Abstract
An outbreak of avian influenza (AI) caused by a low-pathogenic H5N2 type A influenza virus began in Mexico in 1993 and several highly pathogenic strains of the virus emerged in 1994-1995. The highly pathogenic virus has not been reported since 1996, but the low-pathogenic virus remains endemic in Mexico and has spread to two adjacent countries, Guatemala and El Salvador. Measures implemented to control the outbreak and eradicate the virus in Mexico have included a widespread vaccination program in effect since 1995. Because this is the first case of long-term use of AI vaccines in poultry, the Mexican lineage virus presented us with a unique opportunity to examine the evolution of type A influenza virus circulating in poultry populations where there was elevated herd immunity due to maternal and active immunity. We analyzed the coding sequence of the HA1 subunit and the NS gene of 52 Mexican lineage viruses that were isolated between 1993 and 2002. Phylogenetic analysis indicated the presence of multiple sublineages of Mexican lineage isolates at the time vaccine was introduced. Further, most of the viruses isolated after the introduction of vaccine belonged to sublineages separate from the vaccine's sublineage. Serologic analysis using hemagglutination inhibition and virus neutralization tests showed major antigenic differences among isolates belonging to the different sublineages. Vaccine protection studies further confirmed the in vitro serologic results indicating that commercial vaccine was not able to prevent virus shedding when chickens were challenged with antigenically different isolates. These findings indicate that multilineage antigenic drift, which has not been observed in AI virus, is occurring in the Mexican lineage AI viruses and the persistence of the virus in the field is likely aided by its large antigenic difference from the vaccine strain.
Wild aquatic birds are believed to be the primordial reservoir of type A influenza viruses. In the natural host, avian influenza (AI) generally causes an asymptomatic infection, whereas in aberrant hosts, including poultry, swine, and humans, clinical disease is often observed. Type A influenza viruses infecting birds are grouped into two broad pathotypes, low-pathogenicity AI (LPAI) and high-pathogenicity AI (HPAI). The highly pathogenic (HP) form of AI is a “List A” disease according to the Office International des Epizooties (World Organization for Animal Health) and causes systemic disease resulting in high mortality. Although 15 hemagglutinin (HA) subtypes of AI viruses have been described, the HP phenotype has only been associated with some strains of the H5 or H7 HA subtype. LPAI viruses are more commonly isolated from poultry, and clinical signs generally range from asymptomatic infection to drops in egg production and mild respiratory disease, although some low-pathogenic (LP) strains can cause higher mortality, usually due to coinfection with secondary pathogens (32). However, some H5 and H7 subtype LPAI viruses can mutate to the HP form of the virus, and several mechanisms of mutation involved in the emergence of HPAI viruses from LP precursor AI virus have been documented (16, 18, 28). This differentiation of viral pathotypes affects influenza control policy. For control of HPAI, eradication by use of a stamping-out policy is typically employed, although financial constraints in some countries preclude this approach. Responses to LPAI in poultry vary from taking no action against the outbreak to active eradication programs, including the use of quarantines, vaccines, and depopulation of infected flocks. Though vaccines have been used as part of a control program against sporadic outbreaks of LPAI in the United States, their use has been limited for several reasons. Efficacy tests using an LPAI challenge have not been fully standardized. Further, the use of AI vaccines to control LPAI may precipitate poultry embargos by trading partners. However, the use of vaccination to help control AI is gaining increased support and more vaccine is being used worldwide (12).
It is well documented that human influenza virus undergoes frequent antigenic drift, which is the accumulation of point mutations in the antigenic domain of the HA protein (3). As a result, viruses with a slightly changed antigenic structure emerge and can escape the host's acquired immunity, whether this immunity is acquired by natural infection or vaccination. Therefore, to maintain optimal protection by vaccination, the presently prevailing strains of influenza virus need to be included in each year's influenza vaccine, requiring yearly reevaluation and frequent changes to the vaccine formulation (1, 6). In contrast, broad subtype-specific immunity to HPAI has been observed following parenteral AI vaccination in chickens (30, 31). In the United States, H7N2 LPAI viruses have been circulating in live-bird markets since 1994 and progressive genetic drift in the HA gene has also been observed (23, 25). However, an existing commercial AI vaccine prepared from a 1997 seed stock H7N2 virus was able to provide protection against an H7N2 virus isolated in 2002 from a turkey in Virginia (35). In another study, which used an HPAI challenge, vaccine strains with HA protein sequences showing as little as 86% similarity to that of the challenge strain still provided good protection in chickens (29). One possible difference between influenza viruses infecting humans and poultry might relate to the different life spans of the hosts. Humans have multiple exposures to influenza viruses, either through natural infection or vaccination, and have a level of population immunity, but because of their short productive lives, poultry are usually naïve to influenza virus, particularly since vaccination for influenza remains uncommon. This likely results in different types of selection pressure on the virus in each host and may partially explain the broader protection provided by AI vaccine in chickens. In this context, long-term use of AI vaccine in Mexico and adjacent countries presents us with a unique opportunity to examine the evolution of AI virus in the presence of vaccine pressure.
The first suspected cases of AI in poultry in Mexico were detected in late 1993, and LPAI (H5N2) virus was first identified in May 1994. By the time influenza was confirmed in Mexican poultry, it had already spread widely, and eventually the LPAI virus mutated to the HPAI (H5N2) form of the virus in two different regions in Mexico, Puebla and Queretaro (36). The Mexican government started using vaccination in 1995 to aid in the control of both HPAI and LPAI viruses. Over 1 billion doses of inactivated emulsified vaccine have been authorized for use from 1995 to 2001, and 459 million doses of recombinant fowl pox-vectored AI vaccine have been authorized from 1998 to 2001 (37). Though HPAI viruses have not been reported since 1996, LPAI virus continues to circulate in Mexico, and genetically related viruses have also circulated in Guatemala and El Salvador since at least 2000 and 2001, respectively (27). Though some of the earlier Mexican H5N2 isolates have been characterized previously (7, 8, 14), there has been no further characterization of field isolates after the introduction of the vaccine. Thus, it was speculated that antigenic drift might have occurred in this rapidly changing RNA virus. In the present study, we gathered 52 Mexican lineage H5N2 viruses isolated from 1993 through 2002 and conducted molecular and antigenic analysis. Our study demonstrates that the Mexican lineage H5N2 viruses have undergone antigenic drift away from the vaccine strain and highlights the effect of vaccine use on the evolution of AI virus.
MATERIALS AND METHODS
Viruses.
The viral isolates used in this study (Table 1) were obtained from the National Veterinary Services Laboratories (NVSL), Animal and Plant Health Inspection Service, U.S. Department of Agriculture, Ames, Iowa. Viruses were received in allantoic fluid after passage in embryonating chicken eggs (ECE). The isolates were passaged one or two additional times at the Southeast Poultry Research Laboratory (SEPRL) to make working stocks of the virus.
TABLE 1.
Mexican lineage H5N2 AI virus isolates analyzed in this study
| Isolate | Pathotype | HA cleavage site sequence | Genotypeb | HI titerc |
|---|---|---|---|---|
| CK/Mexico/232/94 (commercial vaccine) | LP | RETR/G | Jalisco | — |
| CK/Hidalgo/232/94 | LP | RETR/G | Jalisco | 640 |
| CK/Mexico/31381-7/94 (1993)a | LP | RETR/G | Jalisco | 256 |
| CK/Mexico/31381-1/94 | LP | RETR/G | Jalisco | 320 |
| CK/Mexico/31381-2/94 | LP | RETR/G | Jalisco | 256 |
| CK/Mexico/31381-3/94 | LP | RETR/G | Jalisco | 320 |
| CK/Mexico/31381-4/94 | LP | RETR/G | Jalisco | 640 |
| CK/Mexico/31381-5/94 | LP | RETR/G | Jalisco | 640 |
| CK/Mexico/31381-6/94 | LP | RETR/G | Jalisco | 320 |
| CK/Mexico/31381-8/94 | LP | RETR/G | Jalisco | 512 |
| CK/Mexico/31382-1/94 | LP | RETR/G | Jalisco | 640 |
| CK/Mexico/26654-1374/94 | LP | RETR/G | Jalisco | 256 |
| CK/Queretaro/26654-1373/94 | LP | RETR/G | Jalisco | 256 |
| CK/Jalisco/14585-660/94 | LP | RETR/G | Jalisco | 640 |
| CK/Hidalgo/26654-1368/94 | LP | RETR/G | Jalisco | 320 |
| CK/Queretaro/7653-20/95 | HP | RKRKRKTR/G | Jalisco | 320 |
| CK/Queretaro/14588-19/95 | HP | RKRKTR/G | Jalisco | 320 |
| CK/Queretaro/22019-853/96 (1995)a | HP | RKRKTR/G | Jalisco | 320 |
| CK/Guanajuato/28159-331/95 | LP | RETR/G | Jalisco | 256 |
| CK/Vera Cruz/28159-398/95 | LP | RETR/G | Jalisco | 256 |
| CK/Hidalgo/28159-460/95 | LP | RETR/G | Jalisco | 512 |
| CK/Chiapas/28159-488/95 | LP | RETR/G | Jalisco | 256 |
| CK/Michoacan/28159-530/95 | LP | RETR/G | Jalisco | 256 |
| CK/Morelos/28159-538/95 | LP | RETR/G | Jalisco | 256 |
| CK/Mexico/28159-541/95 | LP | RETR/G | Jalisco | 256 |
| CK/Jalisco/28159-600/95 | LP | RETR/G | Jalisco | 640 |
| CK/Mexico/15407/97 | LP | RETR/G | Jalisco | 512 |
| CK/Puebla/8623-607/94 | HP | RKRKTR/G | Puebla | 160 |
| CK/Puebla/8624-604/94 | HP | RKRKTR/G | Puebla | 160 |
| CK/Puebla/14585-622/94 | HP | RKRKTR/G | Puebla | 256 |
| CK/Puebla/14586-654/94 | HP | RKRKTR/G | Puebla | 160 |
| CK/Mexico/37821-771/96 | LP | RETR/G | A | 320 |
| CK/Chiapas/15406/97 | LP | KETR/G | A | 160 |
| CK/Chiapas/15408/97 | LP | KETR/G | A | 160 |
| CK/VeraCruz/232-6169/98 | LP | KETR/G | A | 80 |
| CK/Puebla/231-5284/98 | LP | KETR/G | A | 80 |
| CK/Morelos/FO22189/98 | LP | KETR/G | A | 128 |
| CK/Jalisco/229-4592/98 | LP | KETR/G | A | 128 |
| CK/Morelos/227-4353/98 | LP | KETR/G | A | 128 |
| CK/Aguascalientes/124-3705/98 | LP | KETR/G | A | 128 |
| CK/Puebla/28159-474/95 | LP | RETR/G | B | 512 |
| CK/Chiapas/15224/97 | LP | RETR/G | B | 128 |
| CK/Chiapas/15405/97 | LP | RETR/G | B | 128 |
| CK/FO/22066/98 | LP | RETR/G | B | 128 |
| CK/Tabasco/234-8289/98 | LP | RETR/G | B | 80 |
| CK/Guatemala/45511-1/00 | LP | RETR/G | B | 40 |
| CK/Guatemala/45511-2/00 | LP | RETR/G | B | 64 |
| CK/Guatemala/45511-3/00 | LP | RETR/G | B | 40 |
| CK/Guatemala/45511-4/00 | LP | RETR/G | B | 64 |
| CK/Guatemala/45511-5/00 | LP | RETR/G | B | 64 |
| CK/El Salvador/102711-1/01 | LP | RETR/G | B | 40 |
| CK/El Salvador/102711-2/01 | LP | RETR/G | B | 32 |
| CK/Guatemala/194573/02 | LP | RETR/G | B | 40 |
Year in parentheses indicates the original year when the sample was collected.
Genotyping is based on the phylogenetic tree of the HA1 gene as shown in Fig. 1.
The HI test was conducted with sera obtained from chickens vaccinated with commercial vaccine. —, not determined.
The commercially available inactivated vaccines in Mexico all use the same seed strain provided by the Mexican government, designated A/CK/Mexico/232/94. The vaccine seed stock we received from NVSL was designated A/CK/Hidalgo/232/94. Because of this nomenclature discrepancy, we sequenced the HA1 subunit of the HA gene, the entire nonstructural (NS) gene, and the 5′ 741 bp of the neuraminidase gene from a commercial oil emulsion vaccine and compared these with the sequences from A/CK/Hidalgo/232/94. These strains were found to be similar, with only five nucleotide changes that resulted in two amino acid changes at positions 169 and 195, which are not associated with antigenic epitopes in HA1, one synonymous change in the NS gene, and no changes in the neuraminidase gene. Based on this close similarity, the commercial vaccine was used to vaccinate birds and produce antisera and A/CK/Hidalgo/232/94 was used as the antigen in hemagglutination inhibition (HI) and virus neutralization (VN) tests and as a challenge virus for the in vivo vaccine protection study described below.
RNA extraction and RT-PCR.
Using an RNeasy mini kit (QIAGEN, Valencia, Calif.), viral RNA from infectious allantoic fluid from ECE was extracted according to a modified protocol as previously described (22). Standard reverse transcription (RT) PCR was carried out with a QIAGEN one-step RT-PCR kit with primers specific for influenza virus. The primer sequences and amplification conditions are available from the authors upon request. The PCR product was separated on an agarose gel by electrophoresis, and amplicons of the appropriate size were subsequently excised from the gel and extracted with a QIAGEN gel extraction kit.
Sequencing and phylogenetic analysis.
Direct sequencing was performed with a PRISM Ready Reaction DyeDeoxy Terminator Cycle Sequencing kit (Perkin-Elmer, Foster City, Calif.) run on a 3700 or 3730 automated sequencer (Perkin-Elmer). The nucleotide sequences were compared initially by using the Megalign program (DNASTAR, Madison, Wis.) with the Clustal V alignment algorithm. Pairwise sequence alignments were also performed in the Megalign program to determine nucleotide and amino acid sequence similarities. Phylogenetic comparisons of the aligned sequences for each gene segment were generated using the maximum parsimony method in a heuristic search using PAUP 4.0b10 software (Sinauer Associates, Inc., Sunderland, Mass.) (33).
Virus titration.
The infectious doses of viruses used for the challenge experiment and for vaccine preparation were determined using ECE as previously described (38). Tracheal swab samples obtained from vaccine experiments were titrated by quantitative real-time RT-PCR assay. In brief, swabs from individual birds were suspended in 1.5 ml of brain heart infusion medium (Difco, Detroit, Mich.). RNA was extracted from fluid containing tracheal swabs by using an RNeasy mini kit (QIAGEN), and quantitative real-time RT-PCR was performed with primers for type A influenza virus as described previously (16a). The 50% egg infective doses (EID50) of virus from the clinical samples were interpolated from the cycle thresholds by using standard curves generated from known amounts of control RNA (101.0 to 106.0 EID50/ml). The details of the procedure and the validation of the assay have been described previously (22).
Production of hyperimmune antisera.
Seven field isolates (Table 2) were selected based on the topology of the HA1 phylogenetic tree (Fig. 1), and antisera were prepared against these isolates and also from a commercial vaccine. The seven field isolates were grown in 10-day-old ECE, and the infectious allantoic fluid was pooled for each virus. Infective titers were determined prior to inactivation with 0.1% betapropiolactone (Sigma, St. Louis, Mo.), and the inactivated virus was used to produce an oil emulsion vaccine as previously described (24). Three-week-old specific-pathogen-free (SPF) chickens were inoculated subcutaneously with either the commercial vaccine or the inactivated oil emulsion vaccines made at SEPRL. The vaccine dose for the seven field viruses contained 107.0 to 108.0 EID50 per 0.5 ml before viral inactivation. Three weeks later, the chickens were booster vaccinated by the same route with the same amount of vaccine. Sera were harvested and inactivated at 56°C for 30 min before being used in the HI and VN procedures.
TABLE 2.
Genetic similarity of the Mexican lineage isolates
| Isolate | Nucleotide sequence similarity (%) to CK/Hidalgo/232/94 at region:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| HA1 (77-1072)a | NA (19-1390) | M (26-1011) | NS (23-873) | NP (13-531) | PA (13-511) | PB1 (13-489) | PB2 (13-701) | |
| CK/Mexico/31381-7/94 | 97.9 | 98.6 | 98.9 | 98.1 | 98.5 | 98.8 | 99.2 | 97.8 |
| CK/Puebla/8623-607/94 | 94.2 | 98.5 | 98.2 | 98.5 | 98.5 | 97.8 | 98.3 | 98.4 |
| CK/Aguascalientes/124-3705/98 | 94.6 | 98.0 | 97.4 | 97.3 | 96.0 | 98.2 | 98.1 | 97.1 |
| CK/Puebla/231-5284/98 | 94.4 | 97.4 | 96.6 | 97.1 | 96.3 | 97.8 | 98.5 | 96.7 |
| CK/Puebla/28159-474/95 | 97.5 | 98.2 | 98.5 | 98.9 | 98.7 | 99.4 | 99.4 | 98.1 |
| CK/El Salvador/102711-1/01 | 92.2 | 92.9 | 95.5 | 96.6 | 97.9 | 93.6 | 96.4 | 93.6 |
| CK/Guatemala/194573/02 | 93.2 | 93.1 | 95.4 | 97.1 | 97.5 | 93.6 | 96.6 | 94.3 |
Numbers in parentheses are the nucleotide positions of the regions compared.
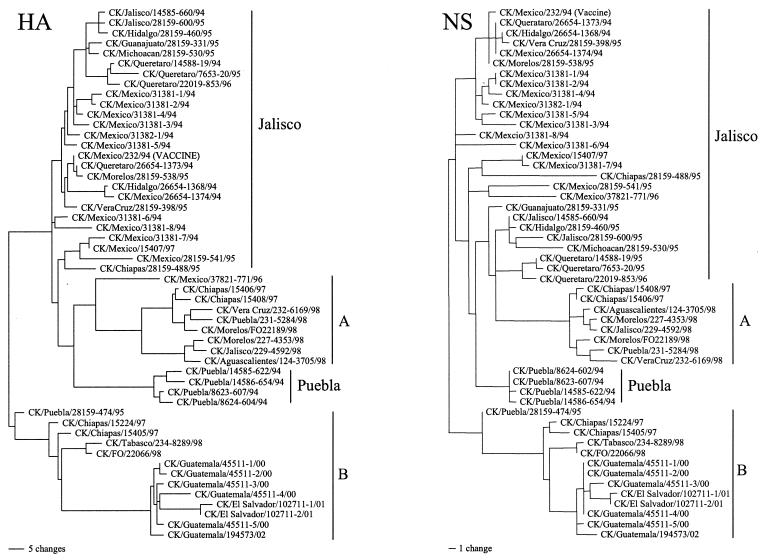
FIG. 1.
Phylogenetic tree based on nucleotide sequences of the HA1 and NS genes from Mexican lineage isolates. The tree, generated by the maximum parsimony method with PAUP 4.0b10, is the result of heuristic search and midpoint rooting.
HI and VN tests.
The HI test was performed as previously described (4). Briefly, titers were determined by using serial twofold dilutions of antisera, 4 HA units of homologous and heterologous antigen, and a 0.5% suspension of chicken erythrocytes per test well. To obtain more accurate HI titers (Table 1), we conducted the HI test with two different starting dilutions of 2 and 5.
The VN test was performed in ECE by using the diluted-serum constant-virus (beta) procedure (9). Briefly, antiserum was mixed with virus for 1 h at room temperature. This mixture was then inoculated into ECE. Three days postinoculation (dpi), allantoic fluid was examined for hemagglutinating activity to determine the presence of the virus. The Reed and Muench formula was used to calculate endpoint titers for homologous and heterologous neutralization (21). Antigenic relatedness values were then calculated using the Archetti and Horsfall formula (2). The isolates and the antisera used in the HI and VN tests are listed in Table 3.
TABLE 3.
Cross-HI and cross-VN test results expressed as percentages of antigenic relationshipa
| Cross-VN test isolate | Result with cross-HI test isolateb
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Vaccine | Mex/94 | Pue/94 | Agu/98 | Pue/98 | Pue/95 | El Sal/01 | Guate/02 | |
| Vaccine | 100 | 71 | 50 | 45 | 17 | 32 | <10 | 12 |
| CK/Mexico/31381-7/94 | 87 | 100 | 79 | 50 | 22 | 45 | 10 | 13 |
| CK/Puebla/8623-607/94 | 49 | 70.2 | 100 | 35 | 22 | 63 | 14 | 18 |
| CK/Aguascalientes/124-3705/98 | 26 | 12 | <10 | 100 | 50 | 25 | 32 | 31 |
| CK/Puebla/231-5284/98 | —c | — | — | — | 100 | 28 | 22 | 27 |
| CK/Puebla/28159-474/95 | — | — | — | — | — | 100 | 17 | 16 |
| CK/El Salvador/102711-1/01 | — | — | — | — | — | — | 100 | 71 |
| CK/Guatemala/194573/02 | 13 | <10 | <10 | 16 | — | — | — | 100 |
The Archetti and Horsfall formula was used to calculate the values (2).
Abbreviated isolate names are used. Full names appear at left.
—, test not done.
Study of vaccine protection against LPAI challenge.
Two-week-old SPF White Rock chickens obtained from flocks maintained at SEPRL were immunized subcutaneously with the commercial vaccine. Two weeks after vaccination, HI titers were determined and the birds were divided into three groups of 10 each, with similar levels of immunity based on HI antibody titers. Three groups of five unvaccinated birds were assigned as challenge controls. We also included one more group of five birds as negative controls. Birds were inoculated intranasally with one of three AI viruses (Table 4) at an infectious titer of 105.0 EID50 per 0.2 ml (dose). Tracheal swabs were collected at 3 and 5 dpi for virus titration.
TABLE 4.
Vaccine protection against LPAI challenge
| Challenge virusa | dpi | Virus titer in vaccinated birds (n = 10) with serum HI antibody titer ofb:
|
Average titer ± SD in unvaccinated birdsc | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 4 | 5 | 5 | 5 | 6 | 6 | 6 | 7 | 7 | |||
| Vaccine strain | 3 | 4.4 | 4.3 | 2.1 | — | — | 2.7 | 3.1 | — | — | — | 4.5 ± 0.3 |
| 5 | 3.2 | 3.3 | 1.3 | — | — | 2.0 | — | — | — | — | 3.1 ± 0.3 | |
| CK/AG/124-3705/98 | 3 | 4.2 | 4.6 | 4.7 | 4.5 | 5.1 | 3.9 | 4.4 | 5.0 | 3.9 | 4.1 | 4.2 ± 0.2 |
| 5 | 3.9 | 3.3 | 2.7 | 1.7 | — | 2.2 | 1.7 | — | 2.1 | 3.9 | 2.4 ± 0.4 | |
| CK/Guatemala/194573/02 | 3 | 5.3 | 5.3 | 4.8 | 5.2 | 4.7 | 4.7 | 4.5 | 4.8 | 4.6 | 4.7 | 4.9 ± 0.3 |
| 5 | 3.9 | 3.6 | 3.4 | 3.7 | 4.4 | 3.9 | 3.1 | 3.4 | 3.7 | 3.1 | 3.4 ± 0.7 | |
AG, Aguascalientes.
The HI titer is expressed as the log2 reciprocal of the endpoint in a twofold dilution of sera. The virus titer is expressed as the log10 EID50 per milliliter. —, no virus detected.
All five birds in this group were positive for virus. SD, standard deviation.
Study of in vivo cross-protection against HPAI challenge.
Two-week-old SPF White Leghorn chickens were immunized subcutaneously with one of three vaccines (Table 5). Birds were booster vaccinated 2 weeks after initial vaccination and intranasally challenged with 107.0 median embryo lethal doses of A/CK/Puebla/8623-607/94 virus per 0.2 ml 2 weeks after booster vaccination. Sera were collected before challenge, and the HI test was conducted with homologous antigen, which is the same virus as the one used for vaccination, and also with the A/CK/Puebla/8623-607/94 antigen. Samples for virus titration were collected at 3 dpi as described above and the chickens were observed daily for illness or death for 10 dpi.
TABLE 5.
Cross-protection against HPAI virus (CK/Puebla/8623/607/94) challenge
| Vaccine group | Postvaccination HI titer ± SDa
|
Postchallenge result
|
||||
|---|---|---|---|---|---|---|
| Homologous Ag | Puebla/94 Ag | Virus isolation (no. of birds positive/no. tested) | Virus titer ± SD (log10 EID50/ml) | Morbidityb | Mortality (no. of birds dead/no. challenged) | |
| CK/Puebla/8623-607/94 | 9.88 ± 1.36 | 9.88 ± 1.36 | 5/16 | 1.45 ± 0.85d | 0/16 | 0/16 |
| CK/Guatemala/194573/02 | 9.94 ± 1.24 | 6.75 ± 1.00 | 14/16 | 3.03 ± 0.91de | 0/16 | 0/16 |
| Commercial vaccine | 8.13 ± 1.36 | 6.94 ± 1.34 | 16/16 | 3.32 ± 0.81de | 0/16 | 0/16 |
| Unvaccinated control | 0 | 0 | 8/8 | 4.20 ± 0.43 | 8/8 | 7/8c |
The HI test was done with homologous antigen (Ag), which is the same virus as the one used for vaccination, and Puebla/94 Ag. The HI titer is expressed as the log2 reciprocal of the endpoint in a twofold dilution of sera.
Values indicate the number of birds showing typical AI clinical signs and the total number challenged.
The mean death time during 10 days of observation was day 6.6.
P < 0.05 compared with the result for the control group.
P < 0.05 compared with the result for the CK/Puebla/8623-607/94-vaccinated group.
Statistical analysis.
The statistical significance of the data was determined by using Student's t test. Results were considered to be statistically significant if the comparison gave a P value of <0.05.
GenBank accession numbers.
The nucleotide sequences presented in this article have been deposited in the GenBank database under accession numbers AY497063 to AY497191.
RESULTS
Viruses.
The viruses in this study were either previously pathotyped or pathotyped in this study using established procedures at the NVSL (7, 8). All of the isolates were of the predicted pathotype based on the HA cleavage site sequence except for one isolate, A/CK/Queretaro/22019-853/96. This isolate, originally isolated in 1995 but characterized in 1996, had multiple basic amino acids in the HA cleavage site consistent with an HPAI virus and also plaqued in chicken embryo fibroblast cells without trypsin, but it killed less than 75% (three of eight) of the birds in a standard intravenous challenge of chickens. The pathotypes of the individual viruses are given in Table 1.
Genetic analysis of H5 HA1.
The coding sequences for the HA1 gene segments from 34 isolates were newly determined. These include six isolates from Guatemala and two isolates from El Salvador. A total of 52 Mexican lineage viruses that were isolated between 1993 and 2002 were compared. The percentages of nucleotide and amino acid similarities among all HA1 sequences ranged from 86.0 to 99.8% and 84.6 to 99.7%, respectively. According to a phylogenetic tree based on the HA1 sequence, at least four distinct clusters of viruses were observed within the Mexican lineage isolates (Fig. 1). Viruses isolated before 1996 were mostly divided into the previously described Jalisco and Puebla sublineages (7), except one isolate, A/CK/Puebla/28159-474/95. The majority of the early isolates belonged to the Jalisco sublineage. However, several clusters of isolates were found within this Jalisco sublineage. Nine viruses isolated in Mexico after 1995 formed a separate clade from the Puebla and Jalisco sublineages and were designated as sublineage A. Viruses isolated in Guatemala and El Salvador were closely related to each other and were part of a fourth Mexican clade of viruses, including the early isolate A/CK/Puebla/28159-474/95, and were designated as sublineage B.
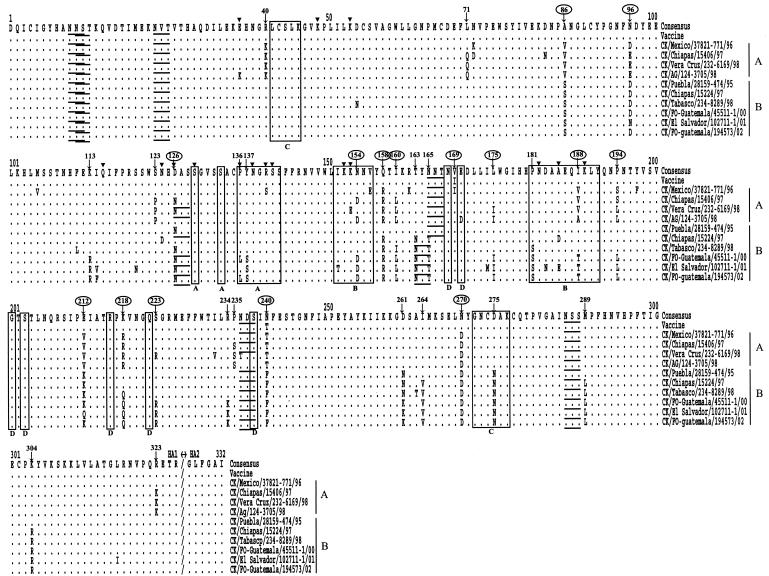
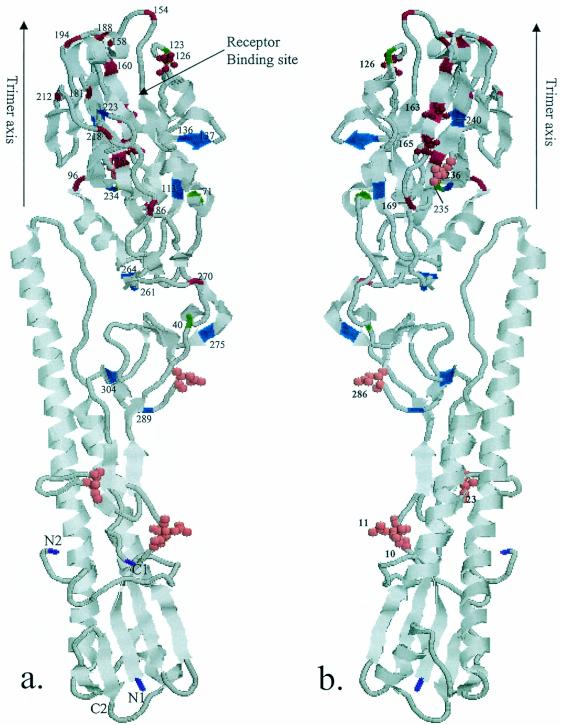
Since phylogenetic analysis demonstrated heterogeneity in the HA gene in the early isolates, we made the consensus sequence of the HA1 gene with 18 viruses isolated before 1995 (Table 1). The sequence of the vaccine strain showed only two amino acid differences from the consensus sequence (Fig. 2). To determine the mutations that may have occurred after the introduction of the vaccine, we compared the sequence of the vaccine strain with sequences from isolates belonging to two recent sublineages (Fig. 2). A total of 20 and 27 amino acid substitution sites, which indicated that mutations had become fixed in a progressive manner from earlier isolates to recent isolates, were detected in sublineages A and B, respectively. Fifteen substitution sites overlapped between the two sublineages. Six substitutions (at positions 136, 137, 154, 181, 188, and 275) occurred in regions previously proposed as antigenic sites A, B, and C in H1 and H3 structures (5, 40). Two amino acid changes (at positions 126 and 136) occurred in the site that was previously demonstrated in an H5 escape mutant (15, 19, 20). The amino acid change from D to N at position 126 resulted in the formation of a glycosylation site that was also observed in an H5 escape mutant. Though many substitutions occurred in regions or sites other than the previously proposed antigenic sites, the majority of changes were located in the distal globular head of HA1. Furthermore, most of the overlapping substitution sites between A and B sublineage viruses were located in the outer surface of the globular head of HA1. The locations of substitution sites are illustrated in the three-dimensional structure of the H5 HA (11) (Fig. 3).
FIG. 2.
Alignment of HA1 amino acid sequences of Mexican lineage isolates with that of the consensus sequence made with 18 early isolates. The underlined residues are potential glycosylation sites, and the residues in the open boxes are previously identified antigenic sites with H1 and H3 molecules. ▾, substitution sites demonstrated in an H5 escape mutant; ↓, amino acid substitution sites in sublineage A and B viruses analyzed in this study. Overlapping sites are circled.
FIG. 3.
Ribbon diagram of the monomer of H5 (A/Duck/Singapore/3/97) HA. Front (a) and back (b) views are shown. Location of amino acid changes in HA1 and potential glycosylation sites of Mexican lineage isolates are labeled. The color scheme is as follows: green, amino acid changes in sublineage A virus; blue, amino acid changes in sublineage B virus; red, overlapping amino acid changes in sublineage A and B viruses; pink, potential glycosylation sites; purple, termini of HA1 (N1 and C1) and HA2 (N2 and C2).
Eight potential glycosylation sites were identified at positions 10, 11, 23, 126, 163, 165, 236, and 286 (Fig. 2 and 3). Although there were some exceptions, the first three sites (at positions 10, 11, and 23) and the last two sites (at positions 236 and 286) were conserved in most of the isolates. As mentioned above, the glycosylation site at position 126 was observed in most of the isolates belonging to sublineages A and B but not in the earlier isolates, including the vaccine strain. In sublineage B viruses, the glycosylation site at position 165 was lost but an additional glycosylation site was observed at position 163. All sequences examined contained conserved sequences in the areas surrounding the proposed receptor-binding site, with the exception of a single amino acid substitution at position 133 (serine to alanine) in A/CK/Mexico/28159-541/95 and at position 223 (serine to arginine) in 10 isolates. The majority of the LP isolates had the RETR/G motif at the HA cleavage site (Table 1), but a single amino acid substitution was observed at the −4 position (arginine to lysine) for the HA1 sequence in isolates of sublineage A, resulting in an unusual KETR/G motif.
By comparing the consensus sequence with sequences of consecutive isolates in both sublineages, the rates of fixation of mutations were calculated to be 12 × 10−3 and 10 × 10−3 substitutions per nucleotide site per year for sublineages A and B, respectively. Evolutionary rates based on amino acid changes were calculated to be 4.2 × 10−3 and 4.0 × 10−3 substitutions per nucleotide site per year, respectively.
Genetic analysis of NS and remaining genes.
The coding sequence for the NS gene segments from 47 isolates were newly determined and a total of 52 Mexican lineage viruses were phylogenetically analyzed. All of the Mexican lineage isolates belonged to subtype B (group B) and assorted into a similar topology, as we observed in the HA tree. The substitution rates for sublineages A and B were calculated to be 6.0 × 10−3 and 12 × 10−3 substitutions per nucleotide site per year, respectively.
The remaining genes (N2, M, NP, PA, PB1, and PB2) of selected isolates from different sublineages were also sequenced and compared with other sequences available in GenBank. All Mexican lineage isolates sequenced were closely related (Table 2) to each other and no evidence of reassortment was observed. All of the isolates examined had stalk deletions in the N2 gene, which is thought to be a characteristic of chicken-adapted AI viruses (17).
One-way serologic test of vaccine antisera to the field isolates.
To determine the cross-reactivity of vaccine antisera to the viruses from different sublineages, the HI test was conducted with sera obtained from chickens vaccinated with commercial vaccine (Table 1). The results of the HI assay demonstrated that there is high cross-reactivity of vaccine antisera to the viruses belonging to the Jalisco sublineage, which was the source of the seed strain for the vaccine. Compared to the HI titer of the sera to the homologous vaccine strain, the HI titers against most of the Jalisco sublineage isolates were similar or within a fourfold difference. The HPAI viruses belonging to the Puebla sublineage demonstrated a fourfold loss in HI activity. In contrast, more than a fourfold difference was observed with several sublineage A viruses, and the sublineage B viruses had as much as a 16-fold difference in HI activity compared to the vaccine strain.
Cross-HI and -VN tests.
To further determine the antigenic relatedness among isolates belonging to different sublineages, eight isolates were selected on the basis of phylogenetic and one-way serologic data. We then conducted cross-HI tests with hyperimmune sera produced in chickens against representative isolates of the different sublineages. The percentage of antigenic relatedness (R value) was then determined with the Archetti and Horsfall method (2) by calculation of homologous and heterologous titer ratios (Table 3). In general, serologically related isolates have R values of more than 50%. The vaccine strain shared more than 50% relatedness only with isolates belonging to the Jalisco and Puebla sublineages. Isolate A/CK/Puebla/28159-474/95, which showed high cross-reactivity with vaccine sera in the one-way HI test, demonstrated only 32% relatedness to the vaccine strain in the cross-HI test. Isolates from Guatemala and El Salvador were antigenically close to each other (71%) but shared very low antigenic relatedness to any of the other isolates examined (less than 32%). To verify these results, we conducted VN tests with five isolates used in the HI test and found results similar to those of the HI tests (Table 3). In general, the VN test was more discriminative than the HI test and hyperimmune sera from vaccine-immunized chickens did not effectively neutralize the sublineage A and B viruses. Therefore, it can be concluded that Jalisco and Puebla sublineage viruses are antigenically similar to the vaccine strain, whereas sublineage A and B viruses are antigenically different from the vaccine strain and each other.
Efficacy of the commercial vaccine against LPAI virus challenge.
To compare the efficacies of the present vaccine against antigenically different viruses, birds were challenged with two viruses from recent sublineages after a single vaccination with commercial vaccine. The vaccine strain was also used for a challenge comparison. Three groups of unvaccinated birds were also challenged with each virus for comparison. The efficacies of the vaccine were compared in terms of the ability to prevent virus replication in the trachea, and the results are summarized in Table 4. In birds challenged with the vaccine strain, large amounts of virus were detected only in the birds that showed low HI titers (1:16 in twofold serial dilution). In most of the birds that had HI titers greater than 32, the vaccine prevented detectable virus shedding, with only a few birds shedding small amounts of virus that were about 2 logs lower than virus detected from unvaccinated and challenged control birds. In contrast, large amounts of virus, comparable to the amount of virus detected in unvaccinated challenged birds, were detected in birds challenged with two recent viruses and the amount of virus shedding was independent of the HI antibody titer of the birds.
Cross-protection against HPAI virus challenge.
To evaluate the effect of the antigenic difference between the vaccine and challenge strains in the protection of chickens against HPAI challenge, three groups of birds were vaccinated with strains that show various degrees of antigenic relatedness to the challenge virus (Table 5). Among birds vaccinated with a homologous strain as the challenge virus, only 5 of 16 shed low amounts of virus (mean titer of 101.45 EID50/ml) at 3 dpi. In contrast, all birds vaccinated with commercial vaccine and most birds (14 of 16) vaccinated with a Guatemala isolate shed higher amounts of virus (mean titers of 103.03 and 103.32 EID50/ml, respectively) from the trachea. The amount of viral shed was approximately 1 log lower than that observed in the unvaccinated and challenged control birds. However, no clinical signs were observed in any of the vaccinated birds and the birds remained healthy during the 10-day observation period. All unvaccinated and challenged control birds demonstrated typical HPAI clinical signs, such as edema of the head and face, cyanosis of unfeathered skin, etc., and seven of eight birds died, with a mean time to death of 6.6 days.
DISCUSSION
The HA glycoprotein of human influenza A virus undergoes antigenic drift that allows the virus to incrementally escape the host's adaptive immunity and requires frequent changes in vaccine formulation for humans (1, 6). In general, when the predicted circulating strain of virus elicits an antibody titer with a fourfold loss in cross-neutralization activity to the present vaccine, then the vaccine is changed. In contrast, AI viruses have not been under constant pressure of vaccines and avian HA subtypes appeared to retain a relatively stable antigenic structure (39). However, vaccine use for poultry has increased, since it is being considered as a more cost-effective approach for control of AI than the traditional stamping-out policy (12). Mexico is one of the first countries to use vaccination to control and eradicate LP as well as HP H5N2 AI viruses in poultry. Though HPAI outbreak has not been reported after the introduction of inactivated vaccine, the Mexican lineage LPAI virus continues to circulate in Mexico and adjacent countries despite 9 years of use of inactivated and fowl pox-vectored vaccines (37). This observation prompted us to address the important question of whether antigenic drift, which has not been observed in AI virus, occurs with the presence of vaccine pressure. Phylogenetic analysis of the HA and NS genes of Mexican lineage AI virus demonstrated the heterogeneity of the early isolates before vaccine was introduced (Fig. 1). In addition to the previously described Puebla and Jalisco sublineage viruses, we found one early isolate (A/CK/Puebla/28159-474/95) that seemed to be the progenitor strain of the new B sublineage which includes isolates from Guatemala and El Salvador. The phylogenetic analysis suggested that the Puebla and Jalisco sublineages have disappeared and have been replaced with new sublineages, A and B. The topology of the tree (Fig. 1) and the pattern of fixation of mutations (Fig. 2) indicate that sublineage A evolved from the Jalisco sublineage. The mutation rates estimated by using linear regression analysis for the A and B lineage viruses were 12 × 10−3 and 10 × 10−3 substitutions per nucleotide site per year, respectively. This mutation rate is higher than those reported for H5 and H7 AI viruses from live-bird markets in the United States, which showed 7.8 × 10−3 and 4.9 × 10−3 substitutions per nucleotide site per year, respectively (23, 26). These results suggest that Mexican lineage viruses are undergoing increased genetic drift that is most likely due to vaccination pressure. However, other variables may be involved, including the fact that more birds are infected yearly in Mexico than in the live-bird markets in the United States, which increases the number of mutations that can be fixed in the population.
In one-way HI tests, we observed high cross-reactivity of vaccine antisera to viruses belonging to the Jalisco sublineage, which includes the vaccine seed strain (Table 1). In contrast, differences of more than fourfold were observed between sublineage A and B viruses. This antigenic difference was further confirmed in cross-HI and cross-VN tests, in which sublineage A and B viruses showed less than 50% antigenic relatedness to the vaccine strain (Table 3). In contrast, HP virus from the Puebla lineage was about 50% related to the vaccine strain and this may partially explain the early disappearance of Puebla lineage HP virus from Mexico after vaccination was introduced. From the above mentioned genetic and serologic findings, it is likely that sublineage A viruses are undergoing antigenic drift away from the vaccine strain, though additional analysis with more recent isolates should be conducted to confirm this conclusion. The sublineage B viruses, possibly originated from A/CK/Puebla/28159-474/95, may have a selective advantage due to their antigenic differences from the vaccine strain and are also drifting further away from the vaccine strain.
In general, in vitro serologic tests assessing similarity are more discriminating than in vivo cross-protection studies (10, 13). Thus, we conducted a vaccine protection test to further determine the antigenic relationship in vivo as well as to assess the efficacies of the presently available commercial vaccine against two recent viruses. The in vivo test further confirmed the serologic data indicating that the commercial vaccine tested was not able to prevent virus shedding when chickens were challenged with antigenically different field isolates (Table 4). Though clinical signs were not observed with these LP viruses in experimental conditions, the shedding of H5 subtype LP viruses is of concern because of the potential for transmission to naïve flocks and the possibility of mutation to the HP form of the virus (7, 14, 16, 18).
Previous studies have demonstrated broad cross-protection against HPAI virus in terms of morbidity and mortality (30, 31). Our study confirms the previous finding that chickens can be protected from clinical disease resulting from HPAI challenge even with a high degree of antigenic difference (Table 5). However, the level of virus shedding in the trachea correlated with the antigenic differences of vaccine and challenge strains. This discrepancy likely reflects the fact that vaccines for AI do not prevent infection but that antibodies in the bloodstream may effectively prevent the systemic phase of disease caused by HPAI viruses.
The H1 and H3 subtype viruses from humans comprise a variety of HA antigenic drift variants, and the antigenic epitopes have been well defined within the H3 molecule's three-dimensional structure (5, 40). In contrast, avian HA subtypes retain a relatively stable antigenic structure, and selection and characterization of monoclonal antibody escape mutants has been used as the only means of identifying antigenic epitopes (15, 19, 20). In this study, we mapped the amino acid changes on the three-dimensional structure of the H5 HA molecule (11) and most of the amino acid substitutions were found in the membrane distal globular head of HA1 (Fig. 3). Between sublineages A and B, we found 15 overlapping substitution sites that were located in the outer surface of the globular head of HA1. Compared to what has been reported previously, six substitutions were found to occur in regions previously proposed as antigenic sites A, B, and C in H1 and H3 structures and two amino acid changes occurred in sites that were previously demonstrated with H5 monoclonal antibody escape mutants (Fig. 2). Though several substitutions occurred in sites other than that previously identified with H5 escape mutants, it is possible that the choice of monoclonal antibodies could have limited the diversity of the selected escape mutants in previous studies. The amino acid change of aspartic acid to asparagine at position 126 that resulted in the formation of a glycosylation site is of interest because glycosylation of viral antigens can mask antigenic epitopes and therefore is an important process in the generation of new viruses. Interestingly, this acquisition of glycosylation was also observed in an H5 escape mutant and in H2 subtype escape mutants at equivalent positions (15, 34). It is plausible that the substitution sites described here may play an important role in drift variation in the field with constant vaccine pressure, and characterization of other H5 subtype antigenic variants will further elucidate the importance of individual substitution sites.
In this study, we demonstrate that presently prevailing Mexican lineage AI viruses, which belong to two different sublineages, are antigenically different from the vaccine strain. Although circumstantial evidence supports the idea that vaccine pressure is driving this phenomenon, other possibilities for explaining the antigenic drift cannot be ruled out. Whatever the cause, the antigenic drift of H5N2 Mexican lineage viruses away from the vaccine strain is of concern for the effectiveness of vaccination strategies within the poultry industry. Vaccination is used not only to prevent clinical disease but also to reduce viral shedding. A reduction in viral shedding reduces the chance of virus spreading from infected vaccinated flocks to uninfected flocks. This is the principle behind barrier or “ring” vaccination around a quarantine zone during an outbreak, which can be a valuable control tool. Vaccination, however, cannot be used alone for the control of AI and must be accompanied by other control measures, including quarantines, controlled depopulation (taking birds to slaughter facilities under tight controls), and increased surveillance. The almost total failure of the present vaccine strain to prevent viral shedding when birds were challenged with recent field isolates was expected because of the extreme difference in cross-HI and -VN activities and greatly reduces the value of vaccination as part of a control and eradication program. Our in vitro and in vivo findings indicate that to maintain optimal protection with our AI vaccines, we should evaluate how well the prevailing isolates match the present AI vaccine formulation and update the vaccine by using criteria similar to those used for human influenza vaccines (1, 6)
Acknowledgments
This work was supported by the U.S. Department of Agriculture (ARS CRIS project 6612-32000-039).
We thank Suzanne Deblois, Joan Beck, and the SAA sequencing facility for technical assistance and Roger Brock for animal care assistance. We also thank Erica Spackman, David Swayne, and Terrence Tumpey for critical reviews of the manuscript.
REFERENCES
- 1.Ada, G. L., and P. D. Jones. 1986. The immune response to influenza infection. Curr. Top. Microbiol. Immunol. 128:1-54. [DOI] [PubMed] [Google Scholar]
- 2.Archetti, I., and F. L. Horsfall. 1950. Persistent antigenic variation of influenza A viruses after incomplete neutralization in ovo with heterologous immune serum. J. Exp. Med. 92:441-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bean, W. J., M. Schell, J. Katz, Y. Kawaoka, C. Naeve, O. Gorman, and R. G. Webster. 1992. Evolution of the H3 influenza virus hemagglutinin from human and nonhuman hosts. J. Virol. 66:1129-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beard, C. W. 1989. Serological procedures, p. 192-200. In H. G. Purchase, L. H. Arp, C. H. Domermuth, J. E. Pearson (ed.), A laboratory manual for the isolation and identification of avian pathotypes. Kendall-Hunt Publishing, Dubuque, Iowa.
- 5.Caton, A. J., G. G. Brownlee, J. M. Yewdell, and W. Gerhard. 1982. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31:417-427. [DOI] [PubMed] [Google Scholar]
- 6.Couch, R. B., and J. A. Kasel. 1983. Immunity to influenza in man. Annu. Rev. Microbiol. 37:529-549. [DOI] [PubMed] [Google Scholar]
- 7.Garcia, M., J. M. Crawford, J. W. Latimer, E. Rivera-Cruz, and M. L. Perdue. 1996. Heterogeneity in the haemagglutinin gene and emergence of the highly pathogenic phenotype among recent H5N2 avian influenza viruses from Mexico. J. Gen. Virol. 77:1493-1504. [DOI] [PubMed] [Google Scholar]
- 8.Garcia, M., D. L. Suarez, J. M. Crawford, J. W. Latimer, R. D. Slemons, D. E. Swayne, and M. L. Perdue. 1997. Evolution of H5 subtype avian influenza A viruses in North America. Virus Res. 51:115-124. [DOI] [PubMed] [Google Scholar]
- 9.Gelb, J., Jr., and M. W. Jackwood. 1998. Infectious bronchitis, p. 169-173. In D. E. Swayne (ed.), A laboratory manual for the isolation and identification of avian pathogens, 4th ed. American Association of Avian Pathologists, Kennett Square, Pa.
- 10.Gelb, J., Jr., C. L. Keeler, Jr., W. A. Nix, J. K. Rosenberger, and S. S. Cloud. 1997. Antigenic and S-1 genomic characterization of the Delaware variant serotypes of infectious bronchitis virus. Avian Dis. 41:661-669. [PubMed] [Google Scholar]
- 11.Ha, Y., D. J. Stevens, J. J. Skehel, and D. C. Wiley. 2001. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc. Natl. Acad. Sci. USA 98:11181-11186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halvorson, D. A. 2002. The control of H5 or H7 mildly pathogenic avian influenza: a role for inactivated vaccine. Avian Pathol. 31:5-12. [DOI] [PubMed] [Google Scholar]
- 13.Hofstad, M. S. 1961. Antigenic and immunological studies of several isolates of avian infectious bronchitis virus. Avian Dis. 5:102-107. [Google Scholar]
- 14.Horimoto, T., E. Rivera, J. Pearson, D. Senne, S. Krauss, Y. Kawaoka, and R. G. Webster. 1995. Origin and molecular changes associated with emergence of a highly pathogenic H5N2 influenza virus in Mexico. Virology 213:223-230. [DOI] [PubMed] [Google Scholar]
- 15.Kaverin, N. V., I. A. Rudneva, N. A. Ilyushina, N. L. Varich, A. S. Lipatov, Y. A. Smirnov, E. A. Govorkova, A. K. Gitelman, D. K. Lvov, and R. G. Webster. 2002. Structure of antigenic sites on the haemagglutinin molecule of H5 avian influenza virus and phenotypic variation of escape mutants. J. Gen. Virol. 83:2497-2505. [DOI] [PubMed] [Google Scholar]
- 16.Kawaoka, Y., C. W. Naeve, and R. G. Webster. 1984. Is virulence of H5N2 influenza viruses in chickens associated with loss of carbohydrate from the hemagglutinin? Virology 139:303-316. [DOI] [PubMed] [Google Scholar]
- 16a.Lee, C. W., and D. L. Suarez. 2004. Application of real-time RT-PCR for the quantification and competitive replication study of H5 and H7 subtype avian influenza virus. J. Virol. Methods 119:151-158. [DOI] [PubMed] [Google Scholar]
- 17.Matrosovich, M., N. Zhou, Y. Kawaoka, R. Webster. 1999. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J. Virol. 73:1146-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perdue, M. L., M. Garcia, J. Beck, M. Brugh, and D. Swayne. 1996. An Arg-Lys insertion at the hemagglutinin cleavage site of an H5N2 avian influenza isolate. Virus Genes 12:77-84. [DOI] [PubMed] [Google Scholar]
- 19.Philpott, M., B. C. Easterday, and V. S. Hinshaw. 1989. Neutralizing epitopes of the H5 hemagglutinin from a virulent avian influenza virus and their relationship to pathogenicity. J. Virol. 63:3453-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Philpott, M., C. Hioe, M. Sheerar, and V. S. Hinshaw. 1990. Hemagglutinin mutations related to attenuation and altered cell tropism of a virulent avian influenza A virus. J. Virol. 64:2941-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reed, L. J., and H. Muench. 1938. A simple method for estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 22.Spackman, E., D. A. Senne, T. J. Myers, L. L. Bulaga, L. P. Garber, M. L. Perdue, K. Lohman, L. T. Daum, and D. L. Suarez. 2002. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 40:3256-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spackman, E., D. A. Senne, S. Davison, and D. L. Suarez. 2003. Sequence analysis of recent H7 avian influenza viruses associated with three different outbreaks in commercial poultry in the United States. J. Virol. 77:13399-13402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stone, H., B. Mitchell, and M. Brugh. 1997. In ovo vaccination of chicken embryos with experimental Newcastle disease and avian influenza oil-emulsion vaccines. Avian Dis. 41:856-863. [PubMed] [Google Scholar]
- 25.Suarez, D. L., M. Garcia, J. Latimer, D. Senne, and M Perdue. 1999. Phylogenetic analysis of H7 avian influenza viruses isolated from the live bird markets of the Northeast United States. J. Virol. 73:3567-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suarez, D. L., and D. A. Senne. 2000. Sequence analysis of related low-pathogenic and highly pathogenic H5N2 avian influenza isolates from United States live bird markets and poultry farms from 1983 to 1989. Avian Dis. 44:356-364. [PubMed] [Google Scholar]
- 27.Suarez, D. L., E. Spackman, and D. A. Senne. 2003. Update on molecular epidemiology of H1, H5, and H7 influenza virus infections in poultry in North America. Avian Dis. 47:888-897. [DOI] [PubMed] [Google Scholar]
- 28.Suarez, D. L., D. A. Senne, J. Banks, I. H. Brown, S. C. Essen, C. W. Lee, R. J. Manvell, C. Mathieu-Benson, V. Moreno, J. Pedersen, B. Panigrahy, H. Rojas, E. Spackman, and D. J. Alexander. 2004. Recombination resulting in virulence shift in avian influenza outbreak, Chile. Emerg. Infect. Dis. 10:693-699. [DOI] [PMC free article] [PubMed]
- 29.Swayne, D. E., J. R. Beck, M. Garcia, and H. D. Stone. 1999. Influence of virus strain and antigen mass on efficacy of H5 avian influenza inactivated vaccines. Avian Pathol. 28:245-255. [DOI] [PubMed] [Google Scholar]
- 30.Swayne, D. E., M. L. Perdue, J. R. Beck, M. Garcia, and D. L. Suarez. 2000. Vaccines protect chickens against H5 highly pathogenic avian influenza in the face of genetic changes in field viruses over multiple years. Vet. Microbiol. 74:165-172. [DOI] [PubMed] [Google Scholar]
- 31.Swayne, D. E., J. R. Beck, M. L. Perdue, and C. W. Beard. 2001. Efficacy of vaccines in chickens against highly pathogenic Hong Kong H5N1 avian influenza. Avian Dis. 45:355-365. [PubMed] [Google Scholar]
- 32.Swayne, D. E., and D. A. Halvorson. 2003. Influenza, p. 135-160. In Y. M. Saif, H. J. Barnes, J. R. Glisson, A. M. Fadly, L. R. McDougald, and D. E. Swayne (ed.), Diseases of poultry. Iowa State University Press, Ames.
- 33.Swofford, D. L. 1998. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 34.Tsuchiya, E., K. Sugawara, S. Hongo, Y. Matsuzaki, Y. Muraki, Z. N. Li, and K. Nakamura. 2001. Antigenic structure of the haemagglutinin of human influenza A/H2N2 virus. J. Gen. Virol. 82:2475-2484. [DOI] [PubMed] [Google Scholar]
- 35.Tumpey, T. M., D. R. Kapczynski, and D. E. Swayne. 2004. Comparative susceptibility of chickens and turkeys to avian influenza A H7N2 virus infection and protective efficacy of a commercial avian influenza H7N2 virus vaccine. Avian Dis. 48:167-176. [DOI] [PubMed] [Google Scholar]
- 36.Villareal-Chavez, C. L., and A. O. Flores. 1998. The Mexican avian influenza (H5N2) outbreak, p. 18-22. In D. E. Swayne and R. D. Slemons (ed.), Proceedings of the Fourth International Symposium on Avian Influenza. United States Animal Health Association, Richmond, Va.
- 37.Villarreal-Chavez, C., and E. Rivera-Cruz. 2003. An update on avian influenza in Mexico. Avian Dis. 47:1002-1005. [DOI] [PubMed] [Google Scholar]
- 38.Villegas, P. 1998. Titration of biological suspensions, p. 248-254. In D. E. Swayne (ed.), A laboratory manual for the isolation and identification of avian pathogens. American Association of Avian Pathologists, Kennett Square, Pa.
- 39.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Mol. Biol. Rev. 56:152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiley, D. C., I. A. Wilson, and J. J. Skehel. 1981. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289:373-378. [DOI] [PubMed] [Google Scholar]