Abstract
Despite recent successes in generating subgenomic RNA replicons derived from genotype 1b strains of hepatitis C virus (HCV) that replicate efficiently in cultured cells, it has proven difficult to generate efficiently replicating RNAs from any other genotype of HCV. This includes genotype 1a, even though it is closely related to genotype 1b. We show here that an important restriction to replication of the genotype 1a H77c strain RNA in normal Huh7 cells resides within the amino-terminal 75 residues of the NS3 protease. We identified adaptive mutations located within this NS3 domain and within NS4A, in close proximity to the essential protease cofactor sequence, that act cooperative to substantially enhance the replication of this genotype 1a RNA in Huh7 cells. These and additional adaptive mutations, identified through a series of iterative transfections and the selection of G418-resistant cell clones, form two groups associating with distinct nonstructural protein domains: the NS3/4A protease and NS5A. A combination of mutations from both groups led to robust replication of otherwise unmodified H77c genomic RNA that was readily detectable by northern analysis within 4 days of transfection into Huh7 cells. We speculate that these adaptive mutations favorably influence assembly of the replicase complex with host cell-specific proteins, or alternatively promote interactions of NS3/4A and/or NS5A with cellular proteins involved in host cell antiviral defenses.
Persistent infection with hepatitis C virus (HCV) is a common cause of chronic liver disease, including chronic hepatitis, cirrhosis, and hepatocellular carcinoma (1, 21). The public health impact of the infection is magnified by the fact that currently available therapy, consisting of alpha interferon typically used in combination with ribavirin, is limited in its ability to provide a sustained response against the most-prevalent genotypes of HCV, 1a and 1b (9, 22). The development of selectable subgenomic genotype 1b HCV replicons and genome length RNAs has provided new opportunities for drug discovery efforts, for assessing in vitro the potential of candidate antiviral agents to inhibit HCV replication, and for studying mechanisms of HCV RNA replication. These RNAs express selectable antibiotic resistance markers and demonstrate a robust replication phenotype in Huh7 cells that were originally derived from a human hepatoma (20). Efficient replication in these cells is often but not always associated with the selection of specific adaptive mutations (3, 14, 15, 19). Some HCV RNAs have been further adapted to growth in HeLa cells and even cells of murine origin (31).
Thus far, however, only RNAs derived from genotype 1b strains of HCV have been adapted to highly efficient replication in Huh7 cells. Efforts to develop replicons derived from other genotypes have generally been met with only limited success. Considering the high genetic variability that is evident among different strains of HCV, the development of replicons from other genotypes is critically needed for drug discovery and in vitro validation of the antiviral activity of novel compounds across a range of viral genotypes (6, 7, 23). In support of this notion, a chimeric replicon containing a genotype 1a polymerase in the background of a 1b RNA was recently found to be less susceptible to alpha interferon treatment in vitro than the parental replicon from which it was derived (12). Moreover, clinical data presented recently suggest that the investigational NS3/4A protease inhibitor BILN 2061 has significantly less antiviral activity against the proteases of non-genotype 1 viruses, compared with genotype 1a and 1b strains of HCV (16). The development of replicons derived from other genotypes would also enable research directed at understanding basic mechanisms of viral RNA replication and the contribution of genetically variable sequences to that process.
Recently, two groups reported the generation of genotype 1a replicons using highly permissive sublines of Huh7 cells. Blight et al. (4) were able to select G418-resistant colonies supporting replication of RNA derived from an infectious molecular clone of the genotype 1a H77 virus. These cell clones were selected following transfection of the RNA into a novel, hyper-permissive Huh7 subline, Huh-7.5, that was generated by curing an established G418-resistant replicon cell line of the subgenomic Con1 replicon RNA that had been used to select it by treatment with alpha interferon (5). An analysis of the sequence of the genotype 1a RNA replicating in these cells identified a critical adaptive mutation located at amino acid position 470 of NS3 (P1496L), within domain II of the NS3 helicase. These RNAs also contained a purposefully introduced mutation in the NS5A protein (S2204I), which had been shown previously to enhance replication of 1b replicons. In a second report, Grobler et al. (11) described a systematic mutational approach leading to a similar conclusion, i.e., that both P1496L and S2204I are necessary for efficient replication of genotype 1a RNA in a highly permissive Huh7 subline which was generated independently, but in a manner similar to the Huh-7.5 cell line (11). Both groups identified other adaptive mutations within the helicase domain of NS3 (S1222T and A1226D), but genotype 1a RNAs containing any of these adaptive mutations were not capable of replication in regular Huh7 cells, indicating a relatively limited range of cellular permisiveness. Gu et al. (12) also recently described replication of a chimeric subgenomic replicon in which most of the polyprotein-coding sequence was genotype 1, but did not identify specific adaptive mutations.
In an effort to establish a more robust genotype 1a replicon, we adopted a somewhat different approach utilizing a previously described secreted alkaline phosphatase (SEAP) reporter system to document viral RNA replication in normal Huh7 cells (28). First, we constructed a chimeric subgenomic replicon in which the RNA region encoding the N-terminal 75 amino acids of NS3 was derived from the genotype 1b Con1 strain and the remainder was derived from the genotype 1a H77c infectious molecular clone (26). We were unable to demonstrate replication of this RNA in a transient-transfection system but were able to select G418-resistant colonies harboring replicating RNA after introduction of the S2204I mutation. We identified a single mutation within the NS4A region of these RNAs that enhanced the replication capacity of the parental RNA. We used a similar approach to identify a second adaptive mutation within the NS3 protease domain that enhanced the replication capacity of a different chimeric replicon that contained genotype 1a NS3/4A sequence within a genotype 1b background. The combination of these two adaptive mutations conferred a relatively robust replication phenotype on genotype 1a RNA, allowing detection of replication in a transient assay in normal Huh7 cells. This replicon was subsequently used in an iterative fashion to identify additional adaptive mutations leading to enhanced replication capacity, ultimately generating a genotype 1a replicon and genome length H77c RNA with extraordinary in vitro replication properties.
MATERIALS AND METHODS
Cells.
Huh7 cells were grown in Dulbecco's modified Eagle's medium (Gibco BRL) supplemented with 10% fetal calf serum, penicillin, and streptomycin. En5-3 is a clonal cell line derived from Huh7 cells by stable transformation with the plasmid pLTR-SEAP (28). These cells were cultured in Dulbecco's modified Eagle's medium (Gibco BRL) supplemented with 10% fetal calf serum, blasticidin (2 μg/ml; Invitrogen), penicillin, and streptomycin. Cell lines were passaged once or twice per week. We used G418 (250 μg/ml) to select colonies from En5-3 cells transfected with replicon RNAs containing 1a sequences.
Plasmids.
The plasmid pBpp-Htat2ANeo was constructed by replacing the BsrGI-XbaI fragment of pBpp-Ntat2ANeo/SI (identical to Ntat2ANeo/SI as described by Yi et al. (28) with the analogous segment of pH77c (GenBank AF011751) (26) engineered to contain a BsrGI site at the corresponding location by QuikChange (Stratagene) mutagenesis. This fragment swap results in the NS3-NS5B sequence in pBpp-Htat2ANeo being identical to that of pH77c, with the exception of the RNA encoding the N-terminal 75 amino acid residues of NS3 that retains the genotype 1b Con1 sequence. Since Bpp-Ntat2ANeo/SI was originally engineered to contain the genotype 1a 5′ nontranslated RNA (5′NTR) sequence (28), the resulting pBpp-Htat2ANeo construct possesses both a genotype 1a 5′NTR and 1a 3′NTR sequence. Overlapping PCR was used to fuse an antigenomic hepatitis delta ribozyme sequence directly to the 3′ end of the genotype 1a 3′NTR, in order to generate a self-cleaving 3′ sequence with the exact 3′-terminal nucleotide of HCV (24). Derivatives of pBpp-Htat2ANeo containing the adaptive mutations K1691R (see Results) or S2204I were created by QuikChange (Stratagene) mutagenesis.
To construct pBpp-H34A-Ntat2ANeo/SI, an EcoRI restriction site was created in pBpp-Ntat2ANeo/SI near the 3′ end of the NS4A coding region by QuikChange mutagenesis. After digestion of the resulting plasmid with BsrGI and EcoRI, the excised HCV segment was replaced with the equivalent sequence from pH77c which had been amplified by PCR using primers pairs containing terminal BsrGI and EcoRI sites, respectively. To construct the plasmid Hpp-H34A-Ntat2ANeo, DNA fragments representing the encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) sequence and the genotype 1a H77c NS3 protein-coding sequence were fused by overlapping PCR. The resulting fragment was digested with KpnI at a site located within the EMCV IRES and BsrGI at the site created within the modified pH77c NS3 region (see above) and then inserted in place of the corresponding fragment in pBpp-H34A-Ntat2ANeo/SI. The adaptive mutation Q1067R or G1188R was introduced into pHpp-H34A-Ntat2ANeo/SI in a similar fashion, using cDNA fragments prepared by reverse transcription-PCR of template RNAs isolated from independent G418-resistant replicon cell lines selected after transfection of En5-3 cells with Hpp-H34A-Ntat2ANeo RNA. pHtat2ANeo/SI was constructed by replacing the BsrGI-XbaI fragment of pHpp-H34A-Ntat2ANeo/SI with that of pBpp-Htat2ANeo/SI. A similar strategy was used to construct pHtat2ANeo/QR/SI, pHtat2ANeo/KR/SI, and pHtat2ANeo/QR/KR/SI. QuikChange (Stratagene) mutagenesis was used to introduce the P1496L, F2080V, and K2040R mutations into replicon constructs derived from pHtat2ANeo/SI.
Modified pH77c plasmids containing adaptive mutations were created by replacing the BsrGI-XbaI fragment with the corresponding fragment from the pHtat2ANeo plasmid derivative containing the indicated mutation, except for the Q1067R mutation which was introduced by QuikChange (Stratagene) mutagenesis. For use as controls, replication-incompetent subgenomic and genome length genotype 1a constructs (Htat2ANeo/QR/VI/KR/KR5A/SI/AAG and H77/QR/VI/KR/KR5A/SI/AAG) were created by replacing residues 2737 to 2739 of NS5B (GDD) with AAG using a similar strategy. Each mutation was confirmed by sequence analysis.
RNA transcription and transfection.
RNA was synthesized with T7 MEGAScript reagents (Ambion), after linearizing plasmids with XbaI. Following treatment with RNase-free DNase to remove template DNA and precipitation of the RNA with lithium chloride, the RNA was transfected into Huh7 cells or En5-3 cells by electroporation. Briefly, 5 μg of RNA was mixed with 2 × 106 cells suspended in 500 μl of phosphate-buffered saline (PBS), in a cuvette with a gap width of 0.2 cm (Bio-Rad). Electroporation consisted of two pulses of current delivered by the Gene Pulser II electroporation device (Bio-Rad), set at 1.5 kV, 25 μF, and maximum resistance. For transient-replication assays, no G418 was added to the medium. Transfected cells were transferred to two wells of a six-well tissue culture plate, and culture medium was removed completely every 24 h and saved at 4°C for subsequent SEAP assay. The cells were washed twice with PBS prior to refeeding with fresh culture medium. Since the culture medium was replaced every 24 h in these transient assays, the SEAP activity measured in these fluids reflected the daily production of SEAP by the cells.
Alkaline phosphatase assay.
SEAP activity was measured in 10-μl aliquots of transfected cell supernatant culture fluids using the Phospha-Light Chemiluminescent Reporter Assay (Tropix) with the manufacturer's suggested protocol reduced in scale. The luminescent signal was read using a TD-20/20 Luminometer (Turner Designs, Inc.).
Sequence analysis of cDNA from replicating HCV RNAs.
HCV RNA was extracted from cells, converted to cDNA and amplified by PCR as described previously (29). First-strand cDNA synthesis was carried out with Superscript II reverse transcriptase (Gibco-BRL); Pfu-Turbo DNA polymerase (Stratagene) was used for PCR amplification of the DNA. The amplified DNAs were subjected to direct sequencing using an ABI 9600 automatic DNA sequencer.
In vitro translation.
In vitro-transcribed RNA, prepared as described above, was used to program in vitro translation reactions in rabbit reticulocyte lysate (Promega). Approximately 1 μg of RNA, 2 μl of [35S]methionine (1,000 Ci/mmol at 10 mCi/ml), and 1 μl of an amino acid mixture lacking methionine were included in each 50-μl reaction mixture. Translation was carried out at 30°C for 90 min. Translation products were separated by SDS-PAGE followed by autoradiography or PhosphorImager (Molecular Dynamics) analysis.
Indirect immunofluorescence.
Cells were grown on chamber slides until 70 to 80% confluent, washed three times with PBS, and fixed in methanol-acetone (1:1, vol/vol) for 10 min at room temperature. A 1:20 dilution of a primary, murine monoclonal antibody to core or NS5A (Maine Biotechnology Services) was prepared in PBS containing 3% bovine serum albumin and incubated with the fixed cells for 1 h at room temperature. Following additional washes with PBS, specific antibody binding was detected with a goat anti-mouse immunoglobulin G fluorescein isothiocyanate-conjugated secondary antibody (Sigma) diluted 1:70. Cells were washed with PBS, counterstained with DAPI (4′,6-diamidino-2-phenylindole), and mounted in Vectashield mounting medium (Vector Laboratories) prior to examination by a Zeiss AxioPlan2 Fluorescence microscope.
Northern analysis for HCV RNA.
We seeded cells transfected with in vitro transcribed RNAs into six-well plates as described above. Total cellular RNA was extracted with Trizol reagent (Gibco-BRL) and quantified by spectrophotometry at 260 nm. Fifteen micrograms of the total RNA extracted from each well was loaded onto a denaturing agarose-formaldehyde gel, subjected to electrophoresis and transferred to positively charged Hybond-N+ nylon membranes (Amersham-Pharmacia Biotec) using reagents provided with the NorthernMax Kit (Ambion). RNAs were immobilized on the membranes by UV cross-linking. The membrane was hybridized with a 32P-labeled antisense riboprobe complementary to the 3′-end of the HCV NS5B sequence (nt 8990 to 9275) derived from pH77C and the hybridized probe was detected by exposure to X-ray film.
RESULTS
Transient replication of 1a replicon containing chimeric NS3-coding sequence.
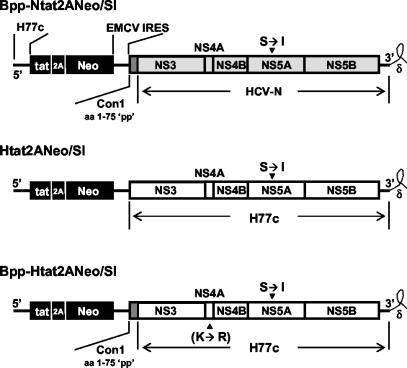
In contrast to genotype 1b HCV, several previous reports suggest that it is difficult to generate subgenomic genotype 1a replicons that are capable of efficient replication in Huh7 cells (3, 13, 14, 17). We encountered similar results with a dicistronic SEAP reporter replicon constructed from the H77c infectious molecular clone (26) that encoded both the human immunodeficiency virus tat protein and neomycin phosphotransferase in the upstream cistron. The organization of this latter replicon, Htat2ANeo/SI (Fig. 1), was similar to that of the efficiently replicating, genotype 1b Bpp-Ntat2ANeo/SI replicon (Fig. 1), referred to previously simply as “Ntat2ANeo/SI” (28). Most of the HCV polyprotein-coding sequence in Bpp-Ntat2ANeo/SI is derived from the genotype 1b HCV-N strain of HCV (2), but the “Bpp” prefix used here and throughout this communication refers to the presence of 225 nt of sequence that are derived from the Con1 strain of HCV at the extreme 5′ end of the polyprotein coding region (“pp” indicates the 5′-proximal protease-coding region [Fig. 1]). In contrast, all of the HCV sequence in Htat2ANeo/SI (Fig. 1) is derived from the genotype 1a H77c virus, including both the 5′ NTR and 3′ NTR sequences. Unlike Bpp-Ntat2ANeo/SI RNA, Htat2ANeo/SI RNA did not transduce the selection of G418-resistant colonies, nor induce secretion of SEAP above that observed with a replication-incompetent NS5B-deletion mutant (ΔGDD) when transfected into En5-3 cells (stably transformed Huh7 cells that express SEAP under control of the human immunodeficiency virus long terminal repeat promoter) (28) in a transient-replication assay (data not shown). This was the case even though the replicon was engineered to contain the genotype 1b adaptive mutation, S2204I, within NS5A (Fig. 1). The absence of apparent replication of Htat2ANeo/SI RNA was striking given the fact that it was derived from a well-documented infectious molecular clone of the H77c strain of HCV (26).
FIG. 1.
Organization of the selectable subgenomic dicistronic HCV replicons, Bpp-Ntat2ANeo/SI (identical to Ntat2ANeo/SI described previously by Yi et al. [28]), Htat2ANeo/SI, and Bpp-Htat2ANeo/SI. The two large open reading frames in each are shown as rectangles, with nontranslated RNA segments shown as lines. “δ” indicates the hepatitis delta ribozyme sequence introduced downstream of the 3′ terminus of the HCV sequence that produces an exact 3′ end. The replicons differ in the source of the sequence encoding the HCV nonstructural proteins in the 3′ open reading frame as well as in the 3′ NTR. The segment of the 3′ open reading frame labeled “pp” (for “proximal protease”) encodes the amino terminus of the NS3 protein (residues 1 to 75): “Bpp” indicates that this region is derived from the genotype 1b Con1 sequence (darkly shaded box). HCV-N sequence is represented as a lightly shaded box; genotype 1b H77c coding sequence is shown as an open box. All three replicons contain the S2204I mutation in NS5A. The location of the K1691R mutation identified in the NS4A sequence of Bpp-Htat2ANeo/SI is indicated.
Recent reports suggest that the EMCV IRES-driven translation of the second cistron in dicistronic, subgenomic RNAs such as those shown in Fig. 1 may be reduced when the translated RNA sequence is derived from genotype 1a virus, rather than genotype 1b (12, 13, 17). However, even when translation of the second cistron is rendered more efficient by replacing the 5′ 225 nt of the genotype 1a NS3 sequence with related sequence from the Con1 genotype 1b virus, replication typically has not been observed when the remainder of the replicon sequence is derived from a genotype 1a virus (13, 17). Gu et al. (12) recently described the successful selection of a replication competent, chimeric replicon in which the 5′ 225 nt of the NS3 coding sequence was derived from genotype 1b virus and the remainder of the second cistron from genotype 1a HCV (construction of chimeric replicons being simplified by a unique BsrGI site within the genotype 1b Con1 virus sequence, 225 nt downstream from the 5′ end of the NS3 region). This replicon also contained 5′NTR sequence derived from genotype 1b virus and had a single base change within the genotype 1a 3′NTR sequence. The results of Gu et al. (12) suggest that the inclusion of the Con1 sequence at the 5′ end of the NS3 region may in some way facilitate replication of the 1a RNA. This hypothesis is strengthened by observations we have made with genotype 1b replicons derived from HCV-N. Those we described previously, including Bpp-Ntat2ANeo/SI RNA, were constructed by ligation of HCV-N sequence to a Con1 replicon at the BsrGI site (13, 14, 28), and thus they contain 5′-proximal NS3 sequence (proximal protease sequence [Fig. 1]) derived from the Con1 virus. Although this chimeric Con1/HCV-N RNA replicates significantly more efficiently than the originally described Con1 replicons, the replacement of the 5′-proximal NS3 sequence in Bpp-Ntat2ANeo/SI with sequence from HCV-N (resulting in Npp-Ntat2ANeo/SI) virtually eliminated its replication phenotype in transient-transfection assays, although it remained possible to select G418-resistant colonies at a low frequency following transfection (data not shown).
To formally assess the ability of the 5′-proximal genotype 1b NS3 sequence to enhance genotype 1a RNA replication, we replaced the 5′ 225 nt of NS3 coding region in Htat2ANeo/SI with the Con1 sequence, generating Bpp-Htat2ANeo/SI (Fig. 1). We also modified the construct by replacing the XbaI restriction site at the 3′ end of the HCV sequence with the hepatitis delta virus ribozyme sequence (24). We have shown previously that the presence of the extraneous 4 nt at the 3′ end of the replicon RNA that results from runoff transcription of XbaI-digested plasmid DNA reduces the replication competence of genotype 1b RNAs by two- to threefold (30). The inclusion of the ribozyme resulted in self-cleaving RNA transcripts capable of generating the exact 3′-terminal HCV RNA sequence. Nonetheless, this modified Bpp-Htat2ANeo/SI RNA still remained incapable of inducing the expression of SEAP in transfected En5-3 cells beyond that observed following transfection of the ΔGDD RNA. Transfection resulted only in an initial burst in SEAP expression due to translation of the input replicon RNA, without the sustained SEAP expression that is indicative of RNA replication (Fig. 2). However, the Bpp-Htat2ANeo/SI RNA was capable of transducing the selection of G418-resistant cell colonies supporting replication of the RNA over a period of 3 to 4 weeks following transfection of the cells.
FIG. 2.
Transient HCV RNA replication assay. Shown is the expression of SEAP by En5-3 cells following transfection with the chimeric 1a replicon Bpp-Htat2ANeo/SI and Bpp-Htat2ANeo/KR/SI, which carries an additional K1691R mutation in NS3 that was identified following selection of G418-resistant cells following transfection with Bpp-Htat2ANeo/SI. As controls, SEAP expression is shown following transfection of cells with the highly replication competent 1b replicon, Bpp-Ntat2ANeo/SI, and a related replication-defective ΔGDD mutant; also shown is SEAP expression by normal En5-3 cells. Cell culture supernatant fluids were collected and replaced with fresh media at 24-h intervals following transfection. Cells were split 5 days after transfection. Samples of media were stored at 4°C until assayed for SEAP activity at the conclusion of the experiment. Results shown represent the mean values obtained from duplicate cultures transfected with each RNA; error bars indicate the range of values. See Table 1 for identification of adaptive mutations in each transcript.
We analyzed the sequence of replicon RNAs extracted from two independent G418-resistant cell clones selected following the transfection of En5-3 cells with Bpp-Htat2ANeo RNA. We determined the presence of a single Lys-to-Arg mutation located within the NS4A region, at residue 1691 (K1691R) of the polyprotein in both cell clones. This residue is located just beyond the 3′ limits of the NS4A cofactor peptide sequence which participates in forming a noncovalent complex with NS3 and enhances its protease activity (25, 27). To determine whether the K1691R mutation facilitated replication of the chimeric genotype 1b/1a RNA in En5-3 cells, we introduced this mutation into the parental Bpp-Htat2ANeo/SI construct, thereby creating Bpp-Htat2ANeo/KR/SI (Table 1 includes a list of all adaptive mutations identified in these studies, as well as the symbols used to indicate their presence in constructs). As shown in Fig. 2, this single mutation significantly enhanced the replication capacity of the RNA, allowing replication to be detected by a sustained increase in SEAP expression following transient transfection of En5-3 cells in the absence of G418 (Fig. 2). Since the level of SEAP production has been shown to correlate closely with intracellular replicon RNA abundance in this reporter system (28, 29), we conclude that K1691R is an adaptive mutation. Interestingly, this mutation has been shown previously to confer an enhanced replication phenotype on Con1 replicons (18), whereas the sequence of HCV-N is naturally Arg at this position (2). Our results stand in contrast to those reported by Gu et al. (12), who identified several mutations within the NS3, NS5A, and NS5B sequences of chimeric genotype 1b-1a RNAs. None of these mutations appeared to enhance the ability of the chimeric RNA to replicate or transduce colony selection.
TABLE 1.
Adaptive mutations identified in genotype 1a H77c replicons
| Symbola | H77c protein and residue | Mutationb |
|---|---|---|
| QR | NS3 41 | Q1067R |
| GR | NS3 162 | G1188R |
| VI | NS3 629 | V1655I |
| KR | NS4A 34 | K1691R |
| KR5A | NS5A 68 | K2040R |
| FV | NS5A 108 | F2080V |
| SI | NS5A 232 | S2204I |
Symbol used to designate presence in RNA transcripts.
Position number refers to H77c polyprotein (GenBank accession no. AF011751).
The 5′ 225 nt of the genotype 1a NS3 sequence down modulate replicon amplification.
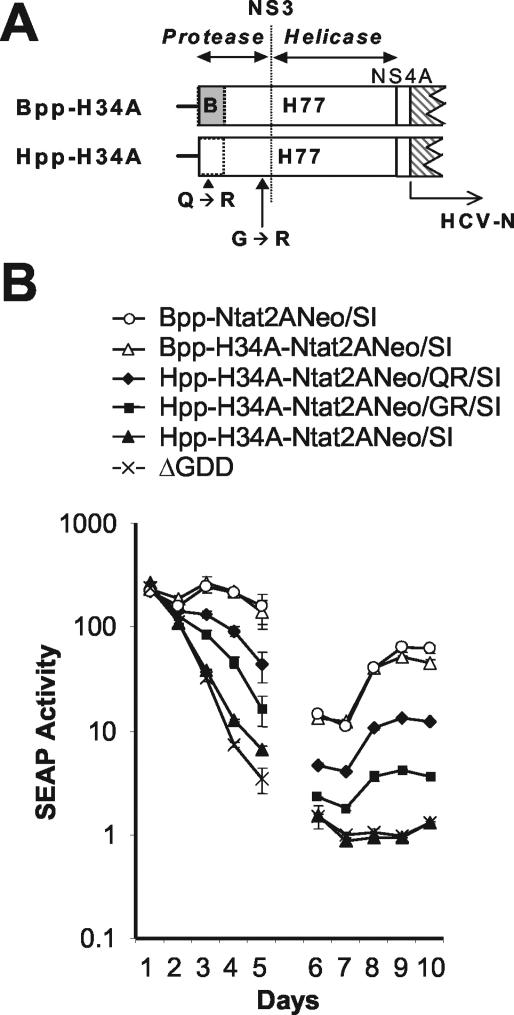
The results described above, as well as those of Gu et al. (12), suggest that the first 225 nt of the genotype 1a NS3 sequence have a negative impact on the replication of subgenomic HCV replicons. This could occur by down modulation of EMCV IRES-directed translation of the nonstructural proteins (13) or by directly influencing replication itself, possibly by influencing an NS3-related function. To address this issue, we sought to identify additional adaptive mutations capable of compensating for the presence of the 5′-proximal genotype 1a protease sequence. We thus constructed additional chimeric replicons containing the entire genotype 1a NS3/4A sequence within the background of Bpp-Ntat2ANeo/SI (Hpp-H34A-Ntat2ANeo/SI [Fig. 3A]). We also constructed a variant of this construct in which the first 225 nt of the NS3/4A sequence was replaced with Con1 sequence (Bpp-H34A-Ntat2ANeo/SI [Fig. 3A]). In both chimeric RNAs, the sequence extending from NS4B to the 3′NTR was derived entirely from the genotype 1b HCV-N strain. While the replicon containing the entire genotype 1a NS3/4A sequence (Hpp-H34A-NtatNeo/SI) did not show evidence of replication in a transient-transfection assay, the variant containing the first 225 nt of the Con1 sequence (Bpp-H34A-NtatNeo/SI) replicated as well as the reference Bpp-Ntat2ANeo/SI replicon (Fig. 3B). This result confirms that the 5′ 225 nt of the genotype 1a NS3 sequence have a negative effect on RNA replication in En5-3 cells, and also indicates that the downstream genotype 1a NS3/4A sequence functions well in this context.
FIG. 3.
(A) Schematic depicting the organization of the 5′ end of the second open reading frame in subgenomic chimeric replicons containing most (Bpp-H34A-Ntat2ANeo/SI) or all (Hpp-H34A-Ntat2ANeo/SI) of the H77 genotype 1a NS34A-coding sequence in the background of the genotype 1b Bpp-Ntat2ANeo/SI. Genotype 1a sequence (H77) is shown as an open box, genotype 1b sequence (Con1 or HCV-N) as a shaded box. “Bpp” indicates the presence of genotype 1b sequence from the Con1 strain of HCV in the 5′-proximal protease coding sequence, whereas “Hpp” indicates that this sequence is derived from the genotype 1a H77 sequence. Approximate locations are shown for the adaptive mutations Q1067R and G1188R, identified in G418-resistant cell clones selected following transfection of Hpp-H34A-Ntat2ANeo/SI. The dotted line indicates the approximate border of the NS3 protease and helicase domains (between residues 1206 and 1207). (B) SEAP activity present in supernatant culture fluids collected at 24 h intervals following transfection of En5-3 cells with various chimeric 1a-1b replicons including Bpp-H34A-Ntat2ANeo/SI, Hpp-H34A-Ntat2ANeo/SI, Hpp-H34A-Ntat2ANeo/QR/SI, and Hpp-H34A-Ntat2ANeo/GR/SI. Control cells were transfected with Bpp-Ntat2ANeo/SI and the replication-defective ΔGDD mutant. See legend to Fig. 2 for further details. See Table 1 for identification of adaptive mutations in each transcript.
Interestingly, despite the lack of detectable RNA replication in the transient assay, we were able to select stable G418-resistant cell clones following transfection of Hpp-H34A-Ntat2ANeo/SI RNA. Sequencing of replicon RNAs derived from two independent cell clones revealed only a single potentially adaptive mutation in each: Q1067R and G1188R, both of which are located within RNA encoding the NS3 protease (Fig. 3A). The Q1067R mutation is of particularly interest, since it is within the 5′ 225 nt of the NS3 region. When introduced into Hpp-H34A-Ntat2ANeo/SI, both the Q1067R and (to a lesser extent) the G1188R mutations enhanced replication of the RNA to a level that was detectable in the transient assay (Fig. 3B), indicating that both are adaptive mutations and capable of compensating, in part, for the presence of the genotype 1a protease sequence. However, neither of these mutations, when introduced into a replicon containing only genotype 1a sequence (Htat2ANeo/SI), was able to enhance replication to the point where it was evident in the transient assay (Htat2ANeo/QR/SI [Fig. 4 and data not shown]).
FIG. 4.
Impact of adaptive mutations on replication competence of the subgenomic genotype 1a replicon, Htat2ANeo/SI. (A) Location of various adaptive mutations within the second open reading frame (derived entirely from the genotype 1a H77sequence): Q1067R and P1496L (described by Blight et al. [4]) (NS3); K1691R (NS4A); and F2080V and S2204I (Blight et al. [3]) (NS5A). Detailed information concerning each mutation is provided in the text. (B) Transient HCV RNA replication assay. SEAP activity in culture supernatants collected at 24-h intervals following electroporation of En5-3 cells with the 1a replicon Htat2ANeo carrying the indicated combinations of the adaptive mutations shown in panel A. Cells were also transfected with genotype 1b Bpp-Ntat2ANeo/SI replicon RNA as a reference. See legend to Fig. 2 for further details. (C) Summary of the replication phenotypes of genotype 1a replicon Htat2ANeo RNAs containing various combinations of adaptive mutations: (−) no detectable replication, (+) modest increase in SEAP expression above background days 3 to 5, and (+++) >10-fold increase in SEAP expression above background 7 days after transfection in the transient replication assay (see panel B). See Table 1 for identification of adaptive mutations in each transcript.
Transient replication of a genotype 1a replicon in normal Huh7 cells.
To determine whether the K1691R and Q1067R mutations might work cooperatively to confer a transient-replication phenotype on the genotype 1a replicon RNA, we introduced both into Htat2ANeo and assessed the ability of the modified RNA to replicate in transfected En5-3 cells. Surprisingly, the combination of the K1691R and Q1067R mutations (in addition to the S2204I mutation in NS5A) conferred a relatively robust replication phenotype on the genotype 1a RNA, such that replication was easily detectable in the transient-transfection assay using the SEAP reporter system (Htat2ANeo/QR/KR/SI [Fig. 4B]). Using an approach similar to that taken in the preceding experiments, we subsequently identified an additional adaptive mutation (F2080V) within the NS5A-coding region (F2080V), when cells transfected with Htat2ANeo/QR/KR/SI RNA were subjected to G418 selection pressure. This mutation resulted in slightly greater replication efficiency when introduced into the genotype 1a replicon containing K1691R and Q1067R in addition to S2204I (Htat2ANeo/QR/KR/FV/SI [Fig. 4B]). However, F2080V had relatively little effect when added to replicons containing only K1691R or Q1067R (in addition to S2204I) (Fig. 4B). Minimally increased secretion of SEAP above the ΔGDD background was observed during the first 5 days after transfection with Htat2ANeo/KR/FV/SI, but this was no longer apparent after 6 days. The replication phenotype of Htat2ANeo/QR/FV/SI was indistinguishable from that of the replication incompetent ΔGDD mutant in this assay (Fig. 4B). These results are summarized in Fig. 4C.
To facilitate a comparison of these results with those reported previously by Blight et al. (4), we introduced the adaptive P1496L mutation identified by this group within the helicase domain of NS3 following transfection of a genotype 1a replicon into the highly permissive Huh7 subline Huh-7.5. Consistent with the previous report, a 1a replicon bearing this mutation P1496L demonstrated only minimal evidence of replication in the transient assay (which utilizes En5-3 cells that are comparable to normal Huh7 cells in terms of their permissiveness for HCV RNA replication) (Htat2ANeo/PL/SI [Fig. 4B]). The addition of the NS5A mutation F2080V failed to noticeably enhance the replication capacity of this RNA (Htat2ANeo/PL/FV/SI [Fig. 4B]). SEAP expression induced by genotype 1a replicons containing both Q1067R and K1691R was approximately 10-fold that induced by replicons containing P1496L. Since SEAP production from En5-3 cells correlates closely with the intracellular abundance of replicon RNA (28), these results suggest that the protease domain mutations make a greater contribution to replication competence of the genotype 1a replicon.
Adaptive mutations within NS3 do not affect EMCV IRES-driven translation of the second cistron.
As mentioned above, previous reports indicate that the EMCV-driven translation of the second cistron is reduced in genotype 1a replicons in comparison to replicons containing the genotype 1b Con1 sequence (12, 13, 17). Although the mechanism is uncertain, the effect appears to be due to the genotype 1a sequence encoding the amino terminus of NS3. Since the adaptive Q1067R mutation is located within this region, we asked whether it or other mutations that enhance 1a replicon amplification do so by improving EMCV IRES-driven translation of the HCV nonstructural proteins. To test this hypothesis, we programmed in vitro translation reactions with genotype 1b and 1a replicon RNAs containing various adaptive mutations, and compared the production of proteins encoded by the second cistron with neomycin phosphotransferase produced from the first cistron. As shown in Fig. 5, the synthesis of NS3 was modestly reduced with replicons containing genotype 1a H77c sequence in the 5′-proximal protease region (compare NS3 abundance in lanes 4 to 8 with that in other lanes). However, it was not increased by any of the adaptive mutations, including Q1067R. This result indicates that the difficulty of establishing replication competent 1a replicons is more likely due to the intrinsic property of the 1a sequence, than to an incompatibility of the HCV and EMCV sequences in this region leading to reduced activity of the EMCV IRES. Nonetheless, the reduced level of translation of the genotype 1a nonstructural proteins that is evident in Fig. 5 may contribute to the poor replication phenotype of these RNAs.
FIG. 5.
Adaptive mutations within the polyprotein do not influence the efficiency of polyprotein translation under control of the EMCV IRES. Shown is an SDS-PAGE gel loaded with products of in vitro translation reactions programmed with RNAs derived from Bpp-Ntat2ANeo (lane 1), Bpp-Htat2ANeo (lanes 2 and 3), Htat2ANeo (lanes 4 to 8), or Bpp-Ntat2ANeo/ΔGDD (lane 9) RNAs carrying various combinations of adaptive mutations (Q1067R, K1691R, F2080V, or S2204I) as indicated. The schematic at the top of the figure indicates the location of these mutations within the polyprotein. Abbreviations: pp, RNA segment encoding the amino-terminal 75 residues of NS3; NS, remainder of the RNA segment encoding the nonstructural proteins. H, genotype 1a H77 sequence; B, genotype 1b Con1; and N, genotype 1b HCV-N sequences. Location of NS3 and Neo product is indicated at the side of gel.
An additional adaptive NS5A mutation further augments replication competence.
Although the F2080V mutation in NS5A provided only a slight additional replication advantage to subgenomic genotype 1a RNAs containing the Q1067R, K1691R, and S2204I mutations (Fig. 4), additional mutations were subsequently identified concurrently near the C terminus of NS3 (V1655I) and within NS5A (K2040R) in RNAs replicating within a G418-resistant cell line selected following transfection with the subgenomic Htat2ANeo/QR/KR/SI replicon. As shown in Fig. 6, both of these mutations enhanced the replication capacity of genotype 1a RNA. Addition of the V1655I mutation resulted in a modest enhancement of Htat2ANeo/QR/KR/SI replication, leading to a replication phenotype slightly better that observed with the addition of the F2080V mutation. In contrast, the addition of the K2040R mutation in NS5A resulted in a dramatic increase in replication competence, rendering the replication phenotype of the genotype 1a RNA equivalent to that of the standard genotype 1b HCV-N replicon used in these studies, Bpp-Ntat2ANeo/SI (Fig. 6B). A genotype 1a replicon containing both of these adaptive mutations in addition to those identified earlier replicated with slightly greater efficiency than this reference genotype 1b RNA in the transient assay (Htat2ANeo/QR/VI/KR/KR5A/SI [Fig. 6B]). These results were confirmed in independent experiments.
FIG. 6.
Impact of additional adaptive mutations on replication competence of the subgenomic genotype 1a replicon, Htat2ANeo/QR/KR/SI (see Fig. 4). (A) Location of various adaptive mutations within the second open reading frame (derived entirely from the genotype 1a H77sequence): Q1067R and V1655I (NS3); K1691R (NS4A); and K2040R (KR5A), F2080V, and S2204I (NS5A). Detailed information concerning each mutation is described in the text. (B) Transient HCV RNA replication assay. SEAP activity in culture supernatants collected at 24 h intervals following electroporation of En5-3 cells with the 1a replicon Htat2ANeo carrying the indicated combinations of the adaptive mutations shown in panel A. Cells were also transfected with genotype 1b Bpp-Ntat2ANeo/SI replicon RNA as a reference. See legend to Fig. 2 for further details. See Table 1 for identification of adaptive mutations in each transcript.
Robust replication of genome length genotype 1a RNA with adaptive mutations.
Encouraged by the above-mentioned results, we assessed the in vitro replication competence of genome-length, genotype 1a H77c RNA engineered to contain the adaptive mutations described above. As with the dicistronic, subgenomic RNAs, we placed the hepatitis delta ribozyme sequence at the 3′ end of the cloned infectious cDNA sequence in pH77c in order to generate RNA transcripts containing an exact HCV 3′ terminus. As these genomic RNAs encoded no selectable marker or reporter protein product, their replication was assessed in transfected Huh7 and En5-3 cells by Northern blot analysis in comparison with related subgenomic RNAs. Subgenomic and genome length replication-incompetent H77 mutant RNAs, in which the GDD motif had been replaced with AAG, served as negative controls for this experiment. For En5-3 cells transfected with the subgenomic RNAs, we also determined levels of SEAP expression.
As expected, the unmodified H77c RNA showed no evidence of replication, even though it has been shown previously to be infectious in chimpanzees when inoculated into liver (Fig. 7, compare lane 7 with the replication-defective 1a genomic RNA in lane 11). The introduction of the Q1067R (NS3) mutation, alone or in combination with S2204I (NS5A), was insufficient to confer a detectable level of replication in Huh7 cells (data not shown). However, when all three mutations were introduced (Q1067R, K1691R and S2204I), the H77c RNA acquired a relatively efficient replication phenotype with readily detectable amplification of the RNA in Northern blots of cell lysates prepared 4 days after transfection of either Huh7 or En5-3 cells (Fig. 7, lane 8). Replication of the genome length RNA was slightly increased by the further addition of the F2080V (NS5A) mutation (Fig. 7, lane 9). However, consistent with the data presented in Fig. 6, the inclusion of both the V1655I mutation in NS3 and the K2040R mutation conferred a substantially more robust replication phenotype on genome length H77c, when present in combination with other adaptive mutations in NS3, NS4A and NS5A (H77c/QR/VI/KR/KR5A/SI [Fig. 7, compare lanes 10 and 11]). This experiment thus confirmed the adaptive effects of these mutations. Northern blotting indicated that the replication capacity of genome length genotype 1a RNAs containing adaptive mutations was significantly greater than the comparable subgenomic, dicistronic genotype 1a replicons, for which the RNA signal 4 days after transfection was low and near the limits of detection in Northern blots (Fig. 7, compare lanes 3 to 6 with lanes 8 to 11). These findings are consistent with those reported previously by Blight et al. (4) and indicate that the inclusion of heterologous sequences in the dicistronic replicons impairs RNA replication competence. Subgenomic replicon RNA was detected unambiguously only in cells transfected with Htat2ANeo/QR/VI/KR/KR5A/SI, the RNA that generated the highest level of SEAP expression (Fig. 7, compare lanes 5 and 6).
FIG. 7.
Northern analysis of HCV RNA abundance 4 days following transfection of normal Huh7 or En5-3 cells with the indicated dicistronic subgenomic and monocistronic genome length HCV RNAs. Lane 1, normal cells; lane 2, the subgenomic replicon, Htat2ANeo/SI; lanes 2 to 5, Htat2ANeo/SI replicon RNAs carrying the indicated combinations of mutations; lane 6, nonreplicating Htat2ANeo/QR/VI/KR//KR5A/SI/AAG; lane 7, genome length H77c RNA; lanes 8 to 10, genome length H77c RNA containing the indicated combinations of mutations; lane 11, genome length H77 RNA containing the lethal NS5B mutation; lanes 12 and 13, subcontrol genomic and genome length synthetic RNA transcripts. See Table 1 for identification of adaptive mutations in each transcript. Blots were probed with a genotype 1a probe derived from the NS5B coding sequence for detection of HCV-specific sequence (top panels); blots were also probed for β-actin message to assess RNA loading (bottom panels). At the top of the figure is shown the En5-3 cell culture supernatant fluid SEAP activity induced by replicating subgenomic RNAs at the time of cell harvest.
As a further measure of the replication competence of these modified genome length H77c RNAs, we also examined transfected En5-3 cells for the presence of core or NS5A proteins using an indirect immunofluorescence method. Introduction of both the K1691R (NS4A) and S2204I mutations resulted in detectable antigen expression 4 days after transfection, albeit only in a very low percentage of cells (less than 0.01% [data not shown]). However, strong expression of both the core and NS5A proteins was observed in approximately 30% of En5-3 cells 4 days after transfection of RNA containing all four adaptive mutations (Fig. 8). Increased replication efficiency of genotype 1a RNAs correlated with a greater proportion of cells supporting the replication of HCV RNA, evidenced by the presence of viral antigen (data not shown).
FIG. 8.
Indirect immunofluorescence detection of core and NS5A proteins 4 days after electroporation of monocistronic genome length RNAs into En5-3 cells. Panels on the right show cells that were transfected with H77c/QR/KR/FV/SI RNA, derived from the genotype 1a infectious molecular cDNA clone pH77c (26), engineered to contain four adaptive mutations: Q1067R (NS3), K1691R (NS4A), and F2080V and S2204I (NS5A). The cells in the panels on the left were transfected with replication-incompetent RNA transcribed from an infectious molecular cDNA clone of HCV-N (2) containing a deletion mutation eliminating the GDD motif within the NS5B polymerase. See Table 1 for identification of adaptive mutations in each transcript.
DISCUSSION
Subgenomic, dicistronic, selectable HCV RNA replicons derived from genotype 1b viruses replicate efficiently in cultured cells (3, 13, 14, 15, 19, 20). These novel RNAs have facilitated the study of HCV RNA replication and substantially accelerated antiviral drug discovery efforts. The Huh7 cell line, derived from a human hepatoma, appears to be uniquely permissive and supportive of the replication of these HCV RNAs, although recent studies suggest that other types of cells may also be permissive for HCV RNA replication (31). However, despite the success of genotype 1b replicons, it has been difficult to generate RNAs that replicate efficiently in any cell type from other genotypes of HCV, including genotype 1a, (3, 13, 14, 17). This surprising observation indicates that significant biological differences exist between genotype 1a and 1b viruses, despite the fact that the nucleotide sequences of genotype 1a viruses are relatively closely related to those of genotype 1b (∼90 to 93% identity). This biological difference raises the likelihood that antiviral agents that are found to be active against the genotype 1b virus may have significantly lesser activity against genotype 1a viruses. Considering these observations and the relatively high genetic variability that exists between different HCV genotypes, the development of cell culture systems supporting replication of viral RNAs from other genotypes will be important for validating in vitro efficacy of candidate antiviral agents across a range of genetically distinct HCV genotypes, as well as developing a better overall understanding of these viruses.
Genotype 1a viruses are the most-prevalent types of HCV in the United States, and like genotype 1 b virus they are relatively refractory to treatment with interferon (9, 22). Thus far, a detectable level of genotype 1a RNA replication has been reported only in specially isolated, highly permissive Huh7 human hepatoma cell sublines (e.g., Huh-7.5 cells) generated by eliminating the replication of genotype 1b RNA replicons from established replicon cell lines using alpha interferon in vitro (4, 11). These previously described genotype 1a RNAs possess cell culture-adaptive mutations that enhance their replication in these special cells, including those selected during the isolation of antibiotic-resistant cell lines containing these 1a replicons (4, 11). However, the published reports suggest that these previously described genotype 1a RNAs do not replicate to a detectable level in standard Huh7 cells, and that their capacity for replication in cultured cells is thus limited. In contrast, we report here genotype 1a HCV RNAs that replicate in a highly efficient manner in normal Huh7 cells.
Our results suggest that the highly efficient replication of genotype 1a RNAs requires at least three adaptive mutations located within the NS3, NS4A, and NS5A proteins. It is evident that these mutations are mutually reinforcing in their ability to enhance the replication of the genotype 1a RNAs, even though they were identified individually under different circumstances. We found that the introduction of the S2204I mutation in NS5A, which is known to promote the replication of genotype 1b virus RNAs in Huh7 cells (3), was not sufficient for subgenomic replicons composed entirely of the genotype 1a sequence to initiate replication in Huh7 cells. However, it made possible the selection of G418-resistant cell colonies following transfection of a chimeric replicon RNA, in which sequence from the infectious molecular clone of the genotype 1a H77c virus encoded all of the nonstructural proteins other than the N-terminal 75 amino acid residues of NS3 which were derived from the genotype 1b Con1 sequence (Bpp-Htat2ANeo/SI [Fig. 1]). The HCV RNAs replicating in these cells contained a single mutation within the NS4A-coding region (K1691R) that enhanced the replication capacity of the original chimeric replicon RNA (Fig. 2). These results suggest that an important restriction to the replication of genotype 1a virus in Huh7 cells may reside within the serine protease domain of NS3, since substitution of the N-terminal domain of the genotype 1a protease with that from the Con1 genotype 1b virus allowed the initiation of replication and the selection of G418-resistant cells. A similar conclusion can be drawn from the results reported by Gu et al. (12). Thus, it is interesting that the adaptive mutation that we identified, K1691R, resides within NS4A very close to the surface of the NS3/4A protease complex that it helps to form (Fig. 9). Exactly what the nature of this protease-related restriction might be, however, remains obscure.
FIG. 9.
Structure of the NS3/4A serine protease/helicase enzyme complex derived from the genotype 1b BK strain of HCV (Protein Database 1CU1) (27), with the locations of adaptive mutations highlighted. (A) Wire diagram of structure showing the NS3 helicase domain in green and the protease domain in magenta. The NS4A cofactor polypeptide is shown in blue in space-filling view, with the NS3 protease active-site residues shown in yellow in space-filling view. Adaptive mutations identified in this study (red arrows) cluster near the protease active site or at sites involved in substrate recognition, including the mutations in the NS3 protease domain at Gln-1067, Gly-1188 (shown in space-filling view as red) and near the carboxyl terminus of NS3 in the helicase domain at Val-1655 (light green). The NS4A adaptive mutation at Lys-1691 (light blue) is just beyond the surface of the protease, at the site of exit of the NS4A strand. Adaptive mutations within the NS3 helicase domain that were identified in other studies, Ser-1222, Ala-1226, and Pro-1496 (4, 11), are shown in copper in space-filling view and are not close to the protease active site. (B) Space-filling view of the structure shown in panel A, in which the adaptive mutations and active site have similar color coding as in panel A. The helicase domain appears in green, and the protease domain is shown in grey. The NS3/4A adaptive mutations identified in this study (Q1067R, G1188R, V1655I, and K1691R) all occur at solvent accessible residues on this side of the molecule. (C) Flip-view of the structure shown in panel B, rotated approximately 180°. The helicase adaptive mutations identified in previous studies are located on the surface of the helicase, distant from the protease active site. (Note that in the HCV-BK sequence, Pro-1496 is Arg, and Lys-1691 is Ser.)
In an effort to better understand this restriction, we constructed a second chimeric replicon containing the complete genotype 1a NS34A sequence within the background of a genotype 1b replicon. This RNA (Hpp-H34A-Ntat2ANeo/SI) did not undergo detectable replication in the transient-transfection system we utilized in these studies (Fig. 3). However, it was capable of transducing the selection of G418-resistant cell colonies following transfection and antibiotic selection. Analysis of the sequence of the HCV RNAs replicating within these cells identified a second, cell culture-adaptive mutation within the N-terminal region of the NS3 protease (Q1067R), providing further evidence that a primary restriction to replication of genotype 1a virus resides within this domain. Yet additional evidence for this comes from the replication phenotype of the Bpp-H34A-Ntat2ANeo/SI replicon, which also contains all of the genotype 1a NS3/4A sequence except for the N-terminal 75 amino acid resides and which demonstrated a robust replication phenotype in the transient-transfection assay. Thus, there appears to be no restriction to replication deriving from inclusion of the genotype 1a NS3 helicase domain, nor for that matter any part of the protease domain except for its N terminus.
Further work demonstrated that the K1691R and Q1067R mutations worked cooperatively: neither by itself was capable of conferring the capacity for efficient replication on a replicon composed entirely of genotype 1a sequence, but a combination of the two (in addition to the genotype 1b S2204I adaptive mutation) resulted in RNA replication that could be readily detected in the transient-transfection assay (Fig. 4). That these mutations should act cooperatively in their effects on replication, as indicated by the data shown in Fig. 4, is consistent with their location in the polyprotein, since the NS4A protease cofactor domain interacts primarily with residues within the N-terminal domain of the NS3 protease (25, 27).
Additional adaptive mutations were identified and verified through an iterative series of experiments involving RNA transfection, isolation of G418-resistant cells, and analysis of the sequence of efficiently replicating genotype 1a RNAs. We also demonstrated that the S2204I mutation did indeed facilitate the replication of the genotype 1a RNA, as its removal from the efficiently replicating subgenomic RNAs shown in Fig. 6 substantially reduced their replication competence in the transient-transfection assay (data not shown). The genotype 1a adaptive mutations we identified are summarized in Table 1. They can be grouped functionally into two groups: K2040R, F2080V, and S2204I, which are all located within NS5A (a common site of genotype 1b adaptive mutations), and Q1067R, G1188R, V1655I, and K1691R, which are all located in or otherwise associated with the protease domain of NS3. While to some extent solvent exposed, both G1188R and Q1067R are close to the active site of the protease (Fig. 9), and would both add a significant charge difference to the active face of the protein. V1655I is particularly interesting. It is located near the extreme C terminus of the NS3 protein, downstream of the helicase domain and close to the protease active site in the crystal structure of the NS3/4A complex (27). In the P3 position of the NS3/4A cleavage site, V1655 is certain to play a role in substrate recognition during the cis-active cleavage of the polyprotein at the NS3/4A junction, and it remains within the substrate-binding pocket in the crystal structure. The potential impact of the K1691R mutation, within NS4A, on the conformation of the protease active site is much less certain, but it is in close proximity to the NS4A cofactor domain, as mentioned above, and intercalation of this domain into the NS3 protease is well known to modulate the activity of the protease.
Significantly, all of these NS3 and NS4A mutations are located at some distance from other genotype 1a adaptive mutations in NS3 that have been described in the literature (Fig. 9). These mutations, located at S1222, A1226, and P1496, are all within the helicase domain of NS3 (4, 11). While on the surface of the protein, they are located on the side opposite the solvent exposed surfaces containing the G1188, V1655, and Q1067 residues (Fig. 9). Thus, we suspect that they facilitate genotype 1a RNA replication by a different mechanism than those mutations that we have identified that cluster near the active site of the protease. At least the P1496L mutation identified by both Blight et al. (4) and Grobler et al. (11) appears to be substantially less active in conferring replication capacity on the genotype 1a H77c RNA. This was demonstrated by the lack of detectable replication of RNA replicons containing this mutation (Htat2ANeo/PL/SI and Htat2ANeo/PL/FV/SI) in the transient-transfection experiment summarized in Fig. 4.
What role could mutations near the active site of the NS3 protease play in promoting the replication of genotype 1a HCV RNA in Huh7 cells? We have excluded the possibility that these mutations work by enhancing translation of the nonstructural proteins under control of the EMCV IRES in the context of the subgenomic replicon, since we observed no difference in translation of these proteins in vitro in reticulocyte lysates programmed with these RNAs (Fig. 5). More importantly, they enhance the replication of genomic H77c RNA lacking any heterologous sequence in Huh7 cells (Fig. 7). These mutations do not seem likely to promote replication by favorably influencing the ability of the protease to process the viral polyprotein, since the polyprotein segment expressed in the Htat2ANeo derivatives is derived entirely from the same H77c genome, and this replicates very efficiently in chimpanzee liver. However, this does remain a formal possibility that needs to be excluded in future studies. We speculate, instead, that these mutations promote interactions of the NS3/4A complex with specific cellular proteins that play a role in assembly of the viral replicase complex, or otherwise influence replication by disabling innate cellular antiviral defenses.
Foy et al. (8) recently demonstrated that expression of the NS3/4A protease effectively blocked activation of interferon regulatory factor 3 (IRF3) in Huh7 cells infected with Sendai virus, thereby preventing the induction of synthesis of beta interferon and other antiviral cytokines. This immuno-evasive action of NS3 was reversed by a specific ketoamide inhibitor of the NS3/4A protease and was dependent upon the protease activity of NS3/4A, indicating that NS3/4A is likely to cleave a cellular protein involved in IRF3 signaling following viral infection. While Foy et al. (8) demonstrated that both genotype 1a and genotype 1b proteases are capable of blocking IRF3 activation, it is intriguing to consider that the adaptive mutations we have identified within NS3/4A may promote its ability to direct such a cleavage, thereby enhancing replication of the virus by lessening cellular antiviral defenses.
The second group of adaptive mutations we identified within NS5A, K2040R, F2080V, and S2204I (Table 1), are likely to function in a fashion similar to NS5A adaptive mutations identified in genotype 1b replicons, which include S2204I. Although their specific mechanism of action is not known, they may either promote the ability of NS5A to assemble a functional replicase complex in Huh7 cells, or perhaps augment the immunomodulatory actions that have been proposed for this viral protein through its interactions with double-stranded RNA stimulated protein kinase R (10). Further studies will be required to determine which if either of these hypotheses is correct, but it is important to point out that the contribution of these adaptive mutations to the replication of the genotype 1a RNA in our studies appears to be additive to that of the NS3/4A mutations (Fig. 4 and 6), not synergistic as shown for the combination of Q1067R and K1691R (Fig. 4).
It remains to be shown whether the adaptive mutations we have identified in this study would have similar effects on RNA transcribed from any other cloned genotype 1a cDNA sequence or whether they may also enhance the ability of RNAs derived from other HCV genotypes to replicate in cultured cells. However, the identification of these adaptive mutations and the construction of genome length H77c RNAs that replicate with extraordinary efficiency (for HCV) in Huh7 cells (Fig. 7) provides new tools for ongoing efforts to establish an in vitro replication system that is fully permissive for all phases of the virus life cycle and that would lead to production of infectious virions. These tools will also be useful for drug discovery efforts, with the caveat that the adaptive mutations in NS3/4A may make the evaluation of protease active-site inhibitors complex, and the results of such tests, potentially difficult to interpret.
Acknowledgments
We are grateful to J. Bukh and R. H. Purcell for providing the pH77c cDNA clone used in these studies.
This work was supported in part by grants and contracts from the National Institute of Allergy and Infectious Diseases (U19-AI40035 and N01-AI25488) and the Advanced Technology Program of the Texas Higher Education Coordinating Board (004952-0027-2001).
REFERENCES
- 1.Alter, M. J. 1997. Epidemiology of hepatitis C. Hepatology 26:62S-65S. [DOI] [PubMed] [Google Scholar]
- 2.Beard, M. R., G. Abell, M. Honda, A. Carroll, M. Gartland, B. Clarke, K. Suzuki, R. Lanford, D. V. Sangar, and S. M. Lemon. 1999. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology 30:316-324. [DOI] [PubMed] [Google Scholar]
- 3.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 4.Blight, K. J., J. A. McKeating, J. Marcotrigiano, and C. M. Rice. 2003. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J. Virol. 77:3181-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brechot, C. 1997. Biology of hepatitis C viruses: clinical implications. Rev. Med. Intern. 18:893-905. [In French.] [DOI] [PubMed] [Google Scholar]
- 7.Farci, P., and R. H. Purcell. 2000. Clinical significance of hepatitis C virus genotypes and quasispecies. Semin. Liver Dis. 20:103-126. [PubMed] [Google Scholar]
- 8.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 9.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 10.Gale, M. J., Jr., M. J. Korth, and M. G. Katze. 1998. Repression of the PKR protein kinase by the hepatitis C virus NS5A protein: a potential mechanism of interferon resistance. Clin. Diagn. Virol. 10:157-162. [DOI] [PubMed] [Google Scholar]
- 11.Grobler, J. A., E. J. Markel, J. F. Fay, D. J. Graham, A. L. Simcoe, S. W. Ludmerer, E. M. Murray, G. Migliaccio, and O. A. Flores. 2003. Identification of a key determinant of hepatitis C virus cell culture adaptation in domain II of NS3 helicase. J. Biol. Chem. 278:16741-16746. [DOI] [PubMed] [Google Scholar]
- 12.Gu, B., A. T. Gates, O. Isken, S. E. Behrens, and R. T. Sarisky. 2003. Replication studies using genotype 1a subgenomic hepatitis C virus replicons. J. Virol. 77:5352-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R. St George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 17.Lanford, R. E., B. Guerra, H. Lee, D. R. Averett, B. Pfeiffer, D. Chavez, L. Notvall, and C. Bigger. 2003. Antiviral effect and virus-host interactions in response to alpha interferon, gamma interferon, poly(i)-poly(c), tumor necrosis factor alpha, and ribavirin in hepatitis C virus subgenomic replicons. J. Virol. 77:1092-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 21.Main, J., B. McCarron, and H. C. Thomas. 1998. Treatment of chronic viral hepatitis. Antivir. Chem. Chemother. 9:449-460. [DOI] [PubMed] [Google Scholar]
- 22.McHutchison, J. G., and M. W. Fried. 2003. Current therapy for hepatitis C: pegylated interferon and ribavirin. Clin. Liver Dis. 7:149-161. [DOI] [PubMed] [Google Scholar]
- 23.Pawlotsky, J. M. 2003. Hepatitis C virus genetic variability: pathogenic and clinical implications. Clin. Liver Dis. 7:45-66. [DOI] [PubMed] [Google Scholar]
- 24.Perrotta, A. T., and M. D. Been. 1996. Core sequences and a cleavage site wobble pair required for HDV antigenomic ribozyme self-cleavage. Nucleic Acids Res. 24:1314-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright-Minogue, J., N. Yao, R. Zhang, N. J. Butkiewicz, B. M. Baroudy, J. Y. Lau, and Z. Hong. 2000. Cross-genotypic interaction between hepatitis C virus NS3 protease domains and NS4A cofactors. J. Hepatol. 32:497-504. [DOI] [PubMed] [Google Scholar]
- 26.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA 94:8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao, N., P. Reichert, S. S. Taremi, W. W. Prosise, and P. C. Weber. 1999. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Struct. Fold Des. 7:1353-1363. [DOI] [PubMed] [Google Scholar]
- 28.Yi, M., F. Bodola, and S. M. Lemon. 2002. Subgenomic hepatitis C virus replicons inducing expression of a secreted enzymatic reporter protein. Virology 304:197-210. [DOI] [PubMed] [Google Scholar]
- 29.Yi, M., and S. M. Lemon. 2003. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 77:3557-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yi, M., and S. M. Lemon. 2003. Structure-function analysis of the 3′ stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA 9:331-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu, Q., J. T. Guo, and C. Seeger. 2003. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J. Virol. 77:9204-9210. [DOI] [PMC free article] [PubMed] [Google Scholar]