Abstract
Hepatitis delta virus (HDV) expresses two essential proteins with distinct functions. The small hepatitis delta antigen (HDAg-S) is expressed throughout replication and is needed to promote that process. The large form (HDAg-L) is farnesylated, is expressed only at later times via RNA editing of the amber/W site, and is required for virion assembly. When HDAg-L is artificially expressed at the onset of replication, it strongly inhibits replication. However, there is controversy concerning whether HDAg-L expressed naturally at later times as a consequence of editing and replication can similarly inhibit replication. Here, by stabilizing the predicted secondary structure downstream from the amber/W site, a replication-competent HDV mutant that exhibited levels of editing higher than those of the wild type was created. This mutant expressed elevated levels of HDAg-L early during replication, and at later times, its replication aborted prematurely. No further increase in amber/W editing was observed following the cessation of replication, indicating that editing was coupled to replication. A mutation in HDAg-L and a farnesyl transferase inhibitor were both used to abolish the ability of HDAg-L to inhibit replication. Such treatments rescued the replication defect of the overediting mutant, and even higher levels of amber/W editing resulted. It was concluded that when expressed naturally during replication, HDAg-L is able to inhibit replication and thereby inhibit amber/W editing and its own synthesis. In addition, the structure adjacent to the amber/W site is suboptimal for editing, and this creates a window of time in which replication can occur in the absence of HDAg-L.
Hepatitis delta virus (HDV) is an infectious RNA agent that exists as a subviral satellite of hepatitis B virus (30). Its genome is a circular single-stranded RNA molecule of 1,679 nucleotides which forms a rod-like imperfect duplex in which roughly 70% of its nucleotides are paired (19). The genome replicates in the nucleus and acts as the template for rolling circle transcription of antigenomic RNA (8). A cis-encoded ribozyme processes the resulting antigenomic multimer into unit-length linear RNA molecules (34). Following self-cleavage, the termini of the resulting monomers are joined by a host-encoded factor, and a circular molecule referred to as the antigenome results (29). This RNA acts as a replication intermediate for genome amplification in an analogous mechanism that also involves rolling circle transcription, self-cleavage, and ligation (20).
In addition to the antigenome, a subantigenomic-length message is also generated during transcription of the genome (14). Initially during replication, this message encodes the small hepatitis delta antigen (HDAg-S), a protein that is required for viral replication (18). Later during replication, some of the messages expressed contain a mutation that changes the HDAg-S UAG (amber) stop codon to a UGG tryptophan (W) codon (24). The protein translated from these messages has an additional 19 amino acids at its C terminus and is referred to as the large hepatitis delta antigen (HDAg-L). This protein is able to interact with the helper virus envelope proteins and is required for virion production (6). Both delta antigens multimerize and form stable ribonucleoprotein (RNP) complexes with the genome and the antigenome (31).
The amber/W conversion that enables HDAg-L expression occurs as a consequence of RNA editing and is mediated by ADAR1-S, the smaller isoform of a host-encoded RNA adenosine deaminase that acts in the nucleus to convert specific adenosines to inosine within imperfect duplex RNA (17, 35). Since the mature delta antigen message lacks the required imperfect duplex structure, only antigenomic RNA is edited (5). How HDAg-L is ultimately expressed from edited antigenomic RNA is not understood. It is possible that nascent antigenomic RNA is first edited, and then following this event, a polyadenylation signal within the rod-structured RNA is recognized and processed to yield an HDAg-L-expressing message that contains inosine. In a second model, the inosine is present only in antigenomic RNA that is ultimately processed to yield the circular replication intermediate. During replication, the inosine is then interpreted as guanosine, and a cytosine is incorporated into the resulting mutant genome. This molecule then serves as the template for a message that contains a UGG codon and expresses HDAg-L. The two models are not mutually exclusive, and it is possible that both mechanisms contribute to the expression of HDAg-L.
The secondary structure immediately adjacent to the edited adenosine plays a critical role in editing. Disruptions of pairing very close to the amber/W site abolish editing, while compensatory mutations that restore pairing also restore editing (3, 32). The minimal structure that can be edited by ADAR1 includes only four base pairs upstream and four base pairs downstream from the edited adenosine (32). However, sequences flanking this essential region may affect the efficiency by which ADAR1 edits this site.
In addition to its essential role in virion assembly, HDAg-L is also reported to be a potent inhibitor of replication. When HDAg-L is expressed artificially from an expression vector at the onset of replication, it can inhibit replication in a dominant manner, and when as little as 1/10 of total viral protein is HDAg-L, replication is inhibited 10-fold (7). This observation raises the question as to how HDV can initiate replication following infection, since HDAg-L comprises roughly a third of the total HDAg in the virion RNP.
It is not understood how HDAg-L inhibits replication when it is present at the onset of replication, and there is evidence to suggest that it may affect the processes of genome and antigenome synthesis differently. When messages expressing both HDAg-S and HDAg-L were cotransfected with genomic RNA, antigenomic RNA was still synthesized, while when the two messages were cotransfected with antigenomic RNA, newly synthesized genomic RNA was not observed (27). Modahl and Lai concluded that HDAg-L specifically inhibits the process of genome synthesis. However, other interpretations of the data are possible; for instance, the authors of that study did not examine whether the transfected genomic versus antigenomic RNAs were circularized and/or imported into the nucleus with comparable efficiency. Nevertheless, their model is attractive because it can explain why HDAg-L present in the virion RNP does not inhibit replication during a natural infection. Once it reaches the nucleus, the HDAg-L containing RNP could synthesize both HDAg-S messages and antigenomes. The resulting HDAg-S could then bind to the newly synthesized antigenomes, and since these RNPs would lack HDAg-L, they would be competent for genome synthesis.
Most recently, workers from the same laboratory used an inducible expression vector to direct HDAg-L synthesis at a time following the onset of HDV replication (26). When HDAg-L synthesis was induced at 3 days following the initiation of replication, no inhibition in the synthesis of either strand was observed. The authors of that study concluded that HDAg-L is able to inhibit replication only when expressed at the onset, and since this is never the case in a natural infection, HDAg-L cannot regulate replication. Again, however, alternate interpretations of the data are possible, and any interpretation is complicated by the RNA transfection methodology employed.
In a natural infection, a single virion is thought to be able to initiate replication within a cell. Hence, the initial concentrations of genome and HDAg-S are extremely low. In the days following infection, replication gives rise to increasing levels of genome, antigenome, and HDAg. In addition, HDV replication controls the relative abundance of all three species. In contrast, in an RNA transfection, very high levels of one RNA strand are delivered at the onset, and this level is often so high that no increase is observed in the level of this species as a consequence of replication. In addition, large amounts of mRNA are often codelivered, and similarly, the level of HDAg may not increase significantly with time. In this setting, by definition, the relative concentrations of genome, antigenome, and HDAg are different from what would occur naturally, as is the time needed to reach maximal levels of replicating RNA. Using such an RNA transfection procedure, Macnaughton and Lai observed that the maximal steady-state levels of replicative species were reached in as few as 4 days posttransfection (26). This very short window of time greatly hampered their ability to express HDAg-L after the onset of replication but still prior to the time when maximal RNA levels were reached. In contrast, when other methods are used to initiate replication, the time needed to reach maximal levels of replicative species can be as much as 8 or 9 days (35). Furthermore, in an RNA transfection, linear molecules that are greater than unit lengths are delivered to cells, and it is usually not possible to monitor the efficiency with which such molecules are either circularized or taken up by the nucleus. Hence, differences observed when replication is initiated with genomic versus antigenomic RNA may result from differences in circularization and/or nuclear import and may not reflect the intrinsic ability of the RNA to serve as the template for RNA-directed synthesis.
Although it is possible to initiate HDV replication when monomeric linear RNA rather than circular RNA is used, the relevance of this observation is not clear (11). These linear RNAs are artificial in that neither their 5′ nor their 3′ termini are generated naturally during HDV replication. Furthermore, there has been no comparison of the efficiency with which these RNAs can initiate replication to that of circular RNA. Given that actual virions contain mainly circular RNA, that HDV replicates via a rolling circle mechanism that generates dimeric and trimeric species, and that ribozyme mutations abolish HDV replication, it is likely, though not certain, that circularization of the template enhances its ability to initiate HDV replication.
We contend that the natural HDV replication cycle is better approximated when cDNA, rather than linear monomeric or multimeric RNA, is used to initiate replication. Since transcription of cDNA must occur in the nucleus, we know a priori that all RNAs are initially delivered to the nucleus with 100% efficiency. Furthermore, although the in vitro generation of circular HDV RNA has not yet been reported, the circularization reaction is known to be very efficient when RNA is transcribed from cDNA in vivo (21, 22, 29). Finally, a very weak promoter, such as the simian virus 40 (SV40) late promoter in the absence of DNA replication and T antigen, can be used to drive synthesis of HDV RNA. In such a setting, the amount of circular RNA and HDAg-S derived from cDNA-directed synthesis can represent a very small fraction of that contributed by RNA-directed synthesis during HDV replication. In such a case, the levels of genome, antigenome, and HDAg-S all increase with time posttransfection, and their relative ratios are controlled by HDV replication.
When such a cDNA-based system is used to initiate HDV replication, and amber/W editing is examined at various times posttransfection, linear kinetics are not observed. Initially, during the first 3 to 4 days of replication, very little amber/W editing and HDAg-L expression are observed (35). If HDAg-L could indeed inhibit replication when expressed as a consequence of editing and replication, then this apparent lag in editing would provide a window of time where replication could proceed in the absence of the inhibitor. From approximately days 4 to 9, RNA editing and HDAg-L expression have been observed to increase in linear fashion (35). However, at very late times posttransfection, a plateau where the levels of amber/W editing and HDAg-L no longer increase with time is observed (35). Typically, when the plateau is reached, HDAg-L constitutes roughly a fifth to a third of the pool of total HDAg, and roughly a similar proportion of the genomes are edited. This observed plateau is also consistent with the HDV replicative cycle. Although amber/W editing is required for HDAg-L expression, in order for HDAg-L to be expressed, the edited antigenomes may need to be copied into mutant genomes. Mutant genomes that are packaged into virions are not infectious, since upon their entry into the cell, they cannot express HDAg-S. Hence, by limiting amber/W editing, the plateau ensures that a majority of genomes are infectious. Although the observed lag and plateau associated with amber/W editing are consistent with the requirements of HDV replication, nothing is known about how either feature is achieved.
Here, we found that a replication-competent HDV mutant possessing increased stability in its rod-like structure downstream from the amber/W site was edited at very high levels compared with the wild-type very early during replication and displayed no lag in editing. This mutant also expressed HDAg-L very early, and its replication aborted prematurely. A premature plateau in editing that coincided with the cessation of replication was also observed. However, under conditions where HDAg-L expressed by the mutant was not functional, replication was restored and the proportion of edited antigenomes reached 80% within 6 days posttransfection. Consistent with these results, when wild-type HDV was replicating in cells in which ADAR1-S was modestly overexpressed, again, editing occurred at high levels very early, no lag was observed, HDAg-L was expressed very early, and replication aborted prematurely. However, under the same conditions when HDV could no longer express functional HDAg-L, replication was restored and very high levels of editing were observed at later times. Based on these results, we propose a model to explain how the regulation of amber/W editing is mediated.
MATERIALS AND METHODS
Plasmids.
pSS15 carries the secreted alkaline phosphatase gene under the control of the cytomegalovirus immediate-early promoter. pDL538 encodes the wild-type replication-competent HDV genomic RNA (21). pCC043 and pCC045 have 6×His mutations and 6×His with compensatory mutations (6×His+compensatory), respectively, in the pDL538 backbone, where the mutations described in Fig. 1A were introduced by site-directed PCR mutagenesis. pSS74 and pSS140 express the wild-type and N-terminally truncated (M39) nonreplicating editing reporter, respectively (32). pSS162 has the D2 M frameshift mutation (18) in the pSS74 background. pSS178 and pSS155 encode wild-type HDAg-expressing nonreplicating reporters with 6×His and 6×His+compensatory mutations, respectively. pSS170, pSS171, and pSS172 encode nonreplicating editing reporters with upstream, downstream, and upstream-downstream mutations, respectively. Upstream, downstream, and upstream-downstream combination mutations as shown in Fig. 1B were introduced by site-directed PCR mutagenesis. pSS202, pSS156, pSS199, pSS200, and pSS201 encode the M39 nonreplicating editing reporter with 6×His, 6×His+compensatory, upstream, downstream, and upstream-downstream combination mutations, respectively. pSS179, pSS168, pSS180, pSS181, and pSS182 encode the D2 M-containing nonreplicating editing reporter with 6×His, 6×His+compensatory, upstream, downstream, and upstream-downstream combination mutations, respectively. pSS174, pSS175, and pSS176 encode replication-competent HDV genomic RNA with upstream, downstream, and upstream-downstream combination mutations in the pDL538 backbone. pCC053 encodes His6-tagged HDAg-L in the background of pDL445 (23). pBOM094 encodes a nonfunctional HDAg-L where the last (214th) amino acid codon was changed from CAG (Q) to TAG (stop) and was made by ligating three fragments, a 2,282-bp fragment from pKW42 (32) digested with SfiI and SalI, a 2,028-bp fragment from pDL444 (23) digested with SfiI and XmaI, and an XmaI and SalI digest of a PCR product that resulted when pKW43 (analogous to pKW42 but with TGG at the amber/W site) was used as the template with oligonucleotides 572 and 573. pSS173 encodes a replication-competent HDV genomic RNA with the same mutation as that in pBOM094 introduced into the pDL538 backbone. pDL701 expresses C-terminally double-hemagglutinin (HA)-tagged ADAR1-S (ADAR1-S-HA) (36).
FIG. 1.
Predicted secondary structures of wild-type and mutant antigenomic RNA. (A) The wild type and 6×His and 6×His+compensatory (6×His+comp.) mutations. Nine substitutions were made to obtain the 6×His mutant, and seven additional mutations are present in the 6×His+compensatory mutant. (B) Mutants with a stabilized secondary structure upstream and downstream of the amber/W site shown in comparison to the wild-type predicted structure. The mutated nucleotides are shown in shaded letters, and the amber/W codon is boxed. Deleted and inserted nucleotides are indicated by Δ and *, respectively.
Tissue culture, transfection, and sample harvesting.
HEK293 and HuH7 cells were cultured as described previously (32, 35). In all transfections, a cDNA that expresses secreted alkaline phosphatase (SEAP) was codelivered so that transfection efficiency could be monitored by kinetic assays of SEAP activity present in the medium. Within each experiment, the resulting SEAP activity varied by less than twofold. For the nonreplicating editing reporters, HEK293 cells were seeded in 35-mm plates at approximately 80% confluence. One day later, the cells were transiently transfected by using Ca3(PO4)2 with 3 μg of the appropriate vector (for vectors expressing wild-type HDAg, 0.3 μg of the reporter vector was diluted with 2.7 μg of pSS43) and 0.3 μg of pSS15. Three days after transfection, cells were harvested for protein with 300 μl of lysis buffer containing 2% sodium dodecyl sulfate and 1% β-mercaptoethanol and for RNA with an RNeasy RNA isolation kit (QIAGEN).
For replication assays, HuH7 cells were transiently transfected with 30 μg of the cDNA vector encoding the appropriate replicon and 3 μg of pSS15 1 day after they were seeded in 100-mm plates. One day posttransfection, the cells were trypsinized and resuspended in 12 ml of Dulbecco's modified Eagle's medium (described above), replated into a 35-mm 6-well plate, and harvested in 300-μl lysis buffer (described above) on the appropriate days thereafter. For the samples shown in Fig. 4, total RNA was then isolated from the same lysis buffer sample as described as follows. The lysed cells were shredded with a 20-gauge tuberculin needle, 150 μl was heated at 95°C for 5 min to be analyzed for protein and the other unheated 150 μl was vortexed and spun down to precipitate the sodium dodecyl sulfate after the addition of 75 μl of 5 M potassium acetate plus 3 M acetic acid mix, and then the residual proteins in the supernatant were removed by phenol-chloroform extraction. The nucleic acids were then precipitated with ethanol, resuspended in water, and then DNase I treated. DNase I was removed by phenol-chloroform extraction, and RNA was then precipitated and resuspended in 100 μl of RNase-free water for Northern blot and reverse transcriptase (RT)-PCR analyses. The RNA samples used in Fig. 2, 3, 5, and 6 were isolated directly from cultured cells with an RNeasy kit as described above. To obtain the samples analyzed in Fig. 6, two 100-mm plates of HuH7 cells were transiently transfected in duplicate with 3 μg of either pDL538 or pSS173, 27 μg of either pDL701 or pSS43, and 3 μg of pSS15. One day later, the cells in the two plates were trypsinized, resuspended and mixed in a total of 24 ml of Dulbecco's modified Eagle's medium, and then seeded in two 35-mm 6-well plates to be used for RNA and protein isolation. The samples analyzed in Fig. 5 were transfected and prepared in an analogous fashion. This method as well as the one used to obtain the samples shown in Fig. 4 and 6 were designed to minimize well-to-well inconsistency due to variation in transfection efficiency. For samples shown in Fig. 5, treatment of cells began at 4 h posttransfection with 2 μM farnesyl transferase inhibitor (FTI) (FTI-277; Calbiochem) in a carrier with final concentrations of 0.2% dimethyl sulfoxide and 400 μM dithiothreitol as described by Bordier et al. (1). The medium containing either FTI with carrier or carrier alone was replaced daily.
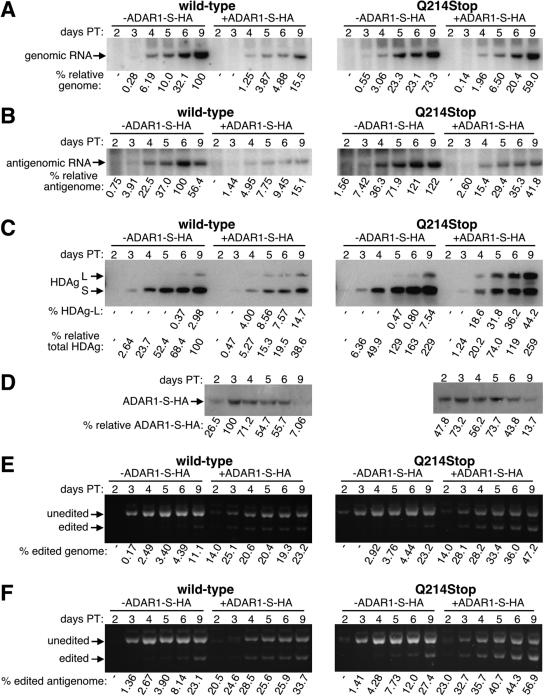
FIG. 4.
Time course analysis of HDV replication, amber/W editing, and HDAg-L expression, with wild-type HDV and the downstream mutant, in the presence and absence of the Q214Stop mutation. HuH7 cells transiently transfected with cDNA that expresses circular genomic RNA were harvested at days 2, 3, 4, 5, 6, and 9, and then the total RNA and protein were analyzed by Northern blot probing for genomic RNA (A), Northern blot probing for antigenomic RNA (B), Western blot probing for HDAg (C), StyI digestion of RT-PCR products that resulted from reverse transcription initiated with a primer complementary to genomic RNA (D), and StyI digestion of RT-PCR products that resulted from reverse transcription initiated with a primer complementary to antigenomic RNA (E). All calculations and abbreviations are as described in the legends to Fig. 2 and 3. Quantification of RNA and protein samples is reported to three significant figures to reflect the precision of the phosphoimager or software used to calculate these values. The amount of total RNA loaded for the Northern blots (A and B) were quantified from ethidium bromide staining of the agarose gels and were as follows (samples are from day 2 to day 9, respectively; the highest sample was given a value of 10). (A) Wild type, 8.72, 9.53, 8.31, 8.98, 8.90, and 7.19; downstream (down) mutant, 4.29, 8.01, 8.42, 8.56, 9.78, and 10; wild-type Q214Stop, 4.33, 5.98, 7.24, 8.53, 7.83, and 8.34; downstream Q214Stop mutant, 5.23, 8.69, 8.52, 8.07, 9.88, and 9.67. (B) Wild type, 7.80, 9.32, 7.47, 8.57, 9.12, and 6.67; downstream mutant, 3.95, 7.32, 6.83, 6.40, 7.73, and 8.59; wild-type Q214Stop, 4.08, 5.57, 5.95, 7.65, 7.36, and 8.72; downstream Q214Stop mutant, 4.99, 9.60, 7.73, 8.41, 9.24, and 10. The amount of total protein loaded into each lane for the Western blots (C) were quantified after Ponceau S staining and were as follows (samples are from day 2 to day 9, respectively; the highest sample was given a value of 10): wild type, 6.18, 6.51, 6.64, 7.76, 8.68, and 9.67; downstream mutant, 7.89, 9.34, 9.21, 10, 9.57, and 8.49; wild-type Q214Stop, 4.95, 6.53, 7.24, 8.09, 8.68, and 9.28; downstream Q214Stop mutant, 7.63, 7.83, 7.57, 8.49, 8.22, and 6.84.
FIG. 2.
Replication and HDAg expression of the 6×His and 6×His+compensatory (6×His+comp.) mutants. Northern blot analysis probing for genomic RNA (A) and antigenomic RNA (B) and Western blot analysis for HDAg (C) with samples obtained from HuH7 cells transiently transfected with cDNA that expressed circular genomic RNA and harvested at 4, 8, and 12 days posttransfection is shown. Normalized expression values of genomic RNA, antigenomic RNA, and HDAg (% relative genome, % relative antigenome, and % relative total HDAg, respectively), where the highest value among the wild-type samples in each set is defined as 100%, are shown. The percentage of HDAg-L was calculated by dividing the HDAg-L signal by the total HDAg signal and multiplying the value by 100. Levels too low to quantify are indicated by −. PT, posttransfection; L, HDAg-L; S, HDAg-S.
FIG. 3.
Editing of nonreplicating amber/W reporters in HEK293 cells. (A) Western blot analysis with wild-type reporters that express functional HDAg. (B) Western blot analysis with reporters that express RNA-binding-deficient HDAg. In this construct, the natural initiator codon has been mutated to alanine, and position 39 has been mutated to methionine. (C) RT-PCR analysis followed by StyI digestion of reporters that do not express HDAg. Lanes: 1, wild-type reporter; 2, 6×His reporter; 3, 6×His+compensatory reporter; 4, upstream reporter; 5, downstream reporter; 6, upstream-downstream combination reporter. The 479-nucleotide unedited RT-PCR product is indicated by a •, while the 393-nucleotide edited product is indicated by *. The percentage of editing was calculated as described in Materials and Methods. All abbreviations are as described in the legend to Fig. 2.
FIG. 5.
Time course analysis of HDV replication, amber/W editing, and HDAg-L expression, with wild-type HDV and the downstream mutant, in the presence and absence of FTI (+FTI and −FTI, respectively). HuH7 cells transiently transfected with cDNA that expresses circular genomic RNA were harvested at days 2, 3, 4, 5, 6, and 9, and then the total RNA and protein were analyzed by Northern blot probing for genomic RNA (A), Northern blot probing for antigenomic RNA (B), Western blot probing for HDAg (C), StyI digestion of RT-PCR products that resulted from reverse transcription initiated with a primer complementary to genomic RNA (D), and StyI digestion of RT-PCR products that resulted from reverse transcription initiated with a primer complementary to antigenomic RNA (E). All calculations and abbreviations are as described in the legends to Fig. 2 and 3. Quantification of RNA and protein samples is reported to three significant figures to reflect the precision of the phosphoimager or software used to calculate these values. The amount of total RNA loaded and transferred for the Northern assay (A and B) were quantified from the blots on which ethidium bromide-stained samples were transferred and were as follows (samples were from day 2 to day 9, respectively; the highest sample was given a value of 10). (A) Wild type (−FTI), 1.62, 3.74, 4.51, 1.78, 5.72, and 2.75; downstream (down) mutant (−FTI), 1.18, 4.56, 10, 5.00, 2.86, and 3.16; wild type (+FTI), 4.31, 5.30, 4.16, 5.57, 4.44, and 4.51; downstream (+FTI) mutant, 2.36, 8.24, 7.87, 5.69, 2.69, and 3.33. (B) Wild type (−FTI), 1.15, 2.05, 4.12, 3.94, 6.30, and 4.63; downstream (−FTI), 2.16, 7.28, 10, 6.46, 3.64, and 4.95; wild type (+FTI), 1.27, 9.48, 8.62, 4.25, 2.40, and 5.34; downstream (+FTI) mutant, 1.00, 4.97, 7.35, 4.45, 3.24, and 4.25 (B). The amounts of total protein loaded into each lane for the Western blots (C) were quantified after Ponceau S staining and are as follows (samples are from day 2 to day 9, respectively; the highest sample was given a value of 10): wild type (−FTI), 5.31, 5.57, 7.59, 8.00, 8.55, and 9.08; downstream (−FTI) mutant, 4.28, 5.66, 7.15, 8.77, 10, and 9.52; wild type (+FTI), 3.91, 5.09, 6.67, 7.94, 7.59, 8.20; downstream (+FTI) mutant, 3.14, 4.65, 5.26, 6.75, 7.76, 7.15.
FIG. 6.
Time course analysis of HDV replication, amber/W editing, and HDAg-L expression, in the presence and absence of the Q214Stop mutation, with and without exogenous ADAR1-S expression. HuH7 cells were transiently transfected with cDNA expressing either wild-type or Q214Stop mutant circular genomic RNA, and the total RNA and protein were analyzed following harvest at days 2, 3, 4, 5, 6, and 9. (A) Northern analysis of genomic RNA. (B) Northern analysis of antigenomic genomic RNA. (C) Western analysis of HDAg. (D) Western analysis of ADAR1-S-HA. (E) StyI digestion of RT-PCR products that resulted from reverse transcription initiated with a primer complementary to genomic RNA. (F) StyI digestion of RT-PCR products that resulted from reverse transcription initiated with a primer complementary to antigenomic RNA. All calculations and abbreviations are as described in the legends to Fig. 2 and 3. Quantification of RNA and protein samples is reported to three significant figures to reflect the precision of the phosphoimager or software used to calculate these values. The amount of total RNA loaded and transferred for the Northern assay (A and B) was quantified from the blots on which ethidium bromide-stained samples were transferred and were as follows (samples are from day 2 to day 9, respectively; the highest sample was given a value of 10). (A) Wild type without ADAR1-S-HA (−ADAR1-S-HA), 1.96, 1.67, 5.93, 4.74, 5.51, and 6.93; wild type with ADAR1-S-HA (+ADAR1-S-HA), 6.61, 3.28, 5.70, 7.11, 4.91, and 7.53; wild- type Q214Stop without ADAR1-S-HA (−ADAR1-S-HA), 3.53, 4.69, 7.26, 8.72, 7.86, and 8.16; wild-type Q214Stop with ADAR1-S-HA (+ADAR1-S-HA), 4.56, 5.88, 6.84, 8.79, 10, and 8.83. (B) Wild type ADAR1-S-HA (−ADAR1-S-HA), 3.51, 2.98, 10, 5.36, 7.35, and 8.27; wild type with ADAR1-S-HA (+ADAR1-S-HA), 5.47, 3.85, 8.51, 7.89, 3.84, and 8.43; wild-type Q214Stop without ADAR1-S-HA (−ADAR-S-HA), 5.33, 6.18, 6.31, 7.40, 7.61, and 6.68; wild-type Q214Stop with ADAR1-S-HA (+ADAR1-S-HA), 3.84, 4.34, 6.41, 5.56, 8.50, and 8.90. The amounts of total protein loaded in each lane for the Western blots (C) were quantified after Ponceau S staining and are as follows (samples are from day 2 to day9; the highest sample was given a value of 10). (A) Wild type without ADAR1-S-HA (−ADAR1-S-HA), 3.02, 2.68, 3.30, 3.86, 4.47, and 6.15; wild type with ADAR1-S-HA (+ADAR1-S-HA), 5.03, 6.27, 7.82, 8.39, 8.09, and 8.72; wild-type Q214Stop without ADAR1-S-HA, 5.46, 6.29, 7.28, 8.35, 8.69, and 10; wild-type Q214Stop with ADAR1-S-HA, 6.10, 6.57, 7.28, 7.43, 7.00, and 7.88.
Western, Northern, and RT-PCR analyses.
RNA samples were analyzed by Northern analysis as described previously (21). Protein samples were subjected to Western analyses for HDAg and ADAR1-S-HA expression as described previously (32). The above-mentioned Western protocols use radiolabeled secondary antibody and have been shown to respond linearly over a broad range of protein concentrations (36). One-fifteenth of total protein and 1/20 of total RNA from each sample was loaded onto the appropriate gels for Western and Northern analyses, respectively. Quantification was achieved by the use of a Storm phosphoimager to detect radioactivity signal and by analysis of the signal using Image Quant version 1.2 software. The amounts of total RNA recovered, loaded into the gel, and transferred to the blot were monitored by quantification of the signal associated with 18S rRNA following staining by ethidium bromide, while the amount of protein recovered, loaded, and then transferred was determined by quantification of the signals associated with all protein bands in each lane after staining with Ponceau S (for values, see Fig. 4, 5, and 6 legends). Before RT-PCR was performed, all RNA samples were subjected to DNase treatment for 1 h, followed by phenol-chloroform extraction. A fraction (1/50) of total isolated RNA was reverse transcribed by using Superscript II (Invitrogen) according to the protocol provided by the manufacturer. For the nonreplicating reporters shown in Fig. 3C, Oli320 (5′TTTTTTTTTTTTTTTTTTTTTTTTTTGAGTGGA3′) was used specifically to reverse transcribe messages polyadenylated at the HDV poly(A) site. For genomic and antigenomic HDV RNA, Oli315 (5′GAAGGTGGATCGAGGGGAGCGCCCGGGG3′) and Oli586 (5′CGGAGGGGGTGCTGGGAACACCGG3′), respectively, were used for reverse transcription. A total of 1/20 of the RT reaction was then amplified by PCR (20 cycles of 94°C for 10 s, 55°C for 30 s, and 68°C for 30 s, followed by 68°C for 5 min) by using Oli320 and Oli586 for the nonreplicating reporter. For the genome and antigenome, 1/20th of the RT reaction was amplified by PCR (20 cycles of 94°C for 10 s and 68°C for 30 s, followed by 68°C for 5 min) by using Oli315 and Oli586. A total of 1/20 of these PCR samples was then reamplified under the same conditions by using Oli587 (5′ CTTCGTCCCCAATCTGCAGGGAGT3′) and Oli733 (5′ GGGACCAGTGGAGCCATGGGATGC3′). By running only 20 cycles for each step, we prevented any component of the PCR from being depleted prior to the completion of the last cycle. If depletion were to occur, heteroduplexes between edited and unedited products could form, such species would not be digested by StyI, and a quantitation error would result. To verify that this error did not occur with our samples, one-fourth of the second PCR sample was added to a fresh 1× PCR mix, and the mixture was treated for a single cycle so that any heteroduplex would be converted to a homoduplex. The results obtained after this step following StyI digestion were nearly identical to those obtained from the PCR products not subjected to the additional cycle. Control PCR samples were also generated in the absence of reverse transcription, and the absence of the 537-bp band was used to verify that DNase I digestion went to completion. High-Fidelity Platinum Taq polymerase (Invitrogen) was used for PCR. Ten microliters of the second PCR samples was digested to completion with 20 U of StyI (New England Biolabs) in 1× NEB buffer 3 in a total volume of 20 μl. The digest was analyzed by electrophoresis on a 1.5% agarose gel, and the gel was stained with ethidium bromide. The fluorescence intensity of ethidium bromide-stained bands at 479 bp (unedited) and 393 bp (edited) was quantified by using Kodak Image Station 440CF and ID image analysis software. Percent editing was calculated in the following manner: % editing = (intensity of 393-bp band/393)/[(intensity of 393-bp band/393) + (intensity of 479-bp band/479)].
RESULTS
Substitution mutations within the C terminus of the HDAg-L open reading frame (ORF) increased amber/W editing.
In a project unrelated to RNA editing, we wished to purify replicating HDV RNPs from cells by using nickel or cobalt chelate chromatography. Toward that end, we constructed a mutant HDV that would express a variant of HDAg-L in which amino acid positions 203 to 208 were replaced with six histidine residues. We speculated that since such a construct would express wild-type HDAg-S, it would not have a replication defect and that the HDV RNP complex could be purified by virtue of the affinity-tagged HDAg-L. We realized that the substitution mutations introduced to generate the six consecutive histidine codons would disrupt the rod-like structure and that this disruption might inhibit HDV replication. We therefore also made a 6×His HDV mutant that contained additional mutations in the opposing side of the rod-like structure to restore that structure. Figure 1A shows the modeled secondary structure of the antigenomic RNA from wild-type HDV, the 6×His, and the 6×His+compensatory mutants.
The mutants were next tested for their ability to replicate in HuH7 cells by using a cDNA that expresses circular genomic RNA from the SV40 late gene promoter. Figure 2A and B show Northern blots detecting genomic and antigenomic RNA, respectively, of the two mutant constructs and of wild-type HDV when samples were harvested at 4, 8, and 12 days posttransfection. The mutant lacking compensatory mutations showed a severe defect in replication, and at day 12, it expressed 200-fold fewer genomes and 50-fold fewer antigenomes than wild-type HDV. Hence, it is likely that the disruption of the secondary structure in this mutant strongly affected its ability to replicate. The genomic RNA expressed by the mutant resulted from both RNA-dependent replication and DNA-dependent transcription from the transfected vector. Since this combined value was 200-fold lower than that attained with wild-type HDV, we concluded that in the latter case, almost all observed signal resulted from HDV replication and virtually none was the result of DNA-dependent transcription. Similarly, Fig. 2C shows Western blot analysis of the same samples. Since the level of HDAg synthesis obtained with the 6×His mutant was almost 1/25 of that obtained with the wild type, we concluded that in the latter case, greater than 95% of the observed viral protein was expressed as a consequence of HDV replication by days 8 and 12.
As shown in Fig. 2, the compensatory mutations restored replication of the 6×His mutant to levels very near that of the wild type, demonstrating that the mutations did not affect replication as long as the rod-like secondary structure was preserved. As expected, we were unable to detect HDAg-L(His6) expressed from the 6×His mutant that lacked compensatory mutations since the replication of this mutant was so impaired. Unexpectedly, however, the construct with the compensatory mutations (6×His+compensatory) expressed nearly fivefold more HDAg-L(His6) than the HDAg-L expressed by wild-type HDV (Fig. 2C). The observed HDAg-L(His6) overexpression from the 6×His+compensatory mutant likely resulted either because the HDAg-L(His6) protein was more stable than was HDAg-L or because the amber/W site within the 6×His+compensatory mutant RNA was edited with higher efficiency than that of the wild-type.
We wondered how the 6×His+compensatory mutant was able to express a very high level of HDAg-L(His6) protein and yet still replicate at wild-type levels. If, when expressed as a consequence of editing and replication, HDAg-L is able to inhibit replication, then it should feed back to inhibit further replication and thereby limit its own expression. Hence, it should be impossible to attain high levels of functional HDAg-L expression from HDV replication. If this model is correct, then the high expression of HDAg-L(His6) protein observed with the 6×His+compensatory mutant could have resulted only if the HDAg-L(His6) protein were defective in its ability to inhibit replication. Alternatively, if HDAg-L (or any variant thereof) is not an inhibitor of replication when expressed naturally from replication, then no defect associated with the HDAg-L(His6) protein need be postulated. The HDAg-L(His6) ORF was next cloned into an expression vector, and the ability of the resulting protein to inhibit HDV replication when present at the onset of replication was compared with that of HDAg-L. Vectors that express HDAg-L, HDAg-L(His6), or no protein were each cotransfected with a vector that expresses circular HDV genomic RNA, and replication was assayed by Northern analysis of the antigenomic strand 6 days posttransfection. Although no antigenomic RNA could be detected in the presence of HDAg-L, abundant signal was observed in the presence of the HDAg-L(His6) protein (data not shown). A Western blot loaded with protein from the same samples was used to confirm that equal levels of HDAg-L and HDAg-L(His6) had been expressed. We concluded that the HDAg-L(His6) protein is defective in its ability to inhibit replication, and this defect could explain how the 6×His+compensatory mutant can express so much of this protein and still replicate at a wild-type level.
We next examined the mechanism responsible for the overexpression of HDAg-L(His6) by the 6×His+compensatory mutant. Such overexpression would result from elevated amber/W editing that could have resulted if either the mutant RNA was a better substrate for editing or the amber/W site within the mutant RNP was more accessible to ADAR1-S. To test such models, we examined the effect of the 6×His+compensatory mutations in the context of an HDV nonreplicating editing reporter (32). Although amber/W editing occurs naturally only in the context of replication, this reporter contains mutations in HDV processing signals that enable the study of editing in the absence of replication.
The mutations shown in Fig. 1A were introduced into an HDAg-S mRNA-derived construct that has additional HDV antigenomic sequences required for editing by ADAR1 and ADAR2. HEK293 cells were transiently transfected with cDNA encoding the reporter, and total protein was harvested at 3 days posttransfection and then subjected to Western analysis. The HDAg expression levels of the wild type and the 6×His and 6×His+compensatory mutants are shown in Fig. 3, lanes 1, 2, and 3, respectively. HDAg-L expression was lower with the 6×His mutant without compensatory mutations than with the wild-type (Fig. 3A, lane 2), showing that disruption of the RNA secondary structure as far as 19 nucleotides downstream from the amber/W site can negatively affect editing. This result was expected, since it was previously observed that a construct lacking sequences beyond the 18th nucleotide downstream from the amber/W site showed decreased editing by endogenously expressed ADAR1 (32).
The construct with 6×His+compensatory mutations (Fig. 3A, lane 3) displayed an almost fourfold increase in HDAg-L expression. This increase could have resulted from an elevated level of amber/W editing either because the mutated RNA was a better substrate for editing or because it was deficient in binding to HDAg-S, and this allowed for greater access by ADAR1. To eliminate any potential effect of the HDAg-RNA interaction on editing, we next tested the wild-type sequence and the two mutants in M39 reporters that express an RNA-binding-deficient form of HDAg in which a methionine initiator was engineered at position 39 of the ORF (32). As shown in Fig. 3B, the 6×His+compensatory mutant was again edited more efficiently than was the wild-type construct.
In order to eliminate all HDAg from the cell, a frameshift mutation that occurs early in the ORF of HDAg and is known to inhibit HDAg expression was also introduced (18). Editing of the amber/W site was monitored by RT-PCR followed by StyI digestion of the PCR product. Figure 3C, lanes 1 through 3, shows the results of the editing assay of these HDAg-free reporters. HDAg has been shown to negatively regulate amber/W editing (28), and as expected, editing levels of all three reporters were higher in the complete absence of HDAg. Similar to what was seen in the presence of HDAg, the mutations disrupting the secondary structure diminished amber/W editing, yet when compensatory mutations were made, an elevated level of editing was observed. We concluded that either the 6×His+compensatory RNA was intrinsically a better substrate for amber/W site editing than was the wild-type substrate or the presence of the 6×His+compensatory RNA structure inside the cell induced additional ADAR activity which resulted in increased amber/W editing. To test for the latter possibility, a 10-fold excess of the M39 reporter that contained either the wild-type amber/W site or the 6×His+compensatory site was cotransfected with the wild-type reporter. In this case, if the 6×His+compensatory site could induce ADAR activity, then editing of the wild-type reporter present in the same cells should also increase. However, we observed that under the two conditions tested, the editing levels associated with the wild-type reporter did not change (data not shown). We concluded that the 6×His+compensatory mutations function only in cis to make the associated amber/W site a better substrate for editing.
Increasing the predicted stability of the RNA secondary structure downstream from the amber/W site enhanced editing.
We noticed that the 6×His+compensatory mutations had moderately changed the predicted local secondary structure downstream from the amber/W site, where three base pairs were made in what was originally a mismatch region. Also, two G · U base pairs were converted to more stable A · U pairs. We hypothesized that these changes may have stabilized the secondary structure downstream from the amber/W site and thereby increased the level of editing. Indeed, the free energy of the RNA structure in the 6×His+compensatory construct, as predicted by the mfold modeling program, had decreased by 4.1 kcal/mol compared to that of the wild-type structure.
To test the hypothesis that increased stability distal to the amber/W site can enhance editing at that site, we constructed two more mutants predicted to have a more stable RNA structure adjacent to the amber/W site. This time, however, we made substitution mutations on the noncoding side of the HDV rod-like structure so that the HDAg ORF remained unaffected. We tested mutants designed to have enhanced stability both upstream and downstream from the amber/W site, as shown in Fig. 1B. Since it has recently been shown that ADAR1-S is responsible for endogenous editing of the amber/W site during replication (35), and the essential cis sequence required for editing by ADAR1 has been previously defined (32), we introduced mutations outside of the essential region. The effects of these mutations on amber/W editing were tested with the three types of nonreplicating reporters: one that expressed wild-type HDAg (Fig. 3A), one that expressed RNA-binding-deficient HDAg, (Fig. 3B), and one that did not express HDAg (Fig. 3C). The upstream mutant showed a modest level of increase in editing, whereas the downstream mutant displayed a more dramatic increase, similar to that observed with the 6×His+compensatory reporter. The highest levels of editing were obtained when the upstream and downstream mutations were combined (compare Fig. 3A and B, lanes 5 and 6). Thus, as was observed with the 6×His+compensatory mutations, stabilizing the predicted RNA secondary structure downstream from the amber/W site caused the RNA to function as a better substrate for editing.
In the context of replication, the downstream mutant expressed elevated levels of HDAg-L at early times and displayed a late replication defect.
Amber/W editing of mutants with a stabilized secondary structure was next examined as it occurs naturally in the context of HDV replication. HuH7 cells were transiently transfected with cDNA that used the SV40 late promoter to express circular genomic RNA. Since only the genome was delivered from the cDNA, any editing of the antigenomic amber/W site must have occurred in the context of HDV replication. Severe defects in replication were observed with the upstream and upstream-downstream mutants (data not shown). Hence, it is likely that the rod-like structure of these mutants was so disrupted that replication could not occur. Since mutants that are completely defective for replication are not very informative, these mutants were not studied further. In contrast, a less severe defect was observed with the downstream mutant, and therefore, this mutant was selected for a more detailed analysis.
Following a single large-scale transfection of HuH7 cells with cDNAs expressing either wild-type HDV or the downstream mutant together with a plasmid that expresses SEAP to monitor transfection efficiency (see Materials and Methods for detail), cells were split into 6-well dishes and then total RNA and protein were harvested at 2, 3, 4, 5, 6, and 9 days posttransfection. The RNA samples were analyzed by using Northern blots probed for either the genome or antigenome (Fig. 4A and B, respectively), and the protein samples were subjected to a quantitative Western blotting procedure that used a polyclonal anti-HDAg antibody and radiolabeled protein A (Fig. 4C). Amber/W conversion was also assayed on both the genomic and antigenomic RNA strands by using RT-PCR followed by StyI digestion, where a strand-specific primer was used to initiate reverse transcription (Fig. 4D and E, respectively). The Northern and Western blots demonstrated that the downstream mutant was defective for replication, and a significant reduction in genomic RNA, antigenomic RNA, and HDAg expression was observed compared with wild-type HDV at all time points (Fig. 4A to C, respectively, left panel).
Despite its defect in replication, the downstream mutant expressed a higher percentage of HDAg-L than did wild-type HDV, especially at early time points (Fig. 4C, left panel, compare the percentages of HDAg-L). This result is consistent with those obtained with the nonreplicating reporters, where the downstream mutant was observed to be a better substrate for editing than wild-type RNA. With wild-type HDV, when HDAg-L expression and amber/W conversion on both viral RNA strands were examined, an initial lag in editing was observed (Fig. 4C to E). In contrast, no such lag was observed with the downstream mutant and, for instance, the proportion of HDAg-L expressed at day 3 was roughly equal to that expressed by wild-type HDV at day 9. With both mutant and wild-type HDV at each time point (12 out of 12 cases), the level of amber/W conversion in the antigenome was higher than that of the genome, which was higher than the proportion of HDAg-L expressed. This was consistent with the editing of the antigenome being a primary event, the copying of the edited antigenome into an amber/W-converted genome being a secondary event, and transcription of these genomes to yield HDAg-L-expressing messages being a tertiary event.
With the downstream mutant, the maximal levels of genomic RNA and HDAg expression were reached by day 4 or 5 (Fig. 4A and C, left panels). In contrast, with wild-type HDV, the maximal levels were not reached until days 6 and 9. We therefore concluded that replication of the downstream mutant aborted prematurely. Note that with the downstream mutant, only small differences in editing were observed with the 4-, 5-, 6-, and 9-day points, as assessed by amber/W conversion on both the antigenome and genome as well as by HDAg-L expression. Hence, at times when little or no replication was occurring, no new editing events occurred. Thus, although the mature antigenomic RNPs of the downstream mutant existed in cells at days 4, 5, 6, and 9, they were not further edited at those times. This result is consistent with the observation that the HDAg-antigenomic RNA interaction inhibits amber/W editing (28). We concluded that the mature circular antigenomic RNP was not a substrate for editing. In contrast to the downstream mutant, with wild-type HDV, where replication was still occurring at the later times as assessed by all three methods, editing levels continued to increase with time, reaching a maximum at the final time point. We therefore concluded that when it occurred in its natural context, amber/W editing was coupled to replication.
HDAg-L regulated amber/W editing and its own synthesis when expressed from the downstream mutant.
Although both the 6×His+compensatory mutant and the downstream mutant expressed elevated levels of their respective versions of HDAg-L, only the downstream mutant displayed a significant defect in replication. One major difference between the downstream mutant and the 6×His+compensatory mutant was that although the former expressed wild-type HDAg-L, the latter expressed a mutant form that was defective in inhibiting HDV replication. Perhaps the downstream mutant expressed too much wild-type HDAg-L too early, and then that protein inhibited further replication. To examine this possibility for both the wild-type- and downstream mutant-expressing constructs, a single point mutation was introduced that had previously been shown to render HDAg-L unable to inhibit replication when it is expressed artificially at the onset of replication. The Q214Stop mutation converts the terminal glutamine codon of HDAg-L to a stop codon, abolishes the CXXQ farnesylation signal at the C terminus, and renders the protein unable to inhibit replication (12). If replication inhibition mediated by wild-type HDAg-L was the cause of the replication defect associated with the downstream mutant, then the introduction of the Q214Stop mutation should rescue that defect.
Since the properties of the Q214Stop protein had previously been evaluated only in the context of an amber/W-converted RNA genome, we first tested whether this protein, when expressed from an expression vector at the onset of replication, could inhibit the replication of wild-type HDV. We compared the trans-dominant inhibitory activity of this protein with that of HDAg-S (negative control) and HDAg-L (positive control). HuH7 cells were cotransfected with a cDNA that expressed replication-competent HDV genomic RNA, an HDAg-S expression vector, and a plasmid that expressed HDAg-L of each type. Total RNA was harvested at 6 days posttransfection and was then subjected to Northern analysis. As expected, wild-type HDAg-L abolished replication, while almost no inhibition was observed with the Q214Stop HDAg-L (data not shown). Western blotting confirmed that the two forms of HDAg-L were expressed at similar levels (data not shown). We therefore concluded that the stability of the Q214Stop HDAg-L protein was similar to that of the wild-type protein but that it was unable to inhibit HDV replication.
After confirming the phenotype of Q214Stop HDAg-L, we carried out a replication time course assay as was done before, with wild-type HDV and the downstream mutant, except that in this case, each harbored the Q214Stop mutation. As shown in the right panels of Fig. 4, when introduced into wild-type HDV, the Q214Stop mutation had no effect on replication or editing. From this, we concluded that there were no cis effects in replication associated with this single nucleotide alteration of the rod-like structure. In addition, since no differences were observed when either functional or nonfunctional HDAg-L was expressed as a consequence of replication, we concluded that this protein does not regulate replication in HuH7 cells when expressed from wild-type HDV, at least during the time frame examined. This result is consistent with previous results obtained with editing-deficient mutants (5, 37, 38).
In contrast to what was observed with wild-type HDV, when the Q214Stop mutation was introduced into the downstream mutant, it rescued the replication defect of that mutant. At later time points (days 6 and 9), the genome levels for the downstream mutant were 29- to 49-fold lower than those of wild-type HDV; however, when HDAg-L (Q214Stop) was expressed, these levels were only 2.0- to 3.4-fold lower (Fig. 4A, compare left and right panels). We concluded that when expressed as a consequence of replication and editing, HDAg-L is able to inhibit replication.
In addition to rescuing the replication defect of the downstream mutant, the Q214Stop mutation also altered the manner in which that mutant was edited. When the downstream mutant expressed functional HDAg-L, its replication aborted prematurely, and no new amber/W editing events were observed after replication had stopped. In contrast, in the absence of functional HDAg-L, replication continued throughout the time course, and genome and HDAg levels reached their maximal values on day 9 (Fig. 4A and C, right panels). Similarly, amber/W editing continued throughout the time course, and by day 9, incredibly high levels of amber/W-converted genome (73.2%) and HDAg-L(Q214Stop) expression (58.2%) were achieved. These levels were much higher than those achieved with the downstream mutant when it could express functional HDAg-L (compare left and right panels of Fig. 4C to E). We therefore concluded that, in the context of HDV replication, amber/W editing occurred only when replication was taking place and that by controlling replication, HDAg-L was able to control amber/W editing and its own expression.
The downstream mutations may have caused other defects not associated with amber/W editing. The additional predicted stability associated with this mutant could have enabled ADAR1 to edit other adenosines or could have induced an antiviral response from a host factor such as the protein kinase PKR. Furthermore, the altered structure could have affected binding by HDAg-S, and the resulting RNP may have been less competent for replication. However, since the downstream Q214Stop mutant displayed only a 2.0-fold defect in its genome replication by day 9, the sum total of all effects unrelated to amber/W editing was rather small. In contrast, since there was a 29-fold defect in replication associated with the downstream mutant in the presence of functional HDAg-L, a 15-fold defect in replication could be directly attributed to the excessive and early editing of the amber/W site that led to functional HDAg-L expression.
The conclusion that HDAg-L can regulate HDV replication and its own expression is based on the observation that the Q214Stop mutation was able to largely rescue the replication defect associated with the downstream mutant. However, it was at least formally possible that the Q214Stop mutation functioned not at the level of HDAg-L activity but rather in cis in the HDV RNA structure to compensate for a defect associated with the rod-like structure of the downstream mutant. We therefore tested whether replication of the downstream mutant could be rescued by another means that did not involve mutation. Deletion of the terminal amino acid from HDAg-L and mutation of Cys211 to serine each renders the resulting protein unable to inhibit HDV replication, and each mutation is predicted to abolish farnesylation of HDAg-L (12, 15). Perhaps HDAg-L inhibits replication only when it is farnesylated. If this were the case, then treatment of cells that harbor replicating HDV with an FTI should prevent the expression of functional HDAg-L that is capable of inhibiting replication. Hence, if the defect in replication associated with the downstream mutant results from its early and elevated expression of functional HDAg-L, FTI treatment of cells should rescue that defect. In contrast, if rescue by the Q214Stop mutation was unrelated to HDAg-L activity, then FTI treatment should not rescue replication of the downstream mutant. Previously, Glenn and coworkers showed that when used at 2 μM, FTI-277 can completely block HDAg-L farnesylation without affecting HDV replication, and we therefore used this dosage of the drug for our study (10).
An analysis of the replication of wild-type and downstream mutant HDV, identical to that shown in Fig. 4, was repeated; however, in this case, replication was analyzed in either the absence or presence of FTI as shown in Fig. 5. In general, Fig. 4 and 5 look remarkably similar to one another. As with the Q214Stop mutation shown in Fig. 4, FTI treatment as shown in Fig. 5 had no effect on the replication and editing of wild-type HDV. This provides further evidence that, in the first nine days of HDV replication in HuH7 cells, HDAg-L does not regulate replication. In contrast, FTI treatment rescued the replication defect associated with the downstream mutant. At 6 days posttransfection, the downstream mutant expressed 4-fold more HDAg (Fig. 5C), 12-fold more antigenomic RNA (Fig. 5B), and 25-fold more genomic RNA (Fig. 5A) in the presence of FTI than in its absence, and the differences were even greater at 9 days posttransfection. Also, as shown in Fig. 4, when functional HDAg-L could not be expressed from the downstream mutant, there was an increase in HDAg-L expression and amber/W editing (Fig. 5C to E). This result provides additional support for the idea that by controlling replication, HDAg-L can also control amber/W editing and its own synthesis. Furthermore, since replication of the downstream mutant in the presence of FTI was equivalent to that of the downstream mutant that also harbored the Q214Stop mutation in the absence of FTI (compare Fig. 4 and 5), we concluded that the Q214Stop mutation functioned only to prevent the trans-inhibitory effect of HDAg-L and that no cis effect on the rod-like structure that influenced replication was observed.
HDAg-L was able to regulate amber/W editing as well as its own synthesis when expressed from wild-type HDV.
Although we observed a dramatic increase in replication, HDAg-L expression, and amber/W editing with the downstream mutant when functional HDAg-L expression was prevented by either mutation or FTI treatment, these same methods produced no detectable difference when tested with wild-type HDV. Hence, if HDAg-L is capable of controlling HDV replication and amber/W editing in the wild-type setting, then too little of that protein in HuH7 cells is expressed too late during replication to have any effect. We next evaluated whether, by overexpressing ADAR1-S, we could increase the level of HDAg-L expression and thereby observe a difference in the replication of wild-type HDV that either could or could not express functional HDAg-L. Previously, Jayan and Casey observed that when ADAR1 and particularly ADAR2 are overexpressed to very high levels, HDV replication is inhibited, and at least some of that inhibition results from the editing of adenosines in addition to that in the amber/W site (16). In an attempt to minimize the editing of additional sites, we expressed ADAR1-S to a more modest level and also used a cDNA that delivered genomic RNA so that antigenomic RNA could be edited only in the context of HDV replication.
A vector that expresses HA-tagged ADAR1-S was cotransfected with both wild-type and Q214Stop mutant HDV-expressing constructs, and a time course analysis was performed. The levels of exogenously expressed ADAR1-S were monitored by Western analysis by using an anti-HA monoclonal antibody (Fig. 6D). In order to compare the levels of ADAR1-S-HA expression between wild-type and Q214Stop samples throughout the time course, the HA signal obtained for the wild-type sample on a given day was divided by that obtained for the Q214Stop sample of the same day, and the average of all six values for the time course was then determined. A value greater than 1 would indicate that ADAR1-S-HA expression was greater in the wild-type samples, while a value less than 1 would indicate the converse. A value of 0.97 was obtained from the gel shown in Fig. 6D, which confirmed that equivalent levels of exogenous ADAR1-S were expressed in both sets of samples.
As shown in Fig. 6C, E, and F, when ADAR1-S was overexpressed in the context of replicating wild-type or Q214Stop HDV, much higher levels of amber/W editing and HDAg-L expression were observed than when ADAR1-S was not overexpressed. Consistent with what has been observed previously, ADAR1-S overexpression also inhibited the replication of wild-type HDV as assessed by the levels of genomes, antigenomes, and HDAg expressed (Fig. 6A to C, left panels). However, in the context of the Q214stop mutant, ADAR1-S overexpression had very little effect on replication (Fig. 6A to C, right panels). We therefore concluded that under the conditions tested, any editing that may have occurred at sites other than the amber/W site had little or no effect on HDV replication. We also concluded that, when expressed from wild-type HDV as a consequence of amber/W editing and replication, functional HDAg-L, but not HDAg-L(Q214Stop), was able to inhibit replication.
When equivalent levels of ADAR1-S were overexpressed, significantly more amber/W conversion and HDAg-L expression were observed with the Q214Stop mutant than with wild-type HDV (Fig. 6C, E, and F, compare right and left panels). These data are fully consistent with those from Fig. 4 and 5 and indicate that by inhibiting replication, HDAg-L inhibited amber/W editing and its own synthesis.
DISCUSSION
During HDV replication, genomes are copied into antigenomes and antigenomes are copied into genomes; however, it is not known how the relative proportion of antigenomic to genomic RNA is controlled. Although this was not the intended topic of study, we nevertheless observed significant differences in the antigenome/genome ratios among various samples. Furthermore, these differences did not fluctuate randomly; rather, they seemed to be controlled temporally. For example, for wild-type HDV in both the presence and absence of FTI (Fig. 5), the steady-state level of antigenomic RNA reached its maximum at day 4, and then the level of this species declined with time. In contrast, for genomic RNA, the maximal steady-state level was not achieved until the final time point at day 9. These differences cannot be attributed to cDNA-directed synthesis since, as was shown in Fig. 2, cDNA-directed synthesis accounted for less than 1% of the genomic RNA that had accumulated at later times. Hence, HDV regulated the synthesis of antigenomic versus genomic RNA differentially with respect to time.
We also observed that during HDV replication, the rate of amber/W site editing was not constant and that the process was apparently regulated. Plots of amber/W conversion of both the genome and antigenome versus time were generated for all of the data obtained with wild-type HDV in Fig. 4 to 6. In all cases, following an initial lag period of 2 to 3 days, a linear increase in amber/W conversion was observed. The associated R value of the linear regression analysis varied from 0.969 to 0.998. In the absence of a lag in editing, the line generated by linear regression should cross the origin (i.e., 0% conversion at time zero). In contrast to this, the six lines generated crossed the x axis (0% conversion) between days 1.8 and 3.3. Thus, during the first few days of HDV replication, there was a lag period in which the rate of editing was very low. Following the lag period, the rate of editing was much higher, and a linear increase in the accumulation of amber/W-converted products with respect to time was observed. In addition, it is known that at very late times, a plateau is reached where very little additional editing is observed (35). Prior to this work, there was no understanding of the mechanisms that regulate HDV amber/W editing or the means by which the lag and plateau are established.
Previously, it was shown that when overexpressed at the onset of replication, HDAg-L functions as a dominant negative inhibitor of replication. In contrast, a recent study proposed a model in which HDAg-L cannot regulate replication when expressed naturally as a consequence of the editing that occurs during replication (26). Thus, it was unclear whether HDAg-L expression can have any effect on the rate of replication and/or editing. Previously, Polson et al. showed that HDAg-S inhibits editing, presumably by binding to the amber/W site and preventing access to ADAR1-S (28). However, since HDAg-S is required for replication and is expressed throughout the replication time course, it is not clear how this protein could be responsible for the regulation of editing that is observed. It was previously observed that the levels of ADAR1-S do not vary during replication; hence, amber/W editing is not regulated by modulation of the level of the editing enzyme (35).
Here, we have directly addressed the mechanism by which amber/W editing is regulated. We found that HDV antigenomic sequences or structures 3′ to the amber/W site were suboptimal for editing by ADAR1. We created two mutants (6×His and the downstream mutant) that were still competent for replication but that served as better substrates for ADAR1. During a replication time course of the downstream mutant, no lag in editing was observed; however, its replication aborted prematurely (Fig. 4A to C). Once replication had aborted, no further amber/W editing occurred (Fig. 4C to E), indicating that the mature circular antigenomic RNP was not a substrate for editing and that editing could occur only when new antigenomic RNA was being synthesized. Hence, it is likely that only nascent antigenomic RNA, prior to its association with HDAg-S, is the substrate for ADAR1-S.
It was also not known how amber/W editing of antigenomic RNA leads to HDAg-L expression. In one model, nascent antigenomic RNA is first edited and then processed by the polyadenylation machinery to yield an inosine-containing message that encodes HDAg-L. This model predicts that HDAg-L expression is not dependent upon fixation of the amber/W mutation into the genome. In this case, the proportion of HDAg-L expressed could exceed the proportion of amber/W-converted genomes. In a second model, the edited nascent antigenomic RNA is processed to yield the mature circular antigenome. This molecule then serves as the template for the synthesis of an amber/W-converted genome that in turn serves as the template for an HDAg-L-expressing message. This second model predicts that the proportion of amber/W-converted antigenomes should exceed the proportion of amber/W-converted genomes and that the proportion of amber/W-converted genomes should exceed the proportion of HDAg-L that is expressed from mRNA. This is exactly what was observed in Fig. 4 to 6, and thus, our data strongly support the second model.
A mutation in HDAg-L that destroyed its farnesylation signal and rendered the protein incapable of inhibiting replication when overexpressed at the onset of replication was used to prevent the HDAg-L expressed during replication from having any affect on that process. This mutation, when introduced into the downstream mutant, rescued the replication defect of that mutant. During a replication time course of this double mutant, not only was no lag in editing observed, but also no plateau was established, and editing continued to increase throughout the time course so that at late times, greater than 70% of the genomes contained the amber/W mutation (Fig. 4, right panels). As an alternative to genetic mutation, we also used an FTI to inhibit HDAg-L function (Fig. 5). Although it was suspected, based on a number of different mutants, that prenylation was required for the trans-dominant inhibitory activity of HDAg-L, the data shown in Fig. 5 were the first to formally prove this point. FTI treatment had the same effect as the Q214Stop mutation and rescued the replication defect of the downstream mutant. Again, in this case, not only was no lag in editing observed but no plateau was established, and editing continued to increase throughout the time course so that by day 9, greater than 88% of the genomes contained the amber/W mutation, and HDAg-L represented 78% of the total HDAg expressed (Fig. 5C to E).
It should be noted that in these experiments, no infectious virus was produced, and what is analogous to only a single round of infection was followed. Although the overediting or nonfunctional HDAg-L double mutant (downstream Q214Stop) could replicate well in a single round, it could not replicate in a natural setting. Since the vast majority of the genomes carry the amber/W mutation and hence cannot express HDAg-S, a protein required for replication even if those genomes could be packaged into virions once they went on to infect new cells, the resulting infection would be nonproductive. Hence, regulation of amber/W conversion of the genome is a crucial mechanism that ensures productive replication over multiple rounds of infection.
In additional experiments, we showed that when ADAR1-S was modestly overexpressed in the presence of replicating wild-type HDV, again, no lag in editing was observed; however, replication aborted prematurely. When ADAR1 was overexpressed with wild-type HDV that expressed nonfunctional HDAg-L, replication was restored, but both the lag and plateau were lost. Again, an overediting phenotype was observed.
From the data summarized above, we have synthesized a model to account for the kinetics of amber/W editing observed during replication. The early lag in editing occurs because the sequence or structure 3′ to the amber/W site has evolved to be suboptimal for editing by ADAR1-S. Editing can occur only when a high concentration of nascent antigenomic RNA is achieved in the cell. Thus, at early times during replication, when the concentration of nascent antigenomic RNA is very low, editing is very inefficient. Later during a replication time course, as the concentration of nascent antigenomic RNA increases, so too does editing efficiency. When either the concentration of ADAR1 (Fig. 6) or the affinity of the amber/W site for that enzyme (Fig. 4 and 5) was increased, the lag was abolished, HDAg-L was expressed prematurely and at elevated levels, and replication aborted. Thus, the lag period that results from the suboptimal nature of the amber/W site provides a window of time where replication can occur in the absence of HDAg-L.
The plateau in editing observed at late times results from two facts. First, the mature antigenomic RNP is not a substrate for editing, while nascent antigenomic RNA not in association with HDAg is a substrate. Thus, the rate of editing is coupled to the rate of replication, and when replication aborts, so too does editing. Second, HDAg-L expressed as a consequence of amber/W editing during replication can indeed inhibit replication. Since replication and editing are coupled, HDAg-L autogenously regulates its synthesis by inhibiting replication and thereby preventing additional editing events. The resulting plateau in editing ensures that the majority of genomes synthesized do not carry the amber/W mutation. These genomes will therefore be competent to initiate a subsequent round of infection.
All of the experiments in this report were conducted with a genotype I clone. Genotype I infections were the first to be discovered, are observed throughout the world, and are thought to represent the most common type of HDV infection (33). More recently, genotypes II and III have been discovered, and the latter genotype is found in Central and South America, often in association with particularly aggressive forms of the disease (4). Interestingly, the amber/W editing that occurs with genotype III is surprisingly different from that which occurs with genotype I. In genotype III, the unbranched rod-like structure is required for replication but is not edited at its amber/W site (2). During RNA replication of this genotype, a second alternate conformation which serves as the substrate for amber/W editing is also thought to form (2). Furthermore, this alternate structure is more efficiently bound by HDAg-L than by HDAg-S (9). Hence, in genotype III, HDAg-L expression is autogenously regulated, and amber/W editing is controlled by a direct feedback mechanism (9). In contrast, in genotype I, the autogenously regulated expression of HDAg-L is achieved through an indirect feedback mechanism. In this case, the same rod-like structure that is required for replication is also the substrate for amber/W editing (5), and we showed here that amber/W editing is coupled to replication. Hence, by controlling replication, genotype I HDAg-L is able to indirectly control amber/W editing and its own expression.
Consistent with previous results, we found that in HuH7 cells, HDAg-L does not regulate the replication of wild-type HDV. Apparently, too little HDAg-L is expressed in these cells, too late during replication, to have any effect on replication. Nevertheless, in these cells, replication does appear to be regulated, and genome levels reach their maximum within 6 to 9 days posttransfection. Furthermore, an editing plateau is still observed in these cells, and in the absence of HDAg-L, runaway replication is not observed. It is presently not known how HDV replication is regulated in HuH7 cells.
It is also not known whether HDAg-L regulates HDV replication in human hepatocytes during a natural infection. As shown in Fig. 6, wild-type HDV replication was regulated by HDAg-L in cells that possessed higher ADAR1 activity. It is not known whether the ADAR1 activity of HuH7 cells is comparable to that of hepatocytes. However, it is known that in tissues of the central nervous system, the ADAR activity in primary cells is much higher than it is in corresponding transformed cell lines derived from those tissues (25). If the same relationship holds for the liver, then hepatocytes would have higher ADAR1 activity than HuH7 cells, and hence, HDAg-L might regulate HDV replication during a natural infection. Also consistent with the notion that editing activity is lower in HuH7 cells than it is in hepatocytes, we note that the proportion of HDAg-L to total HDAg that was expressed by wild-type HDV in HuH7 cells shown in Fig. 4 to 6 ranged from 3 to 9% at the final time point. This level of expression is far lower than what is observed with infected liver samples, where the proportion can be as high as 50% (13).
If HDAg-L does indeed control HDV replication during a human infection, then our results have bearing with respect to a future therapy that might be used to treat infections. Only farnesylated HDAg-L is able to package HDV RNPs into helper virus enveloped particles, and FTIs have been shown to be effective at abolishing HDV virion production both in tissue culture and in an animal model (1, 10). Here, we showed that FTI treatment also abolishes the ability of HDAg-L to antagonize HDV replication. Hence, if an FTI were used to treat an infected patient, it is possible that an increase in intracellular HDV replication would initially result, and such an increase might elicit an acute response. We recommend that in future FTI-based trials, the intrahepatic levels of HDV RNAs should be carefully monitored before, during, and after treatment.
Acknowledgments
We thank Brendan O'Malley for providing pBOM094 and John Coffin, Claire Moore, and Andrew Camilli for their advice and comments.
This work was supported by grant R01-AI40472 from the National Institutes of Health and by the Raymond and Beverly Sackler Research Foundation.
REFERENCES
- 1.Bordier, B. B., P. L. Marion, K. Ohashi, M. A. Kay, H. B. Greenberg, J. L. Casey, and J. S. Glenn. 2002. A prenylation inhibitor prevents production of infectious hepatitis delta virus particles. J. Virol. 76:10465-10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casey, J. L. 2002. RNA editing in hepatitis delta virus genotype III requires a branched double-hairpin RNA structure. J. Virol. 76:7385-7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casey, J. L., K. F. Bergmann, T. L. Brown, and J. L. Gerin. 1992. Structural requirements for RNA editing in hepatitis delta virus: evidence for a uridine-to-cytidine editing mechanism. Proc. Natl. Acad. Sci. USA 89:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casey, J. L., T. L. Brown, E. J. Colan, F. S. Wignall, and J. L. Gerin. 1993. A genotype of hepatitis D virus that occurs in northern South America. Proc. Natl. Acad. Sci. USA 90:9016-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casey, J. L., and J. L. Gerin. 1995. Hepatitis D virus RNA editing: specific modification of adenosine in the antigenomic RNA. J. Virol. 69:7593-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, F. L., P. J. Chen, S. J. Tu, C. J. Wang, and D. S. Chen. 1991. The large form of hepatitis delta antigen is crucial for assembly of hepatitis delta virus. Proc. Natl. Acad. Sci. USA 88:8490-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao, M., S. Y. Hsieh, and J. Taylor. 1990. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J. Virol. 64:5066-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, P. J., G. Kalpana, J. Goldberg, W. Mason, B. Werner, J. Gerin, and J. Taylor. 1986. Structure and replication of the genome of the hepatitis delta virus. Proc. Natl. Acad. Sci. USA 83:8774-8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, Q., G. C. Jayan, and J. L. Casey. 2003. Differential inhibition of RNA editing in hepatitis delta virus genotype III by the short and long forms of hepatitis delta antigen. J. Virol. 77:7786-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glenn, J. S., J. C. Marsters, Jr., and H. B. Greenberg. 1998. Use of a prenylation inhibitor as a novel antiviral agent. J. Virol. 72:9303-9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glenn, J. S., J. Taylor, and J. M. White. 1991. Replication of monomeric genomic delta RNA in cultured cells. Prog. Clin. Biol. Res. 364:293-297. [PubMed] [Google Scholar]
- 12.Glenn, J. S., and J. M. White. 1991. trans-dominant inhibition of human hepatitis delta virus genome replication. J. Virol. 65:2357-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Govindarajan, S., S. Hwang, and M. M. Lai. 1993. Comparison of the presence of two forms of delta antigen in liver tissues of acute versus chronic delta hepatitis. Prog. Clin. Biol. Res. 382:139-143. [PubMed] [Google Scholar]
- 14.Hsieh, S. Y., M. Chao, L. Coates, and J. Taylor. 1990. Hepatitis delta virus genome replication: a polyadenylated mRNA for delta antigen. J. Virol. 64:3192-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang, S. B., and M. M. Lai. 1994. Isoprenylation masks a conformational epitope and enhances trans-dominant inhibitory function of the large hepatitis delta antigen. J. Virol. 68:2958-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayan, G. C., and J. L. Casey. 2002. Increased RNA editing and inhibition of hepatitis delta virus replication by high-level expression of ADAR1 and ADAR2. J. Virol. 76:3819-3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayan, G. C., and J. L. Casey. 2002. Inhibition of hepatitis delta virus RNA editing by short inhibitory RNA-mediated knockdown of ADAR1 but not ADAR2 expression. J. Virol. 76:12399-12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo, M. Y., M. Chao, and J. Taylor. 1989. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 63:1945-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo, M. Y., J. Goldberg, L. Coates, W. Mason, J. Gerin, and J. Taylor. 1988. Molecular cloning of hepatitis delta virus RNA from an infected woodchuck liver: sequence, structure, and applications. J. Virol. 62:1855-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuo, M. Y., L. Sharmeen, G. Dinter-Gottlieb, and J. Taylor. 1988. Characterization of self-cleaving RNA sequences on the genome and antigenome of human hepatitis delta virus. J. Virol. 62:4439-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazinski, D. W., and J. M. Taylor. 1994. Expression of hepatitis delta virus RNA deletions: cis and trans requirements for self-cleavage, ligation, and RNA packaging. J. Virol. 68:2879-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazinski, D. W., and J. M. Taylor. 1995. Intracellular cleavage and ligation of hepatitis delta virus genomic RNA: regulation of ribozyme activity by cis-acting sequences and host factors. J. Virol. 69:1190-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazinski, D. W., and J. M. Taylor. 1993. Relating structure to function in the hepatitis delta virus antigen. J. Virol. 67:2672-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo, G. X., M. Chao, S. Y. Hsieh, C. Sureau, K. Nishikura, and J. Taylor. 1990. A specific base transition occurs on replicating hepatitis delta virus RNA. J. Virol. 64:1021-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maas, S., S. Patt, M. Schrey, and A. Rich. 2001. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl. Acad. Sci. USA 98:14687-14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macnaughton, T. B., and M. M. Lai. 2002. Large hepatitis delta antigen is not a suppressor of hepatitis delta virus RNA synthesis once RNA replication is established. J. Virol. 76:9910-9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Modahl, L. E., and M. M. Lai. 2000. The large delta antigen of hepatitis delta virus potently inhibits genomic but not antigenomic RNA synthesis: a mechanism enabling initiation of viral replication. J. Virol. 74:7375-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polson, A. G., H. L. Ley III, B. L. Bass, and J. L. Casey. 1998. Hepatitis delta virus RNA editing is highly specific for the amber/W site and is suppressed by hepatitis delta antigen. Mol. Cell. Biol. 18:1919-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reid, C. E., and D. W. Lazinski. 2000. A host-specific function is required for ligation of a wide variety of ribozyme-processed RNAs. Proc. Natl. Acad. Sci. USA 97:424-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rizzetto, M., M. G. Canese, J. L. Gerin, W. T. London, D. L. Sly, and R. H. Purcell. 1980. Transmission of the hepatitis B virus-associated delta antigen to chimpanzees. J. Infect. Dis. 141:590-602. [DOI] [PubMed] [Google Scholar]
- 31.Ryu, W. S., H. J. Netter, M. Bayer, and J. Taylor. 1993. Ribonucleoprotein complexes of hepatitis delta virus. J. Virol. 67:3281-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato, S., S. K. Wong, and D. W. Lazinski. 2001. Hepatitis delta virus minimal substrates competent for editing by ADAR1 and ADAR2. J. Virol. 75:8547-8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shakil, A. O., S. Hadziyannis, J. H. Hoofnagle, A. M. Di Bisceglie, J. L. Gerin, and J. L. Casey. 1997. Geographic distribution and genetic variability of hepatitis delta virus genotype I. Virology 234:160-167. [DOI] [PubMed] [Google Scholar]
- 34.Sharmeen, L., M. Y. Kuo, G. Dinter-Gottlieb, and J. Taylor. 1988. Antigenomic RNA of human hepatitis delta virus can undergo self-cleavage. J. Virol. 62:2674-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong, S. K., and D. W. Lazinski. 2002. Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc. Natl. Acad. Sci. USA 99:15118-15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong, S. K., S. Sato, and D. W. Lazinski. 2003. Elevated activity of the large form of ADAR1 in vivo: very efficient RNA editing occurs in the cytoplasm. RNA 9:586-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong, S. K., S. Sato, and D. W. Lazinski. 2001. Substrate recognition by ADAR1 and ADAR2. RNA 7:846-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, T. T., V. V. Bichko, W. S. Ryu, S. M. Lemon, and J. M. Taylor. 1995. Hepatitis delta virus mutant: effect on RNA editing. J. Virol. 69:7226-7231. [DOI] [PMC free article] [PubMed] [Google Scholar]