Abstract
Using a self-replicating reporting replicon of West Nile (WN) virus, we performed a mutagenesis analysis to define the structure and function of the 3′-terminal 6 nucleotides (nt) (5′-GGAUCUOH-3′) of the WN virus genome in viral replication. We show that mutations of nucleotide sequence or base pair structure of any of the 3′-terminal 6 nt do not significantly affect viral translation, but exert discrete effects on RNA replication. (i) The flavivirus-conserved terminal 3′ U is optimal for WN virus replication. Replacement of the wild-type 3′ U with a purine A or G resulted in a substantial reduction in RNA replication, with a complete reversion to the wild-type sequence. In contrast, replacement with a pyrimidine C resulted in a replication level similar to that of the 3′ A or G mutants, with only partial reversion. (ii) The flavivirus-conserved 3′ penultimate C and two upstream nucleotides (positions 78 and 79), which potentially base pair with the 3′-terminal CUOH, are absolutely essential for viral replication. (iii) The base pair structures, but not the nucleotide sequences at the 3rd (U) and the 4th (A) positions, are critical for RNA replication. (iv) The nucleotide sequences of the 5th (G) position and its base pair nucleotide (C) are essential for viral replication. (v) Neither the sequence nor the base pair structure of the 6th nucleotide (G) is critical for WN virus replication. These results provide strong functional evidence for the existence of the 3′ flavivirus-conserved RNA structure, which may function as contact sites for specific assembly of the replication complex or for efficient initiation of minus-sense RNA synthesis.
West Nile (WN) virus is an emerging flavivirus that has caused frequent epidemics since 1996 (51). WN virus is primarily transmitted by mosquitoes among birds; humans and other mammals are incidental hosts (8). Since its introduction to the northeastern United States in 1999, WN virus has caused significant human, equine, and avian disease in North America. Recent studies have shown that, besides natural transmission by mosquitoes, WN virus can also be transmitted in humans through blood transfusion (50), organ transplantation (13), breast feeding (12), intrauterine exposure (10), and laboratory-acquired infection (11). A number of approaches are being pursued to develop a WN virus human vaccine (20, 25, 41, 52, 66, 69, 77) and an antiviral therapy (2, 22, 26, 40, 47, 57, 62).
WN virus belongs to the genus Flavivirus in the family Flaviviridae. Many flaviviruses are significant human pathogens, including dengue (DEN) virus, yellow fever (YF) virus, St. Louis encephalitis (SLE) virus, Japanese encephalitis (JE) virus, Murray Valley encephalitis (MVE) virus, the tick-borne encephalitis (TBE) virus complex, and WN virus (8). Flavivirus virions are spherical in shape, with a diameter of approximately 50 nm. The nucleocapsid, about 30 nm in diameter, consists of capsid protein and genomic RNA and is surrounded by a lipid bilayer in which the viral envelope and membrane proteins are embedded (15). The virion contains a single plus-sense RNA genome of approximately 11 kb in length, which consists of a 5′ untranslated region (UTR), a single long open reading frame, and a 3′ UTR (54). The flavivirus genome contains a 5′ cap followed by the conserved dinucleotide AG; the 3′ end of the genome terminates with the conserved dinucleotide CUOH (6, 7, 18, 29, 43, 44, 54, 71, 72). The 5′ UTR and 3′ UTR of the genomic RNA are approximately 100 and 500 to 700 nucleotides (nt) in length, respectively. The encoded polyprotein is co- and posttranslationally processed by viral and cellular proteases into three structural proteins (capsid [C], premembrane [prM] or membrane [M], and envelope [E]) and seven nonstructural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5) (15). The nonstructural proteins are primarily involved in the replication of viral RNA, as components of a replication complex. NS1 and its interaction with NS4a are required for RNA replication (35, 37, 48). The hydrophobic NS2a was shown to function during assembly and/or release of infectious flavivirus particles (32, 38). NS2b forms a complex with NS3 and is a required cofactor for the serine protease function of NS3 (3, 14, 16, 23). NS3 is a multifunctional protein that has the activities of a serine protease, a 5′-RNA triphosphatase, a nucleoside triphosphatase, and an RNA helicase (33, 70, 73). The functions of the membrane-associated NS4a and NS4b are not known. NS5 has the functions of an RNA-dependent RNA polymerase (RdRp) (1, 24, 65) and a methyltransferase involved in the methylation of the 5′ RNA cap structure (31). During flavivirus replication, a replication complex is formed cotranslationally on the 3′ UTR of the genomic RNA. The plus-sense RNA is replicated into minus-sense RNA, which remains base paired with the plus-sense RNA as a replicative form (RF). The RF then serves as a recycling template for asymmetric synthesis of more plus-sense RNA (36).
The 3′-terminal nucleotides of the flavivirus genomic RNA can form highly conserved secondary and tertiary structures (6, 7, 43, 53, 54, 58, 68). The conserved 3′-terminal structure as well as some short conserved sequences within the 3′ UTR of the genomic RNA may function as cis-acting replication signals, to allow interaction with viral and possibly cellular proteins during the initiation of minus-sense RNA synthesis. In support of this hypothesis, deletions or mutations of the 3′ UTR abolish the replication of full-length genomic RNAs and subgenomic replicons in DEN (19, 45, 46, 80), YF (5, 19, 46), Kunjin (KUN) (27, 30), WN (39, 59, 60, 76), and TBE (44) viruses. Host and viral proteins (including NS3 and NS5) were reported to interact specifically with the 3′ UTR of the flavivirus genome (4, 17, 21, 34, 64, 65).
RdRp activities of recombinant NS5 have been reported for a number of flaviviruses. Recombinant NS5 of DEN-1 virus exhibited a low RdRp activity when RNA templates representing the 3′-terminal 629 and 3,200 nt of the DEN-1 virus genome were used, as well as with a nonviral RNA template (65). Recombinant NS5 of KUN virus could in vitro synthesize full-length RNA from an RNA template of either specific KUN virus replicon (9 kb) or a nonspecific Semliki Forest virus replicon (8.3 kb) (24). Replication complexes purified from DEN-2 virus-infected cells, as well as recombinant DEN-2 virus NS5, exhibited a template requirement for the interaction between 5′ and 3′ ends of the DEN-2 virus subgenomic RNA (1, 79). More recently, it was shown that recombinant NS5 of DEN-2 virus had a strong preference for an RNA template containing a flavivirus-conserved 3′-terminal U, whereas mutation of the flavivirus-conserved 3′ penultimate C did not affect the template activity (49). In contrast, a study with KUN virus replicon showed that mutations of the 3′ penultimate C are completely lethal for replication, whereas substitution of the 3′-terminal U dramatically reduced viral RNA replication (27). Despite the discrepancies between the findings of the latter two studies, the results concordantly suggest that the 3′-terminal nucleotides of the flavivirus genome play a critical role in viral replication.
In this report, we used a WN virus replicon containing a Renilla luciferase reporter to define the requirement of sequence and base-pairing structure of the 3′-terminal 6 nt (5′-GGAUCUOH-3′) of the genome in viral replication. Each of the 3′-terminal 6 nt was mutated to all alternative nucleotides, and, wherever possible, potential base pair structures were restored in the context of mutant 3′-terminal sequences by compensatory mutations. The mutational effects on viral translation and replication were examined by using the luciferase signals. The results suggest that the individual nucleotide identity or the base pair structure of the 3′-terminal 6 nt does not significantly regulate WN virus translation but discretely modulates RNA replication. The essential RNA elements identified in this study may function as recognition signals for efficient and specific initiation of minus-sense RNA synthesis at the 3′ end of WN virus genomic RNA.
MATERIALS AND METHODS
Preparation of plasmids.
Recombinant DNA and cloning procedures were performed by standard methods (55), with modifications as previously described (59, 60). A 196-bp DNA fragment designated as “HDVr” (for hepatitis delta virus ribozyme) was fused to the exact 3′ end of the original WN virus replicon (Rep) and a Renilla luciferase-reporting replicon (RlucRep), resulting in Rep-HDVr and RlucRep-HDVr, respectively (Fig. 1 and 2). The 196-bp HDVr fragment contained a 67-bp HDVr sequence followed by a 129-bp simian virus 40 (SV40) polyadenylation (SV40PolyA) sequence. The 67-bp HDVr was designed to cleave the RNA transcript to yield a precise 3′ end regardless of 3′-terminal sequence (56). The 129-bp SV40PolyA sequence was included for experiments not described in this report.
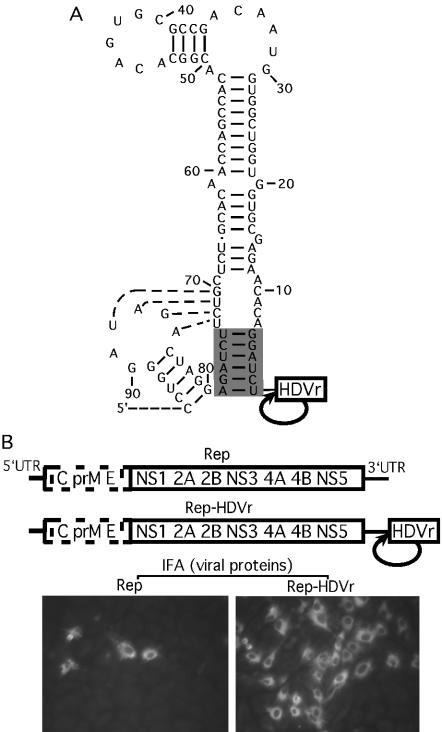
FIG. 1.
Enhancement of replication by the engineering of an HDVr at the 3′ end of WN virus replicons. (A) A stem-loop structure formed by the 3′-terminal sequence of the WN virus genome. The base pairs involved in a putative pseudoknot interaction (58) are indicated by dotted lines. The 3′-terminal 6 nt and their potential base pairs are shaded. An HDVr is engineered immediately at the 3′ end of the WN virus replicon RNA to yield a precise 3′ end through the HDVr-mediated cleavage. The RNA sequence is numbered from the 3′ end. (B, top) WN replicons with (Rep-HDVr) and without (Rep) the 3′ HDVr. All replicons contained an in-frame deletion from nt 190 to 2379 (GenBank accession no. AF404756) spanning three structural genes, C-prM-E (dotted open box). (B, bottom) IFA of BHK cells transfected with Rep or Rep-HDVr at 72 h posttransfection. WN virus immune mouse ascites fluid and Texas red-conjugated goat anti-mouse immunoglobulin G antibody were used as the primary and secondary antibodies for IFA, respectively.
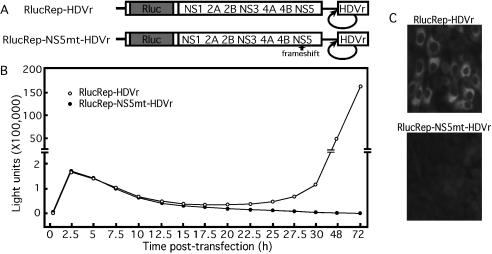
FIG. 2.
The Rluc-reporting replicon containing an HDVr at its 3′ end can be used to monitor viral translation and RNA replication. (A) Rluc-reporting replicon containing a 3′ HDVr (RlucRep-HDVr). Rluc reporter is fused in-frame with the open reading frame of the replicon in the position where the structural region was deleted. RlucRep-NS5mt-HDVr contains a single-nucleotide frameshift upstream of the active site of the RdRp domain of NS5. (B) Time course of the Rluc activity in BHK cells transfected with RlucRep-HDVr or RlucRep-NS5mt-HDVr. (C) IFA of BHK cells transfected with RlucRep-HDVr or RlucRep-NS5mt-HDVr at 72 h posttransfection.
To construct the cDNA clone of RlucRep-HDVr, the 3′-terminal 693 bp of WN virus genomic cDNA (indicated as 3′ UTR), HDVr, and SV40PolyA were fused together through an overlap PCR, resulting in 3′ UTR-HDVr-SV40Poly. Individual DNA fragments of 3′ UTR-HDVr and HDVr-SV40Poly were first PCR amplified from a full-length infectious cDNA clone of WN virus (60) and plasmid pcDNA3.1/Zeo (Invitrogen, Carlsbad, Calif.), using primers F-WN10347 and R-HDVr-3′ UTR and F-HDVr-SV40PolyA and R-Xba-SV40PolyA, respectively (Table 1). Fragments of 3′ UTR-HDVr and HDVr-SV40Poly were then fused to yield 3′ UTR-HDVr-SV40Poly. The 3′ UTR-HDVr-SV40Poly was cloned into the cDNA plasmids of Rep (39, 59) at two unique sites of SacII (at nt 10822 within the 3′ UTR) and XbaI (at the 3′ end of genomic cDNA), resulting in pRep-HDVr (Fig. 1B). Similarly, the PCR fragment of 3′ UTR-HDVr-SV40Poly was cloned into the cDNA plasmids of RlucRep and RlucRep-NS5mt (containing a frameshift to inactivate RdRp) (39) at two unique sites of MluI (at nt 10432 within the 3′ UTR) and XbaI, resulting in pRlucRep-HDVr and pRlucRep-NS5mt-HDVr, respectively (Fig. 2A).
TABLE 1.
Primers used in this study
| Primera | Sequence |
|---|---|
| F-WN10347b | TTCACTAAAGAGATATGAAGACACAACT |
| F-WN10501b | AGAAAGTCAGGCCGGGAAGTTC |
| F-WN10729b | ACTGGGTTAACAAAGGCAAACCAACG |
| R-HDVr-3′UTR | GTAGCCCAGGTCGGACCGCGAGGAGGTGGAGATGCCATGCCGACCCAGATCCTGTGTTCTCGCACCA |
| F-HDVr-SV40PolyA | CTCCTCGCGGTCCGACCTGGGCTACTTCGGTAGGCTAAGGGAGAAGaacttgtttattgcagcttataat |
| R-Xba-SV40PolyA | caggtctagaagacatgataagatacattga |
| F-T7-WN10318b | AATTTAATACGACTCACTATAGGAGATGAGAAGTATGTGGATTACATG |
The primers were named according to their polarity (F, forward sense primer; R, reverse antisense primer), restriction site (underlined lowercase), T7 promoter (underlined uppercase), WN sequence (regular uppercase), HDVr (boldface uppercase), SV40PolyA sequence (regular lowercase), and a tail sequence to facilitate XbaI digestion (italicized lowercase).
The number indicates the nucleotide position of the WN virus genome (GenBank accession no. AF404756) where the 5′ end of the oligonucleotide starts.
All mutagenesis was performed in the pRlucRep-HDVr construct (Fig. 3, 5, and 6). A standard PCR-mediated site-directed mutagenesis was used to prepare mutant fragments of 3′UTR-HDVr-SV40Poly containing the designed nucleotide changes. The resulting mutant, HDVr-SV40Poly, was cloned into pRlucRep-HDVr to replace the wild-type counterpart through unique sites of MluI and XbaI as described above. For construction of replicons containing double mutations, two rounds of PCR-directed mutagenesis were performed in the wild-type RlucRep-HDVr. All constructs were verified by restriction enzyme digestions followed by DNA sequencing.
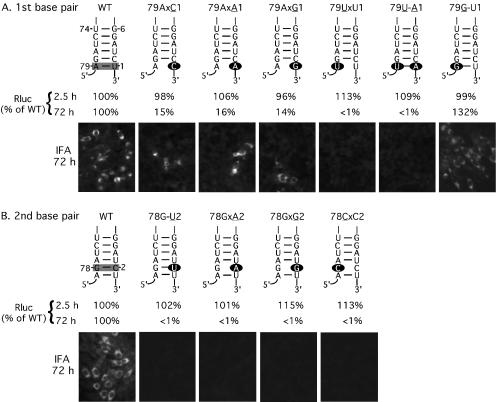
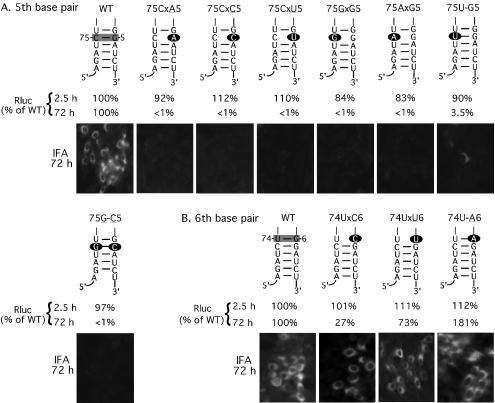
FIG. 3.
Mutagenesis of the 3′-terminal nucleotide (A) and the penultimate nucleotide (B) of the WN virus genome in the RlucRep-HDVr system. Depicted are replicons containing single or double mutations within the 3′-terminal 6 nt (numbered 1 to 6 from the 3′ end of the genome) and their putative base-pairing nucleotides (nt 74 to 79). Specific base pairs analyzed in panels A and B are shaded in the wild-type (WT) structure. For mutant replicons, the mutated nucleotides are indicated as solid ovals. The mutant replicons are named after their mutated position, mutated nucleotides (underlined), blockage of base pair (indicated by an “x”), and potential base pair (indicated by a —). BHK cells transfected with wild-type or mutant replicons were monitored for Rluc activity at 2.5 and 72 h posttransfection. The Rluc activities of mutant replicons are presented as percentages of the corresponding wild-type replicon signal. Representative results of IFAs, performed at 72 h posttransfection, are also presented. Results of one of three representative experiments are shown.
FIG. 5.
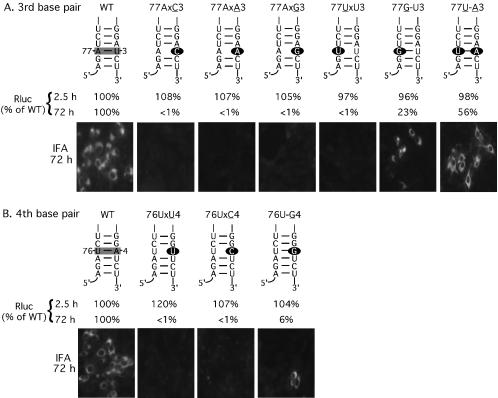
Mutagenesis of the 3′-terminal 3rd (A) and 4th (B) nucleotides of the WN virus genome in viral translation and replication. See the legend to Fig. 3 for details.
FIG. 6.
Mutagenesis of the 3′-terminal 5th (A) and 6th (B) nucleotides of the WN virus genome in viral translation and replication. See the legend to Fig. 3 for details.
RNA transcription, transfection, immunofluorescence assays, and luciferase assays.
Replicon cDNAs of all constructs were driven by a T7 promoter for in vitro transcription. RNA transcription for replicons without a 3′ HDVr (Rep and RlucRep) was performed as previously described (59, 60). RNA transcription for replicons with a 3′ HDVr (Rep-HDVr, RlucRep-HDVr, RlucRep-NS5mt-HDVr, and all RlucRep-HDVr-derivative mutants) was performed similarly, except that no mung bean nuclease treatment of the XbaI-linearized plasmid was required prior to RNA transcription (59, 60). RNA transfection, indirect immunofluorescence assays (IFA), and luciferase assays were performed as previously described (39). The transfection efficiencies of the wild type and various mutant replicons (containing a Renilla luciferase reporter) were normalized by cotransfection of an mRNA containing a firefly luciferase gene (39).
Sequencing of recovered replicon RNAs.
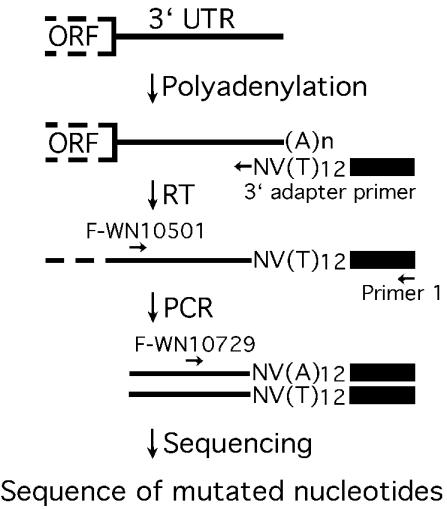
For each viable mutant replicon, a six-well plate was seeded with 3 ml of transfected BHK-21 cell suspension per well as previously described (39). At 72 h posttransfection, cytosolic RNA was extracted with an RNeasy kit (QIAGEN, Valencia, Calif.). As outlined in Fig. 4, extracted RNA (40 μg) was subjected to 3′ polyadenylation by using cloned Escherichia coli poly(A) polymerase (PAP) (Ambion, Austin, Tex.) in a 100-μl reaction (1× PAP buffer, 2.5 mM MnCl2, 1.4 mM ATP, and 8 U of PAP) for 1 h at 37°C. After the PAP reaction, the RNA was cleaned up with the RNeasy kit. The PAP-treated RNA (1 μg) was reverse transcribed at 42°C for 30 min with a 3′ adapter primer (2 μM) and the Superscript III One-Step reverse transcription (RT)-PCR system with Platinum Taq DNA polymerase (Invitrogen, Carlsbad, Calif.). The 3′ adapter primer (5′-GCGAGCACAGAATTAATACGACTCACTATAGGT12VN-3′) contained an adapter sequence (underlined) and a 12-T tract, followed by a V (A, C, or G) and an N (A, C, G, or T). The VN sequence was added to the 3′ end of the 12-T tract to anchor the 3′ adapter primer to the 5′ end of the poly(A) tract of the template RNA during the RT (Fig. 4). Following RT, reactions mixtures were placed on ice, supplemented with primer F-WN10501 (Table 1) and primer 1 (5′-GCGAGCACAGAATTAATACGACT-3′, representing the 5′-terminal 23 nt of the 3′ adapter primer), and subjected to PCR amplification. RT-PCRs were resolved on a 1.5% agarose gel. DNA bands of the expected size (572 bp) were gel purified with a QIAquick gel extraction kit (QIAGEN) and sequenced with primer F-WN10729 (Table 1).
FIG. 4.
Sequencing of viable replicons by 3′ RACE. Replicon RNA recovered at 72 h posttransfection was polyadenylated and amplified through RT-PCR. A 3′ adapter primer containing a 12-T tract was used for RT, and virus-specific primer F-WN10501 and adapter-specific primer 1 were used for PCR. The resulting RT-PCR products were directly subjected to DNA sequencing by a virus-specific primer, F-WN10729. ORF, open reading frame.
HDVr-mediated RNA cleavage.
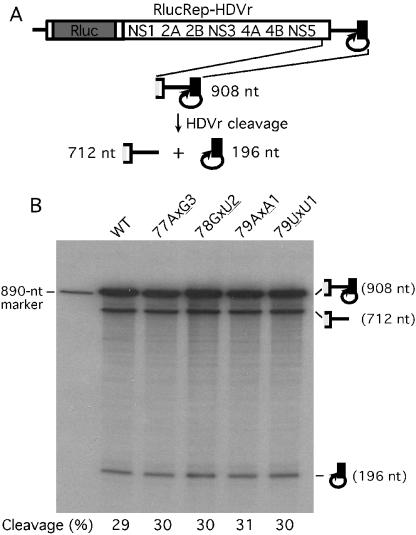
To test whether mutation of nucleotides upstream of the HDVr affected ribozyme cleavage efficiency, we in vitro transcribed a 908-nt RNA containing the 3′-terminal 712 nt of the WN virus genome and the 196 nt of the HDVr-SV40PolyA sequence (see Fig. 7A). The DNA template was prepared by PCR with primers F-T7-WN10318 (containing a T7 promoter sequence followed by the WN virus sequence) and R-Xba-SV40PolyA (Table 1). Individual cDNA plasmid of the wild type or mutant pRlucRep-HDVr was used as a template for PCR. A MEGAscript T7 kit (Ambion) was used to synthesize the 32P-labeled 908-nt RNA according to the manufacturer's protocol with an addition of 1 μl of [α-32P]UTP (3,000 Ci/mmol; Amersham, Piscataway, N.J.) in a standard 20-μl reaction mixture. After incubation of the reaction for 1 h at 37°C, an equal amount of sample from each reaction was analyzed on a 5% sequencing gel. The gel was dried. Individual RNA bands (including the 908-nt uncleaved RNA and two cleaved RNAs of 712 and 196 nt; Fig. 7B) were quantified with a Molecular Dynamics (Sunnyvale, Calif.) PhosphorImager analyzer. The HDVr cleavage efficiency was calculated as the sum of the intensities of the cleaved RNA bands divided by the sum of the intensities of the cleaved and uncleaved RNA bands.
FIG. 7.
In vitro comparison of the HDVr-mediated cleavage efficiencies among wild-type and mutant replicons. (A) Diagram of the HDVr cleavage assay. RNA representing the 3′-terminal 908 nt of the RlucRep-HDVr is in vitro transcribed. The HDVr-mediated cleavage processes the 908-nt RNA precursor into the 712- and 196-nt RNA products. (B) Sequencing gel analysis of the HDVr cleavage assay. 32P-labeled RNAs containing the specified mutations were compared in terms of their cleavage efficiencies by HDVr. The expected sizes of the RNA precursor and products were observed as depicted in panel A. An 890-nt RNA derived from the WN virus sequence (75) was used as an RNA marker. The cleavage efficiency was quantified through a PhosphorImager and is presented as the percentage of the sum of the intensities of the cleaved RNA bands divided by the intensities of the sum of the cleaved and uncleaved RNA bands.
Thermodynamic prediction of RNA secondary structure.
Secondary structures formed by the 3′-terminal nucleotides of various flaviviruses (see Fig. 8) were calculated through minimization of free energy by using the M-Fold program in the GCG package (Genetics Computer Group, Madison, Wis.).
FIG. 8.
Comparison of the 3′ sequences and helix structures formed by various flavivirus genomic RNAs. (A) Summary of nucleotide sequence and base-pairing structure essential for WN virus RNA replication (shaded). (B) 3′-terminal helix structures formed by genomic RNAs of KUN virus (29), MVE virus (GenBank accession no. NC_000943), JE virus (GenBank accession no. AF486638), four serotypes of DEN virus (63), YF virus (54), and TBE virus (43, 44). Essential elements identified in WN virus are shaded in equivalent locations within each helix structure.
RESULTS
Improvement of replication efficiency by engineering an HDVr at the 3′ end of the replicon RNA.
The HDVr-mediated cis-cleavage has been well documented to prepare an RNA molecule with a precise 3′ end (56). To examine the mutational effects of the 3′-terminal nucleotides of WN virus genome on viral replication, we engineered an HDVr at the 3′ end of our original replicon (Rep), resulting in Rep-HDVr (Fig. 1). The original replicon contained an in-frame deletion from nt 190 to 2379 (GenBank accession no. AF404756), spanning three structural genes, C-prM-E (dotted open box; Fig. 1B). Transfection of BHK cells with either Rep or Rep-HDVr revealed IFA-positive cells, indicating that both replicons are replication competent (Fig. 1B). Moreover, five- to eightfold-more IFA-positive cells were observed in transfections with the HDVr-containing replicon than in transfections with the non-HDVr-containing replicon (Fig. 1B), suggesting that the HDVr at the 3′ end of the replicon improves replication efficiency. These results agreed well with a previous report that engineering of an HDVr into a KUN virus replicon substantially improves its replication efficiency (67). The HDVr-mediated enhancement of replicon replication was not likely due to its effect on generation of an authentic 3′ end of the WN virus genome. All of our replicons (with or without HDVr) were prepared to have the exact 3′ terminus of the viral genomic RNA. In the case of the non-HDVr-containing replicons, the authentic 3′ termini of the replicons were achieved through a specific enzyme treatment of the DNA template prior to in vitro transcription (see details in reference 59). As previously reported in KUN virus replicons (67), the increase in replication efficiency of the HDVr-containing replicon of WN virus is likely due to its higher stability (see below).
A luciferase-reporting replicon containing an HDVr at its 3′ end can potentially be utilized to monitor the mutational effect of the 3′-terminal nucleotides on viral translation and RNA replication.
We previously developed a Renilla luciferase-reporting replicon (RlucRep) in which the Rluc gene was in-frame substituted for viral structural genes. This RlucRep can be reliably used to quantify and differentiate viral translation and RNA replication and is ideal for large-scale mutagenesis (39). To facilitate the functional analysis of the 3′-terminal nucleotides of the genome, we engineered an HDVr at the 3′ end of the RlucRep, resulting in RlucRep-HDVr (Fig. 2A). To examine whether addition of the HDVr to the 3′ end of the RlucRep would change the Rluc expression pattern during replication, we performed a time course analysis of the Rluc activity after transfection of BHK cells with the RlucRep-HDVr RNA. Two distinct Rluc signal peaks were observed: one at 2.5 to 10 h and another after 22.5 h posttransfection (Fig. 2B). In contrast, only the 2.5- to 10-h Rluc peak was observed in cells transfected with a replication-deficient replicon that contains a nucleotide insertion upstream of the active site of the viral polymerase NS5 (RlucRep-NS5mt-HDVr; Fig. 2). IFA analysis of cells at 72 h posttransfection showed that viral protein expression was detected only in cells transfected with the replication-competent RlucRep-HDVr, not in cells transfected with the polymerase-defective RlucRep-NS5mt-HDVr (Fig. 2C). These results suggest that the Rluc signals at 2.5 to 10 h posttransfection reflect translation of the input replicon RNA, while the Rluc activity after 22.5 h posttransfection represents RNA replication. Compared with the original RlucRep that lacks an HDVr (39), the current RlucRep-HDVr exhibited a similar pattern of Rluc expression posttransfection. However, two major differences were observed. First, at 48 to 72 h posttransfection, the Rluc signals derived from the RlucRep-HDVr were 20- to 50-fold higher than those derived from the RlucRep (data not shown), suggesting that the HDVr enhances the replication level of the replicon. Second, the negative slope of the first Rluc peak derived from the RlucRep-HDVr (from 2.5 h tracing to 22.5 h posttransfection) was flatter than that derived from the RlucRep (from 2 to 10 h posttransfection) (data not shown), indicating that the HDVr-mediated enhancement of viral replication is most likely due to an increase in RNA stability.
The 3′-terminal U is optimal for viral replication, whereas the identity of its potential base-pairing partner at nucleotide position 79 plays an essential role in RNA replication.
To examine the role of the flavivirus-conserved 3′-terminal U of the WN virus genome in viral replication, we engineered point mutations into the RlucRep-HDVr to replace the wild-type U with a C, A, or G, resulting in the mutants 79AxC1, 79AxA1, and 79AxG1 (mutated nucleotides underlined; blockage of potential base pairing indicated by an “x”; Fig. 3A). Equal amounts of wild type and mutant replicon RNAs were transfected into BHK cells. Rluc activities at various time points posttransfection were quantified: Rluc signals at 2.5 and 5 h posttransfection were measured for estimation of the level of viral translation, while Rluc activities at 48 and 72 h posttransfection were quantified for assessment of the level of RNA replication. For simplicity, only Rluc activities collected at 2.5 and 72 h posttransfection are presented. As shown in Fig. 3A, replacement of the 3′-terminal U with a C, A, or G did not affect the Rluc signal at 2.5 h posttransfection. In contrast, at 72 h posttransfection, Rluc signals from cells transfected with the mutant replicons were about 15% of the corresponding wild-type signal. IFA data collected at 72 h posttransfection correlated well with the Rluc results (Fig. 3A). To examine whether the mutated 3′-terminal nucleotide was retained during replication, we sequenced the mutant replicons recovered at 72 h posttransfection. As outlined in Fig. 4, the recovered replicons were polyadenylated and amplified through RT-PCR with a 3′ adapter primer containing a 12-T tract for RT and a virus-specific primer, F-WN10501, and adapter-specific primer 1 for PCR. The resulting RT-PCR products were directly sequenced by a virus-specific primer, F-WN10729. Strikingly, the 3′-terminal nucleotide of mutant 79AxA1 and 79AxG1 completely reverted to the wild-type U, whereas the 3′-terminal nucleotide of mutant 79AxC1 only partially reverted to the wild-type U (Table 2). The above reversion pattern was consistently observed for each mutant replicon in two independent experiments. The results suggest that the 3′-terminal U is optimal for WN virus replication and that replacement of the wild-type U with a pyrimidine C, but not purine A or G, may support viral replication.
TABLE 2.
Summary of mutagenesis results
| Replicon | Translation (%) | Replication (%)a | Reversionb |
|---|---|---|---|
| 1st bp | |||
| 79A-U1 (WT) | 100 | 100 | |
| 79AxC1 | 98 | 15 | Partial (C or U) |
| 79AxA1 | 106 | 16 | Yes |
| 79AxG1 | 96 | 14 | Yes |
| 79UxU1 | 113 | <1 | ND |
| 79U-A1 | 109 | <1 | ND |
| 79G-U1 | 99 | 132 | No |
| 2nd bp | |||
| 78G-C2 (WT) | 100 | 100 | |
| 78G-U2 | 102 | <1 | ND |
| 78GxA2 | 101 | <1 | ND |
| 78GxG2 | 115 | <1 | ND |
| 78CxC2 | 113 | <1 | ND |
| 3rd bp | |||
| 77A-U3 (WT) | 100 | 100 | |
| 77AxC3 | 108 | <1 | ND |
| 77AxA3 | 107 | <1 | ND |
| 77AxG3 | 105 | <1 | ND |
| 77UxU3 | 97 | <1 | ND |
| 77G-U3 | 96 | 23 | No |
| 77U-A3 | 98 | 56 | No |
| 4th bp | |||
| 76U-A4 (WT) | 100 | 100 | |
| 76UxU4 | 120 | <1 | ND |
| 76U-C4 | 107 | <1 | ND |
| 76U-G4 | 104 | 6 | No |
| 5th bp | |||
| 75C-G5 (WT) | 100 | 100 | |
| 75CxA5 | 92 | <1 | ND |
| 75CxC5 | 112 | <1 | ND |
| 75CxU5 | 110 | <1 | ND |
| 75GxG5 | 84 | <1 | ND |
| 75AxG5 | 83 | <1 | ND |
| 75U-G5 | 90 | 3.5 | No |
| 75G-C5 | 97 | <1 | ND |
| 6th bp | |||
| 74U-G6 (WT) | 100 | 100 | |
| 74UxC6 | 101 | 27 | No |
| 74UxU6 | 111 | 73 | No |
| 74U-A6 | 112 | 181 | No |
The translation and replication levels of mutant replicons are expressed as the replicons' Rluc activities as a percentage of the wild-type (WT) level at 2.5 and 72 h posttransfection, respectively.
For reversion analysis, mutant replicons were extracted at 72 h posttransfection, polyadenylated, and subjected to 3′ RACE analysis (see details in Materials and Methods). ND, not determined.
Since the above mutations changed both the nucleotide identity and the potential base pair structure of the 3′-terminal U, we asked whether changes in only one or in both of the two features were responsible for the observed reduction in RNA replication. To examine the base-pairing effect on viral replication, we prepared mutant replicon 79U-A1 (base pair indicated by a dash) in which the wild-type 79A-U1 was switched, while the base pair structure was retained. Transfection of cells with replicon 79U-A1 showed that the Rluc signal at 2.5 h posttransfection was comparable to that derived from the wild-type replicon, whereas the Rluc activity at 72 h posttransfection was reduced to less than 1% of the wild type. As a negative control, the Rluc activity from cells at 72 h posttransfection with the replication-defective replicon RlucRep-HDVr-mtNS5 was also less than 1% of the activity of the wild type. These results indicate that switching the wild-type 79A-U1 to 79U-A1 is lethal for viral replication. The observation that mutant 79U-A1 was more replication defective than mutant 79AxA1 (less than 1% of the wild type versus 16% of the wild type) indicated that the nucleotide identity at position 79 might be more critical than the base pair structure. To examine the importance of the nucleotide identity at position 79, we generated replicon 79UxU1, in which the wild-type A at position 79 was mutated to a U. The 79UxU1 RNA was completely replication defective, as indicated by the Rluc signals collected at 72 h posttransfection (less than 1% of the wild type). To further analyze the critical role of 79A in WN replication, we prepared mutant 79G-U1, in which the wild-type 79A was replaced with a G and the base pair structure was maintained. Transfection of cells with mutant 79GxU1 showed that the Rluc signal at 2.5 h posttransfection was comparable to that derived from the wild-type replicon. Remarkably, the Rluc activity derived from the 79G-U1-transfected cells at 72 h posttransfection was 132% of the wild-type level. For all constructs, IFA data at 72 h posttransfection correlated well with the Rluc results (Fig. 3A). The results suggest that (i) the nucleotide identity at position 79 is essential for viral replication and (ii) mutations of the 3′-terminal U do not seem to substantially affect viral translation (as indicated by the Rluc signals collected at 2.5 h posttransfection).
The 3′ penultimate C and its potential base-pairing nucleotide at position 78 of the WN virus genome are absolutely essential for RNA replication.
To study the role of the flavivirus-conserved penultimate C in WN virus replication, we prepared three mutant replicons, in which the wild-type penultimate C was replaced with a U, A, or G, resulting in 78G-U2, 78GxA2, and 78GxG2. Among those, mutant 78G-U2 maintained the potential base pair structure between nt 2 and 78. As shown in Fig. 3B, transfection of cells with wild-type and mutant replicons showed similar levels of Rluc signal at 2.5 h posttransfection. In contrast, all mutant-transfected cells exhibited a background level of Rluc activity at 72 h posttransfection, each of which was less than 1% of the wild-type level. In accordance, no IFA-positive cells were detected at 72 h posttransfection, while a substantial number of IFA-positive cells were observed in the wild-type replicon-transfected cells (Fig. 3B). Next, we examined the role of nucleotide G at position 78 in viral replication. Replicon 78CxC2 was prepared, in which the penultimate C was preserved, while the base pair between nt 78 and 2 was abolished (Fig. 3B). Results from both Rluc signal and IFA indicated that replicon 78CxC2 was completely replication defective. These results suggest that (i) the flavivirus-conserved penultimate C is essential for WN replication; (ii) maintenance of the potential base pair structure between nt 2 and 78 is not sufficient for viral replication (as indicated by mutant replicon 78G-U2); and (iii) nucleotide identity at position 78 may play an essential role in viral RNA replication.
Base pair structure, but not nucleotide identity, at the 3rd position from the 3′ end of the genome is essential for WN virus RNA replication.
To dissect the function of the 3rd nucleotide U, we initially changed the wild-type U to a C, A, or G, resulting in the replicons 77AxC3, 77AxA3, and 77AxG3, respectively (Fig. 5A). Transfection of these mutant replicons into BHK cells yielded a nonreplicative level of Rluc activity (less than 1% of the wild-type replicon at 72 h posttransfection) and a negative IFA signal. To examine the effect of base-pairing structure on viral replication, we mutated the nucleotide A at position 77 to a U or G, resulting in replicons 77UxU3 and 77G-U3, respectively. Rluc and IFA results showed that mutant 77UxU3, in which both the nucleotide and its potential base pairing structure at position 77 were changed, was nonreplicative (Fig. 5A). In contrast, mutant 77G-U3, in which the mutation maintained the potential base pairing structure, exhibited a substantial level of replication (23% of the Rluc activity of the wild type at 72 h posttransfection). The results indicated that base pairing structure between nt 3 and 77 is essential for RNA replication. To further examine this hypothesis, we generated mutant 77U-A3, in which the wild-type 77A-U3 was flipped to maintain the base pairing structure. Replicon 77U-A3 was also replication competent, as indicated by a Rluc activity that was 56% of the wild type at 72 h posttransfection, and by a significant level of IFA-positive cells (Fig. 5A). Overall, these results demonstrated that the base pairing structure at the 3rd position from the 3′ end of the genome is essential for viral RNA replication.
Base pair structure at the 4th position from the 3′ end of the genome plays a critical role in WN virus replication.
The wild-type A at the 4th position from the 3′ end of the replicon was mutated to a G, U, or C, resulting in the replicons 76U-G4, 76UxU4, and 76UxC4, respectively (Fig. 5B). Among them, only mutant 76U-G4 retained the potential base pairing structure. Rluc activities from cells at 72 h posttransfection indicated that mutants 76UxU4 and 76UxC4 were lethal, displaying less than 1% of the wild-type replication level. On the other hand, mutant 76U-G4 exhibited about 6% of the wild-type replication level (Fig. 5B). These results implicate that the base pairing structure at the 4th position from the 3′ end of the genome is critical for viral replication.
Nucleotide identities of the 5th position from the 3′ end of the genome and its potential base pair partner are essential for viral replication.
To analyze the function of the 5th nucleotide of the 3′ end of the genome, we prepared replicons 75CxA5, 75CxC5, and 75CxU5 in which the wild-type G at position 5 was mutated to an A, C, or U, respectively. As indicated by the Rluc activity and IFA results, none of the mutants was replication competent (Fig. 6A). Next, we mutated the wild-type C at position 75 to a G, A, or U, resulting in replicons 75GxG5, 75AxG5, and 75U-G5, respectively. Among them, mutants 75GxG5 and 75AxG5 showed no replication activity. In contrast, mutant 75U-G5, which maintained the potential base pair structure, exhibited a marginal level of replication (3.5% of the Rluc signal of the wild type at 72 h posttransfection) (Fig. 6A). To further test the function of the base pairing structure at this position, we prepared mutant 75G-C5, in which the wild-type 75C-G5 was flipped. To our surprise, transfection of cells with replicon 75G-C5 exhibited no replication activity (Fig. 6A). The nonreplicative phenotype of mutant 75G-C5 was verified by three independently prepared constructs. These results suggest that nucleotide G at position 5 and nucleotide C at position 75, but not necessarily the base pairing structure between the two positions (as indicated by the nonreplicative mutant 75G-C5), are essential for WN virus RNA replication. In addition, a pyrimidine U at position 75 could marginally support viral replication (as implicated by the marginal replication level of mutant 75U-G5).
Neither sequence nor structure at the 6th position from the 3′ end of the genome is important for WN virus replication.
We replaced the wild-type G at the 6th position from the 3′ end of the genome with a C, U, or A, resulting in the replicons 74UxC6, 74UxU6, and 74U-A6, respectively (Fig. 6B). Rluc signals collected at 72 h posttransfection indicated that all three mutants were replication competent. Remarkably, mutant 74U-A6, which maintained the base-pairing structure, exhibited a replication level that was about 81% higher than the wild-type level. Mutants 74UxC6 and 74UxU6, in which the base pair structure was abolished between positions 74 and 6, showed replication activities that were 27 and 73% of the wild-type level, respectively. The relative replication levels among the replicon variants (as indicated by the Rluc activity) were in good agreement with the IFA results (Fig. 6B). These results suggest that neither sequence nor structure at the 6th position from the 3′ end of the genome is critical for WN virus replication.
Sequencing analysis of viable mutant replicons.
Point mutations introduced into the WN virus replicon are likely to be reverted to the wild-type sequence if the mutant replicons are replication competent. This was clearly demonstrated in replicons that contained mutations at the 1st position of the 3′ end of the genome (Table 2). To examine whether mutations at other positions within the 3′-terminal nucleotides were retained in replicons after replication, we recovered each viable mutant replicon at 72 h posttransfection and sequenced the 3′-terminal 250 nt of the recovered RNA by 3′ rapid amplification of cDNA ends (RACE) (Fig. 4). The sequencing results showed that, except for the mutations at the 1st nucleotide position, all mutations engineered at other positions of the 3′-terminal nucleotides were retained without any secondary mutation (Table 2). However, we could not exclude the possibility that a compensatory mutation or mutations exist outside the sequenced region of the recovered replicons.
The 3′-terminal 6 nt and their base pair structure do not significantly affect WN virus translation.
Although mutations of the sequence or the base-pairing structure of the 3′-terminal 6 nt exhibited discrete effects on viral RNA replication, the Rluc activities of all mutants at 2.5 h posttransfection ranged from 83 to 132% of the wild-type replicon Rluc activity (Table 2). These results suggest that the 3′-terminal nucleotides and their helix structure do not significantly regulate WN virus translation.
The difference in RNA replication among the various replicons is not due to a difference in HDVr cleavage.
We were concerned that mutations of the 3′-terminal nucleotides of the WN replicon might affect the cleavage efficiency of the HDVr, thus influencing the observed levels of replication among various replicons. To exclude this possibility, we compared the in vitro cleavage efficiencies of the HDVr among several representative replicons. Since the reporting replicon alone (RlucRep) and the HDVr-containing fragment were 9,774 and 196 nt, respectively, in length, it would be technically difficult to resolve the intact RlucRep-HDVr (9,970 nt) and the cleaved product of RlucRep (9,774 nt) on an agarose or a polyacrylamide gel. To overcome this difficulty, we in vitro synthesized shorter RNAs containing the 3′-terminal 712 nt of the WN virus genome (including the complete 3′ UTR of 634 nt) and the 196 nt of the HDVr-containing fragment (Fig. 7A). The HDVr-mediated RNA cleavage occurred simultaneously with the synthesis of the RNAs through in vitro transcription (data not shown). Analysis of such 32P-labeled RNAs (immediately after in vitro transcription) on a sequencing gel showed three RNA species of the expected sizes: an uncleaved precursor (908 nt) and two cleaved products (712 and 196 nt). An 890-nt RNA derived from the WN virus sequence (75) was used as an RNA size marker (Fig. 7B). PhosphorImager quantification of the gel showed similar cleavage efficiencies among RNAs derived from wild-type, 77AxG3, 78GxU2, 79AxA1, and 79UxU1 replicons: approximately 29 to 31% of total RNA was processed. These data agree well with a previous study that demonstrated that the nucleotide sequence upstream of the HDVr cleavage site does not affect the cleavage efficiency of the RNA substrate (56). These results suggest that the observed differences in RNA replication among various replicons are not due to differences in HDVr cleavage efficiency.
DISCUSSION
The flavivirus genome terminates with a conserved 3′-terminal stem-loop structure from which minus-sense RNA synthesis is initiated. Although previous studies have shown that flavivirus 3′ UTRs are essential for replication (5, 19, 30, 39, 44-46, 59, 60, 76, 80), little is currently known about how these sequences and RNA structures specifically contribute to the regulation of viral translation and RNA replication. The objective of this study was to analyze the specific function of the 3′-terminal 6 nt that are involved in formation of the helix structure located at the 3′ end of the WN virus genome. Nucleotide identity and base pair structure were systematically mutated in a self-replicating reporting replicon. The results suggest that neither the nucleotide sequence nor base pair structure of any of the 3′-terminal 6 nt significantly regulates viral translation, but discretely modulates RNA replication (Table 2).
Based on the Rluc activity at 72 h posttransfection, replacement of the conserved 3′-terminal U seemed to reduce the replication level of the WN virus replicon to approximately 15% of the wild-type level (with a complete or partial reversion of the mutated nucleotide to the wild-type U; see below), whereas mutations of the conserved 3′ penultimate C to any other nucleotide completely abolished viral replication (Fig. 3). Similar results were recently reported in a KUN virus replicon (27). However, in the KUN virus replicon, replacement of the 3′-terminal U reduced the RNA replication to about 70% of the wild-type level, as estimated by Northern blot hybridization at 69 h posttransfection (27). The discrepancy in the extent of mutational effects of the 3′ U on viral replication between the WN and KUN viruses (15% versus 70% of the wild type) is most likely due to the different reporting systems used for the two viruses (Rluc enzyme activity versus Northern blot). In our study, we sequenced the viable replicons recovered at 72 h posttransfection (Table 2). Interestingly, a purine A or G replacement of the wild-type 3′ U completely reverted to the wild-type U, whereas a pyrimidine C substitution only partially reverted to the wild-type U. These results suggest that although the 3′-terminal U is optimal for flavivirus replication, replacement of the wild-type U with a pyrimidine C (but not purine A or G) could support viral replication. In contrast, the conserved penultimate C is absolutely required for flavivirus replication. The critical role of the 3′-terminal CUOH in RNA replication was also implicated in flavivirus plus-sense RNA synthesis. Since the flavivirus genome starts with a conserved 5′ dinucleotide AG, the 3′ end of the minus-sense RNA also terminates with the exact dinucleotide CUOH, which is assumed to be the initiation site for plus-sense RNA synthesis. The importance of the 3′ CUOH of the minus-sense RNA was recently demonstrated in KUN virus. Point mutations of the 3′ CUOH of the minus-sense RNA completely blocked the replication of the KUN virus replicon (27).
Using a recombinant NS5 of DEN-2 virus in an in vitro RdRp assay, Nomaguchi and colleagues have recently shown that purified NS5 exhibits a preference for the 3′-terminal nucleotide of the RNA template in the order of U, A∼G, and C, from highest to lowest (49). Surprisingly, substitution of the penultimate C with a U did not affect the template activity for de novo RNA synthesis in the in vitro RdRp assay (49). The difference between the in vitro (recombinant DEN-2 virus RdRp) and in vivo results (KUN and WN virus replicons) may result from the incomplete nature of the in vitro reaction. The DEN-2 virus RdRp assay involved a 770-nt subgenomic RNA, an excess of nucleotide triphosphates, and purified NS5 in the absence of the other viral nonstructural proteins and cellular components (49). Similarly, a discrepancy between the efficiency of transcription initiation of RNA templates with various 3′-terminal nucleotides in vitro (61) and in vivo (78) was observed in hepatitis C virus. However, we cannot exclude the possibility that the difference between the in vitro DEN-2 virus results and the in vivo KUN/WN virus data may truly reflect a difference in template requirement for RNA initiation among various groups of flaviviruses (see below). In vivo mutagenesis of a DEN virus infectious clone or replicon will be required to distinguish between the above two possibilities.
We extended our studies to regions beyond the flavivirus-conserved 3′-terminal dinucleotides. RNA elements within the 3′-terminal 6-bp helix that are essential for WN replication are summarized (shaded) in Fig. 8A.
(i) First and 2nd base pairs.
Besides the 3′-terminal CUOH (described above), the dinucleotides located at positions 78 and 79, which form potential base pairs with the 3′-terminal CUOH of the genome, are also important for viral replication (Fig. 3 and 4). Maintenance of the base pair structure alone at these two positions is not sufficient to support viral replication, as suggested by the results that the base pair-retaining replicons 79U-A1 and 78G-U2 were replication deficient. However, the base pair-retaining replicon 79G-U1 exhibited a replication level that was 32% higher than the wild-type level, suggesting that a purine at position 79 is competent for viral replication. This conclusion is supported by a phylogenetic comparison, which revealed that either an A (KUN, MVE, or JE virus, four serotypes of DEN, YF, and WN viruses) or a G (TBE virus) is located at the equivalent position among different members of flaviviruses (Fig. 8B).
(ii) Third and 4th base pairs.
The base pair structure, but not the nucleotide sequence at these positions is critical for RNA replication. This is clearly demonstrated by the results that, as long as the base pair structure was maintained at these positions (77G-U3, 77U-A3, and 76U-G4), the replicons were replication competent (Fig. 5).
(iii) Fifth base pair.
We mutated either of the nucleotides involved in formation of the 5th base pair to all possible alternative nucleotides. We also flipped the wild-type 75C-G5 to 75G-C5 (Fig. 6A). Among all the variants, only mutant 75U-G5, which maintained the base pair structure, retained a marginal replication activity (3.5% of the wild-type level). In contrast, the base pair-maintaining mutant 75G-C5 was replication defective. These results suggest that the nucleotide identities of G and C at the 5th and the 75th positions, respectively, but not necessarily the base pair structure, are essential for viral replication.
(iv) Sixth base pair
Neither the sequence nor the base pair structure of the 6th nucleotide (G) is critical for WN virus replication. Interestingly, mutation of the wild-type 74U-G6 to 74U-A6 substantially increased the replication level (181% of the wild-type level). Overall, the above data provide strong functional evidence for the existence of an RNA structure at the 3′ terminus of the WN virus genome that until now has been supported only by computer modeling and RNase probing (7, 58).
Phylogenetic comparison among various members of the genus Flavivirus showed that KUN, MVE, and JE viruses (JE virus serocomplex) share a 3′-terminal helix identical to WN virus and, therefore, contain all of the essential sequence and structural elements identified for WN virus (Fig. 8B). All four serotypes of DEN virus possess an identical 3′-terminal helix within the DEN virus serocomplex but differ from those of the JE virus serocomplex. Compared with the JE virus serocomplex, the helix from the DEN virus serocomplex contains a bulge rather than a base pair at the 4th position. Although YF virus and the TBE virus complex share a 3′ helix structure similar to that of the JE virus serocomplex, both YF and TBE viruses contain a G-C base pair rather than the 75C-G5 base pair of WN virus at the 3′ 5th position (Fig. 8B). The outlined differences between WN virus and other flaviviruses were found to be lethal for WN virus replication. These results indicate that although flaviviruses have retained similar sequences and structural elements at the 3′-terminal helix of their genomes, variations specific to particular viral groups have occurred during evolution. These variations may modulate viral replication among different species of hosts during the transmission cycle. In support of this notion, mutagenesis of a DEN-2 virus infectious clone containing fragments of the WN virus 3′-terminal stem-loop showed that retention of the 3′-terminal 17-bp hairpin of DEN-2 virus was essential for the viability of chimeric DEN-2 virus. Moreover, one 7-bp substitution with WN virus sequence in this domain resulted in a mutant DEN-2 virus that grew well in monkey kidney cells but was severely restricted in mosquito cells (80).
Our results, along with a recent report from KUN virus (27), fit well with a model for initiation of RNA replication that is built upon a crystal structure of bacteriophage φ6 RdRp complexed with an oligonucleotide template and two nucleotide triphosphates (NTPs) complementary to the 3′-terminal 2 nt of the template (9). According to this model, the 3′ terminus of the RNA is initially fed into a template channel of the RdRp and is temporarily locked in a specific binding pocket (the “S” pocket) that is too small to accommodate a 3′-terminal purine. If this is the case for WN virus RdRp, an RNA template with a 3′-terminal A or G may not fit into the S pocket, resulting in a complete reversion to the wild-type 3′ U of the mutant replicons (79AxA and 79AxG; Table 2) at 72 h posttransfection. We noticed that, for flavivirus RdRp, the templates of both plus- and minus-sense RNAs universally terminate with the 3′ U. In contrast, for φ6 RdRp, the template terminates with a 3′ pyrimidine: either U for plus-sense RNA or C for minus-sense RNA. Because of the 3′-terminal difference between the flavivirus and bacteriophage φ6 templates, the flavivirus RdRp may have evolved to constrain its S pocket specifically for a pyrimidine U rather than a C, whereas the φ6 RdRp has evolved to retain its S pocket to accommodate both types of pyrimidine. As a result, WN virus RNA template containing a 3′ C may fit, but not snugly, into the stringent S pocket of its RdRp, leading to a partial reversion to the wild-type 3′ U (Table 2). The essential role of the 3′ penultimate C of the template was indicated by a sequence analysis that the template channel of φ6 RdRp prefers a C at the 3′ penultimate position. However, the interactions with the φ6 RdRp that confer this specificity are currently unclear (9). During the formation of the initiation complex, the NTP binding site of the RdRp positions the first incoming GTP to base pair with the 3′ penultimate C of the template. After the initial GTP binding to the 3′ penultimate C, the polymerase ratchets backwards, leading to the release of the 3′-terminal nucleotide of the template from the S pocket. The NTP binding site of the RdRp then positions the second NTP to base pair with the 3′-terminal nucleotide of the template, followed by the formation of the first phosphodiester bond (9).
Although the initiation model described above could explain the importance of the 3′-terminal 2 nt of the template, the molecular basis for the requirements of other RNA elements identified in this study remains enigmatic. Since the flavivirus replication complex contains multiple viral proteins (28, 36, 37, 42, 74) and potentially cellular proteins as well (4, 34, 64), RNA elements other than the 3′-terminal 2 nt are required for specific assembly of the replication complex or for efficient initiation of RNA synthesis at the 3′ end of the viral template. Future studies using genetic, biochemical, and crystallographic approaches are required to define the mechanism of the initiation of flavivirus RNA replication.
Acknowledgments
We are grateful to Paul Masters for helpful discussions during the course of this work. We thank Michael Lo for help during the early phase of this work. We also thank the Molecular Genetics Core and the Cell Culture Facility at the Wadsworth Center for sequencing and oligonucleotide synthesis and for maintenance of BHK cells, respectively.
This work was in part funded by the National Institute of Allergy and Infectious Disease, National Institutes of Health, under contracts no. N01-AI-25490 and 1U01AI061193-01.
REFERENCES
- 1.Ackermann, M., and R. Padmanabhan. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 276:39926-39937. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, J. F., and J. J. Rahal. 2002. Efficacy of interferon alpha-2b and ribavirin against West Nile virus in vitro. Emerg. Infect. Dis. 8:107-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arias, C. F., F. Preugschat, and J. H. Strauss. 1993. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology 193:888-899. [DOI] [PubMed] [Google Scholar]
- 4.Blackwell, J. L., and M. A. Brinton. 1997. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J. Virol. 71:6433-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bredenbeek, P. J., E. A. Kooi, B. Lindenbach, N. Huijkman, C. M. Rice, and W. J. Spaan. 2003. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 84:1261-1268. [DOI] [PubMed] [Google Scholar]
- 6.Brinton, M. A., and J. H. Dispoto. 1988. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology 162:290-299. [DOI] [PubMed] [Google Scholar]
- 7.Brinton, M. A., A. V. Fernandez, and J. H. Dispoto. 1986. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology 153:113-121. [DOI] [PubMed] [Google Scholar]
- 8.Burke, D. S., and T. P. Monath. 2001. Flaviviruses, p. 1043-1126. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott, William & Wilkins, Philadelphia, Pa.
- 9.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. 2002. Intrauterine West Nile virus infection—New York, 2002. Morb. Mortal. Wkly. Rep. 51:1135-1136. [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2002. Laboratory-acquired West Nile virus infections—United States, 2002. Morb. Mortal. Wkly. Rep. 51:1133-1135. [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. 2002. Possible West Nile virus transmission to an infant through breast-feeding—Michigan, 2002. Morb. Mortal. Wkly. Rep. 51:877-878. [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 2002. Update: investigations of West Nile virus infections in recipients of organ transplantation and blood transfusion—Michigan, 2002. Morb. Mortal. Wkly. Rep. 51:879. [PubMed] [Google Scholar]
- 14.Chambers, T. J., A. Grakoui, and C. M. Rice. 1991. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J. Virol. 65:6042-6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 16.Chambers, T. J., A. Nestorowicz, S. M. Amberg, and C. M. Rice. 1993. Mutagenesis of the yellow fever virus NS2B protein: effects on proteolytic processing, NS2B-NS3 complex formation, and viral replication. J. Virol. 67:6797-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, C.-J., M.-D. Kuo, L.-J. Chien, S.-L. Hsu, Y.-M. Wang, and J.-H. Lin. 1997. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J. Virol. 71:3466-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cleaves, G. R., and D. T. Dubin. 1979. Methylation status of intracellular Dengue type 2 40S RNA. Virology 96:159-165. [DOI] [PubMed] [Google Scholar]
- 19.Corver, J., E. Lenches, K. Smith, R. A. Robison, T. Sando, E. G. Strauss, and J. Strauss. 2003. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J. Virol. 77:2265-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis, B. S., G.-J. J. Chang, B. Cropp, J. T. Roehrig, D. A. Martin, C. J. Mitchell, R. Bowen, and M. L. Bunning. 2001. West Nile virus recombinant DNA vaccine protects mouse and horse from virus challenge and expresses in vitro a noninfectious recombinant antigen that can be used in enzyme-linked immunosorbent assays. J. Virol. 75:4040-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Nova-Ocampo, M., N. Villegas-Sepulveda, and R. M. del Angel. 2002. Translation elongation factor-1alpha, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology 295:337-347. [DOI] [PubMed] [Google Scholar]
- 22.Engle, M. J., and M. S. Diamond. 2003. Antibody prophylaxis and therapy against West Nile virus infection in wild-type and immunodeficient mice. J. Virol. 77:12941-12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falgout, B., R. H. Miller, and C.-J. Lai. 1993. Deletion analysis of dengue virus type 4 nonstructural protein NS2B: identification of a domain required for NS2B-NS3 protease activity. J. Virol. 67:2034-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guyatt, K. J., E. G. Westaway, and A. A. Khromykh. 2001. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J. Virol. Methods 92:37-44. [DOI] [PubMed] [Google Scholar]
- 25.Hall, R. A., D. J. Nisbet, K. B. Pham, A. T. Pyke, G. A. Smith, and A. A. Khromykh. 2003. DNA vaccine coding for the full-length infectious Kunjin virus RNA protects mice against the New York strain of West Nile virus. Proc. Natl. Acad. Sci. USA 100:10460-10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jordan, I., T. Briese, N. Fischer, J. Y. Lau, and W. I. Lipkin. 2000. Ribavirin inhibits West Nile virus replication and cytopathic effect in neural cells. J. Infect. Dis. 182:1214-1217. [DOI] [PubMed] [Google Scholar]
- 27.Khromykh, A., N. Kondratieva, J.-Y. Sgro, A. Palmenberg, and E. G. Westaway. 2003. Significance in replication of the terminal nucleotides of the flavivirus genome. J. Virol. 77:10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khromykh, A. A., P. L. Sedlak, and E. G. Westaway. 1999. trans-Complementation analysis of the flavivirus Kunjin ns5 gene reveals an essential role for translation of its N-terminal half in RNA replication. J. Virol. 73:9247-9255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khromykh, A. A., and E. G. Westaway. 1994. Completion of Kunjin virus RNA sequence and recovery of an infectious RNA transcribed from stably cloned full-length cDNA. J. Virol. 68:4580-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koonin, E. V. 1993. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J. Gen. Virol. 74:733-740. [DOI] [PubMed] [Google Scholar]
- 32.Kummerer, B. M., and C. M. Rice. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 76:4773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, H., S. Clum, S. You, K. E. Ebner, and R. Padmanabhan. 1999. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J. Virol. 73:3108-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, W., Y. Li, N. Kedersha, P. Anderson, M. Emara, K. Swiderek, G. T. Moreno, and M. A. Brinton. 2002. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 76:11989-12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindenbach, B. D., and C. M. Rice. 1997. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 71:9608-9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the virus and their replication, p. 991-1042. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 37.Lindenbach, B. D., and C. M. Rice. 1999. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J. Virol. 73:4611-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, W. J., H. B. Chen, and A. A. Khromykh. 2003. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J. Virol. 77:7804-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lo, M. K., M. Tilgner, K. A. Bernard, and P.-Y. Shi. 2003. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 77:10004-10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lo, M. K., M. Tilgner, and P.-Y. Shi. 2003. A potential high-throughput assay for screening inhibitors of West Nile virus replication. J. Virol. 77:12901-12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lustig, S., U. Olshevsky, D. Ben-Nathan, B. E. Lachmi, M. Malkinson, D. Kobiler, and M. Halevy. 2000. A live attenuated West Nile virus strain as a potential veterinary vaccine. Viral Immunol. 13:401-410. [DOI] [PubMed] [Google Scholar]
- 42.Mackenzie, J. M., A. A. Khromykh, M. K. Jones, and E. G. Westaway. 1998. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 245:203-215. [DOI] [PubMed] [Google Scholar]
- 43.Mandl, C. W., H. Holzmann, C. Kunz, and F. X. Heinz. 1993. Complete genomic sequence of Powassan virus: evaluation of genetic elements in tick-borne versus mosquito-borne flaviviruses. Virology 194:173-184. [DOI] [PubMed] [Google Scholar]
- 44.Mandl, C. W., H. Holzmann, T. Meixner, S. Rauscher, P. F. Stadler, S. L. Allison, and F. X. Heinz. 1998. Spontaneous and engineered deletions in the 3′ noncoding region of tick-borne encephalitis virus: construction of highly attenuated mutants of a flavivirus. J. Virol. 72:2132-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Men, R., M. Bray, D. Clark, R. M. Chanock, and C.-J. Lai. 1996. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 70:3930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molenkamp, R., E. A. Kooi, M. A. Lucassen, S. Greve, J. C. P. Thijssen, W. J. M. Spaan, and P. J. Bredenbeek. 2003. Yellow fever virus replicons as an expression system for hepatitis C virus structural proteins. J. Virol. 77:1644-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morrey, J., D. Smee, R. Sidwell, and C. Tseng. 2002. Identification of active antiviral compounds against a New York isolate of West Nile virus. Antiviral Res. 55:107-116. [DOI] [PubMed] [Google Scholar]
- 48.Muylaert, I. R., R. Galler, and C. M. Rice. 1997. Genetic analysis of the yellow fever virus NS1 protein: identification of a temperature-sensitive mutation which blocks RNA accumulation. J. Virol. 71:291-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nomaguchi, M., M. Ackermann, C. Yon, S. You, and R. Padmanbhan. 2003. De novo synthesis of negative-strand RNA by dengue virus RNA-dependent RNA polymerase in vitro: nucleotide, primer, and template parameters. J. Virol. 77:8831-8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pealer, L., A. Marfin, L. Petersen, R. Lanciotti, P. Page, S. Stramer, M. Stobierski, K. Signs, B. Newman, H. Kapoor, J. Goodman, and M. Chamberland. 2003. Transmission of West Nile virus through blood transfusion in the United States in 2002. N. Engl. J. Med. 349:1236-1245. [DOI] [PubMed] [Google Scholar]
- 51.Petersen, L. R., and J. T. Roehrig. 2001. West Nile virus: a reemerging global pathogen. Emerg. Infect. Dis. 7:611-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pletnev, A. G., R. Putnak, J. Speicher, E. J. Wagar, and D. W. Vaughn. 2002. West Nile virus/dengue type 4 virus chimeras that are reduced in neurovirulence and peripheral virulence without loss of immunogenicity or protective efficacy. Proc. Natl. Acad. Sci. USA 99:3036-3041. (Erratum, 99:7184.) [DOI] [PMC free article] [PubMed]
- 53.Proutski, V., E. A. Gould, and E. C. Holmes. 1997. Secondary structure of the 3′ untranslated region of flaviviruses: similarities and differences. Nucleic Acids Res. 25:1194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rice, C. M., E. M. Lenches, S. R. Eddy, S. J. Shin, R. L. Sheets, and J. H. Strauss. 1985. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science 229:726-733. [DOI] [PubMed] [Google Scholar]
- 55.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 56.Schurer, H., K. Lang, J. Schuster, and M. Morl. 2002. A universal method to produce in vitro transcripts with homogeneous 3′ ends. Nucleic Acids Res. 30:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi, P. Y. 2002. Strategies for the identification of inhibitors of West Nile virus and other flaviviruses. Curr. Opin. Investig. Drugs 3:1567-1573. [PubMed] [Google Scholar]
- 58.Shi, P. Y., M. A. Brinton, J. M. Veal, Y. Y. Zhong, and W. D. Wilson. 1996. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry 35:4222-4230. [DOI] [PubMed] [Google Scholar]
- 59.Shi, P. Y., M. Tilgner, and M. K. Lo. 2002. Construction and characterization of subgenomic replicons of New York strain of West Nile virus. Virology 296:219-233. [DOI] [PubMed] [Google Scholar]
- 60.Shi, P.-Y., M. Tilgner, M. K. Lo, K. A. Kent, and K. A. Bernard. 2002. Infectious cDNA clone of the epidemic West Nile virus from New York City. J. Virol. 76:5847-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shim, J. H., G. Larson, J. Z. Wu, and Z. Hong. 2002. Selection of 3′-template bases and initiating nucleotides by hepatitis C virus NS5B RNA-dependent RNA polymerase. J. Virol. 76:7030-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shimoni, Z., M. J. Niven, S. Pitlick, and S. Bulvik. 2001. Treatment of West Nile virus encephalitis with intravenous immunoglobulin. Emerg. Infect. Dis. 7:759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shurtleff, A. C., D. W. Beasley, J. J. Chen, H. Ni, M. T. Suderman, H. Wang, R. Xu, E. Wang, S. C. Weaver, D. M. Watts, K. L. Russell, and A. D. Barrett. 2001. Genetic variation in the 3′ non-coding region of dengue viruses. Virology 281:75-87. [DOI] [PubMed] [Google Scholar]
- 64.Ta, M., and S. Vrati. 2000. Mov34 protein from mouse brain interacts with the 3′ noncoding region of Japanese encephalitis virus. J. Virol. 74:5108-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tan, B. H., J. Fu, R. J. Sugrue, E. H. Yap, Y. C. Chan, and Y. H. Tan. 1996. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology 216:317-325. [DOI] [PubMed] [Google Scholar]
- 66.Tesh, R. B., J. Arroyo, A. P. Travassos Da Rosa, H. Guzman, S. Y. Xiao, and T. P. Monath. 2002. Efficacy of killed virus vaccine, live attenuated chimeric virus vaccine, and passive immunization for prevention of West Nile virus encephalitis in hamster model. Emerg. Infect. Dis. 8:1392-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Varnavski, A. N., P. R. Young, and A. A. Khromykh. 2000. Stable high-level expression of heterologous genes in vitro and in vivo by noncytopathic DNA-based Kunjin virus replicon vectors. J. Virol. 74:4394-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wallner, G., C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. The flavivirus 3′-noncoding region: extensive size heterogeneity independent of evolutionary relationships among strains of tick-borne encephalitis virus. Virology 213:169-178. [DOI] [PubMed] [Google Scholar]
- 69.Wang, T., J. F. Anderson, L. A. Magnarelli, S. J. Wong, R. A. Koski, and E. Fikrig. 2001. Immunization of mice against West Nile virus with recombinant envelope protein. J. Immunol. 167:5273-5277. [DOI] [PubMed] [Google Scholar]
- 70.Warrener, P., J. K. Tamura, and M. S. Collett. 1993. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J. Virol. 67:989-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wengler, G. 1981. Terminal sequences of the genome and replicative-form RNA of the flavivirus West Nile virus: absence of poly(A) and possible role in RNA replication. Virology 113:544-555. [DOI] [PubMed] [Google Scholar]
- 72.Wengler, G., and E. Castle. 1986. Analysis of structural properties which possibly are characteristic for the 3′-terminal sequence of the genome RNA of flaviviruses. J. Gen. Virol. 67:1183-1188. [DOI] [PubMed] [Google Scholar]
- 73.Wengler, G., and G. Wengler. 1991. The carboxy-terminal part of the NS 3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology 184:707-715. [DOI] [PubMed] [Google Scholar]
- 74.Westaway, E. G., J. M. Mackenzie, M. T. Kenney, M. K. Jones, and A. A. Khromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wong, S. J., R. H. Boyle, V. L. Demarest, A. N. Woodmansee, L. D. Kramer, H. Li, M. Drebot, R. A. Koski, E. Fikrig, D. A. Martin, and P.-Y. Shi. 2003. An immunoassay targeting nonstructural protein 5 to differentiate West Nile virus infection from dengue and St. Louis encephalitis virus infections, and from flavivirus vaccination. J. Clin. Microbiol. 41:4217-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamshchikov, V. F., G. Wengler, A. A. Perelygin, M. A. Brinton, and R. W. Compans. 2001. An infectious clone of the West Nile flavivirus. Virology 281:294-304. [DOI] [PubMed] [Google Scholar]
- 77.Yang, J. S., J. J. Kim, D. Hwang, A. Y. Choo, K. Dang, H. Maguire, S. Kudchodkar, M. P. Ramanathan, and D. B. Weiner. 2001. Induction of potent Th1-type immune responses from a novel DNA vaccine for West Nile virus New York isolate (WNV-NY1999). J. Infect. Dis. 184:809-816. [DOI] [PubMed] [Google Scholar]
- 78.Yi, M., and S. M. Lemon. 2003. Structure-function analysis of the 3′ stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA 9:331-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.You, S., and R. Padmanabhan. 1999. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 274:33714-33722. [DOI] [PubMed] [Google Scholar]
- 80.Zeng, L., B. Falgout, and L. Markoff. 1998. Identification of specific nucleotide sequences within the conserved 3′-SL in the dengue type 2 virus genome required for replication. J. Virol. 72:7510-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]