Abstract
Background
The aim of this study was to investigate the effect of hypoxia on the level of microRNA-18a (miR-18a) and hypoxia-inducible factor 1A (HIF1A) expressed by choroidal endothelial cells through cytological analysis.
Material/Methods
After culturing choroidal endothelial cells (CECs) under normoxia or hypoxia, microRNAs, expressed in different ways, were screened by using GeneChip microRNA array, and the results were confirmed by real-time PCR. The bioinformatics approach was used to screen target genes of target miRNA. In addition, CECs were transfected with target miRNA mimic or inhibitor and then expression levels of targeted miRNA and genes were observed.
Results
The GeneChip microRNA Array detected 14 miRNAs that were expressed differently. Among these miRNAs, 12 miRNAs were identified as being upregulated and 2 miRNAs as being down-regulated. MiR-18a was most significantly down-regulated. Bioinformatics analysis showed that HIF1A was the target gene of miR-18a. In CECs transfected with miR-18a mimic, there was a remarkable decrease in gene expression level and protein level of HIF1A, and thereby, significant reduction in proliferation and migration of CECs. In CECs transfected with miR-18a inhibitor, there was a significant increase in gene and protein expression levels of HIF1A, and enhanced proliferation and migration of CECs.
Conclusions
We conclude that miR-18a affects the function of CECs by inhibiting the expression of HIF1A.
Keywords: Choroidal Neovascularization, Hypoxia-Inducible Factor 1, Macular Degeneration, MicroRNAs
Background
Age-related macular degeneration (AMD) is a distressing disease which may lead to visual damage in elderly people. This vision impairment is chronic and progressive. Choroidal neovascularization (CNV) is the major reason for vision loss due to AMD [1]. Pathologically, endothelial function is damaged when choroidal endothelial cells (CECs) are exposed to traumatic factors such as hypoxia, causing the secretion imbalance of proangiogenic and anti-angiogenic factors, which finally trigger CNV [2].
Hypoxia plays important roles in the pathogenesis of CNV [3] that may involve over-expression of vascular endothelial growth factor (VEGF) and hypoxia-induced factor A (HIF-1A). These factors act as the transcription regulators, which may induce genes expression related to angiogenesis, erythropoiesis, and glucose metabolism to regulate the response of cells to hypoxia. HIF-1 may also induce proliferation and migration of CECs, which is the important pathological mechanism of CNV [4].
In cellular proliferation and differentiation, apoptosis, gene regulation, and disease development, microRNAs (miRNAs) have been found to play critical roles. miRNA, a kind of non-coding RNA consisting of 22 nucleotides, is capable of binding to 3′ untranslated or coding regions of target mRNA sequence by base-pairing, promoting the degradation of target mRNA or inhibiting protein expression to regulate the expression of target genes [5]. In this study, we used GeneChip microRNA Array technology to explore the effect of hypoxia on miRNAs expression profiles, as well as the expression of miR-18a and HIF1A, providing theoretical guidance and experimental evidence for the mechanism by which miRNAs regulate CNV.
Material and Methods
Reagents and equipment
Ham F12 medium, fetal bovine serum, endothelial cell growth factor (ECGF), 0.25% trypsin-0.1% EDTA (Ethylenediaminetetraacetic acid), and penicillin streptomycin double antibody were obtained from Gibco (USA). Trypsin, methyl thiazolyl tetrazolium (MTT), dimethyl sulfoxide (DMSO), heparin, and insulin were purchased from Sigma (USA). Restriction enzyme (Hea III) and DL2000 DNA marker were provided by Takara (Dalian, China). Equipment used in this study included ELIASA (ELX800, Bio-Tek, USA), inverted microscope (IX71, Olympus, Japan), and real-time PCR (7500, ABI, USA).
Human CECs culture
Human CECs (purchased from CHI Scientific, Inc.) were thawed in Ham F12 medium supplemented with 200 g/L fetal bovine serum, 100 mg/L ECGF, 100 mg/L heparin, 0.3 IU/mL insulin, and 1 g/L penicillin-streptomycin double antibody, and then cultivated in culture bottles coated with 2 g/L gelatin, at 37°C in 5% CO2. Medium was changed every 3–4 days. When 80% of the cells fused, they were digested for 1 min with 0.25 trypsin-0.1% EDTA, and passage was done by 1: 2. Cells of 3–6 passages were used for further experiments.
Establishment of hypoxia model
When the cells entered into log phase after culturing in an incubator at 37°C with 5% CO2, some of them continued to grow under this condition, and the other cells were transferred to a hypoxic incubator (1% O2, 5% CO2, and 94% N2). These cells were harvested after 24 h.
MiRNA extraction and GeneChip microRNA array analysis
Cells, under normoxia and hypoxia, were collected and their miRNA was extracted by mirVana™ miRNA Isolation Kit (Invitrogen, USA). One microgram of miRNA was labeled by biotin using FlashTag Biotin RNA Labeling Kit (Genisphere, USA). GeneChip miRNA2.0 Array (Affymetrix, USA) was used to analyze miRNA expression profiles. According to GeneChip instructions, hybridization, washing, image scanning, and data normalization were successively conducted. MiRNAs with P<0.01 and multiple value ≥20 or ≤−20 were identified as miRNAs which expressed differently.
Target gene prediction of miR-18a
Online bioinformatics software, including miRanda, DIANA-microT, PicTar, TargetScan, miRBase Target, and MICRORNA.ORG were used to predict the possible target genes of miR-18a. The selected target genes, which possessed species conservatism with 2–7 bases of its 3′ untranslated region (UTR), underwent complete complementary pairing with the sequence of miR-18a.
Transfection of miR-18a mimic or inhibitor
Transfection of miR-18a mimic or inhibitor was performed according to the instructions of Lipofect AMINE 2000 Kit (Invitrogen, USA). An amount of 50 nmol/L miR-18a mimic (5′-UAAUACUGCCGCCUGGUAAUGAUGGA-3′) or inhibitor (5′-UCACAACCAAAUCCUAGAAAGAGUAGA-3′) (Dharmacon, USA) was transfected into CECs, and the cells were harvested after 72 h. Untransfected cells were used as control.
Gene and protein expression of HIF1A
Cells under normoxia and hypoxia, as well as transfected with miR-18a mimic and inhibitor, were collected and total RNA was extracted by QiazolTM Lysis reagent Kit (Oiagen, USA). Total RNA was reversely transcribed into cDNA and PCR amplification was performed. For HIF1A, the forward primer was 5′-CGTTCCTTCGATCAGTTGTC-3′ and the reverse primer was 5′-TCAGTGGTGGCAGTGGTAGT-3′. β-actin served as the internal reference, together with the forward primer of 5′-TGTTACCAGGGAGGAGCAGT-3′ and the reverse primer of 5′-TGCCCTTCCTTTCCTGTGT-3′. All primers were synthesized by TaKaRa (Dalian, China) and 2−ΔΔCt method was used to calculate the relative expression of HIF1A.
After collection of cells under normoxia and hypoxia, as well as those transfected with miR-18a mimic and inhibitor, total protein was extracted by RIPA buffer and quantified by BCA method. Then 100 μg of total proteins were loaded onto 10% polyacrylamide SDS gels (SDS-PAGE), separated by electrophoresis before transferring onto PVDF membranes. The membrane was blocked with 5% skimmed milk for 1 h, after which it was incubated with anti-HIF1A primary antibody (1:500, Abcam) overnight at 4°C. Second antibody hybridization, membrane washing, and ECL visualization were performed regularly. β-actin was used as the internal reference and the relative protein expression of HIF1A was also calculated. Each experiment was carried out independently in triplicate.
Proliferation and migration of CECs after transfection
Proliferation of CECs was detected by MTT method: Cells transfected with miR-18a mimic or inhibitor were cultivated in 96-well plate before incubated for 72 h at 37°C with 5% CO2. Then, 20 μL of MTT (5 mg/mL) was added to each well, after which cells were cultured for another 4 h. After discarding culture medium, 150 μL DMSO was added to each well. Cells were oscillated for 5 min to fully dissolve crystals. The absorbance value was determined at 570 nm.
Migration of cells was detected by scratch test: Cells transfected with miR-18a mimic or inhibitor were cultivated in 96-well plate before incubating to cell confluence at 37°C with 5% CO2. The well was scratched by sterile tip for 200 uL pipet and the cells were washed with phosphate-buffered saline (PBS) before adding to serum-free medium. Then, the cells were observed by inverted microscope after 24 h at 37°C in 5% CO2, and the migration distance was calculated.
Statistical analysis
Statistical analysis was carried out by SPSS 13.0 software package (SPSS, Inc., Chicago, IL). After conducting normal distribution testing, normally distributed data were expressed as mean ± standard deviation (X±SD). Two-tailed Student t-test or Wilcoxon signed ranks test was used to evaluate differences between groups, which were considered statistically significant at P<0.05 in all cases.
Results
GeneChip miRNA array analysis
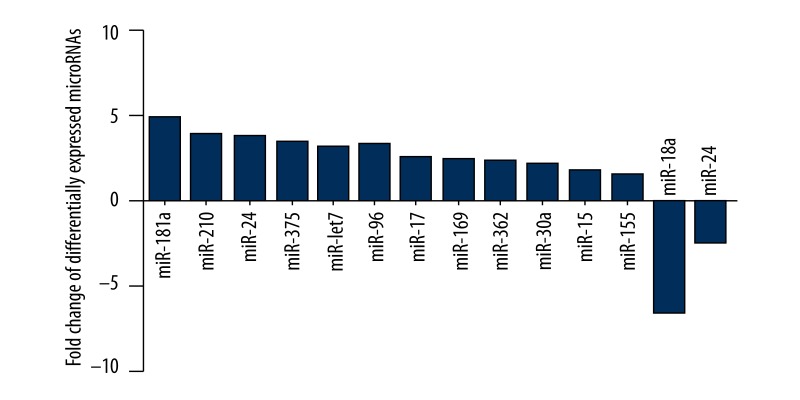
GeneChip miRNA array was applied to detect different miRNAs in CECs under normoxia or hypoxia. As a result, 14 different miRNAs (P<0.01, multiple value ≥20 or ≤−20) were screened out. Compared with cells under normoxia culture, 12 miRNAs were upregulated in CECs under hypoxia culture. The up-regulated miRNAs were miR-181a, miR-210, miR-24, miR-375, miR-lct7, miR-96, miR-17, miR-169, miR-362, miR-30a, miR-15, and miR-155. Two miRNAs, 1 miR-18a and 1 miR-24, were down-regulated. According to our results, miR-18a was the most significantly down-regulated (Figure 1).
Figure 1.
List of microRNAs expressed in different ways by comparing the choroidal endothelial cells under normoxic and hypoxic conditions.
Target gene prediction of miR-18a
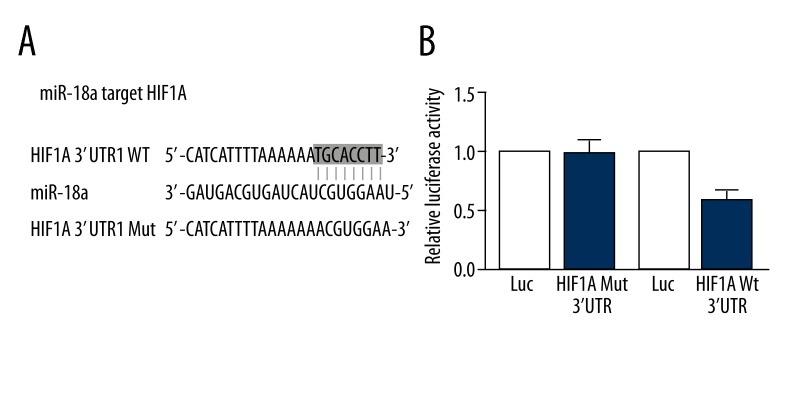
The intersection result obtained from the above-mentioned software was considered as the possible target gene. MiR-18a possessed species conservatism, and there were 8 bases (ACGUGGAA) in its seed zone. These bases were completely and complementarily paired with the 3′ untranslated region (UTR) of HIF1A (TGCACCTT) (Figure 2). Moreover, the binding energy between HIF1A and miR-18a was relatively low. Bioinformatics analysis indicated that HIF1A was the possible target gene of miR-18a.
Figure 2.
(A) Sequences of miR-18a, wild-type seed sequence in 3′UTR of HIF1A. (B) Luciferase assay showing that miR-18a mimic can significantly suppress the activity of luciferase containing the wild-type 3′UTR of HIF1A instead of the one containing mutant 3′UTR of HIF1A.
Expression of miR-18a and HIF1A in CECs under normoxia or hypoxia
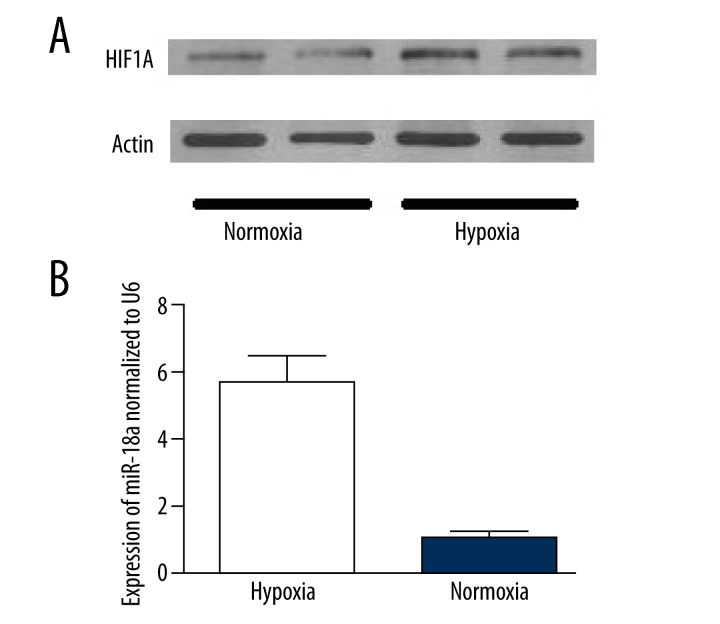
Effects of normoxia or hypoxia on expressing miR-18a and HIF1A in CECs are illustrated in Figure 3. According to our results, the protein level of HIF1A was very low under normoxia, while hypoxia increased the expression of HIF1A and the difference was statistically significant (P<0.05) between both groups. Hypoxia obviously inhibited the expression of miR-18a in CECs, which was 1/6 of that under normoxia, and there was also a significant (P<0.01) difference between both groups.
Figure 3.
(A) The different protein expression levels of HIF1A between the choroidal endothelial cells under normoxic and hypoxic conditions. (B) The different expression levels of miR-18a of choroidal endothelial cells under normoxic and hypoxic conditions.
Expression of HIF1A, proliferation and migration of CECs after miR-18a mimic transfection
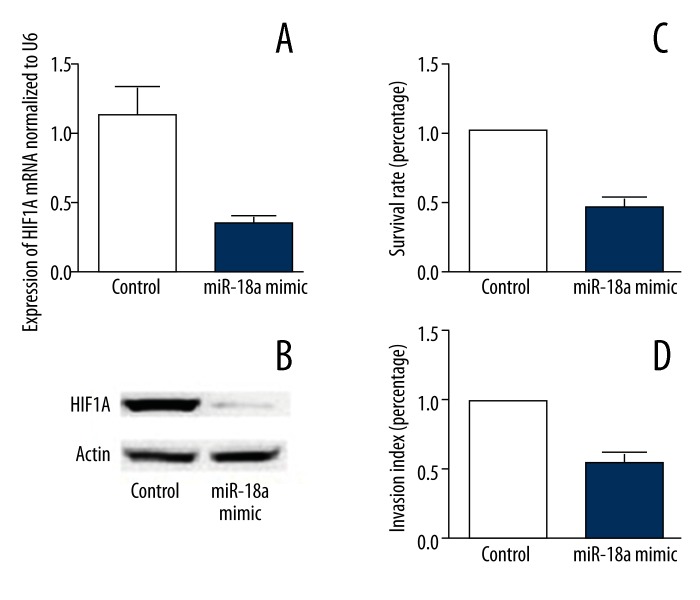
Compared with control cells, gene expression and protein levels of HIF1A dramatically decreased in CECs transfected with miR-18a mimic, indicating that overexpression of miR-18a in CECs significantly inhibited the expression of its target gene, HIF1A. Additionally, our findings showed that proliferation and migration of CECs were remarkably reduced after overexpression of miR-18a (Figure 4).
Figure 4.
(A) miR-18a mimic significantly decreases the mRNA expression level of HIF1A in CECs. (B) miR-18a mimic significantly decreases the protein expression level of HIF1A in CECs. (C) miR-18a mimic significantly suppresses the survival rate of CECs. (D) miR-18a mimic significantly suppresses the migratory ability of CECs.
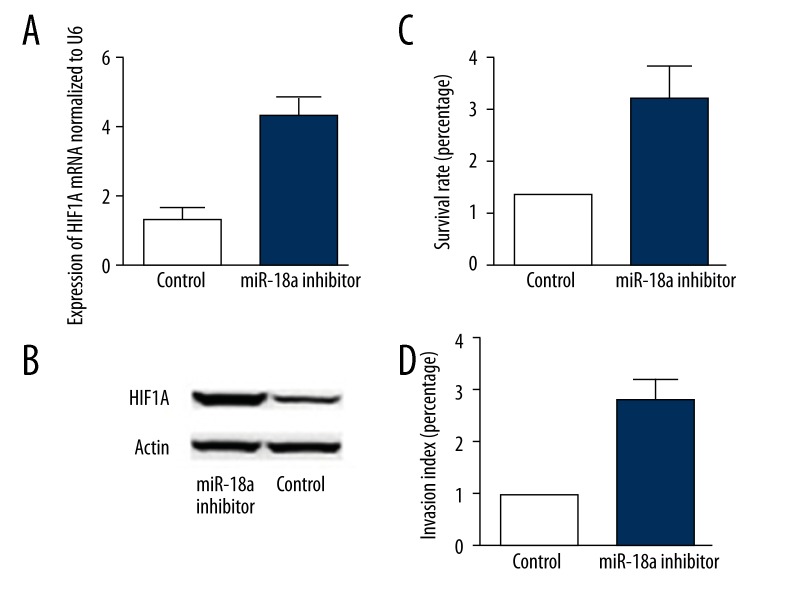
Expression of HIF1A as well as proliferation and migration of CECs after miR-18a inhibitor transfection
Overexpression of miR-18a inhibitor in CECs significantly enhanced gene expression and protein levels of HIF1A compared with control cells, suggesting that inhibition of miR-18a in CECs enhanced expression of HIF1A. Furthermore, inhibition of miR-18a in CECs significantly increased proliferation and migration of cells (Figure 5).
Figure 5.
(A) miR-18a inhibitor significantly increases the mRNA expression level of HIF1A in CECs. (B) miR-18a inhibitor significantly increases the protein expression level of HIF1A in CECs. (C) miR-18a inhibitor significantly improves the survival rate of CECs. (D) miR-18a inhibitor significantly promotes the migratory ability of CECs.
Discussion
Basically, CNV occurs in the macular area and is the pathological basis of many ocular fundus diseases [1]. The vascular wall of CNV is thin and fragile. It can easily lead to hemorrhage, which results in local inflammation accompanied by lymphocytes and macrophages infiltration as well as the secretion of inflammatory factors [6]. After that, new vessels are fiberized and the formed fibrous scars cause irreversible visual impairment.
Hypoxia plays an important role in development of CNV. Ischemia and hypoxia caused by abnormal blood flow, oxidative stress, and local inflammatory response can stimulate the release of various angiogenic growth factors and promote the formation of new blood vessels to increase oxygen supply, which might be the critical reason for CNV [2]. Previous studies demonstrated that the choroidal blood flow in AMD patients was significantly decreased compared with that in healthy people of equivalent age. In CNV patients, local blood circulation was problematic, and ocular pulsatile blood flow was slower compared with that in normal people, suggesting that hypoxia due to decreased choroidal blood flow might be the initial factor for development of CNV [7]. In addition, many diseases that accompany CNV could be due to degeneration and thickening of the Bruch membrane, which results in oxygen diffusion disorder in chorioid, pigment epithelium, and the retina, which suggests that hypoxia might participate in the pathological process of CNV [8].
HIF-1 is crucial in inducing the creation of new vessels [3]. HIF-1 consists of two subunits, A and B. HIF-1B, a structural subunit, expresses stability and is not affected by oxygen content. Hypoxia leads to an increase in the expression level of HIF-1A, which combines with subunit B to form activated HIF-1. It acts as a transcription initiation factor, triggering the expression of various genes related to angiogenesis, erythropoiesis, and glucose metabolism to regulate the response of cells to hypoxia. Target genes such as VEGF, basic fibroblast growth factor (bFGF), glucose transporter, EGF, and erythropoietin are closely related to the generation of new blood vessels. VEGF is the most potent angiogenic stimulator, and HIF-1 specifically binds to its enhancer sequence to strengthen the transcription of VEGF and stabilize the mRNA of VEGF, thereby increasing the activity of VEGF [4]. The inhibition of HIF-1A expression under hypoxia can effectively reduce the expression of VEGF in retinal pigment epithelial cells, inhibiting neovascularization [9]. Additionally, pigment epithelium-derived factor (PEDF) can help to inhibit neovascularization through VEGF. Although hypoxia can cause a decrease in the expression of PEDF, it indirectly up-regulates VEGF expression and promotes the development of CNV [10].
It has been suggested that many miRNAs participate in regulating vascular endothelial function. For instance, miR-221 and miR-222 can inhibit the proliferation and migration of vascular endothelial cells. MiR-23, miR-27b, and miR-130a can promote angiogenesis, and MiR-155, miR-21, and miR-126 can take part in the regulation of vascular inflammation [11]. Inhibition of miR-23 and miR-27 can block angiogenesis in vitro, and cause retinal development disorder in vivo after birth. miR-23 and miR-27 are correlated to pathological vasculogenesis in laser-induced CNV model mice, while MiR-23 and miR-27 can promote vasculogenesis by inhibiting anti-angiogenesis factors Sprouty2 and Sema6A [12]. Moreover, miRNAs participate in regulating the response of vascular endothelial cells against VEGF. MiR-126 can directly inhibit Sprouty-dependent protein as well as the regulatory subunit of PI3K to enhance the response of vascular endothelium to VEGF [13]. Results derived from inhibition of the VEGF pathway are similar to those in miR-126 knockout. The above-detailed findings suggest that miRNAs play important roles in regulating vascular integrity and vasculogenesis. In ischemic retinas of mice, expression levels of miR-31, miR-150, and miR-184 were significantly decreased. Intraocular injection of miR-31, miR-150, and miR-184 precursors obviously reduced CNV and neovascularization in ischemic retinas [14].
Conclusions
In this study, there was significant decrease in expression of miR-18a under hypoxic condition, while the expression of HIF-1A remarkably increased in CECs, suggesting a correlation between hypoxia and miR-18a. Our results further proved that overexpression of miR-18a mimic in CECs significantly reduced mRNA and protein levels of HIF-1A. The proliferation and migration of CECs transfected with miR-18a mimic also decreased. However, overexpression of miR-18a inhibitor in CECs significantly increased mRNA and protein levels of HIF-1A, along with an increase in the proliferation and migration of CECs. This study did not explore the role of miR-18a in CNV, although the association with CECs was detected. Further studies are needed to investigate how miR-18a contributes to regulation of CNV and pathogenesis of AMD.
Footnotes
Source of support: Departmental sources
Conflict of interest statement
We declare that we have no conflict of interest.
References
- 1.Zarbin MA. Current concepts in the pathogenesis of age-related macular degeneration[J] Arch Ophthalmol. 2004;122(4):598–614. doi: 10.1001/archopht.122.4.598. [DOI] [PubMed] [Google Scholar]
- 2.Brown DM, Heier JS, Ciulla T, et al. Primary endpoint results of a phase II study of vascular endothelial growth factor trap-eye in wet age-related macular degeneration. Ophthalmology. 2011;118(6):1089–97. doi: 10.1016/j.ophtha.2011.02.039. [DOI] [PubMed] [Google Scholar]
- 3.Vadlapatla RK, Vadlapudi AD, Mitra AK. Hypoxia-inducible factor-1 (HIF-1): A potential target for intervention in ocular neovascular diseases. Curr Drug Targets. 2013;14(8):919–35. doi: 10.2174/13894501113149990015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin CH, Li CH, Liao PL, et al. Silibinin inhibits VEGF secretion and age related macular degeneration in a hypoxia dependent manner through the PI 3 kinase/Akt/mTOR pathway. Br J Pharmacol. 2013;168(4):920–31. doi: 10.1111/j.1476-5381.2012.02227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie T, Fan Y, Wang F. The correlation between microRNA and ophthalmic disease. Recent Advances in Ophthalmology. 2009;29(5):391–94. [Google Scholar]
- 6.Doyle SL, Campbell M, Ozaki E, et al. NLRP3 has a protective role in age-related macular degeneration through the induction of IL-18 by drusen components. Nat Med. 2012;18(5):791–98. doi: 10.1038/nm.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riva CE, Geiser M, Petrig BL. Ocular blood flow assessment using continuous laser Doppler flowmetry. Acta Ophthalmol. 2010;88(6):622–29. doi: 10.1111/j.1755-3768.2009.01621.x. [DOI] [PubMed] [Google Scholar]
- 8.Klein R, Cruickshanks KJ, Nash SD, et al. The prevalence of age-related macular degeneration and associated risk factors. Arch Ophthalmol. 2010;128(6):750–58. doi: 10.1001/archophthalmol.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arjamaa O, Nikinmaa M, Salminen A, et al. Regulatory role of HIF-1α in the pathogenesis of age-related macular degeneration (AMD) Ageing Res Rev. 2009;8(4):349–58. doi: 10.1016/j.arr.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Kudelka MR, Grossniklaus HE, Mandell KJ. Emergence of dual VEGF and PDGF antagonists in the treatment of exudative age-related macular degeneration. 2013;8(5):475–84. doi: 10.1586/17469899.2013.840095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79(4):581–88. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Q, Gallagher R, Ufret-Vincenty R, et al. Regulation of angiogenesis and choroidal neovascularization by members of microRNA-23 27 24 clusters. Proc Natl Acad Sci USA. 2011;108(20):8287–92. doi: 10.1073/pnas.1105254108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fish JE, Santoro MM, Morton SU, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15(2):272–84. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen JK, Yang X, Xie B, et al. MicroRNAs regulate ocular neovascularization. Mol Ther. 2008;16(7):1208–16. doi: 10.1038/mt.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]