il6st, Gzmk, and Hspb3 gene coexpression network in the mechanism of freezing reaction in mice. J Neurosci Res. 2011;89:267–273. doi: 10.1002/jnr.22545. [DOI] [PubMed] [Google Scholar]
- Kulikov AV, Bazovkina DV, Kondaurova EM, Popova NK. Genetic structure of hereditary catalepsy in mice. Genes Brain Behav. 2008;7:506–512. doi: 10.1111/j.1601-183X.2008.00387.x. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7:e1001057. doi: 10.1371/journal.pcbi.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DT, Fretier P, Jiang C, Vincent SR. Nitric oxide signaling via cGMP-stimulated phosphodiesterase in striatal neurons. Synapse. 2010;64:460–466. doi: 10.1002/syn.20750. [DOI] [PubMed] [Google Scholar]
- Malmanger B, Lawler M, Coulombe S, Murray R, Cooper S, Polyakov Y, Belknap J, Hitzemann R. Further studies on using multiple-cross mapping (MCM) to map quantitative trait loci. Mamm Genome. 2006;17:1193–1204. doi: 10.1007/s00335-006-0070-2. [DOI] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Modica-Napolitano JS, Lagace CJ, Brennan WA, Aprille JR. Differential effects of typical and atypical neuroleptics on mitochondrial function in vitro. Arch Pharm Res. 2003;26:951–959. doi: 10.1007/BF02980205. [DOI] [PubMed] [Google Scholar]
- Mott R, Talbot CJ, Turri MG, Collins AC, Flint J. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci U S A. 2000;97:12649–12654. doi: 10.1073/pnas.230304397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci U S A. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oti M, van Reeuwijk J, Huynen MA, Brunner HG. Conserved co-expression for candidate disease gene prioritization. BMC Bioinformatics. 2008;9:208. doi: 10.1186/1471-2105-9-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NV, Hitzemann RJ. Detection and mapping of quantitative trait loci for haloperidol-induced catalepsy in a C57BL/6J × DBA/2J F2 intercross. Behav Genet. 1999;29:303–310. doi: 10.1023/a:1021653732147. [DOI] [PubMed] [Google Scholar]
- Qian Y, Hitzemann B, Yount GL, White JD, Hitzemann R. D1 and D2 dopamine receptor turnover and D2 messenger RNA levels in the neuroleptic-responsive and the neuroleptic nonresponsive lines of mice. J Pharmacol Exp Ther. 1993;267:1582–1590. [PubMed] [Google Scholar]
- Rasmussen E, Cipp L, Hitzemann R. Identification of quantitative trait loci for haloperidol-induced catalepsy on mouse chromosome 14. J Pharmacol Exp Ther. 1999;290:1337–1346. [PubMed] [Google Scholar]
- Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- Severson JA, Randall PK, Finch CE. Genotypic influences on striatal dopaminergic regulation in mice. Brain Res. 1981;210:201–215. doi: 10.1016/0006-8993(81)90894-5. [DOI] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article 17. [DOI] [PubMed] [Google Scholar]
- Zhao W, Langfelder P, Fuller T, Dong J, Li A, Hovarth S. Weighted gene coexpression network analysis: state of the art. J Biopharm Stat. 2010;20:281–300. doi: 10.1080/10543400903572753. [DOI] [PubMed] [Google Scholar]
- Zhu J, Chu CT. Mitochondrial dysfunction in Parkinson's disease. J Alzheimers Dis. 2010;20(Suppl. 2):S325–334. doi: 10.3233/JAD-2010-100363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.