Abstract
VP16 is an essential structural protein of herpes simplex virus. It plays important roles in immediate-early transcriptional regulation, in the modulation of the activities of other viral components, and in the pathway of assembly and egress of infectious virions. To gain further insight into the compartmentalization of this multifunctional protein we constructed and characterized recombinant viruses expressing VP16 linked to the green fluorescent protein (GFP). These viruses replicate with virtually normal kinetics and yields and incorporate the fusion protein into the virion, resulting in autofluorescent particles. De novo-synthesized VP16-GFP was first detected in a diffuse pattern within the nucleus. Nuclear VP16-GFP was progressively recruited to replication compartments, which coalesced into large globular domains. By 10 to 12 h after infection additional distinct foci containing VP16-GFP could be seen, almost exclusively located at the periphery of the replication compartments. At the same time pronounced accumulation was observed in the cytoplasm, first in a diffuse pattern and then accumulating in vesicle-like compartments which were concentrated in an asymmetric fashion reminiscent of the Golgi. Inhibition of DNA replication resulted in prolonged diffuse nuclear distribution with minimal cytoplasmic accumulation. Treatment with brefeldin disrupted the cytoplasm vesicular pattern, resulting in redistributed large foci. Time-lapse microscopy demonstrated various dynamic features of infection, including the active induction of very long cellular projections (up to 100 μM). Vesicular clusters containing VP16 were transported within projections to the termini, which developed bulbous ends and appeared to embed into the membranes of adjacent uninfected cells.
The herpesvirus tegument is defined as the unstructured proteinaceous layer between the viral capsid and the envelope incorporating, in the case of herpes simplex virus (HSV), at least 15 or more virus-encoded proteins in differing copy number and accounting for approximately 50% of the volume of the virion (8, 17). Although only a subset of HSV tegument proteins is absolutely essential for replication in tissue culture, it is becoming clear that these proteins play important and diverse structural and functional roles during infection. Recent data on the pathway of maturation of HSV indicate that the virus acquires a primary envelope at the inner nuclear membrane but that this is followed by fusion at the outer nuclear envelope and reenvelopment in a Golgi-related compartment (12, 26, 40). However, the site(s) and mechanism(s) of recruitment of tegument proteins remain unclear.
One of the most-abundant tegument proteins is VP16 (18), the product of the UL48 gene (6) of HSV, and it is one of the subset of essential tegument proteins (43). VP16 plays important roles in the transcriptional regulation of immediate-early (IE) genes, in the modulation of the activities of other viral components, and in the assembly and egress of infectious virions. VP16 is assembled in approximately 1,000 to 1,500 copies per virion and as a component of the infecting virion acts to stimulate transcription of the immediate-early genes from the infecting genome (6, 29, 30). In its role as an activator of IE transcription, VP16 forms a multicomponent complex with at least two cellular proteins, Oct-1 and HCF, which is targeted to specific sites upstream of IE promoters on the infecting virus genome (21, 29, 31, 39, 44). The highly acidic C-terminal domain of VP16 then promotes transcription through recruitment of host RNA polymerase II and associated initiation components (7, 15, 33). Recombinant viruses which are defective in this activity of VP16, either through impaired complex formation or deletions within the activation domain, show reduced levels of IE transcription and significantly impaired replication (1, 37, 45). While this activity of VP16 acts to boost the onset of lytic cycle gene expression, it is not essential for virus replication. VP16 is, however, absolutely required for assembly of infectious virus (43). VP16-null mutants can be propagated on a complementing cell line since the endogenously synthesized protein can be incorporated into virions. In noncomplementing cells there was a relatively moderate effect on gene expression and overall DNA replication, and although capsids were formed there appeared to be a defect in DNA packaging. Virus maturation was severely affected and no infectious progeny virions could be detected (43). More recent work, accounting for a pleiotropic effect of VP16 deletion on vhs (virion host shutoff) activity has suggested that the defect in maturation of viruses lacking VP16 is in a step subsequent to primary envelopment at the inner nuclear membrane (28). Similar results have been observed with a VP16-null mutant of pseudorabies virus (PRV), in which virus replication is severely compromised at a step involving maturation in the cytoplasm (13). However, unlike the situation with HSV, null mutants of VP16 in PRV are viable and replicate, although with some 1,000-fold decrease in yield (13). Recently, interactions between VP16 and several other tegument proteins have been demonstrated. VP16 can be coimmunoprecipitated with vhs protein using anti-vhs antibodies, and vhs can inhibit complex formation between VP16/Oct-1/HCF on IE promoters (36). It was also shown that VP16 suppresses the translational shutoff function of vhs (22). The original VP16-null mutant, 8MA, exhibited a dramatic increase in vhs shutoff function which resulted in translational arrest during infection. Cells expressing VP16 were resistant to the shutoff function of superinfecting virions, and the translational defect in 8MA could be rescued in the derivative strain 8MA/ΔSma, in which the vhs function is inactivated (22). Thus, VP16 appears to modulate vhs activity, and VP16 binding by vhs appears also to be important for the latter's incorporation into the tegument. In virus-infected cells VP16 was also shown to copurify in a highly enriched fraction with another tegument protein, VP22 (10). A direct interaction was demonstrated in vitro, and it was further shown that, in transient transfections, coexpression of both proteins resulted in a profound reorganization from their normal independent locations to a novel macromolecular assembly in the cytoplasm which contained both proteins (10). The transcriptional activity of VP16 has been reported to be influenced by the tegument proteins UL46 and UL47 (46), and copurification between UL46 and VP16 has also been observed (20). VP16 has also been reported to interact with other virion components. In chemical cross-linking experiments, VP16 was reported to be cross-linked into complexes of high molecular weight in association with gB as well as gD and gH (47).
To gain further insight into the organization and compartmentalization of this essential multifunctional protein we report the construction and analysis of recombinant viruses expressing the VP16 linked to the green fluorescent protein (GFP). These viruses replicate with virtually normal kinetics and yields and incorporate the fusion protein into the virion. In analysis of the earliest stages of infection, we found no evidence for the association of VP16-GFP on infecting virions congregating around the centrosome. De novo-synthesized VP16-GFP appeared in a diffuse pattern in the nucleus at times when VP22 is mainly cytoplasmic. Between 5 and 7 h after infection the majority of nuclear VP16-GFP was recruited to replication compartments, but distinct domains lacking replication proteins and containing VP16 formed on the periphery of the replication compartments as infection progressed. Pronounced accumulation was then observed in cytoplasmic vesicle-like compartments which were concentrated in an asymmetric fashion reminiscent of the Golgi. Treatment with brefeldin disrupted the vesicular pattern, resulting in distributed large foci of the protein. VP16-GFP could be used to track the induction of very long cellular projections from infected cells, which appeared to develop bulbous ends and drive into the membranes of adjacent uninfected cells. These results are discussed with respect to the involvement of VP16 in virion maturation and its associations in particular with VP22.
MATERIALS AND METHODS
Cells, virus, and infections.
Vero, BHK-21, and COS-7 cells were grown in Dulbecco's modified minimal essential medium containing 10% newborn calf serum. HSV type 1 (HSV-1) strain 17 was used as the parental virus. Titrations were routinely performed in Vero cells or BHK-21 cells. Construction of the recombinant VP16-GFP viruses was performed by cotransfection of purified virus DNA together with the appropriate transfer vector as described below. Purification of virus particles was performed after low multiplicity of infection (MOI) of BHK-21 cells. Assays for transactivation by VP16-GFP fusion proteins were performed in COS cells as previously described (16).
Construction of HSV-1 VP16-GFP viruses.
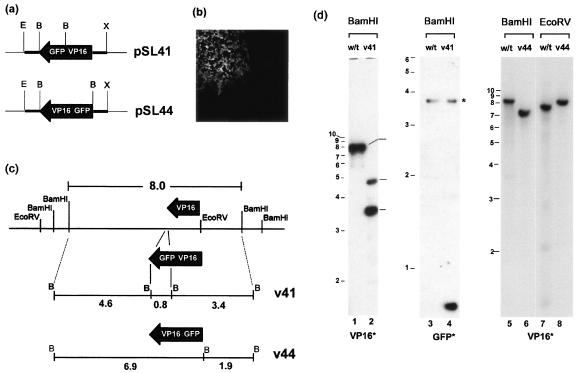
The main features of the construction of the VP16-GFP and GFP-VP16 viruses are illustrated in Fig. 1. The transfer plasmid vector pSL41 (Fig. 1a) was generated as follows. The VP16 open reading frame (ORF) (UL48 gene) was amplified by PCR as a BglII/BamHI DNA fragment using as a template the BamHI F fragment of the viral genome and was inserted into the unique BamHI site of pGE122 (kindly provided by G. Elliott), generating pSL39. pGE122 contains approximately 200 bp of each of the 5′ and 3′ flanking sequences of the UL48 gene, amplified by PCR as an XbaI/BamHI and a EcoRI/BamHI fragments, respectively. A BamHI fragment encompassing the GFP ORF was cloned from the commercial vector pEGFP.N (Clontech) into the BamHI site of pSL39, in frame with the VP16 ORF yielding pSL41. This plasmid encodes VP16-GFP and contains the native promoter and leader sequences at the 5′ end and the native nontranslated and polyadenylation sequences at the 3′ end. A similar strategy was used to construct the transfer vector, pSL44 which contains GFP fused at the N terminus of VP16 in a vector containing the same native 5′ and 3′ sequences as above.
FIG. 1.
(a) Schematic diagram of the transfer plasmids pSL41 and pSL44 which contain the genes for VP16-GFP and GFP-VP16, respectively, in each case flanked by 200 bp of the 5′ and 3′ regions of the native VP16 gene, constructed as described in Materials and Methods. These plasmids were cotransfected with purified HSV-1 [17] DNA, and autofluorescent plaques (b) were isolated and plaque purified. (c) A schematic representation of the VP16 region and the native BamHI and EcoRV sites is illustrated, together with the predicted structure after recombination into the locus. (d) Diagnostic Southern blotting of DNA from stocks of plaque-purified virus was performed to confirm recombination. DNA samples from HSV wt, v41, or v44 viruses are indicated at the top of each lane together with the diagnostic enzyme, and the radiolabeled probes, either VP16 or GFP, are indicated at the bottom. The profile of fragments precisely reflects that expected in panel c. The band marked with an asterisk is likely due to a weak hybridization because of the presence of a short region from HSV thymidine kinase gene in the commercial vector (pEGFPN) used to make the GFP probe.
The construction of VP16-GFP recombinant viruses was performed essentially as described before for the construction of a GFP-VP22 virus (11). Vero cells (106 cells in a 60-mm-diameter dish) were cotransfected with 3 μg of pSL41 or pSL44 and 1 μg of viral DNA. After a complete cytopathic effect (CPE) was observed (approximately 5 days), infected cells were harvested, titrated on Vero cells, and examined live for GFP fluorescence. Green plaques were picked, titrated, and subject to three to five rounds of plaque purification, the resulting viruses being labeled HSV-1 v41 and HSV-1 v44, respectively.
Purification of viral DNA.
Vero cells were infected with HSV-1 (17), [v41] or [v44] at an MOI of 0.02. When complete CPE was observed, infected cells were lysed by cycles of freeze-thawing and clarified by centrifugation, and supernatants were applied to a 2-ml 40% sucrose-NTE cushion (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 150 mM NaCl) and centrifuged for 2 h at 25,000 rpm at 4°C in a 641TH rotor. The virus pellet was resuspended in 800 μl of NTE, sonicated, and transferred to a fresh tube. DNase I and RNase A (10 μl each of 10-mg/ml stock solutions) were added, the solution was made to 10 mM MgCl2, and the sample was incubated at 37°C for 2 h. Following inactivation of DNase by the addition of 10 mM EDTA, viral particles were lysed in 1% sodium dodecyl sulfate (SDS) and digested in the presence of proteinase K (400 μg/ml) for 2 h at 37°C. Proteins were extracted, once with 1 volume of phenol and twice more with 0.5 volume of phenol and 0.5 volume of chloroform-isoamyl alcohol (24:1). DNA was precipitated with ethanol, washed in 70% ethanol, dried, and resuspended in water.
Diagnostic Southern blotting of virus DNA.
DNA was digested with either BamHI or EcoRV and fractionated by electrophoresis in 0.6% gels. Following transfer to Hybond-N membranes, the blots were probed by standard methods. The probe for VP16 was an isolated fragment encompassing the VP16 ORF plus approximately 30 and 200 bp of the 5′ and 3′ flanking DNA, respectively. The probe for GFP was the commercial plasmid pEGFP.N (Clontech). Probes were labeled by random priming using [α-32P]dCTP and denatured just before addition to the hybridization solution. Prehybridization, hybridization washing, and autoradiography were all performed by standard methods.
Purification of virus particles.
Viral particles were purified from the medium of infected BHK-21 cells essentially as described in detail elsewhere (11), except that the cells were initially infected at an MOI of 0.02 PFU/cell. After initial pelleting, virus preparations were applied to 5 to 15% Ficoll gradients and the bands were harvested by side puncture of the tube. Virus particles were then pelleted and resuspended in phosphate-buffered saline.
SDS-PAGE and Western blotting.
Mock-infected or infected cells (1× 106 to 2 × 106 cells) were washed in cold phosphate-buffered saline, and total lysates were prepared by adding 250 μl of SDS loading buffer. Samples were briefly sonicated prior to electrophoresis. Equal amounts of total proteins were fractionated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred onto Hybond-C membranes. Primary antibodies for immunodetection were diluted as follows: anti-IE110k (monoclonal antibody [MAb] 11060; 1:10,000), anti-IE175k (MAb 10176; 1:5,000), anti-HSV-thymidine kinase (tk; 1:2,000), anti-VP16 (MAb LP1, 1:4,000; or polyclonal rabbit antibody POS1, 1:2,000), anti-VP22 (polyclonal rabbit antibody AGV30; 1:50,000), antiactin (MAb AC-40; 1:5,000; Sigma). Target proteins were visualized by enhanced chemiluminescence (Pierce).
Live-cell microscopy.
Cells were plated in two-well chambered coverglasses, infected as indicated in the figure legends, and incubated at 37°C in a humidified incubator at 5% CO2. The chambered slides were then transferred at various intervals to the stage of an inverted confocal microscope (LSM410; Carl Zeiss Ltd.) for analysis, imaged, and replaced in the incubator for further analysis. Images were routinely acquired using a Plan-apochromat ×63 oil immersion objective lens, NA 1.4, and zoom factors ranging from 1 to 8 of the LSM410 acquisition software. The images for the time-lapse animation (see Fig. 9) were acquired with Plan-Neofluar ×40 oil immersion lens, NA 1.3. Scale bars are indicated on the images. For long-term analysis and time-lapse microscopy, cells were plated onto 42-mm coverslips within 60-mm-diameter culture dishes. The coverslips were then transferred to a Bachhoffer POC-chamber (Carl Zeiss Ltd.), and the chamber placed in a Saur heated frame covered with a Perspex lid through which a constant supply of 5% CO2 was delivered. For each time point a Z-series of images was collected and stacked using the XYZT module of the LSM410 confocal software, to produce an individual Z stack for each time point. The series were then imported into QuickTime for presentation as an animated movie. Other images were annotated using Adobe Photoshop.
FIG. 9.
Analysis of VP16-GFP movement and accumulation within infected cells by time lapse. This figure represents the first frame of a QuickTime movie. Vero cells were plated on 40-mm coverslips which were assembled into a Bachhoffer POC-chamber (Carl Zeiss Ltd.) which could be maintained at constant temperature and with a controlled CO2 environment on the confocal microscope. Cells were imaged using the XYZT module of the LSM410 imaging software, collecting 10 individual sections separated by 1.5 μm at each time point, stacking the sections to give one extended depth of focus and compiling the individual images into a QuickTime movie series. Several movies were compiled. This series summarizing some of the key features discussed in the text was taken at 72 h after an original infection at an MOI of 0.001. Each of 150 frames was taken at 5 min covering a total time span of 12 h. Bar, 50 μm.
RESULTS
Construction of recombinant HSV expressing VP16-GFP proteins.
VP16, encoded by the UL48 gene of HSV-1, is an essential structural component and a key regulatory protein, involved in the activation of virus immediate-early gene expression. Previous analysis has demonstrated that other tegument proteins can be modified by fusion to GFP and that such fusion proteins are incorporated normally into the virion (11, 19). However, unlike the previous candidate proteins, VP16 is an essential structural protein and plays an additional important role in activation of IE expression. Since there was a possibility that orientation of GFP with respect to VP16 may influence one or both functions, we constructed VP16-GFP fusions in which the GFP component was present at either the N- and C-terminal ends. Prior to the construction of viruses containing the VP16 fusion proteins we also examined, by transient-transfection assays, whether the presence of GFP fused to the C terminus of VP16 would affect the ability of the protein to transactivate in vivo. The results showed that VP16-GFP could activate transcription of an IE reporter gene as efficiently as native VP16 (data not shown), consistent with previous analysis of various proteins showing that the activation domain of VP16 can function in chimeric proteins and in an internal position.
To replace the native VP16 gene with that encoding VP16-GFP or GFP-VP16, purified HSV-1(17) DNA was cotransfected with the transfer vectors, pSL41 or SL44, respectively, as described in detail in Materials and Methods. These vectors contain the appropriate fusion construct flanked by approximately 200 bp of the native 5′ promoter-leader sequence and 3′ untranslated sequence (Fig. 1a). After a complete CPE was observed, virus was harvested and screened for intrinsically fluorescent plaques, a typical example of which is shown in Fig. 1b. Plaques were isolated, and the virus was titrated and then plaque purified a further three to five times to generate the resulting strains HSV-1 v41 (for VP16-GFP) and HSV-1 v44 (for GFP-VP16). To ensure that recombination had occurred as expected, DNA was purified from each of the strains and compared with that of the parental wild-type (wt) strain. Incorporation of GFP-VP16 or VP16-GFP into the virus genome was examined by probing BamHI or EcoRV digests and monitoring changes in the 8.0-kb BamHI F fragment and overlapping 7.4-kb EcoRV I fragments (Fig. 1c). Incorporation of the fusion construct in place of the native VP16 gene should result in an increase in size of the EcoRV I fragment from 7.4 kb to approximately 8.2 kb. The expected result was observed for both HSV-1 v44 (Fig. 1d, cf. lanes 7 and 8) and HSV-1 v41 (data not shown). The extra BamHI site incorporated in the construction of the vectors pSL41 and pSL44 (Fig. 1a) also aided in the diagnostic tests. Thus, as expected for HSV-1 v41 using a VP16 labeled probe, the 8.0-kb BamHI F fragment was lost and two new fragments of 4.6 and 3.4 kb were observed (Fig. 1d, cf. lanes 1 and 2). (Note that the lower detection of the 4.6-kb fragment was due to the use of a purified, isolated probe for VP16 which encompassed only a short region 3′ of the gene.) Correspondingly a new GFP containing a 0.8-kb BamHI fragment was observed for HSV-1 v41 when the blots were probed with a GFP-specific probe (Fig. 1d, lanes 3 and 4). For HSV-1 v44, the normal BamHI F fragment was replaced with an approximately 7.0-kb fragment (Fig. 1, cf. lanes 5 and 6). This result indicated that the extra BamHI site on the 5′ side of the GFP-VP16 gene had been incorporated, but not the one on the 3′ side (which would have produced the 4.6-kb fragment).
Expression of VP16-GFP and replication of recombinant viruses.
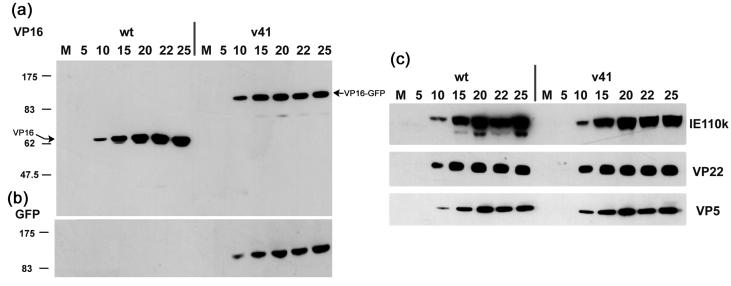
To examine expression of the VP16-GFP fusion proteins, Vero cells were infected with either the wt, HSV-1 v41, or HSV-1 v44 at an MOI of 10 PFU/cell, the cells harvested at intervals thereafter and total cell lysates analyzed by SDS-PAGE and Western blotting. The time course of accumulation of VP16, detected using the MAb LP1, is shown in Fig. 2a. For HSV-1 v41, native VP16 was not observed and instead was replaced with the accumulation of a band of approximately 95 kDa. The same band was detected when probing the blots with anti-GFP antibody while as expected no reaction was observed for the wt virus (Fig. 2b). Similar results were obtained for HSV-1 v44 (data not shown). The results indicate that in the recombinant strains, expression of native VP16 had been replaced with expression of VP16-GFP fusion proteins. Expression kinetics and overall accumulation of VP16-GFP in the v41 and v44 recombinant strains were similar to those of native VP16 in the parental strain. We also compared expression of additional virus proteins including IE110, VP22, and VP5, representing different temporal classes. No significant differences were observed between wt and recombinant viruses in either the levels or kinetics of accumulation between the viruses (Fig. 2c and data not shown).
FIG. 2.
Vero cells were infected with wt or v41 strains at an MOI of 10 or mock-infected (m), and cultures were harvested at the times indicated (hours postinfection). Equivalent amounts of total cell lysates were separated by SDS-PAGE, transferred to nitrocellulose and probed with anti-VP16 (a) or anti-GFP (b) antibodies. Expression of native VP16 was absent and replaced with a VP16 fusion protein of approximately 95 kDa which was also detected with anti-GFP antibody. (c) Samples from the same time course were probed for expression of the immediate-early protein IE110k (ICP0), and the structural proteins VP22 and VP5. No significant differences in the abundance or kinetics of accumulation were observed between the wt of recombinant viruses.
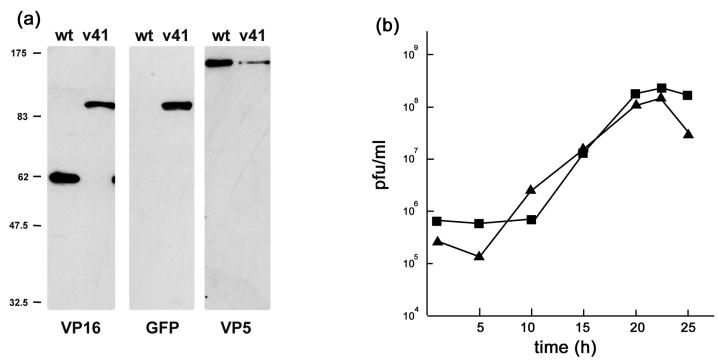
To examine incorporation of the VP16-GFP fusion proteins into virions, extracellular virus particles were pelleted and purified on Ficoll gradients as previously described, and protein profiles were analyzed by Western blotting with anti-VP16 and anti-GFP antibodies after separation by SDS-PAGE. The results show that native VP16, present in the wt virus, was absent from the profile of the recombinant virus, and replaced with a band migrating at approximately 95 kDa (Fig. 3a). The 95-kDa band was detected with both anti-VP16 and anti-GFP antibodies. Note that in this case, as shown with the control anti-VP5 antibody, less material from the HSV-1 v41 preparation was loaded on the gel, accounting for the lower abundance of VP16-GFP compared to VP16 in the recombinant and wt viruses. From this and additional analysis we found no evidence for any significant difference in the abundance of VP16 versus VP16-GFP in virion preparations.
FIG. 3.
(a) Incorporation of VP16-GFP into virions. Gradient-purified virions from wt or v41 infected cells were separated by SDS-PAGE, blotted, and probed with antibodies to VP16, GFP, or VP5 as indicated. VP16 was present in v41 virions as a 95-kDa protein and when corrected for VP5 loading was incorporated in similar amounts as native VP16 into wt virions. (b) Single-step growth curves for wt and v41 viruses. Vero cells were infected at an MOI of 10, and cells and media were harvested and titrated on Vero cells. Results are plotted as total PFU/ml for wt (squares) and v41 virus (triangles). The results indicate that the recombinant v41 replicates with similar efficiency to wt virus.
VP16 is an essential structural protein of HSV-1 (43) being recruited into the tegument compartment at approximately 1,500 to 2,000 molecules per virion. While little difference was observed at the level of protein synthesis it remained possible that fusion of the GFP moiety onto VP16 may have affected virus assembly. To examine this possibility we performed single-cycle growth analysis comparing each of the recombinant viruses to the wt after high MOI (10 PFU/cell). No significant differences were observed in the total yield or the rate of replication for either strain compared to the parent virus (Fig. 3b and data not shown).
Localization of VP16-GFP: compartmentalization within the nucleus.
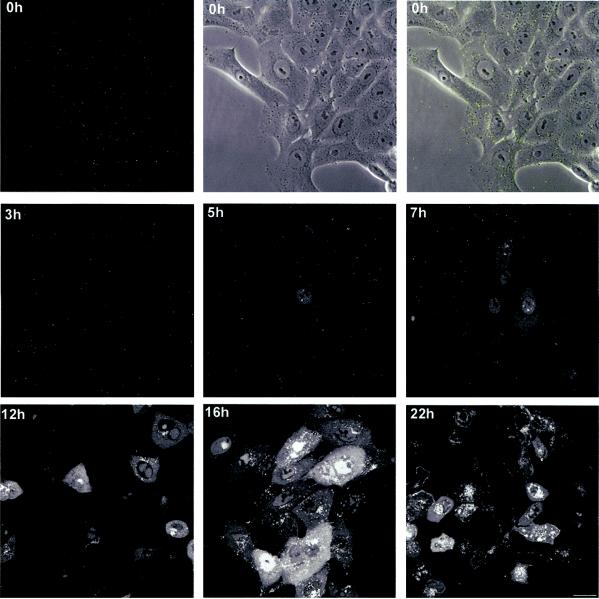
We next examined the localization of VP16-GFP in live infected cells. Vero cells were plated in chambered coverslips, infected with HSV-1 v41 at an MOI of 10 PFU/cell, and examined by confocal microscopy at various intervals thereafter. In this case the cultures were replaced in the incubator and typical images recorded for each time point (Fig. 4). Images were collected in live cells by sectioning though the depth of the cells (at 1-μm intervals) and then stacking the sections to yield an extended depth of focus covering the entire field. Immediately after infection, well-resolved fluorescent particles could be seen over the cells (Fig. 4 0 h, separate fields for fluorescence, phase, and overlay). No significant grouping of these particles could be discerned at the earliest stages of infection, and by 3 h after infection only a general decrease in the number of fluorescent particles could be seen. Between 3 and 5 h after infection VP16-GFP was now detected mostly in a diffuse pattern within the nucleus (Fig. 4, 5 h). At around 5 h, VP16-GFP could be observed beginning to accumulate in bright punctate foci with numbers ranging from one to approximately eight per cell, but mostly within the range one to four. At intermediate times from 7 to 12 h after infection, these nuclear foci had expanded and coalesced into large globular domains within the nucleus which likely represented replication compartments (see below). Between 12 and 16 h these further expanded resulting in intense nuclear accumulations. At around 7 to 12 h, VP16-GFP could be observed in a homogeneous diffuse cytoplasmic pattern, beginning by 12 h after infection to accumulate in vesicular like foci within the cytoplasm, and increasing in intensity at late stages with additional accumulation at the boundaries between cells (see below).
FIG. 4.
Localization of VP16-GFP in virus infected cells. Vero cells, grown in chambered coverslips were infected with HSV-1 v41 at an MOI of 10 for 1 h, washed in buffer, and reincubated at 37°C in medium containing 2% calf serum. At various times thereafter the cultures were removed from the incubator and images captured in live cells as described in Materials and Methods. Representative images are show wherein the extended depth of field was captured by Z-sectioning through the depth of the cell and then stacking the sections using the appropriate Zeiss LSM software module. The time point at 0 h (i.e., 1 h after addition of virus), was imaged both for GFP and for phase contrast, both fields being combined in the top right hand panel. Bar; 25 μm.
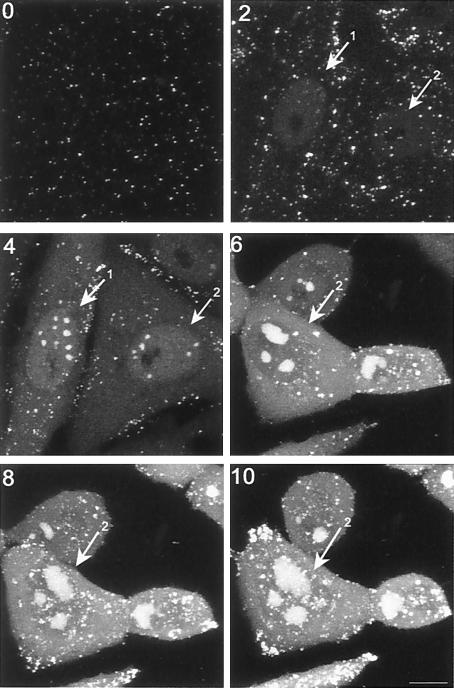
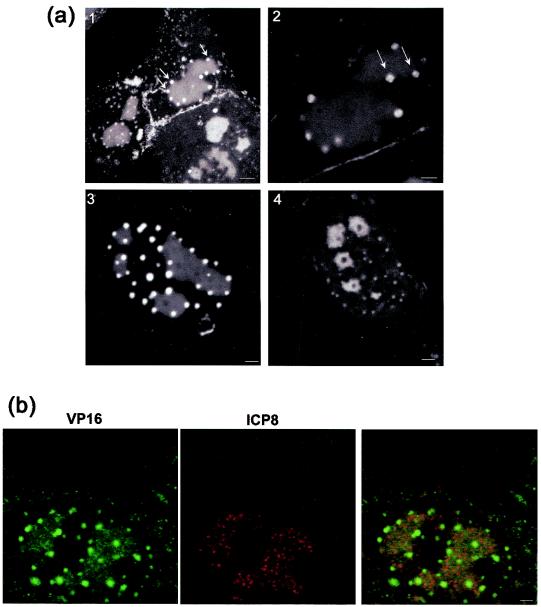
To analyze the pattern of accumulation in more detail, we examined individual infected cells throughout infection. Typical results examining infection up to 10 h postinfection are shown on Fig. 5. Within the cell labeled 1, weak diffuse nuclear GFP-VP16 was observed, in this case by 2 h, while discrete nuclear foci were observed by 4 h on the background of diffuse localization (4 h, cell 1,2). Two hours later these foci had coalesced into three large intense domains (6 h, cell 2). (Note that the imaged field for time points 6, 8, and 10 had shifted slightly to the right). Interestingly a distinct aspect of the nuclear pattern began to emerge by around 8 to 10 h, wherein foci of GFP-VP16 began to appear on the periphery of the large globular domains. While more difficult to visualize in the total extended depth of field images such as those in Fig. 4 and 5, the new foci were readily observed by imaging individual sections through cells (Fig. 6a, panel 1). Foci of GFP-VP16 can be seen accumulating almost exclusively at the periphery of the globular domains. While the large globular domains could be irregular in size and shape, we noted that the GFP-VP16 foci at the periphery were for the most part homogeneous, spherical, and of approximately similar size within any individual cell. This may indicate that the domains progressively increase in size, in a uniform way within the infected cell. We also note that it was occasionally possible to visualize substructure within these foci, such as the ones indicated in the higher-magnification image (panel 2), where smaller areas of increased intensity could be discerned. Generally, these foci appeared at approximately 10 to 12 h after infection, with their number appearing to be related to the number of large globular domains. For example, in panel 3, three to four large globular domains could be observed, each with the additional GFP-VP16 peripheral foci. We occasionally observed foci that appeared not to be on the periphery of a globular domain, and although this may indicate the occasional dissociation of foci from the globular domains, this more likely reflects the optical sectioning used for optimal imaging. Finally we also noted that by sectioning through live cells, we were able to occasionally observe that the larger globular domains representing replication foci (see below) were present around areas devoid of GFP-VP16 (panel 4). This could represent internal areas of the replication foci that were specifically devoid of GFP-VP16, or areas of the cell, such as the PML domains, around which replication compartments matured (see discussion). We note that general features of VP16 recruitment into replication compartments and into the peripheral smaller foci was also observed by immunofluorescence studies of cells infected with wt HSV-1 (data not shown).
FIG. 5.
Progression of localization of GFP-VP16 in individual infected cells. Vero cells grown on gridded coverslips so that individual specific cells could be tracked, were infected with HSV v44 at an MOI of 10. The cells were infected and replaced in the incubator, and live cell images were recorded using the same settings and laser attenuation at various times thereafter (time indicated in hours postinfection on each panel). This time course highlights key aspects of the nuclear phases of localization where VP16 first appears in a general diffuse pattern, concentrating in subnuclear foci which fuse into large compartments, and additionally is recruited to numerous smaller foci which begin to form around 8 to 10 h after infection. Note that the imaged field for the 6, 8, and 10 h time points had shifted slightly to the right. Bar, 10 μm.
FIG. 6.
GFP-VP16 localization in replication compartments. (a) Aspects of GFP-VP16 localization in the nucleus were studied in more detail using Z-sectioning and higher magnification imaging. Vero cells were infected and live cells imaged as described above. In panel 1 distinct small foci directly impinging on the replication compartments can be readily observed (arrows). They are spherical and relatively homogeneous in size distribution. Occasionally (panel 2), it appeared that these novel foci had themselves substructures resembling coalescing microfoci, but this was generally difficult to discern. The majority of the smaller GFP-VP16 foci appeared to be associated with the perimeter of the larger replication foci (panel 3). By sectioning cells through the larger foci, areas devoid of GFP-VP16 could be identified, as shown in panel 4. Bar; panel 1, 5 μm; panels 2 to 4, 2 μm. (b) VP16 colocalizes with ICP8 in replication compartments, but ICP8 is not recruited into the adjacent foci. To confirm that the larger coalesced compartments containing GFP-VP16 represented replication compartments, infected cells were fixed and stained with antibody to ICP8 or to GFP (as indicated). With this antibody ICP8 is observed in a microspeckled pattern in the compartments, and there was clear colocalization with VP16. However, ICP8 was absent from the VP16-containing foci which formed on the perimeter of the replication foci. Bar, 2 μm.
To confirm that the progression of VP16 from an initially nuclear diffuse pattern to accumulation in foci represented accumulation into replication foci, we examined localization between VP16 and ICP8 in fixed cells (Fig. 6b). As expected the large globular foci containing GFP-VP16 colocalized with ICP8. However, the adjacent VP16-positive domains which developed around the perimeter of the replication foci were devoid of ICP8, indicating selective recruitment of VP16 into these additional foci.
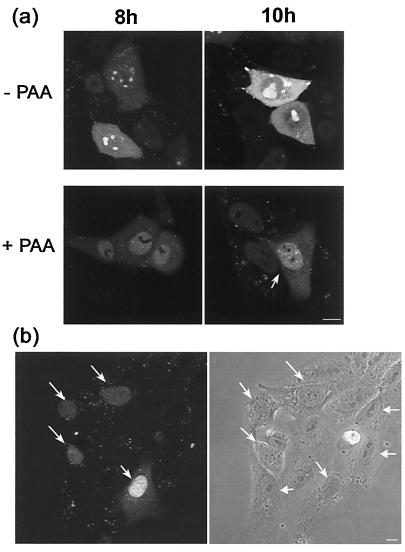
We also examined the localization of GFP-VP16 in the absence and presence of phosphonoacetic acid (PAA) an inhibitor of virus DNA synthesis. The results (Fig. 7a) show that in the presence of PAA, GFP-VP16 accumulated in a diffuse pattern that was maintained for prolonged times, without recruitment into nuclear foci. While the localization of nuclear GFP-VP16 in foci which coalesce into large globular domains is clearly similar to the pattern expected of replication foci, the exclusively diffuse nuclear pattern in the absence of DNA synthesis does not exactly reflect that of ICP8. In the absence of DNA synthesis ICP8 accumulates in quite numerous, distinct prereplicative compartments, a subset of which progress to replication compartments (4, 24). It may therefore be that VP16 is not recruited to prereplicative compartments containing proteins such as ICP8 and only associates with replication compartments due to a replication-dependent interaction.
FIG. 7.
Effect of inhibition on DNA replication on localization of GFP-VP16. (a) Vero cells plated in chambered coverslips were infected with HSV-1 v44 at an MOI of 10, the inoculum replaced 1 h later and the cells further incubated in the presence or absence of PAA at a concentration of 400 μg/ml as indicated. At various time points (indicated in hours) the localization of GFP-VP16 was examined by confocal microscopy in live cells as discussed in the text. (b) Disruption of normal nucleolar organization early in infection. Vero cells infected with HSV-1 v44 at an MOI of 10 analyzed very early after infection (2 h) show de novo-synthesized VP16 in a diffuse nuclear pattern (arrows). When examined by phase microscopy these same cells show a distinctly altered gross nucleolar morphology comprising an untangled fibrillar type pattern (diagonal arrows) readily distinct from the more compact, phase dense lobular morphology in those cells not yet expressing VP16 (horizontal arrows). Bar, 10 μm.
Fate of input VP16.
We wished to examine whether at the level of detection by GFP fluorescence in live cells, we could track the fate of input VP16 from the infecting particles. At an input MOI of 10 PFU/cell and with an estimate of 1,500 to 2,000 molecules per virion (18, 23), there should on average be several thousands molecules of VP16 deposited within the cell immediately after infection. Although it has been previously reported (38) that infecting particles congregate around the microtubule organizing center, this analysis used capsid components as the detection system. Throughout this work we found no evidence for such progressive localization of VP16-GFP fluorescent particles (Fig. 3 and 4 and data not shown). Although not definitive evidence, this may indicate that VP16 dissociates from the capsid upon entry, a conclusion previously reached from studies of the transcriptional activity of VP16 wherein, using a temperature-sensitive virus, VP16 was able to activate a resident nuclear target gene despite the accumulation of capsids on the outside of the nuclear pore (2). From examination of cells after infection in the presence of cycloheximide we were unable to obtain any firm evidence for the fate of input VP16. No diffuse VP16 could be observed in the absence of de novo protein synthesis (data not shown) indicating that the diffuse nuclear pattern seen beginning between 3 and 5 h was indeed due to de novo-synthesized GFP-VP16 rather than the slow accumulation of protein to detectable levels. We note as an aside that during these studies we observed that at early stages of infection, when GFP-VP16 was just beginning to accumulate in the nucleus and some infected cells were not yet positive, those cells expressing VP16 could be identified by phase microscopy by virtue of a reorganized nucleolus (Fig. 7b). This occurred between 2 and 4 h, well before any other morphological effect was evident. Nucleolar disruption from a defined dense pattern to a loose, tangled appearance (Fig. 7b) could be used to predict those cells which were expressing discernible levels of protein at the very earliest stages. Some form of nucleolar sparing of newly synthesized viral proteins remains during HSV infection, indicative of some remaining nucleolar organization. However, there is clearly significant reorganization of the nucleolus very early in infection. While not the subject of investigation here, further analysis of this process and its requirements are under way using GFP-tagged resident nucleolar proteins.
VP16 compartmentalization within the cytoplasm.
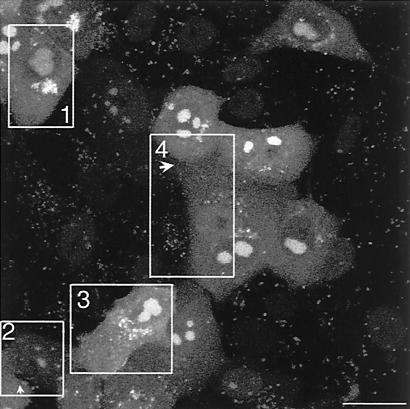
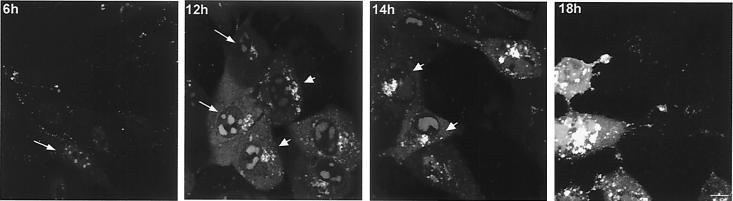
As indicated above, by 6 to 10 h GFP-VP16 began to appear in the cytoplasm, initially in a diffuse pattern with no specific localization and progressively into small vesicular foci. Further aspects of cytoplasmic localization are shown in Fig. 8 and 9. By 12 h GFP-VP16 showed pronounced accumulation in perinuclear clusters reminiscent of the Golgi (Fig. 8, 12 h). Later abundant larger foci and intense aggregates accumulated (Fig. 8, 14 to 18 h). Among the advantages of probing protein localization using GFP fusion proteins are the absence of a requirement for cell fixation and processing and the ability to perform time-lapse analysis yielding a dynamic picture of compartmentalization and movement in individual cells. Features of late infection were examined with the use of time-lapse analysis wherein coverslips in a sealed chamber were maintained at 37°C in a heated platform at 5% CO2 and images collected at desired intervals, as described previously (11). The results of one such analysis showing several features of infection are shown in the accompanying video (Fig. 9 represents the first frame [video available at http://www.mcri.ac.uk/herpesvirus/video/]). Within the cell marked box 1, VP16-GFP has accumulated in perinuclear clusters. Progression of infection in the cell (box 2) lags somewhat behind. In both cells intense clusters can be seen with smaller satellite foci moving within the cell. Large sections of the clustered foci can be seen to detach from the main body and move rapidly across the cell, often being reabsorbed back into the main cluster. In both these examples the vast majority of cytoplasmic VP16-GFP is seen as a single large vesicular cluster. Frequently, as seen in cell (box 3), the perinuclear clusters accumulate and as individual foci expand, they also disperse into a more random pattern throughout the cell. It may be that the pericentric clustering of VP16-GFP represents its accumulation in a Golgi-associated compartment which has remained intact in the cell, while dispersal may represent alteration to the underlying cellular compartments, a process which has been previously described (5).
FIG. 8.
VP16-GFP accumulation in the cytoplasm. Vero cells were infected and processed as for Fig. 4. From approximately 10 h onwards VP16-GFP begins to accumulate in the cytoplasm, with a homogeneous diffuse pattern underlying progressive accumulation into vesicular clusters which accumulate pericentrically around the nucleus. As infection progresses VP16-GFP clusters accumulate at the periphery of cells and in projections as discussed in the text. Bar, 10 μm.
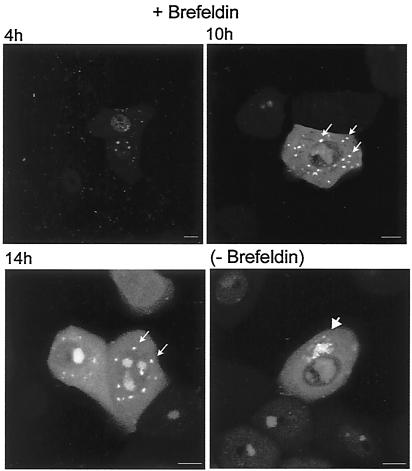
We examined whether the pattern of VP16 localization would be altered under conditions where the Golgi would be disrupted, i.e., in the presence of brefeldin A. Infected cells were treated with brefeldin at 3 h after infection and the localization of GFP-VP16 monitored thereafter. The results (Fig. 10) indicate that while little effect was observed on the accumulation of GFP-VP16 in the nucleus or the globular replication compartments, a distinct effect was observed in the pattern of accumulation in the cytoplasm. Instead of accumulation in perinuclear vesicular foci, GFP-VP16 was found in comparatively large, discrete foci, which were less numerous and randomly dispersed throughout the cytoplasm, showing no obvious localized clustering (Fig. 10, 10 to 14 h). Taken together, the asymmetric clustering seen as infection progresses and the results of brefeldin treatment are consistent with the interpretation that VP16 associates with a perinuclear Golgi-associated compartment. However, the effect of brefeldin could be an indirect one and other interpretations of the cytoplasmic localization are possible. Further work, in conjunction with cell lines expressing tagged Golgi markers is under way to analyze late cytoplasmic compartmentalization in detail.
FIG. 10.
Effect of brefeldin A on the localization of GFP-VP16. Vero cells were infected with HSV-1 v44 at an MOI of 10 and incubated in the presence or absence of brefeldin A at 1 μg/ml. Live infected cells were analyzed at various times thereafter as indicated. While the nuclear pattern of GFP-VP16 was not significantly altered, within the cytoplasm, GFP-VP16 accumulated in larger distributed spherical foci, which did not cluster pericentrically within the cell, in contrast to the vesicular clustering seen in the absence of the drug. Bars, 10 μm.
An additional feature of infection can be observed in the video of the infection within box 2. As infection progresses, the adjacent cell retracts and a long process emanating from this cell extends towards the cell on its right. Bright foci of GFP-VP16 accumulate at the tip of the process which appears to embed and then spread onto the adjacent cell membrane. Later this material either disperses throughout the membrane, is endocytosed or otherwise is lost. We frequently observe this activity and examine it here at the morphological level in more detail. A good example of the induction in infected cells of this type of long single process is shown in box 4. A cell towards the bottom of the box shows the perinuclear clustering described above. As infection progresses an elongated process develops which progresses towards the adjacent cell. Vesicular material within the body of the cell moves towards the tip, but can also be seen to halt or reverse direction. A single image showing these extended processes is illustrated in Fig. 11. The processes can be up to 100 μm in length and frequently terminate with a bulbous-like morphology likely reflecting the movement of cytoplasm and vesicles towards the end (see cell at left hand side of panel). A higher magnification image of the boxed section of Fig. 11 is shown in panel b, and shows that the tips of these processes make intimate contact with adjacent cells. The VP16-GFP containing material over this cell, which is not itself yet expressing endogenous VP16, originates from the infected cell at its lower left side and accumulates there due to the extended process developed from this infected cell and the trafficking of VP16-GFP vesicular material along it (see also cell box 4 in the movie above). We also observed that these termini could subsequently detach from the originating cell, resulting in the association of VP16-GFP containing membrane components and vesicles with the recipient cell (see Discussion). Generally the induction of these processes was more readily observed under conditions of low cell density and asynchronous infection where not all cells were initially infected. Their induction was also not restricted to cell lines. Similar observations on the induction of elongated processes and trafficking of VP16-GFP material were observed in primary keratinoctyes (data not shown).
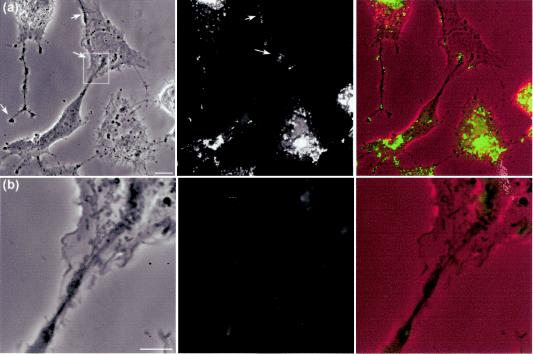
FIG. 11.
HSV infection induces long extensions along which VP16-GFP clusters accumulate. The demonstration of long cellular projections induced by HSV infection is best seen when cells are plated at a relatively low density. Vero cells plated at low density were infected with HSV-1 v41 as before and analyzed by phase microscopy and for VP16-GFP localization. In the combined images (right-hand panel) the phase image is placed in the red channel of the color image, while the VP16-GFP signal is in the green channel. (a) In the top panel long extensions can be seen which can have a bifurcated or bulbous end (arrow), while the right-hand cell shows a similar extension impinging upon a cell not yet expressing VP16. The arrows point to VP16-GFP material which overlies this uninfected cell but originates from the lower infected cell in the panel. Bar, 10 μm. (b) Enlargement of the boxed section of panel a, as discussed in the text. Bar, 5 μm.
DISCUSSION
VP16 is an essential structural protein of HSV. It is incorporated into the tegument compartment of the virion and plays important roles in the transcriptional regulation of immediate-early genes, in the modulation of the activities of other viral components and in the pathways for assembly and egress of infectious virions. To gain insight into the organization and compartmentalization of this multifunctional protein we constructed and analyzed recombinant viruses expressing VP16 linked to the green fluorescent protein. We have previously used this approach to examine other tegument proteins, but unlike these components VP16 is an essential protein required for replication, and incorporation of GFP may have had a deleterious effect on replication and assembly. However, viruses HSV-1 v41 and v44, respectively, encoding fusion proteins with GFP at the C terminus or N terminus of VP16, replicated with similar kinetics to wt virus and incorporated the fusion proteins into the virion in similar abundance.
VP16 nuclear localization.
Newly synthesized VP16 accumulated firstly in a diffuse pattern in the nucleus. Between 5 and 7 h after infection, nuclear VP16-GFP was recruited to replication compartments within the cell, which coalesced into large globular domains. We previously demonstrated a direct physical complex in virus infected cells between VP16 and one of the other major tegument proteins VP22 (10). We also showed in studies of a recombinant virus that expresses VP22-GFP, that VP22 was overwhelming located in the cytoplasm of infected cells. Thus, the association between VP16 and VP22 reflected in their copurification seems unlikely to take place in the nucleus or to be involved with VP16 nuclear entry, but more likely reflects a cytoplasmic complex between the two proteins (see below).
The pattern of initial progressive accumulation in the nucleus resembles that seen for several proteins including not only replication proteins such as ICP8, DNA polymerase or origin binding protein (24, 25), but also regulatory proteins including ICP4 (32) and other tegument proteins including VP13/14 (9). As for VP13/14, the association of VP16 with maturing replication compartments may not represent any direct involvement in DNA replication itself. Nevertheless DNA synthesis was partially reduced in VP16-null mutants and it remains possible that its accumulation reflects a direct role in DNA replication (28, 43). We note that the nuclear pattern of this phase of VP16 may not simply recapitulate that of e.g., ICP8 since the latter has been reported to accumulate into punctate prereplication sites in the presence of inhibitors of DNA synthesis, while VP16 remains quite homogeneously diffuse under such conditions. Recruitment into the replication compartments may therefore require a DNA synthesis-dependent component or event and indeed in the absence of DNA synthesis VP16 remained in a diffuse homogeneous nuclear pattern, albeit with minimal accumulation in a diffuse cytoplasmic pattern.
By 10 to 12 h after infection additional distinct foci containing VP16-GFP could be seen, almost exclusively located at the periphery of the replication compartments. These foci were clearly distinct from the large replication compartments and lacked ICP8, but nevertheless appeared to be in close contact with the perimeter of the compartments. Recent results indicate that another tegument protein VP13/14 together with a minor population of VP22 are recruited into similar foci located on the periphery of the large DNA replication compartments (9, 19). These additional compartments could represent some assembly intermediate, for example a site where assembly may take place, or alternatively as a repository from where VP16 or other proteins are sampled, e.g., after appropriate interaction into subcomplexes, for incorporation into maturing capsids at a different site. We note that foci on the periphery of what were likely to be replication compartments have been previously observed in immunofluorescence studies (42). In that study, VP16 was detected in the nucleus but little was observed accumulating in the peripheral foci, termed assemblons. It remains to be seen whether the foci observed in that study are similar to those we show here in which VP16 accumulates.
From their analysis of replication of VP16-null mutants, Mossman et al. reported that no enveloped capsids were assembled in the cytoplasm, and they concluded that VP16 was required for viral egress downstream of the initial envelopment site (28). This is consistent with conclusions from studies on a PRV mutant lacking its VP16 homologue, where unenveloped capsids accumulated in the cytoplasm. The authors proposed that PRV VP16 may serve to link tegument budding into cytoplasmic membranes, with maturing cytoplasmic capsids (13). However, there was a notable difference in the effects of deletion of VP16 in PRV and HSV. In PRV the nuclear phase of capsid formation and primary envelopment at the inner nuclear membrane was unaltered quantitatively or qualitatively, in the absence of VP16. In contrast, in HSV, deletion of VP16 resulted in a significant reduction in total levels of nuclear capsid formation, and moreover those capsids which underwent primary envelopment showed a distinctly different morphology compared to wt capsids undergoing primary envelopment, being smaller, more uniform and lacking the spiked appearance associated with glycoprotein recruitment (28). These primary enveloped capsids were also found exclusively between the inner and outer nuclear membranes, with no capsid envelopment observed in the cytoplasm. One conclusion consistent with these data is that VP16 is required in the nucleus for the formation of a primary enveloped particle which has the capacity to fuse properly with the outer nuclear membrane in the pathway to assembly. This role would be in addition to its cytoplasmic role. It would be reasonable to propose that this nuclear role of VP16 would be reflected in its recruitment into the assembling primary particle, thus, e.g., directing the particle to suitable nuclear exit sites, or interacting with viral glycoproteins on the inner side of the inner nuclear membrane. It may therefore be that the sites of VP16 accumulation we observe on the perimeter of replication compartments, rather than the replication compartments themselves, represent sites of incorporation of VP16 onto the outside of assembled capsids.
However, a requirement for VP16 in the nucleus for proper assembly does not necessarily mean that it is assembled into the particle in this compartment. In this case VP16 within the peripheral nuclear compartments, may be required for the recruitment and assembly of proteins or complexes which are inserted into the capsid, for the interaction with or alteration of the host nuclear lamina or membrane, or for the selective transport or accumulation of virus glycoproteins at the inner nuclear membrane, the absence of which would result in a particle unable to progress further out of the nucleus in the exit pathway. We note in this context that differences in the functional roles of the PRV and HSV VP16 may be partly reflected in their localization since the PRV protein accumulated abundantly in the cytoplasm, with only a very minor population in the nucleus (13), while in this work VP16 first accumulated in the nucleus and was clearly present in replication compartments and the novel adjacent compartments. In addition we note that while PRV VP16 has not been observed to be associated with nuclear capsids by immunoelectronmicroscopy (13), at least a proportion of nuclear capsids were reported to show some VP16 labeling, although it was unclear whether this reflected true recruitment onto capsids (27).
Nevertheless it remains the case that the accumulation of GFP-VP16 in nuclear compartments, whether the main replication compartments, or the adjacent domains, may not necessarily reflect the route of its recruitment into virions. It remains possible that VP16 has some additional nuclear function or even that the nuclear accumulation is a reflection of its nuclear role in transcription of immediate-early genes (even though it is the de novo-synthesized VP16 and not the input VP16 which is being tracked).
VP16 cytoplasmic accumulation.
VP16 was first observed in the cytoplasm, initially in a diffuse pattern, then accumulating in vesicle-like compartments which were frequently concentrated in an asymmetric fashion reminiscent of the Golgi. Time-lapse confocal microscopy illustrated the dynamic nature of the vesicular accumulation, whereby large sections of the clusters could rapidly detach from and be readsorbed into the main pericentric focus. On an individual cell basis, while the perinuclear clusters aggregate and expand as individual foci, they also dispersed into a random vesiculated pattern throughout the cell. While there could be several explanations for this, it may be that differences in the organization or dissociation of the Golgi apparatus in individual cells (5) could account for such heterogeneity. Studies are now under way to establish cell lines expressing Golgi components fused to fluorescent proteins in which to study dynamic association with Golgi components on an individual cell basis.
As indicated above, at relatively early times in the cytoplasm VP16-GFP appeared mainly diffuse and in the presence of inhibitors of DNA synthesis, cytoplasmic VP16-GFP remained in an exclusively diffuse pattern for prolonged times. The recruitment of VP16-GFP into vesicular compartments could be the result of its interaction with membrane-associated proteins synthesized as infection progressed. Alternatively such recruitment could reflect the association with membrane compartments of emerging virions which had incorporated VP16 in a different compartment, including the nucleus. No direct binding of VP16 to membranes has been reported and the acidic pI of the protein may make this unlikely. However, VP16 directly associates with VP22 (10), a protein which has been reported to bind membranes (3) and, at least for the PRV VP22, to interact with the cytoplasmic tails of virus-encoded glycoproteins (14). VP16 of HSV was also reported to interact with gB, gD, and gH in biochemical cross-linking studies of structural proteins (47).
Treatment with brefeldin disrupted the coalesced vesicular pattern in the cytoplasm, resulting in redistributed large foci. Studies of the effect of brefeldin indicate that its effect is comparatively rapid, and using GFP-fused to Golgi resident proteins, (35) the redistribution of Golgi proteins to the endoplasmic reticulum has been demonstrated. However, the resulting pattern of localization of Golgi-associated cellular proteins after brefeldin treatment does not resemble the accumulation of VP16-GFP. The simplest interpretation of these results is that treatment with brefeldin disrupts the Golgi and thus the normal destination of cytoplasmic VP16-GFP, and in the absence of the Golgi, VP16-GFP accumulates or associates in another compartment with a more disperse punctate location. Alternatively since virus infection per se perturbs normal Golgi structure, it may be that brefeldin treatment of infected cells results in Golgi distribution to the larger punctate foci reflected in VP16 accumulation. However, other interpretations of the effect of brefeldin remain possible. Studies of live infected cells expressing Golgi associated fluorescent proteins and fluorescent VP16 should help address whether VP16 associates with specific cytoplasmic compartments and the dynamic nature of its cytoplasmic compartmentalization.
As previously reported in analysis of viruses expressing VP22-GFP fusion proteins, (11, 41), we observed that infected cells frequently formed very pronounced plasma membrane protrusions which establish intimate contact with adjacent cells. Time-lapse analyses indicate that this was not a passive effect of a virus induced CPE but an active process induced by infection which was observed not only in cell lines but also in infected primary keratinocytes. These extensions contained GFP-VP16, which could be seen to accumulate in vesicular clusters at the termini and initially progressed with a bulbous terminal structure that spread out upon contact with adjacent cells. Previous reports have demonstrated the induction of similar very long protrusions induced by Vaccinia virus infection (34). While there may be similarities with our observations in HSV-infected cells, analysis in vaccinia-infected cells indicated that these protrusions were initially formed from the induction of broader lamellipodia which compacted to finer projections. This does not appear to be the case with the HSV-induced projections. We also observed that frequently these GFP-containing extensions appeared to be taken up by adjacent cells, likely requiring some detachment or membrane fission followed by endocytosis. Clearly HSV can spread from cell to cell, by a route independent of the production of extracellular virus. The active induction of cell extensions which terminate in or are engulfed by adjacent cells may contribute to cell-to-cell spread. Work is now in progress in the attempt to show that this process results in a productive infection in the absence of the normal route of infection by extracellular virus, (e.g., in the presence of neutralizing antibody), and to examine activation of cellular pathways potentially involved in the reorganization, such as the RhoGTPase-actin network.
Given that VP16, in addition to playing structural role, also plays an important role in the transcriptional induction of the lytic stages of replication, both in acute-stage infection and reactivation, future studies of GFP-VP16 trafficking in neurons in culture and in vivo may also be informative for understanding its role and regulation at this stage of infection.
REFERENCES
- 1.Ace, C. I., T. A. McKee, J. M. Ryan, J. M. Cameron, and C. M. Preston. 1989. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J. Virol. 63:2260-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brignati, M. J., J. S. Loomis, J. W. Wills, and R. J. Courtney. 2003. Membrane association of VP22, a herpes simplex virus type 1 tegument protein. J. Virol. 77:4888-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burkham, J., D. M. Coen, and S. K. Weller. 1998. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J. Virol. 72:10100-10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campadelli, G., R. Brandimarti, C. Di Lazzaro, P. L. Ward, B. Roizman, and M. R. Torrisi. 1993. Fragmentation and dispersal of Golgi proteins and redistribution of glycoproteins and glycolipids processed through the Golgi apparatus after infection with herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 90:2798-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 180:1-19. [DOI] [PubMed] [Google Scholar]
- 7.Cousens, D. J., R. Greaves, C. R. Goding, and P. O'Hare. 1989. The C-terminal 79 amino acids of the herpes simplex virus regulatory protein, Vmw65, efficiently activate transcription in yeast and mammalian cells in chimeric DNA-binding proteins. EMBO J. 8:2337-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dargan, D. 1986. The structure and assembly of herpesviruses. Viral Struct. 5:359-437. [Google Scholar]
- 9.Donnelly, M., and G. Elliott. 2001. Fluorescent tagging of herpes simplex virus tegument protein VP13/14 in virus infection. J. Virol. 75:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 73:4110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flint, S. J., L. W. Enquist, R. M. Krug, V. R. Racaniello, and A. M. Skalka. 2000. Principles of virology. ASM Press, Washington, D.C.
- 13.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greaves, R., and P. O'Hare. 1989. Separation of requirements for protein-DNA complex assembly from those for functional activity in the herpes simplex virus regulatory protein Vmw65. J. Virol. 63:1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greaves, R. F., and P. O'Hare. 1990. Structural requirements in the herpes simplex virus type 1 transactivator Vmw65 for interaction with the cellular octamer-binding protein and target TAATGARAT sequences. J. Virol. 64:2716-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haarr, L., and S. Skulstad. 1994. The herpes simplex virus type 1 particle: structure and molecular functions. APMIS 102:321-346. [DOI] [PubMed] [Google Scholar]
- 18.Heine, J. W., R. W. Honess, E. Cassai, and B. Roizman. 1974. Proteins specified by herpes simplex virus XII. The virion polypeptides of type 1 strains. J. Virol. 14:640-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutchinson, I., A. Whiteley, H. Browne, and G. Elliott. 2002. Sequential localization of two herpes simplex virus tegument proteins to punctate nuclear dots adjacent to ICP0 domains. J. Virol. 76:10365-10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato, k., T. Daikoku, F. Goshima, H. Kume, K. Yamakii, and Y. Nishiyama. 2000. Synthesis, subcellular localisation and VP16 interaction of the herpes simplex virus type 2 UL46 gene product. Arch. Virol. 145:2149-2162. [DOI] [PubMed] [Google Scholar]
- 21.Kristie, T. M., and P. A. Sharp. 1993. Purification of the cellular C1 factor required for the stable recognition of the Oct-1 homeodomain by the herpes simplex virus alpha-trans-induction factor (VP16). J. Biol. Chem. 268:6525-6534. [PubMed] [Google Scholar]
- 22.Lam, Q., C. A. Smibert, K. E. Koop, C. Lavery, J. P. Capone, S. P. Weinheimer, and J. R. Smiley. 1996. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 15:2575-2581. [PMC free article] [PubMed] [Google Scholar]
- 23.Leslie, J., F. J. Rixon, and J. McLauchlan. 1996. Overexpression of the herpes simpex virus type 1 tegument protein VP22 increases its incorporation into virus particles. Virology 220:60-68. [DOI] [PubMed] [Google Scholar]
- 24.Lukonis, C. J., J. Burkham, and S. K. Weller. 1997. Herpes simplex virus type 1 prereplicative sites are a heterogeneous population: only a subset are likely to be precursors to replication compartments. J. Virol. 71:4771-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukonis, C. J., and S. K. Weller. 1996. Characterization of nuclear structures in cells infected with herpes simplex virus type 1 in the absence of viral DNA replication. J. Virol. 70:1751-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mettenleiter, T. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miranda-Saksena, M., R. A. Boadle, P. Armati, and A. L. Cunningham. 2002. In rat dorsal root ganglion neurons, herpes simplex virus type 1 tegument forms in the cytoplasm of the cell body. J. Virol. 76:9934-9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Hare, P., and C. R. Goding. 1988. Herpes simplex virus regulatory elements and the immunoglobulin octamer domain bind a common factor and are both targets for virion transactivation. Cell 52:435-445. [DOI] [PubMed] [Google Scholar]
- 30.Post, L. E., S. Mackem, and B. Roizman. 1981. Regulation of alpha genes of herpes simplex virus: expression of chimeric genes produced by fusion of thymidine kinase with alpha gene promoters. Cell 24:555-565. [DOI] [PubMed] [Google Scholar]
- 31.Preston, C. M., M. C. Frame, and M. E. Campbell. 1988. A complex formed between cell components and an HSV structural polypeptide binds to a viral immediate early gene regulatory DNA sequence. Cell 52:425-434. [DOI] [PubMed] [Google Scholar]
- 32.Randall, R. E., and N. Dinwoodie. 1986. Intranuclear localization of herpes simplex virus immediate-early and delayed-early proteins: evidence that ICP 4 is associated with progeny virus DNA. J. Gen. Virol. 67:2163-2177. [DOI] [PubMed] [Google Scholar]
- 33.Sadowski, I., J. Ma, S. Triezenberg, and M. Ptashne. 1988. GAL4-VP16 is an unusually potent transcriptional activator. Nature 335:563-564. [DOI] [PubMed] [Google Scholar]
- 34.Sanderson, C. M., M. Way, and G. L. Smith. 1998. Virus-induced cell motility. J. Virol. 72:1235-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sciaky, N., J. Presley, C. Smith, K. J. Zaal, N. Cole, J. E. Moreira, M. Terasaki, E. Siggia, and J. Lippincott-Schwartz. 1997. Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J. Cell Biol. 139:1137-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smibert, C. A., B. Popova, P. Xiao, J. P. Capone, and J. R. Smiley. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68:2339-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smiley, J. R., and J. Duncan. 1997. Truncation of the C-terminal acidic transcriptional activation domain of herpes simplex virus VP16 produces a phenotype similar to that of the in1814 linker insertion mutation. J. Virol. 71:6191-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sodeik, B., M. W. Ebersold, and A. Helenius. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136:1007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stern, S., and W. Herr. 1991. The herpes simplex virus trans-activator VP16 recognizes the Oct-1 homeo domain: evidence for a homeo domain recognition subdomain. Genes Dev. 5:2555-2566. [DOI] [PubMed] [Google Scholar]
- 40.Tomishima, M. J., G. A. Smith, and L. W. Enquist. 2001. Sorting and transport of alpha herpesviruses in axons. Traffic 2:429-436. [DOI] [PubMed] [Google Scholar]
- 41.van Leeuwen, H., G. Elliott, and P. O'Hare. 2002. Evidence of a role for nonmuscle myosin II in herpes simplex virus type 1 egress. J. Virol. 76:3471-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward, P. L., W. O. Ogle, and B. Roizman. 1996. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J. Virol. 70:4623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinheimer, S. P., B. A. Boyd, S. K. Durham, J. L. Resnick, and D. R. O'Boyle II. 1992. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J. Virol. 66:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson, A. C., K. LaMarco, M. G. Peterson, and W. Herr. 1993. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell 74:115-125. [DOI] [PubMed] [Google Scholar]
- 45.Yang, W. C., G. V. Devi-Rao, P. Ghazal, E. K. Wagner, and S. J. Triezenberg. 2002. General and specific alterations in programming of global viral gene expression during infection by VP16 activation-deficient mutants of herpes simplex virus type 1. J. Virol. 76:12758-12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, Y., D. A. Sirko, and J. L. McKnight. 1991. Role of herpes simplex virus type 1 UL46 and UL47 in alpha TIF-mediated transcriptional induction: characterization of three viral deletion mutants. J. Virol. 65:829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, Q., and R. J. Courtney. 1994. Chemical cross-linking of virion envelope and tegument proteins of herpes simplex virus type 1. Virology 204:590-599. [DOI] [PubMed] [Google Scholar]