Abstract
Hantaviruses are tripartite negative-sense RNA viruses and members of the Bunyaviridae family. The nucleocapsid (N) protein is encoded by the smallest of the three genome segments (S). N protein is the principal structural component of the viral capsid and is central to the hantavirus replication cycle. We examined intermolecular N-protein interaction and RNA binding by using bacterially expressed Sin Nombre virus N protein. N assembles into di- and trimeric forms. The mono- and dimeric forms exist transiently and assemble into a trimeric form. In contrast, the trimer is highly stable and does not efficiently disassemble into the mono- and dimeric forms. The purified N-protein trimer is able to discriminate between viral and nonviral RNA molecules and, interestingly, recognizes and binds with high affinity the panhandle structure composed of the 3′ and 5′ ends of the genomic RNA. In contrast, the mono- and dimeric forms of N bind RNA to form a complex that is semispecific and salt sensitive. We suggest that trimerization of N protein is a molecular switch to generate a protein complex that can discriminate between viral and nonviral RNA molecules during the early steps of the encapsidation process.
Hantaviruses are enveloped tripartite negative-sense RNA viruses and make up one genus of the Bunyaviridae family (21, 22). The three genomic RNA segments, L, M, and S, encode an RNA-dependent RNA polymerase, the envelope glycoproteins (G1 and G2), and nucleocapsid (N) protein, respectively. Hantaviruses are of significant medical importance because they can cause either hemorrhagic fever with renal syndrome or hantavirus pulmonary syndrome characterized by lung damage and cardiac dysfunction (19). The hantavirus replication cycle starts with binding of the G1 and G2 proteins to β3 integrin (8, 9) or other cell surface receptors, followed by virus entry and uncoating. After entry, L-protein-mediated primary transcription of minus-strand RNA results in plus-stranded mRNA in the cytoplasm, apparently with an orthomyxovirus-like cap-snatching mechanism (7, 16, 11). Following viral mRNA translation, transcription shifts from mRNA to plus-strand complementary RNA (cRNA) and de novo minus-strand viral RNA synthesis concomitant with the formation of ribonucleoprotein structures (7, 11, 20). The ribonucleoproteins appear to be composed of viral RNA, N protein, and presumably viral polymerase and accumulate on the cytoplasmic side of intracellular membranes, possibly through interaction with the G1 ands G2 proteins (10, 15). Additional evidence suggests that hantavirus assembly occurs at the plasma membranes of infected cells (10, 18).
Central to the hantavirus replication cycle is the viral N protein. The activity and biological function of N protein in the viral life cycle are proposed to depend on differential interaction with minus-strand viral RNA, plus-stranded cRNA, and the mRNA. Cell culture-based experiments indicate that encapsidation requires full-length viral RNA or cRNA molecules since no viral mRNA molecules are observed in N proteins (12). The termini of full-length La Crosse virus (genus Bunyavirus) RNA may undergo base pairing, resulting in the formation of intramolecular “panhandle” structures (17). Since the S, M, and L segments of other members of the Bunyaviridae family, including the hantaviruses, also contain termini that could base pair, it is likely that all members of this virus family also form panhandles. Unlike viral RNA, mRNA in the Bunyaviridae family has a truncated 3′ terminus and a 5′ terminus that is derived from cellular RNA by way of cap snatching. Since mRNAs lack the 3′-terminal nucleotides, panhandle formation mediated by the terminal nucleotides would not be expected to occur. The presence or absence of this terminal structure might play a role in the recognition and encapsidation of viral RNA during particle assembly.
On the basis of in vitro studies it appears that the 5′ termini of S-segment RNA may be necessary and sufficient for interaction with N protein (23, 24). The RNA binding domain of N protein has also been mapped to the central conserved region, amino acids 175 to 217 (25). Bacterially expressed N protein has been shown to associate into dimers, trimers, and large aggregates. Biochemical and genetic studies have suggested that the C-terminal half plus the N-terminal 40 amino acid residues of N protein are responsible for intermolecular N-protein association (1, 2). A recent analysis of protein-protein interaction with truncated N mutants, in conjunction with a mammalian two-hybrid assay, indicates that N-terminal amino acids 1 to 43 are involved in, and C-terminal amino acids 393 to 398 (VNHFHL) are absolutely crucial for, the homotypic interactions in N-protein homodimers and homotrimers in Tula hantavirus (14).
The mechanism for selective encapsidation of viral RNA and cRNA by the N protein in hantaviruses and other members of family Bunyaviridae is poorly understood. Here we show that bacterially expressed Sin Nombre virus (SNV) N protein assembles into di- and trimeric forms. Of the three forms, the trimeric form is the most stable. Interestingly, N-protein trimers specifically recognize the panhandle structure formed by the complementary sequences in the 3′ and 5′ termini in the S-segment RNA molecule, whereas a mixed population of mono-, di-, and trimeric forms binds RNA with lower affinity. We suggest that N-protein trimerization and its specific recognition of panhandle structures may be an intermediate step in the discrimination between viral and nonviral RNA molecules during early steps of the encapsidation process in the family Bunyaviridae.
MATERIALS AND METHODS
Oligonucleotides and enzymes.
The PCR primers used were from Sigma Genosys. The restriction enzymes and T4 DNA ligase used were from New England Biolabs. Hot Mastertaq polymerase was from Eppendorf. Moloney murine leukemia virus reverse transcriptase, RNase out, random primers, DNase I, and T7 transcription reagents were from Invitrogen. All other chemicals were purchased from Sigma.
Hantavirus N-protein purification.
On the basis of previous work, fusion of a glutathione S-transferase (GST) tag with hantavirus N protein does not appear to eliminate RNA-N-protein interaction (13). To generate an SNV N expression plasmid the N gene isolate SN77734 (3, 4) was amplified by PCR with 3′ and 5′ primers containing flanking EcoRI and HindIII sites. The amplified N gene was cloned into pGEX-UBP (5) that had been previously digested with the same two enzymes to create the expression plasmid pGEX-SNV N. GST-tagged N protein was purified as follows. Transformed cells were grown in 500 ml of Luria-Bertani medium containing 100 μg of ampicillin per ml for 3 to 4 h to an optical density at 26 nm of 0.6. Five hundred microliters of 1 M isopropyl-β-d-thiogalactopyranoside (IPTG) was added, and cells were grown for another 2 h at room temperature. Cells were harvested by centrifugation at 2,800 × g for 25 min. The resulting bacterial pellet was washed with 60 ml of 1× phosphate-buffered saline (PBS) buffer. The washed pellet was sonicated in 15 ml of solubilization buffer {50 mM sodium phosphate (pH 8.0), 300 mM NaCl, 20 mM β-mercaptoethanol, lysozyme (1 mg/100 ml), 10 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), protease cocktail inhibitor} and centrifuged at 16,000 × g for 15 min. The supernatant, which contained most of the solubilized N protein, was dialyzed overnight against 2 liters of ice-cold dialysis buffer (1× PBS, 1% Triton X-100). After dialysis, the material was centrifuged again at 16,000 × g for 20 min to remove insoluble N protein. The supernatant was incubated with 100 μl of glutathione Sepharose 4B beads that had been washed twice with 750 μl of ice-cold 1× PBS buffer. The mixture was incubated at room temperature for 3 to 4 h and centrifuged at 2,300 × g for 5 min. Bound N protein remained associated with the glutathione beads and could not be efficiently recovered by incubation of the beads with 10 mM reduced glutathione or by rapid incubation of the beads with buffer containing 8 M urea. However, it was possible to remove the protein by incubation in buffer containing guanidine HCl. Rapid treatment with 8 M urea resulted in removal of most of the contaminating bacterial protein. Thus, the beads containing bound GST-N protein were washed twice with 1 ml of 1× PBS buffer (pH 8.0) and then washed quickly with 750 μl of 1× PBS buffer (pH 8.0) containing 8 M urea to remove contaminating protein, followed by a single wash with 1 ml of 1× PBS buffer (pH 8.0). The remaining GST-N protein was recovered by incubating the beads overnight in 200 μl of denaturation buffer (50 mM sodium phosphate [pH 8.0], 0.5 M NaCl, 20 mM β-mercaptoethanol, 10 mM CHAPS, 6 M guanidine hydrochloride) and centrifuged at 2,300 × g for 30 min. The soluble, denatured N protein was renatured by stepwise dialysis against 1× PBS to allow gradual removal of the guanidine hydrochloride. The resulting protein was used in RNA binding or sedimentation experiments.
Preparation of RNA substrates.
RNA from SNV strain 77734-infected cells was used to generate full-length S-segment DNA that could be used for in vitro transcription to produce full-length S-segment RNA. The S-segment gene containing intact, full-length SNV S-segment genomic RNA was amplified by PCR with 3′ and 5′ primers containing an appropriately positioned T7 promoter. Terminal primers corresponding to SNV strain NMR11 were used for amplification since the termini of that virus had been previously sequenced (6). The amplified full-length S-segment gene was used directly in T7 transcription reaction mixtures to produce the full-length S-segment genomic RNA (designated S SNV RNA) (Fig. 1).
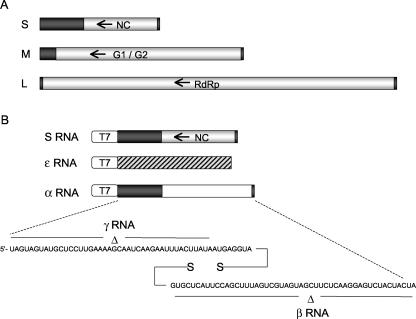
FIG. 1.
Hantavirus RNAs. The three negative-sense viral RNAs are depicted in panel A. The S segment encodes the N protein (NC), the M segment encodes viral envelope proteins G1 and G2, and the L segment encodes the RNA-dependent RNA polymerase (RdRp). The coding regions are indicated, and the flanking noncoding regions are shown in black. Arrows indicate gene orientation. Synthetic RNAs derived by in vitro transcription are shown in panel B. DNA template construction is described in the text. S RNA is initiated via correct juxtaposition of a T7 promoter to the S-segment nucleic acid. Truncation of the template was achieved by cleavage with the type I restriction enzyme BsmBI. A BsmBI site was placed distal to the S-segment transcription unit such that cleavage would result in a 5′ template nucleotide in the template DNA strand that ends at a site corresponding to the 3′-terminal nucleotide of the viral RNA. As described in the text, ɛ RNA is a nonviral RNA synthesized with T7 polymerase (indicated by diagonal markings). α RNA contains foreign RNA in place of the NC gene (indicated in white). The terminal nucleotides present in both S RNA and α RNA are shown at the bottom of panel B. γ RNA contains a 42-nucleotide deletion at the 5′ end of the genome, and β RNA contains a 42-nucleotide deletion at the 3′ end of the genome, as indicated.
Several additional DNAs suitable for the synthesis of a hantavirus-like RNA by T7 RNA polymerase were constructed by cloning in several steps. RNA from SNV-infected cells was isolated, and the 5′ noncoding region of the S segment was amplified by reverse transcription (RT)-PCR with the primers 5′-CCGGTACGCGTCCAGGCGTCTCGTATTTAGTAGTATGCTCCTTGAAAAG and 5′-GGATCTAGATAAGTGGGGCAGTAATCAACTTAT. The PCR product was digested with MluI and XbaI and cloned into pEGFP (Clontech) that had been cut with the same two enzymes to generate pEGF-5′-S. This intermediate plasmid was amplified with the following primers to generate double-stranded DNA with a T7 promoter juxtaposed to the 5′ noncoding region of the S segment, the green fluorescent protein gene in the antisense orientation relative to the S-segment sequence, the 3′ noncoding region of S, and a flanking BsmBI restriction site: 5′-CCCCCGCTAGCTAATACGACTCACTATAGTAGTAGTATGCTCCTTGAAAAGCAA and 5′-GCATGCGCGCCGTCTCCTAGTAGTAGACTCCTTGAGAAGCTACTACGACTAAAGCTGGAGGTCGCCACCATGGTGAGCAAGG. Following cleavage with NheI and BssHII, this DNA was cloned into pIREShyg2 that had been cut with XbaI and BssHII, resulting in plasmid pT7-S.
pT7-S was linearized with BsmBI and used as the template for in vitro transcription with T7 polymerase to synthesize an RNA molecule 2,353 nucleotides long containing the 730 noncoding nucleotides from the 5′ termini, 42 noncoding nucleotides from the 3′ termini of the SNV S segment, and 1,580 nonviral (green fluorescent protein gene) nucleotides in place of the coding region (designated α RNA) (Fig. 1). To generate RNA lacking the 3′ 42 nucleotides of the S segment, pT7-S was linearized with NcoI prior to transcription with T7 polymerase. This RNA was designated β RNA. For the preparation of SNV S-segment RNA lacking the terminal 5′ viral sequences, we amplified the sequence including nucleotides 2575 to 4132 of pT7-S with a 5′ primer containing a flanking T7 promoter. The amplified sequence was used directly as the template in in vitro T7 transcription reactions. The resulting transcript was designated γ RNA. We used XmnI-digested pGEM-11Zf(−), which contains a T7 promoter, as the template for in vitro synthesis of a nonviral RNA molecule 1,980 nucleotides in length (ɛ RNA).
For the synthesis of “minipanhandle” RNA, we used the following primers in PCRs with plasmid pT7-S as the template: 5′-ATATGATCAGCATGCTAATACGACTCACTATAGTAGTAGTATGCTCCTTGAAAAGCAATCAAGATTTTTTTTAGTCGTAGTAGCTTCTCAAGGAGTCTACTA and 5′-GGATCGGGAGATCTCCCGATCCCCTATGGT-3′. The amplified sequence was digested with BsmBI prior to transcription with T7 RNA polymerase. Minipanhandle RNA had 32 nucleotides from both the 5′ and 3′ termini of genomic S-segment RNA separated by six uracil residues to facilitate the formation of an intramolecular panhandle.
Radiolabeled transcripts were produced from linearized plasmids with a [α-32P]CTP T7 transcription kit (MBI Fermentase). Purification of RNA transcripts was performed with Trizol reagent (QIAGEN). Purified RNA was stored at −20°C in 25-μl aliquots for up to 2 weeks.
Sucrose density gradient centrifugation.
Sucrose gradients were used to examine the oligomeric forms of N protein and to characterize N protein in the presence of RNA. One-hundred-microliter samples containing 100 μM purified N protein were layered onto linear gradients containing 10 to 60% (wt/vol) sucrose in binding buffer (40 mM HEPES [pH 7.4], 80 mM NaCl, 20 mM KCl, 1 mM dithiothreitol) (23) and centrifuged at 30,000 rpm in an SW40 rotor at 4°C for 22 h. Fractions of 0.5 ml were collected from the bottom of the gradient. The protein molecular mass markers bovine serum albumin (BSA; 82 kDa), alcohol dehydrogenase (150 kDa), and β-amylase (200 kDa) were similarly fractionated. A 20-μl sample of each fraction was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). N protein was detected by Western blot analysis with anti-N-protein antiserum.
Filter binding assays.
RNA molecules were synthesized by in vitro transcription with T7 RNA polymerase in the presence of [α-32P]CTP as described above. All binding reactions were carried out with binding buffer at a constant concentration of RNA (1 pM) and increasing concentrations of N protein. The reaction mixture was incubated at room temperature for 45 min and filtered through nitrocellulose filters under vacuum. The filters were washed with 10 ml of binding buffer and dried, and the amount of RNA retained was quantified with a scintillation counter. Dissociation constants were calculated by fitting the experimental points into either sigmoidal or hyperbolic equations with the program Origin 6 (Microcal). The apparent dissociation constant (Kd) corresponds to the concentration of N protein required to obtain half saturation, assuming that the complex formation obeys a simple bimolecular equilibrium. We assumed that the plateau in the binding profile represents complete binding of RNA to allow the calculations at half saturation.
RT-PCR.
RNA was isolated from fractions of sucrose gradients with Trizol reagent (Invitrogen) in accordance with the manufacturer's instructions. Moloney murine leukemia virus reverse transcriptase (Invitrogen) was used for first-strand cDNA synthesis with random primers in accordance with the manufacturer's instructions. Two microliters of cDNA generated by RT-PCR was used as the template for a PCR to amplify a 500-bp region in the 5′ terminus of the S-segment SNV RNA gene with specific primers.
RNase digestion.
One-hundred-microliter samples containing 50 μM N-protein mixture and 0.5 μM α RNA in binding buffer were run in a 10 to 60% sucrose gradient as described above. One-hundred-microliter samples from fractions 6 and 7 of sucrose gradients, which contained α RNA-N-protein complexes, were digested with 1 μl of an RNase A-T1 mixture containing 40 μg of RNase A per μl and 95 U of RNase T1 per μl. Control experiments were done with the same amount of RNA in the absence of N protein under identical reaction conditions. After RNase digestion, undigested RNA was purified with Trizol reagent detected by RT-PCR.
RESULTS
Hantavirus N protein is a stable trimer.
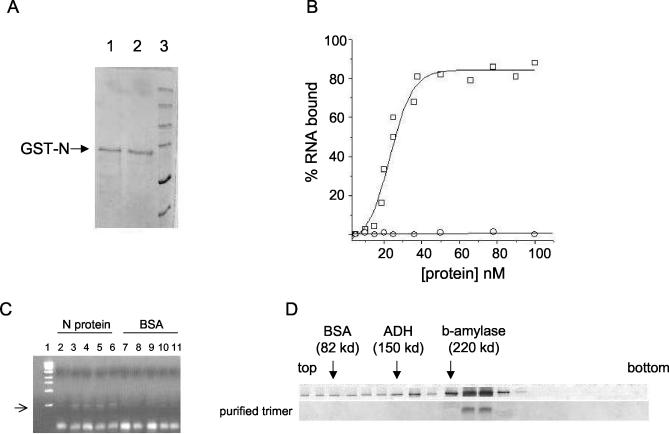
Escherichia coli cells containing the expression vector pGEX-SNV N were induced with IPTG to express the GST-tagged N protein, and the GST fusion protein was isolated and purified (see Materials and Methods). Figure 2A depicts an SDS-PAGE analysis of purified N protein under reducing and nonreducing buffer conditions. This protein was largely free of detectible heterologous bacterial protein and had an electrophoretic mobility consistent with the expected mass of the peptide (74 kDa), as evidenced by comigration relative to that of protein molecular weight markers. Similar results were obtained when treated or untreated samples were run on separate gels to mitigate the potential problem of diffusion of the reducing agent (data not shown). To determine whether this expressed N protein was able to stably interact with RNA, indicative of an expected intrinsic activity, we carried out a preliminary RNA-filter binding experiment with viral S-segment RNA generated by in vitro transcription with T7 RNA polymerase. Figure 2B shows the binding profile for the association of N protein with radioactively labeled S-segment RNA. As a control in this initial experiment, we carried out a parallel binding experiment with a protein that does not stably interact with RNA (BSA). On the basis of these data, it is evident that S-segment RNA interacted with N protein. To further verify that N protein stably interacts with this RNA, we carried out RNase digestion experiments to see whether S-segment RNA is partially protected from RNase digestion in the presence of N protein. Figure 2C shows the digestion of S-segment RNA in the presence of increasing concentrations of either N protein or BSA. These data indicate that N protein partially protected the S-segment RNA from RNase digestion while BSA did not.
FIG. 2.
Analysis of SNV N protein expressed in E. coli. (A) SDS-PAGE analysis of purified N protein in reducing buffer (lane 1) and nonreducing buffer (lane 2). Protein markers are shown in lane 3. (B) RNA binding profile of purified N protein. Increasing concentrations of either N protein (□) or BSA (○) were incubated with S RNA as described in the text, and RNA-protein complexes were isolated and quantified by nitrocellulose filter binding with 32P-labeled RNA. (C) RNase digestion of S RNA in the presence of either N protein (lanes 2 to 6) or BSA (lanes 7 to 11) in binding buffer at room temperature for 1 min. N protein or BSA was incubated with S RNA for 1 h at room temperature and then digested for 1 min at room temperature with 1 μl of a mixture of RNases A and T1 at 40 μg/μl and 95 U/μl, respectively. After digestion RNA was isolated and purified by Trizol reagent (see Materials and Methods). RT-PCR was used to generate cDNA with random primers, and 2 μl of cDNA was used as the template in a PCR with primers specific to a 500-nucleotide-long region in the 5′ noncoding terminus of the S segment. The amplified DNA was run on a 1% agarose gel. Lane 1 contains a DNA size marker. No N protein or BSA (lane 2), 10 μM N (lane 3), 20 μM N (lane 4), 30 μM N (lane 5) 40 μM N (lane 6), 10 μM BSA (lane 7), 20 μM BSA (lane 8), 30 μM BSA (lane 9), 40 μM BSA (lane 10), and 70 μM BSA (lane 11) were used. (D) Ten to 60% sucrose gradient and Western blot analysis of bacterially expressed N protein. The top gradient contains unfractionated GST-N protein, and the bottom gradient shows purified trimeric N protein. Protein standards (BSA, alcohol dehydrogenase (ADH), and β-amylase) were run in parallel with the N samples, and the migration of those standards is indicated by arrows.
To assess the subunit composition of the purified N protein, we carried out sucrose gradient centrifugation. N-protein samples at a concentration of 100 μM were run on 10 to 60% sucrose gradients and detected by Western analysis with anti-N polyclonal antibody. N protein was found in a continuous set of samples that ranged from fractions 10 to 23 from the bottom of the gradient. However, N was most abundant in fractions 12 and 13 (Fig. 2D). On the basis of cosedimentation with molecular weight standards, the abundant N protein in fractions 12 and 13 is likely trimeric, the protein in fractions 16 and 17 is probably dimeric, and the protein in fractions 20 and 21 is monomeric. As described previously, the mobility of N protein was the same following incubation in either the presence or the absence of reducing agents (Fig. 2A). This indicates that the dimer and trimer species are apparently formed and maintained through noncovalent interactions.
To examine potential interconversion of the mono-, di-, and trimeric N-protein species, we pooled the appropriate fractions corresponding to these three forms of N protein, dialyzed the samples against 1× PBS, and resedimented the N species on 10 to 60% sucrose gradients. The purified dimers and monomers resedimented to fractions 16 and 17 and fractions 12 and 13, indicating progressive accumulation of the trimeric species (data not shown). In contrast, the purified trimeric species retained trimeric form and apparently constitutes a more stable molecular species (Fig. 2D). Such purified trimer preparations remained stable at 4°C for 2 to 3 days but by 7 to 8 days formed high-molecular-weight insoluble aggregates (data not shown).
Trimeric N protein binds specifically to the S RNA panhandle.
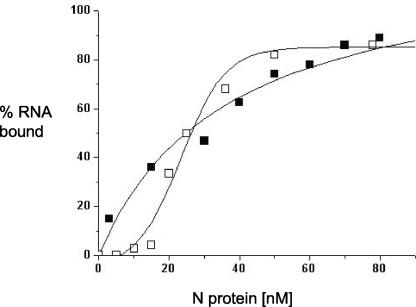
To measure the RNA binding affinity of N protein we carried out binding assays with an array of RNA molecules related to the hantavirus S segment. In these experiments radioactively labeled RNA was incubated with increasing concentrations of either unfractionated N protein or purified trimeric N prepared on sucrose gradients. Protein-RNA complexes were then filtered through nitrocellulose membranes, and the amount of protein-bound RNA retained was measured. Representative binding profiles with full-length S-segment RNA are shown in Fig. 3. These data indicate that both purified trimeric N and unfractionated N protein have a dissociation constant of about 25 nM with full-length S-segment RNA (Table 1). In contrast, the interaction between N protein and nonviral RNA (ɛ RNA) of approximately the same length and the overall nucleotide composition of the S segment were markedly reduced, particularly for purified trimeric N protein (Table 1).
FIG. 3.
Binding profile for the association of unfractionated N protein (□) and purified trimeric N (▪) with 32P-labeled S-segment RNA in binding buffer at room temperature. Binding experiments were carried out as described in the text.
TABLE 1.
Dissociation constants for the association of purified N-protein trimer and a mixture of mono-, di-, and trimers with different types of RNA molecules under different conditions
| System |
Kd (nM) at NaCl concn (mM) of:
|
||
|---|---|---|---|
| 80 | 160 | 320 | |
| S segment RNA + N-protein mixture | 25 ± 2 | 27 ± 3 | 30 ± 2 |
| S segment RNA + N-protein trimer | 24 ± 1 | 23 ± 2 | 24 ± 1 |
| α RNA + N-protein mixture | 42 ± 3 | 22 ± 4 | 42 ± 2 |
| α RNA + N-protein trimer | 25 ± 5 | 30 ± 3 | 45 ± 1 |
| β RNA + N-protein mixture | 150 ± 6 | 280 ± 4 | 485 ± 6 |
| β RNA + N-protein trimer | 525 ± 9 | NDa | ND |
| γ RNA + N-protein mixture | 600 ± 10 | ND | ND |
| γ RNA + N-protein trimer | 802 ± 9 | ND | ND |
| Mini pan RNA + N-protein mixture | 186 ± 3 | 193 ± 5 | 203 ± 2 |
| Mini pan RNA + N-protein trimer | 39 ± 1 | 22 ± 2 | 35 ± 5 |
| ɛ RNA + N-protein mixture | 165 ± 2 | 240 ± 5 | 350 ± 1 |
| ɛ RNA + N-protein trimer | 577 ± 8 | ND | ND |
ND, not determined.
These initial binding data were obtained with buffer containing 80 mM NaCl. To examine the effect of higher salt concentrations on the binding affinity between full-length S RNA and N, additional RNA-protein binding experiments were carried out at 160 and 320 mM NaCl. As shown in Table 1, even at these higher salt concentrations both unfractionated and purified, trimeric N had dissociation constants with S-segment RNA similar to those observed at 80 mM NaCl. These data are consistent with the specific recognition of viral RNA by both trimeric N protein and a mixed population of N proteins. Moreover, the binding affinity between N and hantavirus RNA is similar to that observed for the interaction between N proteins and the corresponding RNAs of other RNA viruses.
We next attempted to identify regions within the S segment that are recognized with high affinity by N. α RNA is a derivative of S-segment RNA that contains an exact substitution of nonviral RNA in place of the open reading frame in the central region of the S segment so that the gene that encodes N is absent but the flanking noncoding sequences of the gene that encodes S are retained (Fig. 1). β RNA is identical to α RNA except that the terminal 3′ noncoding region of the S segment is deleted (Fig. 1). Finally, γ RNA is also derived from α RNA but the terminal 5′ noncoding sequences of the viral RNA are deleted (Fig. 1). The interaction between the mixed population of N or purified, trimeric N with α RNA was only marginally reduced compared to that observed for the full-length S segment (Table 1). Moreover, carrying out binding reactions with higher salt concentrations again indicated that the N-α RNA interaction was stable. These data indicate that the coding region of the S segment is not necessary for specific N-RNA interaction. In contrast, RNAs that lacked either the 5′ or 3′ noncoding regions of the S segment (β RNA or γ RNA) reduced the binding affinity of trimeric N to a level similar to, or less than, that observed for nonviral RNA (ɛ RNA), indicating that the terminal noncoding regions of the S segment are required for specific interaction with the N trimer.
Since both the terminal noncoding regions of S RNA appeared to be required for specific, salt-stable N recognition, this raises the question of the nature of the RNA substrate recognized by N. The termini of the viral RNA contain nucleotides that could facilitate the formation of a panhandle structure through intramolecular hydrogen bonding, and this secondary structure could serve as the specific recognition site for N. Alternatively, both the 5′ and 3′ ends of the RNA could be necessary for specific recognition by N but not in panhandle configuration. To try to distinguish between these two possibilities we carried out filter binding experiments with a short minipanhandle RNA containing the terminal nucleotides of the S segment (Fig. 4). On the basis of thermodynamic considerations, this short RNA would be highly likely to form a panhandle structure composed of the same base-paired nucleotides that would mediate hypothetical panhandle formation in full-length S-segment RNA and alternative structures would be thermodynamically unfavorable. Formation of the minipanhandle structure would result in a free-energy change of about −29 kcal. Prior to filter binding this small RNA was heat denatured and allowed to slowly refold. Examination of the dissociation constants between N and this RNA (Table 1) leads to three primary observations. First, the minipanhandle RNA was sufficient for specific binding by N compared with either full-length S-segment RNA or α RNA. Second, the purified trimeric form of N was required for specific binding to this RNA, exhibiting a binding affinity significantly higher than that of the mixed population of N. Third, complexes consisting of trimeric N and the minipanhandle RNA were resistant to higher salt concentrations, indicative of specificity and in concordance with the binding affinities of full-length S-segment RNA or α RNA.
FIG. 4.
Viral panhandle RNA. At the top is shown the secondary structure of minipanhandle RNA used as a binding substrate for the SNV N-protein interaction. Mfold was used to analyze multiple alternative RNA secondary structures. The structure shown is the most probable on the basis of thermodynamic considerations. At the bottom is the terminal structure likely to be present in full-length S-segment RNA and in α RNA. The same structure formed by the terminal nucleotides in the S RNA and α RNA is shown at the bottom. Analysis of 20 alternative theoretical secondary structures indicated that the panhandle is likely to form in each. In addition, the shaded nucleotides are those deleted in γ or β RNA. The shaded nucleotides near the 5′ end are deleted in γ RNA, and those in the 3′ end are deleted in β RNA.
Mono- and/or dimeric N protein interacts with RNA nonspecifically.
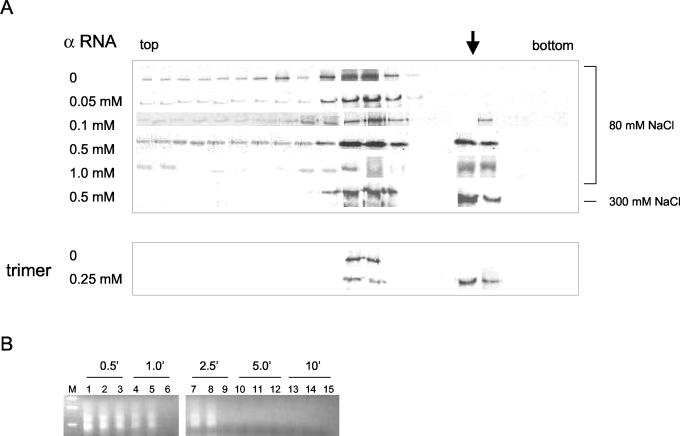
To verify the filter binding results and further explore the interaction between N and RNA we attempted to examine protein-RNA interaction by sedimentation in sucrose gradients. We incubated a constant concentration of unfractionated N protein in binding reaction mixtures with increasing concentrations of α RNA. Following incubation the samples were then analyzed by sedimentation and the distribution of N protein in the gradient was determined by Western analysis with anti-N antibody. With increasing RNA concentrations some of the N protein sedimented more rapidly through the gradient (Fig. 5). The peak of this more rapidly sedimenting material was centered in fractions 5 and 6 from the bottom of the gradient. Further, detection of α RNA by RT-PCR indicated that most of the RNA was also present in the same fractions as that of the rapidly sedimenting N protein (data not shown). When sedimented alone, α RNA migrated to fraction 9 in the gradient (data not shown). These data are consistent with the idea that the rapidly migrating N protein is composed of N-protein-RNA complexes.
FIG. 5.
Sucrose gradient analysis of N-protein-viral RNA complexes. (A) Following sedimentation through sucrose gradients N protein was detected by Western analysis with anti-N antibody as in Fig. 2D. In gradients 1 through 6, 50 μM unfractionated N was incubated with increasing amounts of α RNA for 2 h at room temperature prior to sedimentation as indicated. All samples were incubated prior to and during centrifugation in 80 mM NaCl, except the sample labeled 300 mM, for which both the binding and sedimentation were carried out in the presence of 300 mM NaCl. In the last two gradients, 25 μM purified trimeric N protein was incubated in the absence or presence of 0.25 μM α RNA for 2 h at room temperature prior to sedimentation and Western analysis. The arrow indicates the position of the N-RNA complexes. (B) The material from fractions 5 and 6, which contained putative N-α RNA complexes, were treated with RNases A and T1 for various times (as indicated), and remaining undegraded α RNA was detected by RT-PCR and electrophoresis on a DNA gel as described in the text. Lanes: M, DNA size markers; 1, 4, 7, 10, and 13, material from fraction 5; 2, 5, 8, 11, and 14, material from fraction 6; 3, 6, 9, 12, and 15, α RNA in sucrose gradient buffer plus 20% sucrose in the absence of N protein.
To see whether the RNA in fractions 6 and 7 might be bound to and partially protected from RNase degradation by N protein, we treated the putative N-protein-RNA complexes with a mixture of RNases A and T1, removed aliquots at various time intervals, and compared the degradation of the RNA in the samples with that of RNA in the absence of N protein. The presence or absence of RNA was determined by RT-PCR with primers specific for the SNV S segment (Fig. 5B). On the basis of this experiment, it appears that N protein inhibited but did not eliminate the degradation of α RNA, again consistent with the idea that N protein is associated with this RNA. Two DNA products arising from PCR are evident in Fig. 5. In control experiments we also observed that PCR amplification of α RNA resulted in two DNA species (data not shown). We speculate that one species arose from alternative annealing of one of the primers to the template during either RT or PCR. Nonetheless, the two RNAs indicate that α RNA is present.
On the basis of the nitrocellulose filter binding assays the affinity between N and full-length S RNA or α RNA was unaffected by higher salt concentrations (Table 1). If N protein migrated more rapidly in the gradient because of association with RNA, then such protein-RNA complexes should similarly be unaffected by higher salt concentrations. Thus, we carried out a binding experiment and sucrose sedimentation in the presence of 300, rather than 80, mM NaCl. As shown in Fig. 5A, the more rapidly sedimenting N protein was still present at this higher salt concentration, consistent with the idea that more rapidly sedimenting material in the gradient is composed of RNA-N complexes.
We also carried out sedimentation analysis to verify the interaction of purified trimeric N with α RNA. We again observed RNA-dependent migration of N protein to a position lower in the gradient (Fig. 5A). Moreover, this rapidly sedimenting N-protein-RNA complex was stable in the presence of 300 mM NaCl (data not shown). Although the filter binding experiments indicated that α RNA interacts with an affinity similar to that of full-length S RNA, we also examined the interaction of N protein with full-length S RNA rather than α RNA. As expected, we observed a fractionation pattern similar that observed with α RNA at both 80 and 300 mM NaCl (data not shown).
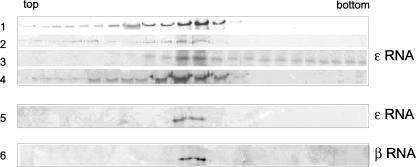
As described previously, ɛ RNA is a nonviral RNA similar in size to the full-length S segment (Fig. 1). We carried out RNA-protein binding experiments with N protein and ɛ RNA and analyzed the resulting material by sedimentation. As with α RNA, incubation of unfractionated N with ɛ RNA resulted in the sedimentation of N to a position in the gradient consistent with the presence of protein-RNA complexes (Fig. 6). However, this rapidly sedimenting N protein appeared to be distributed throughout fractions near the bottom of the gradient and only in the presence of relatively large amounts of ɛ RNA (Fig. 6, gradient 3). Moreover, in contrast to α RNA, binding and sedimentation in the presence of 300 mM NaCl eliminated rapidly sedimenting N protein (Fig. 6, gradient 4).
FIG. 6.
Sucrose gradient analysis of complexes formed between N protein and nonviral RNA or lacking the 3′ terminus of the S segment. Binding reactions, sucrose gradient analysis, and detection of N by Western blotting were performed as described in the legend to Fig. 5. Fifty micromolar N protein was incubated with ɛ or β RNA prior to analysis on sucrose gradients. 1, no RNA; 2, 0.1 μM RNA; 3, 0.5 μM RNA; 4, 0.5 μM RNA with binding and sedimentation carried out in the presence of 300 mM NaCl; 5, purified trimeric N protein with ɛ RNA; 6, purified trimeric N protein with β RNA.
When ɛ RNA was incubated with the purified N-protein trimer there was no evidence of RNA binding, even at the lower salt concentration (Fig. 6, gradient 5). These data are consistent with a relatively low-affinity nonspecific RNA interaction with unfractionated N protein and are in concordance with the filter binding data. β RNA, which lacks the 3′-terminal nucleotides, appeared to be similarly unable to stably interact with trimeric N protein, as evidenced by the absence of rapidly sedimenting N (Fig. 6, gradient 6). These sedimentation data, and the filter binding data, indicate that both the 5′ and 3′ ends are required for stable association with the N trimer. Since rapidly sedimenting N was detected only in preparations in which the mono- and dimeric N protein was present, it appears that the recognition of RNA by the monomer and dimer is disparate from that by the trimer. RNA-protein interaction is less specific and salt sensitive for N preparations composed of a mixed population than for purified trimeric N.
DISCUSSION
The differential binding activity of purified trimeric N protein relative to that of a mixed population of N-protein species leads to a model with potential significance for Bunyaviridae RNA encapsidation (Fig. 7). In particular, the purified trimer specifically recognizes RNA containing the preformed panhandle structure derived from antiparallel hydrogen bonding of the terminal nucleotides of the S segment. It seems likely that this form of N is required for recognition of viral RNA during the encapsidation process. N preparations containing monomeric and/or dimeric N interacts with RNA with reduced specificity relative to that of the trimer and exhibit a salt-sensitive interaction with RNA that is not dependent on the presence of a panhandle. However, from the binding data it appears that there is a higher-affinity interaction between the mixed N population and RNA at low salt concentrations. Further, this interaction appears to require the presence of the 5′-terminal region of the S segment, and this is in concordance with previous observations of the in vitro interaction between Hantaan virus N protein and viral RNA (24).
FIG. 7.
Association of N protein with viral RNA. The monomeric and dimeric forms of N require the 5′ terminus of the S segment for binding. However, this association is sensitive to increased ionic strength. Interaction of trimeric N protein is specific for RNA containing the 5′ and 3′ termini and is resistant to high salt concentrations.
The binding curve observed with increasing concentrations of trimeric N is parabolic, whereas that formed with the mixed population is sigmoidal (Fig. 3). This is consistent with the idea that in the presence of the monomer and dimer, interaction with RNA is cooperative or that the prior binding of N to RNA influences the association between additional N molecules and RNA. To determine the basis of this differential interaction it is necessary to generate additional data to calculate cooperativity parameters. However, since mono- and dimeric N protein has the capacity to form trimeric N, it is possible that such protein-protein interaction can occur following the interaction of a monomer or a dimer with RNA. In contrast, in the case of the purified trimer it appears that the association with RNA is a straightforward bimolecular interaction, consistent with a lack of subsequent protein-protein interaction.
A mixed population of N molecules contains trimeric species as well as monomers and dimers, yet the RNA binding property of the monomers and dimers is apparently either dominant over that of the trimer or significantly affects the properties of trimeric N. For example, the interaction between purified trimeric N protein and minipanhandle RNA is of relatively high affinity, but the interaction is of lower affinity for the mixed population of N even though the trimer is present. It is possible that the monomeric and dimeric forms of N protein competitively interfere with binding of the trimer. It is difficult to assess the exact proportion of mono-, di-, and trimeric forms of N protein in mixed populations since over time trimeric N accumulates. It will be interesting to examine the stoichiometry of the various forms of N protein in virus-infected cells. It is possible also that the various forms of N protein play different roles during virus nucleic acid replication. Such an association may function in bringing the 5′ and 3′ ends into juxtaposition to facilitate panhandle formation. Recent experiments with N-protein preparations lacking the GST tag indicate that these peptide preparations also preferentially form trimers and bind with specificity to the S RNA panhandle (data not shown). Thus, the N-terminal GST tag present on the N-protein preparations does not to overtly affect intermolecular interaction with other N peptides or RNA.
A minipanhandle RNA containing only the viral RNA termini is sufficient for recognition by trimeric N and the termini of the viral S segment are necessary for high-affinity interaction between N and RNA. In contrast, the internal segment of the RNA had little effect on the association with N. However, it is likely that for longer RNAs, the internal sequence and higher-order structure will influence the likelihood that the terminal panhandle structures may form. If panhandle formation is indeed important in the replication of Bunyaviridae RNA, then there would be selection for either intrinsic features of the virus RNA or a protein-driven mechanism to ensure that the termini of the RNA have the opportunity to hydrogen bond.
Bunyaviridae N protein plays a key role in the viral replication cycle. During the assembly of virus particles, minus-strand viral RNA molecules are specifically packaged relative to either of the plus-strand viral RNA species (cRNA or mRNA). This indicates an operational mechanism for the selective discrimination among the three types of RNA molecules. Full-length plus-strand RNA can also potentially form panhandle structures, although the stability of such structures would vary somewhat from that formed by corresponding viral RNA. It will be interesting to compare the interaction of N protein with these two alternative substrates, and such experiments will likely help in understanding the role of the panhandle structures and N protein in virus replication. Since trimeric N specifically recognizes the RNA molecules composed of the 3′ and 5′ noncoding viral sequences, panhandle formation may be required for selective encapsidation of viral RNA molecules during the viral replication cycle.
Acknowledgments
We thank Brian Hjelle for encouragement and the generous contribution of numerous reagents. We thank Dave Peabody, Jesse Summers, and Brian Hjelle for critical reviews of the manuscript and for discussions.
This work was supported by grant R21-AI053400 from the National Institutes of Health.
REFERENCES
- 1.Alfadhli, A., Z. Love, B. Arvidson, J. Seeds, J. Willey, and E. Barklis. 2001. Hantavirus nucleocapsid protein oligomerization. J. Virol. 75:2019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfadhli, A., E. Steel, L. Finlay, H. P. Bachinger, and E. Barklis. 2002. Hantavirus nucleocapsid protein coiled-coil domains. J. Biol. Chem. 277:27103-27108. [DOI] [PubMed] [Google Scholar]
- 3.Botten, J., K. Mirowsky, D. Kusewitt, M. Bharadwaj, J. Yee, R. Ricci, R. M. Feddersen, and B. Hjelle. 2000. Experimental infection model for Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus). Proc. Natl. Acad. Sci. USA 97:10578-10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Botten, J., K. Mirowsky, C. Ye, K. Gottlieb, M. Saavedra, L. Ponce, and B. Hjelle. 2002. Shedding and intracage transmission of Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus) model. J. Virol. 76:7587-7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callahan, M. A., M. A. Handley, Y.-H. Lee, K. J. Talbot, J. W. Harper, and A. T. Panganiban. 1998. Functional interaction of human immunodeficiency virus type 1 Vpu and Gag with a novel member of the tetratricopeptide repeat protein family. J. Virol. 72:5189-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chizhikov, V. E., C. F. Spiropoulou, S. P. Morzunov, M. C. Monroe, C. J. Peters, and S. T. Nichol. 1995. Complete genetic characterization and analysis of isolation of Sin Nombre virus. J. Virol. 69:8132-8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn, E. F., D. C. Pritlove, H. Jin, and R. M. Elliott. 1995. Transcription of a recombinant bunyavirus RNA template by transiently expressed bunyavirus proteins. Virology 211:133-143. [DOI] [PubMed] [Google Scholar]
- 8.Gavrilovskaya, I. N., E. J. Brown, M. H. Ginsberg, and E. R. Mackow. 1999. Cellular entry of hantaviruses which cause hemorrhagic fever with renal syndrome is mediated by β3 integrins. J. Virol. 73:3951-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gavrilovskaya, I. N., M. Shepley, R. Shaw, M. H. Ginsberg, and E. R. Mackow. 1998. β3 integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc. Natl. Acad. Sci. USA 95:7074-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldsmith, C. S., L. H. Elliott, C. J. Peters, and S. R. Zaki. 1995. Ultrastructural characteristics of Sin Nombre virus, causative agent of hantavirus pulmonary syndrome. Arch. Virol. 140:2107-2122. [DOI] [PubMed] [Google Scholar]
- 11.Hutchinson, K. L., C. J. Peters, and S. T. Nichol. 1996. Sin Nombre virus mRNA synthesis. Virology 224:139-149. [DOI] [PubMed] [Google Scholar]
- 12.Jin, H., and R. M. Elliott. 1993. Characterization of Bunyamwera virus S RNA that is transcribed and replicated by the L protein expressed from recombinant vaccinia virus. J. Virol. 67:1396-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaukinen, P., V. Koistinen, O. Vapalahti, A. Vaheri, and A. Plyusnin. 2001. Interaction between molecules of hantavirus nucleocapsid protein. J. Gen. Virol. 82:1845-1853. [DOI] [PubMed] [Google Scholar]
- 14.Kaukinen, P., A. Vaheri, and A. Plyusnin. 2003. Mapping of the regions involved in homotypic interactions of Tula hantavirus N protein. J. Virol. 77:10910-10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Persson, R., and R. F. Pettersson. 1991. Formation and intracellular transport of a heterodimeric viral spike protein complex. J. Cell Biol. 112:257-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plotch, S. J., M. Bouloy, I. Ulmanen, and R. M. Krug. 1981. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23:847-858. [DOI] [PubMed] [Google Scholar]
- 17.Raju, R., and D. Kolakofsky. 1989. The ends of La Crosse virus genome and antigenome RNAs within nucleocapsids are base paired. J. Virol. 63:122-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ravkov, E. V., S. T. Nichol, and R. W. Compans. 1997. Polarized entry and release in epithelial cells of Black Creek Canal virus, a New World hantavirus. J. Virol. 71:1147-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmaljohn, C., and B. Hjelle. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 3:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmaljohn, C. M. 1996. Molecular biology of hantaviruses. Plenum Press, New York, N.Y.
- 21.Schmaljohn, C. S., and J. W. Hooper. 2001. Bunyaviridae: the viruses and their replication, p. 1581-1602. In K. A. Howley (ed.), Virology, fourth ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 22.Schmaljohn, C. S., and C. B. Jonsson. 2001. Replication of hantaviruses, p. 15-32. In S. A. Nichol (ed.), Hantaviruses. Springer-Verlag, Berlin, Germany.
- 23.Severson, W., L. Partin, C. S. Schmaljohn, and C. B. Jonsson. 1999. Characterization of the Hantaan nucleocapsid protein-ribonucleic acid interaction. J. Biol. Chem. 274:33732-33739. [DOI] [PubMed] [Google Scholar]
- 24.Severson, W. E., X. Xu, and C. B. Jonsson. 2001. cis-acting signals in encapsidation of Hantaan virus S-segment viral genomic RNA by its N protein. J. Virol. 75:2646-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu, X., W. Severson, N. Villegas, C. S. Schmaljohn, and C. B. Jonsson. 2002. The RNA binding domain of the Hantaan virus N protein maps to a central, conserved region. J. Virol. 76:3301-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]