Abstract
Purified recombinant viral replicases are useful for studying the mechanism of viral RNA replication in vitro. In this work, we obtained a highly active template-dependent replicase complex for Cucumber necrosis tombusvirus (CNV), which is a plus-stranded RNA virus, from Saccharomyces cerevisiae. The recombinant CNV replicase showed properties similar to those of the plant-derived CNV replicase (P. D. Nagy and J. Pogany, Virology 276:279-288, 2000), including the ability (i) to initiate cRNA synthesis de novo on both plus- and minus-stranded templates, (ii) to generate replicase products that are shorter than full length by internal initiation, and (iii) to perform primer extension from the 3′ end of the template. We also found that isolation of functional replicase required the coexpression of the CNV p92 RNA-dependent RNA polymerase and the auxiliary p33 protein in yeast. Moreover, coexpression of a viral RNA template with the replicase proteins in yeast increased the activity of the purified CNV replicase by 40-fold, suggesting that the viral RNA might promote the assembly of the replicase complex and/or that the RNA increases the stability of the replicase. In summary, this paper reports the first purified recombinant tombusvirus replicase showing high activity and template dependence, a finding that will greatly facilitate future studies on RNA replication in vitro.
Plus-stranded RNA viruses, which constitute the largest group among plant and animal viruses, replicate in infected cells by using the viral replicase complex. The replicase complex consists of virus-coded proteins, such as the RNA-dependent RNA polymerase (RdRp), auxiliary proteins, and possibly host-derived proteins, and the RNA template (1, 4, 5, 20, 27). To study the mechanism of viral RNA replication, functional replicases are purified from virus-infected hosts (3, 10, 12, 16, 23, 26, 38, 41, 42, 53, 55) or from heterologous systems, including Escherichia coli (17, 19, 21, 24, 44, 45), yeast (40), insect (22, 24, 58), Xenopus (13), and mammalian cells (14, 24). The advantage of the heterologous systems is that expression of the replicase proteins can be achieved without dependence on virus replication, thus facilitating mutational analysis of the replicase genes. These studies have established that the RdRp of several viruses, including Turnip crinkle virus, Tobacco etch virus, Bamboo mosaic virus, Hepatitis C virus, Bovine viral diarrhea virus (17, 19-22, 44, 45), etc., are active when expressed without other virus-coded auxiliary proteins. On the contrary, RdRps for several other viruses, such as Brome mosaic virus (BMV) and Alfalfa mosaic virus (AMV), required the presence of several factors, such as the RdRp, a viral auxiliary protein, and the viral RNA, in order to be functional in vitro (40, 54). In summary, viral replicase systems, which are very useful to dissect the protein (trans-acting) and RNA (cis-acting) factors that control virus replication, have been developed only for a limited number of plus-stranded RNA viruses.
Tombusviruses are small plus-stranded viruses that belong to supergroup 2 viruses, such as hepatitis C virus, Flaviviruses, and Pestiviruses, based on the similarity among their RdRp sequences (25, 50). The genome of tombusviruses codes for five genes involved in replication (termed p33 and p92; Fig. 1), cell-to-cell movement (p22), encapsidation (p41), and suppression of gene silencing (p19; Fig. 1A) (reviewed in reference 50). The two overlapping replicase genes are essential for replication of the genomic RNA (gRNA) in plant cells (28, 34, 52). While the p92 has the RdRp signature motifs in its unique C terminus, the function(s) of p33 is presently unknown. Mutagenesis of the RNA-binding site (an arginine-proline-rich motif, termed RPR motif [43]) in p33 was found to affect gRNA replication (34), subgenomic RNA (sgRNA) synthesis, and RNA recombination (35), suggesting that p33 is a multifunctional protein. Similar mutations within the RPR motif in p92 also affected RNA replication, suggesting that the overlapping (prereadthrough) domain in p92 is essential for its function in infected cells (34).
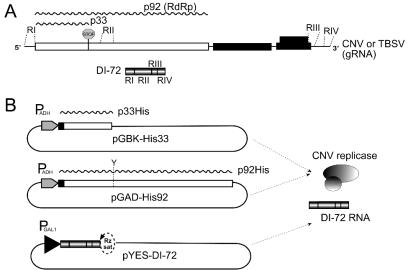
FIG. 1.
Expression of CNV replicase proteins in yeast. (A) Genome organization of CNV, the closely related TBSV RNA encoding five open reading frames, and TBSV-derived DI-72 RNA. The p33 and p92 replicase genes are shown as white boxes (the stop sign represents translational termination codon at the end of p33), and the remaining three genes are shown as black boxes. Regions of TBSV present in DI-72 RNA are marked on the gRNA. (B) Plasmids used to express the CNV p33 and p92 replicase proteins and DI-72 RNA in yeast cells. pGBK-His33 and pGAD-His92 have a constitutive PADH promoter, and DI-72 RNA is expressed from a galactose-inducible (PGAL1) promoter. Black boxes in pGBK-His33 and pGAD-His92 constructs represent the 6× His tag. The translation termination codon of p33 was replaced with a tyrosine (Y) codon, allowing p92 expression from the pGAD-His92 plasmid. There is a satTRSV(−) ribozyme (Rz sat) at the 3′ end of DI-72.
Tombusviruses are popular model viruses to study virus replication due to the presence of small defective interfering (DI) RNAs, which are deletion derivatives of the viral gRNA (Fig. 1A). These DI RNAs do not code for essential genes, allowing for convenient testing of the role of essential and/or regulatory cis-acting elements, such as promoters (15, 32, 33), replication enhancers (29, 31, 47), and a replication silencer (39). Another advantage of tombusviruses is the availability of yeast as a model host to study replication of DI RNAs in vivo (30, 36).
Efficient assays based on partially purified RdRp have been developed for Tomato bushy stunt virus (TBSV) and Cucumber necrosis virus (CNV), which were obtained from infected plants (23). These preparations were then used to confirm the role of cis-acting sequences and structures in TBSV DI RNA replication and recombination in vitro (8, 29, 31-33, 39). The disadvantage of the above preparations is that replication of tombusvirus is necessary to obtain enough RdRp activity for biochemical characterization. Therefore, development of a heterologous system to express and purify highly active recombinant tombusvirus replicase would be useful to define the role of the replication proteins and RNA templates in replication. To obtain such a system in this study, we used yeast cells, which coexpressed p33 and p92, as well as the DI RNA template from plasmids. Characterization of the obtained CNV replicase preparation from yeast revealed that it had properties similar to those of the plant-derived preparation. The yeast-derived preparation was capable of de novo initiation on exogenous plus- and minus-stranded DI RNA templates. In addition, we found that the generation of highly active replicase in yeast required the coexpression of a DI RNA template together with the essential p33 and p92 replicase proteins. Overall, the development of the recombinant CNV replicase assay will be of great help in defining the role of virus- and possibly host-derived proteins and the RNA template in tombusvirus replication.
MATERIALS AND METHODS
Construction of expression plasmids in yeast cells.
To express the CNV p92 gene with an N-terminal 6× His tag, we generated pGAD-His92 (Fig. 1B). This was done by the following steps. (i) We eliminated the HindIII site in pGADT7 (position 2280; BD Biosciences) by partial digestion with HindIII, blunted the ends with Klenow polymerase, and performed religation. This modification yielded pGAD(H−) vector. (ii) The CNV p92 gene was amplified by PCR using primers #424 (5′-CGACGGATCCGATACCATCAAGAGGATGCTGTG) and #952 (5′-CCCGCTCGAGTCATGCTACGGCGGAGTCAAGGA) from p92Y, in which the stop codon of the p33 gene was replaced with a tyrosine codon (28, 52). The obtained 2.5-kb PCR fragment was digested with BamHI and XhoI. (iii) In addition, we generated the 116-bp HindIII-BamHI fragment encoding a 6× His tag at the 5′ end (the actual sequence is 5′-AAGCTTACCATGGGGGGTTCTCATCATCATCATCATCATGGTATGGCTAGCATGACTGGTGGACAGCAAATGGGTCGGGATCTGTACGACGATGACGATAAGGTACCCGGATCC) from the pYES2/NT-C vector (Invitrogen). (iv) Finally, the 2.5-kb and 116-bp fragments were cloned simultaneously into HindIII-XhoI-digested pGAD(H−) vector to generate pGAD-His92.
For the construction of pGBK-His33 (Fig. 1B) to express the CNV p33 gene with an N-terminal 6× His tag, we gel isolated the 1.5-kb HindIII (position 6544)-HindIII (position 738) fragment from HindIII-digested pGBKT7 (BD Biosciences). The CNV p33 sequence was amplified with PCR using primers #424 and #992B (5′-GAGCTGCAGCTATTTCACACCAAGGGA). The obtained 0.9-kb PCR fragment was digested with BamHI and PstI and was gel isolated. In addition, the 116-bp HindIII-BamHI fragment encoding a 6× His tag fragment was obtained as described above. The generated 1.5-kb, 0.9-kb, and 116-bp fragments were ligated together, followed by reamplification of the ligated product with PCR using primers #953 (5′-GATCCTTTTGTTGTTTCCGGGTGTACAATA) and #992B. The obtained PCR product was digested with Bsp1407I and PstI and was inserted into Bsp1407I- and PstI-digested pGBKT7 vector.
To modify the original RNA transcription vector (30) that produced yDI-72 RNA with a ∼120-nucleotide 5′ leader, which is a plasmid-borne nonviral sequence, we constructed a new expression vector based on the high-copy-number pYES2/NT-A plasmid carrying a URA3-selectable marker (Invitrogen) and the GAL1 promoter. First, pYC2/CT (Invitrogen), which was originally used to express yDI-72 RNA in yeast (30), was modified to remove most of the plasmid-borne leader sequence, leaving only 4 bp between the start site of transcription and a HindIII site in pYC2/CTm. This was done with primers #1080 (5′-CAGGCAAGCGATCCGTCCGCCGGCGAACGT) and #1145 (5′-CCCGAAGCTTACTTTTATTACATTTGAATAAGAAGTAAT) in a PCR in the presence of pYC2/CT template. The obtained PCR product was digested with NaeI and HindIII enzymes, gel purified, and ligated into pYC2/CT between NaeI (position 4479) and HindIII (position 501) sites, resulting in the pYC2/CTm plasmid. After the above modification of the expression vector, we inserted DI-72 sequence in order to facilitate the expression of plus- and minus-stranded DI-72 RNA. This was made by a two-step PCR. First, we used primers #542 (5′-GCCCGAAGCTTGGAAATTCTCCAGGATTTC) and #157 (5′-GGGCTGCATTTCTGCAATGTTCC) for plus-stranded DI-72 and primers #719 (5′-GCCCGAAGCTTGGGCTGCATTTCTGCAATGTTC) and #20 (5′-GGAAATTCTCCAGGATTTCTC) for minus-stranded DI-72 in the presence of DI-72SXP template (56). The obtained PCR products were separately gel isolated and ligated with gel-isolated PCR products that included the ribozyme sequence from the minus-stranded satellite of Tobacco ringspot virus (6, 7), which was obtained with primers #1067 (5′-AGTCCTGTTTCTTGCCAAACAGAGAAGGGCACCAGAGAAA) and #1068 (5′-GGTAATATACCACAACGTGTGTTTCTCTGGTGCCCTTCTC). The ligation products were PCR amplified with primers #542 and #1069 (5′-CCGGTCGAGCTCTACCAGGTAATATACCACAACGTGTGT) for plus-stranded DI-72 and with primers #719 and #1069 for minus-stranded DI-72. The obtained PCR products were digested with HindIII and SacI enzymes, gel isolated, and ligated into the similarly treated pYC2/CTm plasmid. Finally, to transfer the cloned plus- and minus-stranded DI-72 sequences to the high-copy plasmid pYES2/NT-A, we treated the above plasmids with NgoMIV and EcoRI and then inserted the gel-isolated fragments into NgoMIV and EcoRI-treated pYES2/NT-A plasmid. The obtained plasmids were termed pYES-DI-72(+)Rz (Fig. 1B) and pYES-DI-72(−)Rz.
Yeast transformation and growth.
Saccharomyces cerevisiae strain INVSc1 (Invitrogen) was used in these studies (30). The yeast cells were cotransformed with all three plasmids [i.e., pGAD-His92, pGBK-His33, and pYES-DI-72(+)Rz] by the LiAc-single-stranded DNA-polyethylene glycol method (30). In control experiments, we replaced one of the three plasmids with the original plasmid lacking viral sequences. After transformation, yeast cells were plated on selective SC medium without uracil, leucine, and tryptophan (SC-ULT−).
For CNV replicase studies, yeast was grown in SC-ULT− medium containing 2% galactose for 24 h at 30°C. The cultures were then diluted 10-fold with fresh SC-ULT− medium with 2% galactose and grown under the same conditions until an optical density at 600 nm of 0.6 to 0.7 (approximately 24 h). Yeast cells were then harvested by centrifugation at 1,100 × g for 5 min, followed by washing the pellet with 20 mM Tris-HCl, pH 8.0, and centrifugation (same as described above). The pelleted cells were resuspended in a small amount (∼1 to 2 ml) of fresh buffer (20 mM Tris-HCl, pH 8.0) and aliquoted, followed by centrifugation at 21,000 × g for 1 min and storage of the pellet at −80°C until further use.
For obtaining the enriched membrane fraction, yeast was grown at 30°C in SC-ULT− with 2% glucose for 24 h, followed by dilution to an optical density at 600 nm of 0.1 in SC-ULT− with 2% galactose. After further incubation at 23°C, yeast samples were taken at the time points described in the legend to Fig. 2.
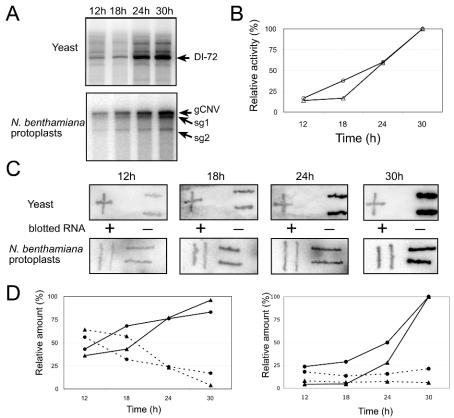
FIG. 2.
Characterization of CNV replicase activity in enriched membrane fractions derived from yeast or N. benthamiana protoplasts. (A) PAGE analysis of the (32P-labeled) in vitro replicase products on the endogenous templates, which are present in the enriched membrane fractions, obtained from yeast (top) and from protoplasts (bottom). Time of incubation, shown on the top of the gel in the time course experiments, started after the addition of galactose to the media for yeast or with the addition of incubation media after electroporation for N. benthamiana protoplasts. Arrows indicate the bands that correspond to products obtained on the various endogenous RNAs. (B) Relative RdRp activity of enriched membrane fractions. For quantification, we measured the intensity of 32P-labeled DI-72 and gCNV RNA bands (see panel A) obtained with yeast- and plant-derived CNV replicase preparations by using a phosphorimager. The gels shown in panel A were exposed for the same time, and 100% value represents the signal obtained after 30 h of incubation in each experiment. Circles and triangles represent data obtained with CNV replicase derived from protoplasts and yeast, respectively. (C) RNA blot showing plus-strand (+) and minus-strand (−) levels in the in vitro CNV replicase products on endogenous templates. Unlabeled T7 RNA polymerase transcripts of DI-72(+) and DI-72(−) (400 ng of each) for yeast-derived samples and gCNV(+) and gCNV(−) for protoplasts-derived samples were blotted on the membrane as shown between the blots. Time points for harvesting the yeast and protoplast samples for isolation of membrane fractions are shown on the top. The blotted RNAs were hybridized with denatured 32P-labeled RNA probes, which were generated by the CNV replicase in vitro on the endogenous templates present in the enriched membrane fractions from yeast or protoplast (see panel A). (D) Relative amounts of plus- and minus-stranded RNAs in the in vitro replicase products. Total replicase products at each time point were taken as 100% (left panel), or the amount of plus-stranded replicase product at the 30 h time point was taken as 100% (right panel). Solid and dotted lines represent plus- and minus-stranded products, while circles and triangles represent data obtained with preparations from protoplasts and yeast, respectively.
Purification of the CNV replicase from S. cerevisiae cells coexpressing the replicase proteins and DI RNA.
Frozen yeast cells were homogenized in liquid nitrogen by grinding in a mortar, followed by transferring the obtained powder to a tube containing the extraction buffer (200 mM sorbitol, 50 mM Tris-HCl [pH 7.5], 15 mM MgCl2, 10 mM KCl, 10 mM β-mercaptoethanol, yeast protease inhibitor mix; Sigma), which was applied in 10-fold excess over the (fresh weight) yeast cells. Unbroken cells were removed by centrifugation at 100 × g for 1.5 min at 4°C. Supernatant was then transferred to a new tube, and the enriched membrane fraction was pelleted via centrifugation at 21,000 × g for 15 min at 4°C. The obtained fraction was considered enriched membrane fraction, which was resuspended in the extraction buffer and used in the in vitro replicase reactions as described below.
To further purify the CNV replicase preparation, the enriched membrane fraction was resuspended in the extraction buffer containing 1.2 M NaCl, followed by gentle rotation for 20 min at 4°C and centrifugation at 21,000 × g for 15 min at 4°C. The obtained pellet was resuspended in the extraction buffer containing 1% Triton X-100 and 5% SB3-10 (caprylyl sulfobetaine) (Sigma) by gentle rotation for 1 h at 4°C, followed by centrifugation at 21,000 × g for 15 min at 4°C. The obtained supernatant was considered the solubilized membrane fraction. This preparation was tested in the in vitro replicase assay as described below.
To prepare the samples for the His tag-based metal affinity purification, we solubilized the enriched membrane fraction in the solubilization buffer (extraction buffer plus 1% Triton X-100, 5% SB3-10, and 0.5 M KCl) as described above. After centrifugation, the supernatant was applied to a column containing ProBond resin (Invitrogen) equilibrated with the solubilization buffer. The column was then rotated for 1 h, followed by washing with two column volumes of the solubilization buffer, washing with the extraction buffer containing 1% Triton X-100 and 5% SB3-10, and then a second wash with the extraction buffer containing 1% Triton-100, 5% SB3-10, and 2 mM imidazole. The recombinant proteins were recovered from the column in the extraction buffer containing 150 mM imidazole, 1% SB3-10, and 0.1% Triton X-100 in a two-step elution (each in a half-column volume). The purity of the obtained recombinant protein-containing preparations was tested by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (51, 57), while the amount of the recombinant proteins in various samples was compared by using Western blotting with monoclonal anti-His tag antibody (Amersham).
In vitro CNV replicase assay.
To test the activity of various preparations, we performed two types of assays. One tested the endogenous template activity present within the CNV replicase preparation, whereas the other assay examined the ability of the CNV replicase preparation to accept exogenous templates. The only difference between the two assays is whether extra templates were added to the reaction mixture or not.
We used 0.5 μg of RNA templates for the CNV replicase reactions. The reaction mixtures contained either 25 μl of recombinant or 25 μl of plant-derived CNV replicase as described previously (23). The RdRp products were phenol-chloroform extracted and analyzed under denaturing conditions (i.e., 5% PAGE containing 8 M urea) (23).
Preparation of protoplast membrane fraction for in vitro RdRp assay.
Nicotiana benthamiana protoplasts were prepared and electroporated as described previously (34). The enriched membrane fraction and the solubilized membrane fraction were prepared from pelleted protoplast cells by using a method described earlier (23).
Preparation of RNA transcripts.
RNA transcripts for the replicase assays and protoplast experiments were prepared by standard in vitro T7 transcription reaction with various DNA templates as described previously (34). The DNA templates used in this work were prepared as described previously (30, 32, 33). Following the T7 RNA polymerase reaction, the RNA transcripts were isolated from 1% agarose gel for the RNA blot, followed by phenol-chloroform extraction and precipitation in 95% ethanol. The RNA transcripts were dissolved in water.
Total RNA extraction from yeast cells and RNA blot analysis.
To extract total RNA from yeast, equal volumes of buffer (50 mM sodium acetate [pH 5.2], 10 mM EDTA, 1% SDS) and water-saturated phenol were added to the pelleted cells (30). Samples were vortexed, incubated for 4 min at 65°C, and then incubated for 2 min on ice and centrifuged at 21,000 × g for 5 min at room temperature. Total RNA was precipitated from the aqueous phase by adding 3 volumes of 95% ethanol with 0.1 volume of 3 M sodium acetate, pH 5.2, and was washed with 70% ethanol. RNA was dissolved in Tris-EDTA buffer and formamide (in 1:1 ratio). Total yeast RNA samples were heated for 5 min at 85°C, electrophoresed in 1% agarose gels, and transferred to Hybond XL membrane (Amersham). In vitro-made RNA transcripts, following 5 min of preincubation with formamide at 85°C, were pipetted to the Hybond XL membrane and cross-linked with UV (Bio-Rad). Hybridization was done with ULTRAhyb solution (Ambion) at 68°C according to the supplier's instructions. The 32P-labeled replicase products were used as probes for hybridization.
Western blot.
Aliquots of replicase proteins purified from yeast cells by using nickel-chelating affinity columns were mixed in 1:1 ratios with SDS-PAGE sample loading buffer (51), heated for 5 min at 85°C, electrophoresed in SDS-8% PAGE gels, and electrotransferred to a polyvinylidene difluoride membrane (Bio-Rad). Nonspecific binding sites on the membranes were blocked with 5% nonfat dry milk solution in Tris-buffered saline (TBS) buffer (51) containing 0.1% Tween 20 (TTBS), and the membranes were washed three times with TTBS buffer and incubated with monoclonal anti-His antibodies (Amersham) for 1 h at room temperature. Following three 10-min washes with TTBS buffer, membranes were incubated for 1 h at room temperature with secondary alkaline phosphatase-conjugated antibody (Sigma). After three washes of membranes with TTBS, protein bands were developed by using 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium (Sigma).
RESULTS
Comparison of CNV replicase activity of the enriched membrane fraction obtained from yeast and plant protoplasts.
To facilitate the detection and purification of CNV replicase proteins from yeast, we modified the yeast expression system described earlier (30). Briefly, we added an N-terminal 6× His tag to both the p33 and p92 proteins, which were expressed constitutively from high-copy-number plasmids (Fig. 1B). Coexpression of the p33 and p92 proteins together with the DI-72 RNA replicon, which is obtained from the galactose-inducible GAL1 promoter (Fig. 1B), results in robust DI-72 RNA replication in yeast cells. The presence of His tags in p33 and p92 does not appear to inhibit DI-72 RNA replication, because the amount of DI-72 plus strand can reach rRNA levels in yeast similar to those in plant cells (see below).
Enriched membrane preparations were obtained from yeast coexpressing p33, p92, and DI-72 as described in Materials and Methods. Briefly, yeast was grown on selective medium in the presence of galactose for 12 to 30 h (in a time course experiment; Fig. 2), followed by disruption of cells and differential centrifugation to obtain the crude, enriched membrane fraction. For comparison we used N. benthamiana protoplasts electroporated with CNV gRNA transcripts (note that we did not use DI RNA template here because the super-competitive DI-72 RNA would reduce gCNV replication, thus resulting in reduced amount of replicase in plant cells), followed by harvesting of cells at various time points (Fig. 2A). The enriched membrane preparations were then obtained from the CNV gRNA-transfected protoplasts by using a method described previously (23). Subsequently, the obtained enriched membrane preparations from yeast and from plant protoplasts were tested in the presence of four ribonucleotides, including 32P-labeled UTP. Under these conditions the CNV replicase complex is expected to complete RNA synthesis on the endogenous templates (i.e., the viral RNA that was copurified with the replicase), which are part of the actively synthesizing replicase complex in cells. Because the replicase products became 32P-labeled during the in vitro reaction, they were analyzed in denaturing gels (Fig. 2A and B). These experiments demonstrated that both the yeast- and plant protoplast-derived CNV replicase complexes synthesized 32P-labeled products, which were either DI-72 RNA-sized (in yeast; Fig. 2A) or gRNA-, sgRNA1-, and sgRNA2-sized (in plant protoplast; Fig. 2A). The amounts of these products increased over time in both hosts, suggesting that more replicase complexes were formed in the cells during longer incubation times. Accordingly, the amount of p33 increased continuously during incubation up to 24 h in N. benthamiana protoplasts (data not shown).
To test the nature of the in vitro-synthesized replicase products, we recovered the 32P-labeled RNAs after the in vitro replicase reaction and used them as probes in RNA blotting. We applied the same amounts of denatured plus- and minus-stranded DI-72 RNA for yeast samples and CNV gRNA for protoplast samples as target RNAs, which were fixed on nylon membranes (Fig. 2C). After hybridization of the membranes with the 32P-labeled RNAs obtained from the in vitro replicase reactions, we measured the ratio of plus- versus minus-strand-specific signals on the RNA blots by using a phosphorimager (23). These experiments demonstrated that the enriched membrane preparations containing the CNV replicase from both yeast and plant protoplasts synthesized more minus-stranded than plus-stranded RNAs at the early time point, while the synthesis was gradually shifted toward plus strands at later time points (Fig. 2D, left panel). This finding was expected based on detection of more abundant plus strands than minus-strands in plant protoplasts at late time points (47). Interestingly, we observed new minus-strand synthesis by the CNV replicase even at late time points in both preparations (Fig. 2D, right panel), suggesting that the minus-strand synthesis had not been shut down in order to favor plus-strand synthesis (see Discussion).
Solubilized CNV replicase preparation from yeast is capable of using exogenous templates in vitro.
The CNV replicase present in either the crude yeast extract or the enriched membrane fraction was unable to utilize an exogenous template (i.e., template added for the in vitro reaction) (Fig. 3A). The exogenous template was RI/III(−), which contains minus-stranded sequence of region I and region III of DI-72 (Fig. 1A). RI/III(−) was chosen because (i) it is a very efficient template {it includes both the cPR promoter and the RIII(−) replication enhancer [29]} in vitro, (ii) it gives smaller replicase products than the endogenous DI-72 template, and (iii) it generates various replicase products (due to de novo initiation from the 3′-terminal promoter and from internal promoter-like sequences or primer extension; see the more detailed description below), which is useful for comparing replicase products obtained with the previously characterized CNV replicase preparation obtained from N. benthamiana. In order to efficiently solubilize the CNV replicase from the enriched yeast membrane preparation, we tried various detergents and their combinations at different concentrations (data not shown). We found that after treatment with 1% Triton X-100-5% SB3-10, the replicase complex remained active because it could synthesize 32P-labeled RNA product by using the endogenous template (Fig. 3A, lane 9, and data not shown). The same preparation, however, was also capable of cRNA synthesis on added (exogenous) template as well (Fig. 3A, lane 9). In summary, we conclude that the solubilized preparation contains a highly active CNV replicase that can be programmed by added templates.
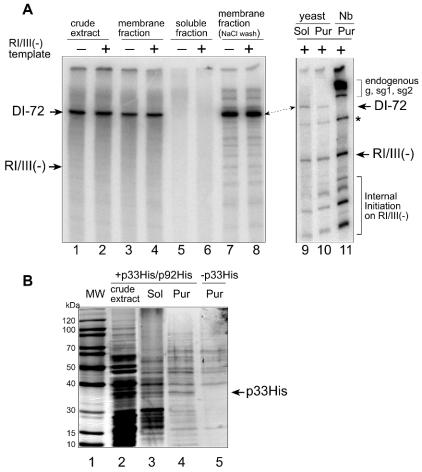
FIG. 3.
(A) Activity of CNV replicase in different preparations obtained from yeast and from N. benthamiana. Each preparation, as shown on the top, was tested in the absence or presence of exogenous RI/III(−) template, which contains the minus-stranded regions I and III of DI-72 (see Fig. 1A) in a standard CNV replicase assay. The positions of the exogenous RI/III (−), the endogenous DI-72 (for the yeast-derived samples), and genomic (g) and subgenomic (sg1 and sg2) templates (plant) are shown by arrows and bracketed lines on the side of the gel. The internal initiation products and the primer extension product are shown with a bracket and an asterisk, respectively. Sol represents solubilized membrane fraction, Pur indicates affinity-purified CNV replicase from yeast, and Nb represents the partially purified CNV replicase obtained from N. benthamiana plants. The replicase products obtained from the endogenous templates representing gCNV, sgRNA1, and sgRNA2 are indicated as g, sg1, and sg2, respectively. (B) SDS-PAGE analysis of proteins present in various CNV replicase preparations from yeast. The SDS-10% PAGE gel was stained with silver, and the position of p33His is marked on the right (p33His was confirmed with Western blotting; data not shown). Lanes 2 to 4 represent samples that were obtained from yeast expressing DI-72(+), p33His, and p92His, while the sample in lane 5 was derived from yeast expressing DI-72(+) and p92His but lacking p33His expression. Other symbols are as described for panel A. MW, molecular size marker.
Affinity-based purification of CNV replicase from yeast.
To purify the recombinant CNV replicase from the solubilized preparation, we utilized the engineered 6× His tag present at the N termini of p33 and p92 for nickel-chelating affinity purification as described in Materials and Methods. We found that the obtained purified CNV replicase preparation showed high polymerase activity (Fig. 3A, lane 10). Characterization of the replicase product by PAGE analysis revealed that the recombinant CNV replicase was able to synthesize cRNA on added RI/III(−) template in the presence of ribonucleotides and 32P-labeled UTP (Fig. 3A, lane 10). An analysis of the CNV replicase preparations via silver-stained SDS-PAGE revealed that the His tag-based purification led to the enrichment of p33 and several other host proteins (Fig. 3B, lane 4) whose functions are presently unknown.
We next wanted to know if the properties of the affinity-purified recombinant CNV replicase were comparable to those of the partially purified CNV replicase (RdRp) preparation obtained from infected N. benthamiana plants (23). First, we tested plus- and minus-stranded DI-73 RNA templates in vitro. DI-73 RNA contains the same sequence as DI-72 (Fig. 1A), plus a 167-nucleotide-long sequence between RIII and RIV, which is missing in DI-72 RNA (56). Importantly, the larger size of DI-73 compared to that of DI-72 allowed us to distinguish between the added exogenous DI-73 RNA and the copurified endogenous DI-72 templates after the in vitro reaction. We found that the yeast-derived purified recombinant and the plant-derived CNV replicase produced comparable products with DI-73(+) and DI-73(−) templates (Fig. 4, lanes 1 to 4). For example, both preparations used the minus-stranded template more efficiently than the plus-stranded DI-73. In addition, both replicase preparations synthesized three different types of RNA products on DI-73(−), such as (i) template-sized cRNA obtained by de novo initiation from the 3′-terminal promoter, (ii) several products that were shorter than the template, due to de novo initiation at internal positions (resembling promoter-like sequences [33]), and (iii) a product that was longer than the template, which was the result of primer extension from the 3′ end of the template (self-priming reaction). All of these in vitro replicase products were characterized in detail before, based on the plant-derived preparation (23, 32, 33).
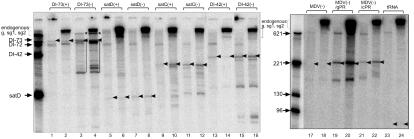
FIG. 4.
Comparison of template usage by the purified CNV replicases obtained from yeast and plant in vitro. Radiolabeled replicase products were analyzed on denaturing 5% PAGE-8 M urea gels. The exogenous RNA templates used in the in vitro replicase reaction are shown on the top of the gel. The products of the CNV replicase affinity purified from yeast (odd lane numbers) and from plants (even lane numbers) are shown. RNA size markers obtained by T7 transcription with [α-32P]UTP are shown on the left side of the gels. Note that the two sets of size markers are different, except for that of DI-72 (621 nucleotides) RNA. Template-sized products are marked with arrowheads, while the primer extension and internal initiation products are marked with asterisks and bracketed lines, respectively. The replicase products obtained from the endogenous templates representing gCNV, sgRNA1, and sgRNA2 are indicated as g, sg1, and sg2, respectively.
Additional templates tested with the purified recombinant CNV replicase were the CNV-derived DI-42, the related Turnip crinkle virus (TCV)-associated satD and satC (53), the Qβ-phage-associated satellite RNA (termed MDV [2]), and the nonviral yeast tRNA (Fig. 4). All these viruses belong to supergroup 2, although TCV and CNV are more closely related (they belong to the Tombusviridae family) than Qβ bacteriophage. The recombinant and the plant-derived CNV replicases were found to use DI-42(−) (lanes 15 and 16) and satC(−) (lanes 11 and 12) templates efficiently, while DI-42(+) (lanes 13 and 14), satC(+) (lanes 9 and 10), satD(+) (lanes 5 and 6), and satD(−) (lanes 7 and 8) RNAs were less efficient templates in vitro (Fig. 4). In contrast, MDV(−) (lanes 17 and 18) and the yeast tRNA (lanes 23 and 24) were not recognized by the CNV replicase as templates (Fig. 4). Overall the template specificities of the recombinant and the plant-derived CNV replicases were very similar.
To test if the recombinant CNV replicase can properly recognize TBSV- and CNV-derived cis-acting sequences, such as minus- or plus-strand initiation promoters, we used constructs MDV(−)/gPR and MDV(−)/cPR, which carried either the gPR promoter (i.e., the minimal minus-strand initiation sequence in TBSV and CNV [32, 33]) or the cPR promoter (i.e., the minimal plus-strand initiation sequence [32]) in addition to the MDV(−) sequences at the 5′ end. Analysis of the in vitro replicase products revealed that the recombinant CNV replicase, similar to the plant-derived preparation, recognized these minimal promoter sequences correctly and efficiently (Fig. 4, lanes 19 to 22).
Isolation of highly active CNV replicase complex requires coexpression of template RNA in yeast.
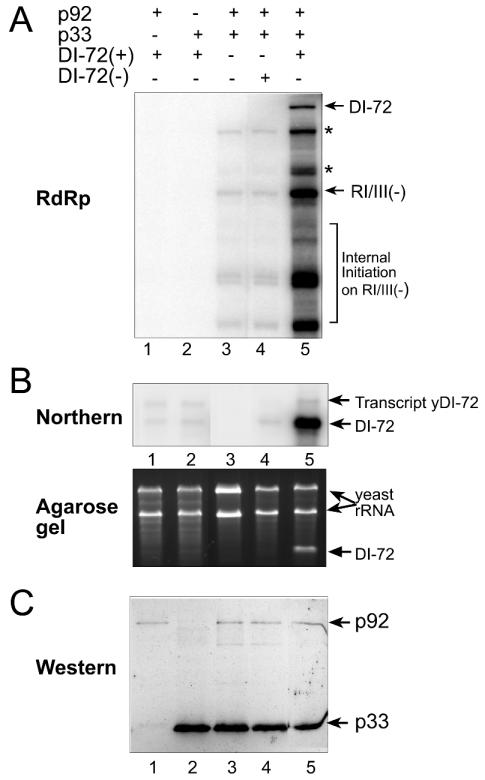
To take advantage of the heterologous CNV replicase system, we coexpressed various combinations of p33, p92, and DI-72 RNA in yeast, followed by testing the replicase activity of the affinity-purified preparations in the presence of an added RNA template [RI/III(−)] as shown in Fig. 5. These experiments demonstrated that neither p33 nor p92, when expressed separately in yeast, had detectable levels of replicase activity in vitro (Fig. 5, lanes 1 and 2). Coexpression of p33 and p92 in the same yeast cells, however, resulted in detectable, albeit very low, levels of replicase activity on the added template in vitro (Fig. 5, lane 3). The observed polymerase activity seems to come from the affinity-purified CNV replicase, because it was capable of producing full-length products which were single-strand specific and RNase resistant (data not shown), and these products were similar to those observed with the plant-derived CNV replicase preparation (Fig. 3A, lane 11).
FIG. 5.
Coexpression of DI-72 RNA in yeast stimulates the activity of the affinity-purified CNV replicase. (A) 32P-labeled RNA products obtained in the in vitro CNV replicase assay with exogenous template [RI/III(−)] were analyzed on 5% PAGE-8 M urea gels. The coexpressed p33, p92, and DI-72(+) or (−) RNAs in yeast cells are shown on the top with plus signs, while yeast lacking one of these components is marked with a minus sign. Additional symbols are as defined in the legend to Fig. 4. (B) Northern blot of total yeast RNA (top) and corresponding ethidium bromide-stained agarose gel (bottom). The total RNA was extracted from the yeast cells coexpressing various components as shown in panel A and was analyzed in 1% agarose gel (bottom) and with Northern blotting (top) using 32P-labeled RI/III/IV(−) RNA probe. Transcripts of yDI-72-sized (before ribozyme cleavage) and DI-72-sized bands are indicated by arrows. (C). Western blot analysis of p33 and p92 proteins, which were affinity purified from yeast cells coexpressing the shown viral components (see panel A), with anti-His antibodies.
Coexpression of p33, p92, and DI-72(+) RNA in the same yeast cells, however, led to the isolation of a CNV replicase preparation with the highest activity as shown in Fig. 5 (lane 5). These observations suggest that the RNA template significantly stimulates (∼40-fold increase) the formation of stable CNV replicase complex in vivo (see Discussion). In contrast, coexpression of DI-72(−) template with p33 and p92 resulted in basal levels of CNV replicase activity (Fig. 5, lane 4), suggesting that the minus-stranded RNA cannot stimulate the formation of the CNV replicase in vivo. Western blot analysis of p33 and the less abundant p92 in the purified replicase preparations revealed that the protein levels were comparable in the active (Fig. 5C, lane 5), less active (lanes 3 to 4), and inactive preparations (lanes 1 to 2), suggesting that there is no correlation between the amount of the individual replicase proteins and the activity of the purified CNV replicase in vitro.
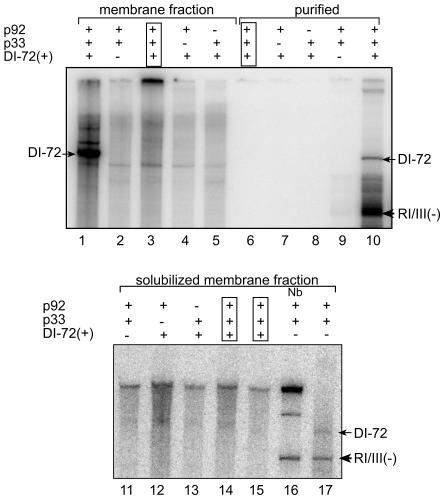
To examine if active CNV replicase can be assembled in vitro from separately expressed components, we used three types of mix-and-match experiments. In one type we mixed (in 1:1 ratio) two separate enriched membrane fractions obtained from two yeast strains, which coexpressed p92/DI-72(+) RNA and p33/DI-72(+) RNA separately prior to the replicase assay (Fig. 6, lane 3). We did not observe endogenous (32P-labeled) DI-72 RNA product in this mix-and-match assay, in contrast to the high-template activity obtained with similar extracts derived from yeast coexpressing p33/p92/DI-72 RNA (Fig. 6, lane 1). The second type of assay included the mixture of purified p33 and p92 components (expressed separately in two yeast strains) and exogenous RI/III(−) template obtained by in vitro transcription. Again, we did not observe detectable levels of replicase activity in this in vitro mix-and-match experiment (Fig. 6, lane 6). The control experiment, which included affinity-purified preparations obtained from yeast coexpressing p33/p92/DI-72 RNA, resulted in 32P-labeled products from the endogenous and exogenous templates (Fig. 6, lane 10).
FIG. 6.
Mix and match in vitro replicase assays. The CNV replicase preparations represent enriched membrane fractions (lanes 1 to 5), affinity-purified proteins (lanes 6 to 10), and solubilized membrane fractions (lanes 11 to 17) obtained from yeast, except for lane 16, which contains products that were generated by CNV replicase obtained from N. benthamiana plants. The CNV replicase assay depicted in lanes 6 to 17 included exogenous RI/III(−) templates. The coexpressed p33, p92, and DI-72(+) RNA in yeast cells are shown on the top as plus signs, while yeast lacking one of these components is marked with minus signs. The boxed lanes represent the mixed preparations as follows. Preparations containing either p33 or p92 (coexpressed with DI-72 RNA) were obtained from separate yeast lines, which were mixed in 1:1 ratio prior to the CNV replicase reaction (replicase products in lanes 3, 6, 14, and 15 are from mixing preparations corresponding to yeast lines shown in lanes 4 and 5; 7 and 8; and 12 and 13, respectively). Note that the CNV replicase preparation in lane 15 was obtained by mixing p92 and p33 preparations in a 1:4 ratio. The CNV replicase products generated on endogenous DI-72 or exogenous RI/III(−) templates are marked. The lane marked Nb represents the replicase product obtained with the plant-derived CNV replicase preparation.
Because it is possible that we might have lost one or more host-derived components from the CNV replicase during purification, thus resulting in inactive replicase in the above mix-and-match experiment, we also used nonpurified solubilized membrane fractions for in vitro studies (Fig. 6, lanes 11 to 17). However, mixing (prior to the replicase assay) solubilized membrane fractions obtained from two separate yeast strains which coexpressed either p92/DI-72(+) or p33/DI-72(+) in 1:1 (Fig. 6, lane 14) or 1:4 ratios (Fig. 6, lane 15), respectively, did not give the predicted 32P-labeled products in the in vitro assays. Based on these mix-and-match experiments with (i) the enriched membrane fractions, (ii) the solubilized membrane fractions, or (iii) the affinity-purified replicase proteins and RNA transcripts, we conclude that the in vitro conditions used did not favor the assembly of functional CNV replicase complex. This result is in contrast with those from the in vivo conditions in yeast coexpressing p33/p92/DI-72(+) RNA, which gave functional CNV replicase. Future studies will be devised to test if cellular membranes or host factors, which might be missing in the above preparations, are needed for the assembly of the CNV replicase complex.
DISCUSSION
TBSV and CNV replicases obtained from infected plants have been used extensively to characterize viral RNA elements that affect plus- and minus-strand synthesis, including promoter (initiation) elements (32, 33), replication enhancers (29, 31), a replication silencer element (39), and template-switching in vitro (8, 9). These elements and processes are important for replication and recombination (including DI RNA formation) of tombusvirus. Despite the usefulness of the above-described plant-derived CNV replicase preparations in providing in vitro data that complement the wealth of in vivo data on tombusvirus (11, 15, 37, 46-48), there is one major drawback in obtaining such preparations: the production of the replicase proteins depends on virus replication. Therefore, the effect of detrimental mutations within the viral RNA or the replicase proteins on replication is difficult to study in the plant-derived in vitro CNV replicase assay. The development of a heterologous expression system for the CNV replicase genes in yeast should, however, circumvent this problem. This is because the recombinant replicase proteins are produced from expression plasmids in yeast even in the absence of viral replication (Fig. 5C). Accordingly, we succeeded in purifying active CNV replicase complex from yeast in the absence of replicating CNV gRNA (Fig. 5A). This recombinant preparation will complement the traditional plant-based and the newly developed protoplast-based (Fig. 2) CNV replicase assays.
Comparison of the recombinant CNV replicase from yeast and the partially purified CNV replicase preparation from plants revealed that both were capable of cRNA synthesis on added templates. We observed three different types of replicase initiation when minus-stranded DI-73 RNA was used as a template: (i) full-length cRNA product (via de novo initiation from the 3′-terminal cPR promoter [32]); (ii) internal initiation from promoter-like sequences (also via de novo initiation (33); and (iii) primer-based initiation (self-priming from the 3′ end) of cRNA synthesis (9). In contrast to the minus-stranded templates, the plus-stranded DI-73 RNA was used inefficiently by both replicase preparations (Fig. 4). Thus, in spite of (i) the different expression strategies for p33 and p92 in plants (CNV infection based) and in yeast (plasmid based), (ii) the different purification methods (i.e., affinity-based purification from yeast and purification via chromatography from plants), and (iii) the different N-terminal sequences in p33 and p92 (i.e., wild-type p33/p92 in the plant-expressed preparations and His-tagged p33/p92 in the yeast-expressed preparations), the properties of the purified recombinant CNV replicase preparations and the plant-derived preparations are comparable under the in vitro conditions used.
The results shown in this work confirm that the functional CNV replicase complex contains both p33 and p92 replicase proteins (Fig. 5). This has been predicted earlier based on data obtained from biochemical and genetic experiments (23, 28, 34, 52). However, it is somewhat surprising that the purified recombinant p92 was not a functional polymerase (RdRp) in vitro when expressed alone in yeast (Fig. 5). This is because p92 contains the same sequence as p33 in its N-terminal overlapping (prereadthrough) domain (Fig. 1A). Apparently, the N-terminal region in p92 cannot fulfill the function(s) provided by the free p33 in the CNV replicase complex. In contrast to the CNV p92, which requires p33 in the functional CNV replicase, the similar TCV p88 RdRp protein or its N-terminally truncated version (when purified from E. coli) are highly active polymerases in vitro in the absence of the smaller replicase protein (i.e., p28 [44]). The reason for this difference between the CNV and TCV RdRp proteins in the requirement for the auxiliary viral protein is not presently known.
In addition to the p33 and p92 replicase proteins, the endogenous DI RNA is also an important factor in the CNV replicase complex. Although the purified CNV replicase containing p92 and p33 was active in the in vitro assay, coexpression of DI-72 RNA in the same yeast cells resulted in a CNV replicase preparation with ∼40-fold enhanced activity (Fig. 5A). By using Western blotting, we excluded the possibility that the increase in replicase activity is due to increased amounts of the replicase proteins in the purified preparations obtained from yeast lacking or coexpressing DI-72 RNA (Fig. 5D). We also demonstrated that the enhanced CNV replicase activity requires the plus-stranded DI-72 RNA, while the minus-stranded DI-72 failed to achieve the same effect (Fig. 5). Based on these observations, we propose that the role of the plus-stranded RNA template is to promote the assembly of the functional replicase in cells by possibly providing an assembly platform. A similar model on the nontemplate role of the RNA has also been proposed for the BMV and the AMV replicases, which are related to each other but are only very distantly related to the CNV replicase (40, 54). A seemingly important difference between the BMV and AMV replicases versus the CNV replicase is that the latter is functional, albeit at a low level, in the absence of the RNA template (Fig. 5).
Although the coexpression of DI-72 RNA is important for isolation of highly active CNV replicase from yeast, we found that, after purification of the replicase complex, the template RNA can be removed partially without the loss of or large reduction in the activity of the CNV replicase (based on exogenous templates; Fig. 4 and data not shown). This suggests that the CNV replicase, once it has been formed in cells, is a stable complex even in the absence (or in the presence of a minute amount) of DI-72 RNA.
The actual role(s) of the RNA template during the assembly of the CNV replicase complex or during other processes, such as recruitment of replicase proteins to the membranous structures (the proposed sites of tombusvirus replication [36, 49]), is presently unknown. Our effort to assemble the CNV replicase in vitro from either purified components (p33/p92 and DI-72 RNA) or by mixing enriched or solubilized membrane fractions failed to yield functional preparation (Fig. 6). It is possible that our in vitro preparations lacked host factors and/or particular membrane surface, which might be essential for the assembly of the functional CNV replicase complex in vivo.
Time-course experiments (Fig. 2) revealed that the CNV replicase present in the enriched membrane fractions obtained from yeast and N. benthamiana protoplasts synthesized both plus- and minus-stranded products in vitro. As expected, the CNV replicase synthesized more plus-stranded products at the late time points than at the early time point (Fig. 2D, left panel). However, we also observed new minus-strand synthesis taking place even at the late time points in the enriched membrane preparations obtained from yeast and plant protoplasts. Based on this observation, we propose that the CNV replicase is involved in minus-strand synthesis during the entire replication process; thus, there is no shut down of minus-strand synthesis in cells, a process that would favor plus-strand synthesis at the late time points, as suggested for the unrelated Tobacco mosaic virus (18). Our observation seems to be valid for both the yeast-based infection-independent and the protoplast-based infection-dependent systems (Fig. 2C). We interpret these results to mean that new CNV replicase complexes, which first synthesize minus-stranded RNA products, are continuously being formed in yeast cells and in plant protoplasts, even at the late time points. In agreement with this model, the amounts of plus- and minus-stranded TBSV (47) and CNV RNAs (data not shown), both gRNA and DI RNA, increase continuously over 24 to 30 h of infection in cucumber and N. benthamiana protoplasts. Also, we found that the amount of p33 increases over time (up to 24 h) in plant protoplasts (data not shown), which suggests continuous production of p33, and possibly p92, replicase protein in these cells.
The above-described model is based on the assumption that the obtained CNV replicase preparations can only complete RNA synthesis on the endogenous templates. Thus, the new 32P-labeled in vitro replicase products would only consist of the complementary strands of the copurified RNAs. However, it is also possible that the CNV replicase present in the enriched membrane fraction can perform sequential plus- and minus-strand synthesis from the endogenous templates. This complete in vitro replication would result in new 32P-labeled replicase products consisting of both strands, even if the copurified RNA consisted of one strand. Although we cannot exclude this model at this time, we do not have any evidence supporting complete replication in our CNV replicase assays (based on either endogenous or exogenous templates). Future experiments will address this possibility.
In summary, in this work we have developed three new in vitro assays with the recombinant CNV replicase. One assay is based on obtaining an enriched-membrane fraction, the second on solubilized membrane preparation, and the third on affinity-purified replicase preparation, which will be useful to further study the mechanism of tombusvirus replication and recombination. In addition, we have demonstrated that the CNV replicase complex contains both p33 and p92 replicase proteins. Also, the activity of the CNV replicase obtained from yeast is greatly enhanced by coexpression of the DI-72(+) strand, suggesting that the viral RNA plays a role in assembly of the CNV replicase.
Acknowledgments
We thank Judit Pogany and Jozsef Gal for critical comments and helpful suggestions.
This work was supported by the NSF (MCB0078152) and by the Kentucky Tobacco Research and Development Center at the University of Kentucky.
Footnotes
This study is Publication No. 04-12-034 of the Kentucky Agricultural Experiment Station.
REFERENCES
- 1.Ahlquist, P., A. O. Noueiry, W. M. Lee, D. B. Kushner, and B. T. Dye. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 77:8181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Axelrod, V. D., E. Brown, C. Priano, and D. R. Mills. 1991. Coliphage Q beta RNA replication: RNA catalytic for single-strand release. Virology 184:595-608. [DOI] [PubMed] [Google Scholar]
- 3.Bates, H. J., M. Farjah, T. A. Osman, and K. W. Buck. 1995. Isolation and characterization of an RNA-dependent RNA polymerase from Nicotiana clevelandii plants infected with red clover necrotic mosaic dianthovirus. J. Gen. Virol. 76:1483-1491. [DOI] [PubMed] [Google Scholar]
- 4.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buck, K. W. 1999. Replication of tobacco mosaic virus RNA. Philos. Trans. R Soc. Lond. B Biol. Sci. 354:613-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buzayan, J. M., P. A. Feldstein, G. Bruening, and F. Eckstein. 1988. RNA mediated formation of a phosphorothioate diester bond. Biochem. Biophys. Res. Commun. 156:340-347. [DOI] [PubMed] [Google Scholar]
- 7.Buzayan, J. M., A. Hampel, and G. Bruening. 1986. Nucleotide sequence and newly formed phosphodiester bond of spontaneously ligated satellite tobacco ringspot virus RNA. Nucleic Acids Res. 14:9729-9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, C. P., and P. D. Nagy. 2003. Mechanism of RNA recombination in carmo- and tombusviruses: evidence for template switching by the RNA-dependent RNA polymerase in vitro. J. Virol. 77:12033-12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, C. P., J. Pogany, and P. D. Nagy. 2002. Mechanism of DI RNA formation in tombusviruses: dissecting the requirement for primer extension by the tombusvirus RNA dependent RNA polymerase in vitro. Virology 304:460-473. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, J. H., M. P. Ding, Y. H. Hsu, and C. H. Tsai. 2001. The partial purified RNA-dependent RNA polymerases from bamboo mosaic potexvirus and potato virus X infected plants containing the template-dependent activities. Virus Res. 80:41-52. [DOI] [PubMed] [Google Scholar]
- 11.Fabian, M. R., H. Na, D. Ray, and K. A. White. 2003. 3′-Terminal RNA secondary structures are important for accumulation of tomato bushy stunt virus DI RNAs. Virology 313:567-580. [DOI] [PubMed] [Google Scholar]
- 12.Gal-On, A., T. Canto, and P. Palukaitis. 2000. Characterisation of genetically modified cucumber mosaic virus expressing histidine-tagged 1a and 2a proteins. Arch. Virol. 145:37-50. [DOI] [PubMed] [Google Scholar]
- 13.Gamarnik, A. V., and R. Andino. 1996. Replication of poliovirus in Xenopus oocytes requires two human factors. EMBO J. 15:5988-5998. [PMC free article] [PubMed] [Google Scholar]
- 14.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Havelda, Z., and J. Burgyan. 1995. 3′ Terminal putative stem-loop structure required for the accumulation of cymbidium ringspot viral RNA. Virology 214:269-272. [DOI] [PubMed] [Google Scholar]
- 16.Hayes, R. J., and K. W. Buck. 1990. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell 63:363-368. [DOI] [PubMed] [Google Scholar]
- 17.Hong, Y., and A. G. Hunt. 1996. RNA polymerase activity catalyzed by a potyvirus-encoded RNA-dependent RNA polymerase. Virology 226:146-151. [DOI] [PubMed] [Google Scholar]
- 18.Ishikawa, M., T. Meshi, T. Ohno, and Y. Okada. 1991. Specific cessation of minus-strand RNA accumulation at an early stage of tobacco mosaic virus infection. J. Virol. 65:861-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kao, C. C., A. M. Del Vecchio, and W. Zhong. 1999. De novo initiation of RNA synthesis by a recombinant flaviviridae RNA-dependent RNA polymerase. Virology 253:1-7. [DOI] [PubMed] [Google Scholar]
- 20.Kao, C. C., P. Singh, and D. J. Ecker. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:251-260. [DOI] [PubMed] [Google Scholar]
- 21.Lai, V. C., C. C. Kao, E. Ferrari, J. Park, A. S. Uss, J. Wright-Minogue, Z. Hong, and J. Y. Lau. 1999. Mutational analysis of bovine viral diarrhea virus RNA-dependent RNA polymerase. J. Virol. 73:10129-10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagy, P. D., and J. Pogany. 2000. Partial purification and characterization of Cucumber necrosis virus and Tomato bushy stunt virus RNA-dependent RNA polymerases: similarities and differences in template usage between tombusvirus and carmovirus RNA-dependent RNA polymerases. Virology 276:279-288. [DOI] [PubMed] [Google Scholar]
- 24.Neufeld, K. L., O. C. Richards, and E. Ehrenfeld. 1991. Purification, characterization, and comparison of poliovirus RNA polymerase from native and recombinant sources. J. Biol. Chem. 266:24212-24219. [PubMed] [Google Scholar]
- 25.O'Reilly, E. K., and C. C. Kao. 1998. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology 252:287-303. [DOI] [PubMed] [Google Scholar]
- 26.Osman, T. A., and K. W. Buck. 1996. Complete replication in vitro of tobacco mosaic virus RNA by a template-dependent, membrane-bound RNA polymerase. J. Virol. 70:6227-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osman, T. A., and K. W. Buck. 1997. The tobacco mosaic virus RNA polymerase complex contains a plant protein related to the RNA-binding subunit of yeast eIF-3. J. Virol. 71:6075-6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oster, S. K., B. Wu, and K. A. White. 1998. Uncoupled expression of p33 and p92 permits amplification of tomato bushy stunt virus RNAs. J. Virol. 72:5845-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panavas, T., and P. D. Nagy. 2003. The RNA replication enhancer element of tombusviruses contains two interchangeable hairpins that are functional during plus-strand synthesis. J. Virol. 77:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panavas, T., and P. D. Nagy. 2003. Yeast as a model host to study replication and recombination of defective interfering RNA of Tomato bushy stunt virus. Virology 314:315-325. [DOI] [PubMed] [Google Scholar]
- 31.Panavas, T., Z. Panaviene, J. Pogany, and P. D. Nagy. 2003. Enhancement of RNA synthesis by promoter duplication in tombusviruses. Virology 310:118-129. [DOI] [PubMed] [Google Scholar]
- 32.Panavas, T., J. Pogany, and P. D. Nagy. 2002. Analysis of minimal promoter sequences for plus-strand synthesis by the cucumber necrosis virus RNA-dependent RNA polymerase. Virology 296:263-274. [DOI] [PubMed] [Google Scholar]
- 33.Panavas, T., J. Pogany, and P. D. Nagy. 2002. Internal initiation by the cucumber necrosis virus RNA-dependent RNA polymerase is facilitated by promoter-like sequences. Virology 296:275-287. [DOI] [PubMed] [Google Scholar]
- 34.Panaviene, Z., J. M. Baker, and P. D. Nagy. 2003. The overlapping RNA-binding domains of p33 and p92 replicase proteins are essential for tombusvirus replication. Virology 308:191-205. [DOI] [PubMed] [Google Scholar]
- 35.Panaviene, Z., and P. D. Nagy. 2003. Mutations in the RNA-binding domains of tombusvirus replicase proteins affect RNA recombination in vivo. Virology 317:359-372. [DOI] [PubMed] [Google Scholar]
- 36.Pantaleo, V., L. Rubino, and M. Russo. 2003. Replication of Carnation Italian Ringspot Virus defective interfering RNA in Saccharomyces cerevisiae. J. Virol. 77:2116-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park, J. W., B. Desvoyes, and H. B. Scholthof. 2002. Tomato bushy stunt virus genomic RNA accumulation is regulated by interdependent cis-acting elements within the movement protein open reading frames. J. Virol. 76:12747-12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plante, C. A., K. H. Kim, N. Pillai-Nair, T. A. Osman, K. W. Buck, and C. L. Hemenway. 2000. Soluble, template-dependent extracts from Nicotiana benthamiana plants infected with potato virus X transcribe both plus- and minus-strand RNA templates. Virology 275:444-451. [DOI] [PubMed] [Google Scholar]
- 39.Pogany, J., M. R. Fabian, K. A. White, and P. D. Nagy. 2003. A replication silencer element in a plus-strand RNA virus. EMBO J. 22:5602-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quadt, R., M. Ishikawa, M. Janda, and P. Ahlquist. 1995. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc. Natl. Acad. Sci. USA 92:4892-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quadt, R., and E. M. Jaspars. 1990. Purification and characterization of brome mosaic virus RNA-dependent RNA polymerase. Virology 178:189-194. [DOI] [PubMed] [Google Scholar]
- 42.Quadt, R., H. J. Rosdorff, T. W. Hunt, and E. M. Jaspars. 1991. Analysis of the protein composition of alfalfa mosaic virus RNA-dependent RNA polymerase. Virology 182:309-315. [DOI] [PubMed] [Google Scholar]
- 43.Rajendran, K. S., and P. D. Nagy. 2003. Characterization of the RNA-binding domains in the replicase proteins of Tomato bushy stunt virus. J. Virol. 77:9244-9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajendran, K. S., J. Pogany, and P. D. Nagy. 2002. Comparison of turnip crinkle virus RNA-dependent RNA polymerase preparations expressed in Escherichia coli or derived from infected plants. J. Virol. 76:1707-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ranjith-Kumar, C. T., J. L. Santos, L. L. Gutshall, V. K. Johnston, J. Lin-Goerke, M. J. Kim, D. J. Porter, D. Maley, C. Greenwood, D. L. Earnshaw, A. Baker, B. Gu, C. Silverman, R. T. Sarisky, and C. Kao. 2003. Enzymatic activities of the GB virus-B RNA-dependent RNA polymerase. Virology 312:270-280. [DOI] [PubMed] [Google Scholar]
- 46.Ray, D., and K. A. White. 1999. Enhancer-like properties of an RNA element that modulates Tombusvirus RNA accumulation. Virology 256:162-171. [DOI] [PubMed] [Google Scholar]
- 47.Ray, D., and K. A. White. 2003. An internally located RNA hairpin enhances replication of Tomato bushy stunt virus RNAs. J. Virol. 77:245-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ray, D., B. Wu, and K. A. White. 2003. A second functional RNA domain in the 5′ UTR of the tomato bushy stunt virus genome: intra- and interdomain interactions mediate viral RNA replication. RNA 9:1232-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubino, L., and M. Russo. 1998. Membrane targeting sequences in tombusvirus infections. Virology 252:431-437. [DOI] [PubMed] [Google Scholar]
- 50.Russo, M., J. Burgyan, and G. P. Martelli. 1994. Molecular biology of Tombusviridae. Adv. Virus Res. 44:381-428. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook, J., T. Maniatis, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Scholthof, K. B., H. B. Scholthof, and A. O. Jackson. 1995. The tomato bushy stunt virus replicase proteins are coordinately expressed and membrane associated. Virology 208:365-369. [DOI] [PubMed] [Google Scholar]
- 53.Song, C., and A. E. Simon. 1994. RNA-dependent RNA polymerase from plants infected with turnip crinkle virus can transcribe (+) and (−) strands of virus-associated RNAs. Proc. Natl. Acad. Sci. USA 91:8792-8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vlot, A. C., L. Neeleman, H. J. Linthorst, and J. F. Bol. 2001. Role of the 3′-untranslated regions of alfalfa mosaic virus RNAs in the formation of a transiently expressed replicase in plants and in the assembly of virions. J. Virol. 75:6440-6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe, T., A. Honda, A. Iwata, S. Ueda, T. Hibi, and A. Ishihama. 1999. Isolation from tobacco mosaic virus-infected tobacco of a solubilized template-specific RNA-dependent RNA polymerase containing a 126K/183K protein heterodimer. J. Virol. 73:2633-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White, K. A., and T. J. Morris. 1994. Recombination between defective tombusvirus RNAs generates functional hybrid genomes. Proc. Natl. Acad. Sci. USA 91:3642-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wray, W., T. Boulikas, V. P. Wray, and R. Hanckock. 1981. Silver staining of proteins in polyacrylamide gels. Anal. Biochem. 118:197-203. [DOI] [PubMed]
- 58.You, S., and R. Padmanabhan. 1999. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 274:33714-33722. [DOI] [PubMed] [Google Scholar]